Abstract
Persisters are cells which evade stresses like antibiotics and which are characterized by reduced metabolism and a lack of genetic alterations required to achieve this state. We showed previously that MqsR and MqsA of Escherichia coli are a toxin-antitoxin pair that influence cell physiology (e.g., biofilm formation and motility) via RNase activity as well as through regulation of toxin CspD. Here, we show that deletion of the mqsRA locus decreases persister cell formation and, consistent with this result, overexpression of MqsR increases persister cell formation. Furthermore, toxins Hha, CspD, and HokA increase persister cell formation. In addition, by overproducing MqsR in a series of isogenic mutants, we show that Hha and CspD are necessary for persister cell formation via MqsR overexpression. Surprisingly, Hfq, a small RNA chaperone, decreases persistence. A whole-transcriptome study shows that Hfq induces transport-related genes (opp genes and dppA), outer membrane protein-related genes (ybfM and ybfN), toxins (hha), and proteases (clpX, clpP, and lon). Taken together, these results indicate that toxins CspD, Hha, and HokA influence persister cell formation via MqsR and that Hfq plays an important role in the regulation of persister cell formation via regulation of transport or outer membrane proteins OppA and YbfM.
Keywords: MqsR, persister formation, CspD, Hha, Hfq
Introduction
Persister cells are a small fraction of bacteria that demonstrate resistance to antibiotics without genetic change [1]. The number of persister cells may reach 1% of stationary-phase cells or biofilm cells for a variety of microorganisms including Pseudomonas aeruginosa [2], Escherichia coli [1], and Candida albicans [3]. However, the genetic mechanism of persister cell formation is still unclear.
Toxin/antitoxin (TA) systems are ubiquitous [4] in bacterial chromosomes and in low copy number plasmids where they appear to stabilize these systems [5]. TA systems typically consist of pairs of genes in an operon which encode a stable toxin that can cause cell death by disrupting an essential cellular process and a labile antitoxin that can bind and form a tight complex with the toxin and neutralize its activity [6]. A number of TA systems have been identified in E. coli including (listed as toxin/antitoxin) MqsR/MqsA [7; 8; 9; 10], MazF/MazE [11], RelE/RelB [12], ChpB/ChpS [13], YoeB/YefM [14], YafQ/DinJ [15], and HipA/HipB [16], and several of these have been linked to biofilm formation [17; 18].
TA systems have also been linked to persister cells [19; 20]. In a whole-transcriptome study, MqsR/MqsA of E. coli were found to be the most induced loci in persister cells [20]. Although deletion of nearly all TA loci has no effect on persister cell formation due to the redundancy of these systems [19; 20], overexpression of HipA toxin increased persistence 10-fold whereas deletion of hipBA decreased persistence by 10- to 100-fold in stationary and biofilm conditions [19]; hence, hipA is the first bona fide persister gene.
Previously, we discovered that MqsR is induced in biofilms [21] and that MqsR is a regulator of motility and quorum sensing that is directly associated with biofilm development as it mediates the ability of autoinducer 2 to increase E. coli biofilm formation [22]. In addition, we found MqsR in the absence of the antitoxin, MqsA, is much more toxic [10], and the three dimensional structure of MqsA revealed it is an RNase similar to RelE and YoeB [9]. MqsR degrades RNA with a GCU motif [7]. Furthermore, MqsA binds DNA via a helix-turn-helix (HTH) C-terminal domain [9]. Unlike most TA pairs including RelE/RelB, YoeB/YefM and MazF/MazE, the MqsR/MqsA TA pair is unique [9] in that (i) the antitoxin is larger than the toxin, (ii) both toxin and antitoxin are basic, (iii) the mqsRA sequences are not homologous to any member of a recognized TA system, and (iv) the antitoxin binds the toxin at its amino terminus and requires a zinc metal for structural stability. Most importantly, the antitoxin MqsA regulates more than its own synthesis as it is the first antitoxin that has been shown to bind directly the mqsRA, spy, mcbR, and cspD promoters as well as to regulate transcription of these loci [10].
Here, we show direct evidence that MqsR influences persister formation since deletion of mqsRA reduces persister cell formation and MqsR overexpression increases persister cell formation. Using whole transcriptome analysis, quantitative real time-polymerase chain reaction (qRT-PCR), and cell survival studies in which selected genes were overproduced in isogenic mutants, we explored the mechanism by which MqsR influences persister cell formation and found that MqsR positively regulates persister cell production via Hha and CspD and that the small RNA chaperone Hfq represses persister production.
Materials and Methods
Bacterial strains, plasmids, and growth conditions
The bacterial strains and plasmids used in this study are listed in Supplemental Table S1. For isogenic mutants and overexpression of specific genes, we used the Keio collection [23] and ASKA library [24], respectively, from the Genome Analysis Project in Japan. All experiments were conducted in Luria-Bertani (LB) medium [25] at 37°C. Kanamycin (50 μg/mL) was used for pre-culturing isogenic knockout mutants and for maintaining pBS(Kan)-based plasmids [26], whereas chloramphenicol (30 μg/mL) was used for maintaining pCA24N-based plasmids [24].
Persister assay
Persister cell formation was performed as described [27] with slight modifications. To determine the number of persister cells with the knockout mutants, cells were inoculated in LB medium and grown to a turbidity of 0.5 at 600 nm. After collecting the cells, the concentration was adjusted to a turbidity of 1 (to obtain 1 to 3 × 108 CFU/mL of viable cells), and these cultures were exposed to 100 μg/mL ampicillin for 2 h. Cells were diluted by 102 to 107 via 10-fold serial dilution steps in 0.85% NaCl solution and applied as 10 μL drops on LB agar with kanamycin or chloramphenicol to determine persister cell viability [28]. For producing proteins using pBS(Kan)- or pCA24N-based plasmids, when the cell density reached a turbidity of 0.5, 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) was added for 2 h, the turbidity was adjusted to 1, and ampicillin was added as above. Two independent cultures were used for each strain.
RNA isolation and whole-transcriptome analysis
The whole-transcriptome analysis was performed using planktonic cells of BW25113 hfq vs. BW25113 by growing cells to a turbidity of 0.5 at 600 nm in LB medium at 37°C, adjusting the turbidity to 1, and exposing the cells to 100 μg/mL ampicillin for 2 h. Total RNA was isolated from cells as described previously [21] using RNALater buffer® (Applied Biosystems, Foster City, CA). cDNA synthesis, fragmentation and hybridizations were as described previously [22]. The E. coli GeneChip Genome 2.0 array (Affymetrix, Santa Clara, CA; P/N 511302) was used. Corroborating the deletion of hfq, the microarray signals of hfq were very low. For the binary microarray comparisons to determine differential genes expression, if the gene with the larger transcription rate did not have a consistent transcription rate based on the 11 probe pairs (P-value less than 0.05), these genes were not used. A gene was considered differentially expressed when the P value for comparing two chips was lower than 0.05 (to ensure that the change in gene expression was statistically significant and that false positives arise less than 5%) and when the expression ratio was higher than the standard deviation (3.5 fold) for all K-12 genes of the microarrays [29].
Quantitative real time-PCR (qRT-PCR)
qRT-PCR was performed using the StepOnePlus™ Real-Time PCR System (Applied Biosystems). The preparation of RNA samples were performed as described previously [21]. For the qRT-PCR experiments, we used RNALater buffer for RNA stabilization and protection during the RNA preparation steps. 50 ng of total RNA was used for the qRT-PCR reaction using the Power SYBR® Green RNA-to-CT™ 1-Step Kit (Applied Biosystems). Primers were designed using Primer3Input Software (v0.4.0) and are listed in Supplemental Table S1. The housekeeping gene rrsG [30] was used to normalize the genes expression data. The annealing temperature was 60°C for all the genes in this study. For the level of expression of the hha, cspD, hokA, and hfq transcripts, overnight cultures of BW25113/pBS(Kan) and BW25113/pBS(Kan)-mqsR were inoculated into LB medium and grown to a turbidity of 1 at 600 nm, and 1 mM IPTG was added for 1 h to induce mqsR. To explore whether mqsR, hha, cspD, and hfq transcription was affected by antibiotic stress, overnight cultures of BW25113 were grown to a turbidity of 0.5, cells were adjusted to a turbidity of 1 and were exposed with or without 100 μg/mL ampicillin for 2 h.
Microarray accession numbers
The differential gene expression data have been deposited in the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) and are accessible through accession numbers GSE18852.
Results
MqsR strongly influences persister production
Previously, we found that overexpression of MqsR is toxic [8], whereas overexpression of MqsA diminished MqsR toxicity [9; 10]. Using E. coli MG1655, MqsR production increased persister cell formation 10,000- to 100,000-fold against a number of antibiotics including ofloxacin and cefotaxime while deletion of mqsR did not reduce the number of persisters [20]. In the current study, we used E. coli BW25113 so that we could take advantage of the single gene knockout Keio library [23] and used ampicillin (100 μg/mL) as a representative antibiotic since non-persister cells die within 15 min at this concentration [19]. Unlike the results with MG1655, deletion of mqsR from BW25113 significantly repressed persister production (6-fold) compared to the wild-type strain (Fig. 1A). Corroborating this result, deletion of mqsR and mqsA also repressed persister production (7-fold) compared to the wild-type strain (Fig. 1A). In addition, producing MqsR (using two different of expression vectors) increased persister cell formation (12- to 16-fold), but producing MqsA or MqsR/MqsA did not affect persister cell formation (Fig. 1B). Taken together, these results show the MqsR/MqsA TA pair have an important role in persister cell formation as it becomes only the second locus that upon deletion reduces persistence.
Fig. 1.
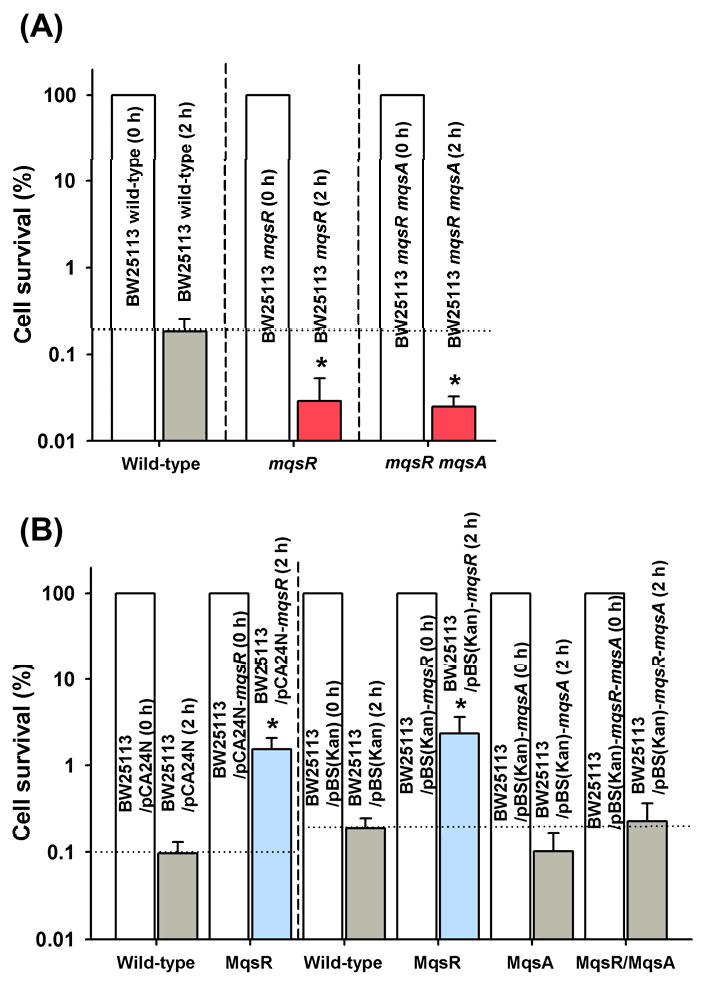
Effect of mqsR and mqsA mutations on persister cell formation. BW25113 wild-type, BW25113 mqsR, and BW25113 mqsR mqsA were grown to a turbidity of 0.5 at 600 nm in LB medium at 37°C, adjusted on a turbidity of 1, and exposed to 100 μg/mL ampicillin for 2 h (A). Persister cell formation of BW25113/pCA24N, BW25113/pCA24N-mqsR, BW25113/pBS(Kan), BW25113/pBS(Kan)-mqsA BW25113/pBS(Kan)-mqsR, and BW25113/pBS(Kan)-mqsR-mqsA grown to a turbidity of 0.5 in LB medium at 37°C, contacted with 1 mM IPTG for 2 hr for induction, adjusted to a turbidity of 1, and exposed to 100 μg/mL ampicillin for 2 h in LB (B). Persister data are the average of two independent cultures, and one standard deviation is shown. The asterisk indicates statistical significance determined using a Student's t-test (p < 0.05).
Hha, CspD, and HokA increase persistence
Previously, using whole-transcriptome studies and nickel-enrichment DNA binding microarrays coupled with cell survival experiments, we identified eight proteins (CspD, HokA, ClpX, ClpP, Lon, YfjZ, RelB, and RelE) that are involved in MqsR toxicity [10]. We also identified that the toxin Hha controls cell death and biofilm dispersal [18]. Since the RelE toxin in the RelE/RelB pair is already linked to persister cell formation [19], we explored the impact of seven of these nine proteins in persister cell formation by performing the persister formation assay while inducing the genes that encode these eight proteins in hosts that lack each of these genes (Fig. 2A).
Fig. 2.
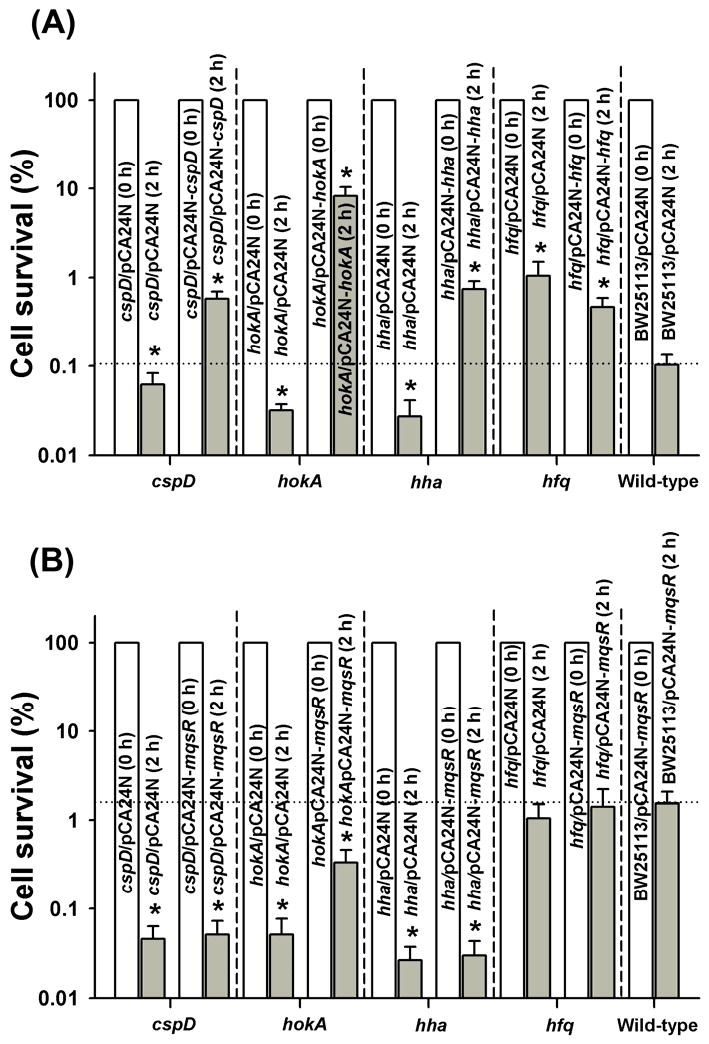
Effect of isogenic mutations on persister cell production. BW25113 hha/pCA24N-hha, BW25113 cspD/pCA24N-cspD, BW25113 hokA/pCA24N-hokA, and BW25113 hfq/pCA24N-hfq were grown to a turbidity of 0.5 in LB medium at 37°C, contacted with 1 mM IPTG for 2 hr for induction, adjusted to a turbidity of 1, and exposed to 100 μg/mL ampicillin for 2 h in LB (A). Effect of hha, cspD, hokA, and hfq isogenic deletions on persister cell formation with MqsR via pCA24N-mqsR (B). Persister data are the average of two independent cultures, and one standard deviation is shown. The asterisk indicates statistical significance determined using a Student's t-test (p < 0.05).
Among the seven proteins, Hha, CspD and HokA increased persister cell formation by 6- to 85-fold (Fig. 2A), whereas ClpX, ClpP, and Lon only slightly increased the number of persisters (2- to 3-fold) (Supplemental Fig. S1). Corroborating these studies, the deletion of cspD (2-fold), hokA (3-fold), and hha (4-fold) significantly repressed the production of persister cells (Fig. 2A); hence, these three toxins directly involved in the persister production.
Hha and CspD are essential for persister cell production via MqsR
Confirming the relationship between Hha and CspD with MqsR and persister cells, deleting hha and cspD abolished persister cell formation upon producing MqsR (Fig. 2B). In contrast, deleting hokA only slightly reduced the ability of MqsR to increase persistence (Fig. 2B). Therefore, toxins Hha and CspD are most directly involved in persister cell formation with MqsR.
Antibiotic stress induces Hha and CspD via MqsR
Using qRT-PCR and BW25113/pBS(Kan)-mqsR, we found that hha (10 ± 1 fold) and cspD (13 ± 1 fold) are induced by overproducing MqsR. Also, under antibiotics stress (100 μg/mL ampicillin for 2 h), mqsR was induced (5 ± 1 fold) in wild-type BW25113. Under these conditions, hha (4 ± 1 fold) and cspD (3 ± 1 fold) were also induced. These results show that persister cells produced by antibiotic stress induce mqsR, and its induction influences production of both the Hha and CspD toxins.
Hfq reduces persistence
Hfq was selected for study here because of its impact on persistence since it an RNA-binding protein that is important for small RNA-mRNA transactions [31] and because MqsR is an RNase. We found that inducing hfq reduced persister cell formation by 2-fold compared to the empty plasmid (Fig. 2A). More importantly, deleting hfq increased persister production 11-fold (BW25113 hfq/pCA24N vs. BW25113/pCA24N for 2 h) (Fig. 2A). Similar phenotypes were observed for the persister assay with strains BW25113 hfq and BW25113 in the absence of plasmids (data not shown). Also, hfq was not induced by stress (qRT-PCR). Hence, Hfq reduces persister cell formation.
Hfq whole-transcriptome study
Since Hfq reduces persistence, we hypothesized that Hfq, as an RNA chaperone, may be strongly associated with persister-related RNA under antibiotic stress. Hence, we investigated the genes controlled by Hfq under antibiotic stress using a whole-transcriptome analysis. Using planktonic cells, hfq deletion under persister conditions repressed 124 genes by more than 3.5-fold and induced 22 genes (Supplemental Table S2). Primarily, the glycolate utilization glc cluster [32] was repressed upon deleting hfq including glcB (23-fold), glcG (21-fold), glcF (23-fold), glcD (15-fold), and glcC (29-fold). Also, motility-related genes including flagella, curli, and chemotaxis were repressed by deleting hfq along with emrY (encoding multi-drug resistance protein Y) and micC (encoding a MicC small RNA regulator of OmpC translation). Deletion of hfq induced hha as well as clpP, clpX, and lon which encode antitoxin-degrading proteases.
Transport proteins encoded by dppA and the opp locus well as genes encoding outer membrane proteins ybfM and ybfN were induced upon deleting hfq (Supplemental Table S2). Therefore, Hfq may decrease persistence by repressing specific transport- or membrane-related proteins. Corroborating these results, producing YbfM and OppA increased persistence by 28- and 12-fold, respectively (Fig. 3).
Fig. 3.
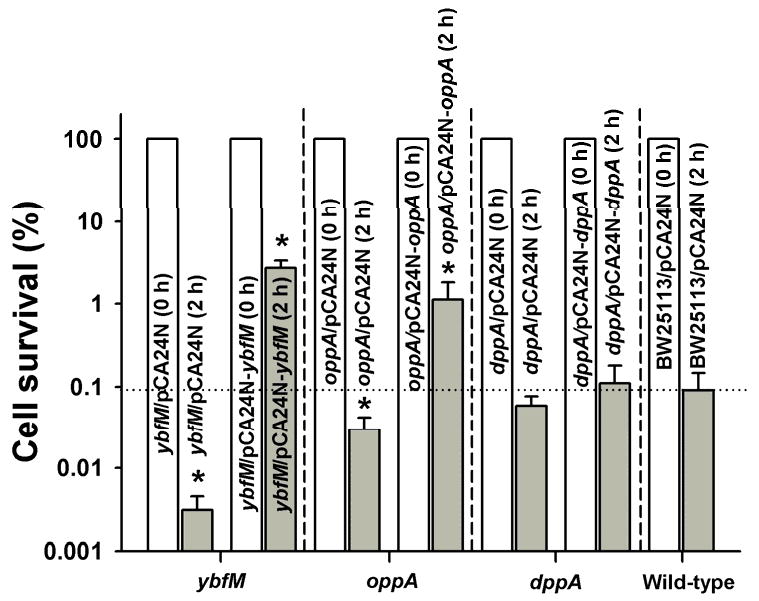
Impact of YbfM, OppA, and DppA on the production of persister cells. BW25113 ybfM/pCA24N-ybfM, BW25113 oppA/pCA24N-oppA, and BW25113 dppA/pCA24N-dppA were grown to a turbidity of 0.5 in LB medium at 37°C, contacted with 1 mM IPTG for 2 h for induction, adjusted to a turbidity of 1, and exposed to 100 μg/mL ampicillin for 2 h. Persister data are the average of two independent cultures, and one standard deviation is shown. The asterisk indicates statistical significance determined using a Student's t-test (p < 0.05).
Discussion
Understanding the genetic basis of persistence may lead to new concepts in anti-microbial therapy. To date, several E. coli proteins such as GlpD, PlsB, DnaJ, and PmrC have been linked to persistence [27; 33]. Here, we linked for the first time Hha, CspD, HokA, Hfq, YbfM, and OppA to persistence as well as show that deletion of mqsRA decreases persistence making it only the second locus that upon deletion, affects persistence. A number of toxins in TA systems that inhibit translation via RNase activity are well-suited for initiating cell dormancy [20]. The role of TA systems on cell physiology is not clear, and nine hypothetical biological functions of TA systems have been proposed [34] including persister formation. Recently, we found one more function for TA systems, that of influencing biofilm formation, as shown with Hha/TomB [18], MqsR/MqsA [8; 21; 22], and the five TA systems of the E. coli Δ5 strain [17]. Among these toxin genes, here we focused on the function of MqsR in persisters.
The lines of evidence that MqsR is important for persister production are (i) mqsR is induced 8-fold in biofilms [21], a mode of growth resistant to antibiotics and which generates persisters [1], (ii) MqsR is a toxin paired with antitoxin MqsA and its toxicity is due to both its RNase activity and regulation of the CspD toxin by interaction with MqsA [10], and toxins have been linked to persister cell formation [20], (iii) MqsR cleaves mRNA at GCU sites and 4,212 open reading frames (99.6% population) of the 4,226 genes of the E. coli genome contain a GCU sequence, hence MqsR is a potent toxin [7], (iv) mqsR is the most highly induced gene in persister cells [20], and (v) production of MqsR increases persistence and deletion of mqsR decreases persistence (Fig. 1). Fig. 4 summarizes our understanding of the genetic mechanism of MqsR in persister cell production.
Fig. 4.
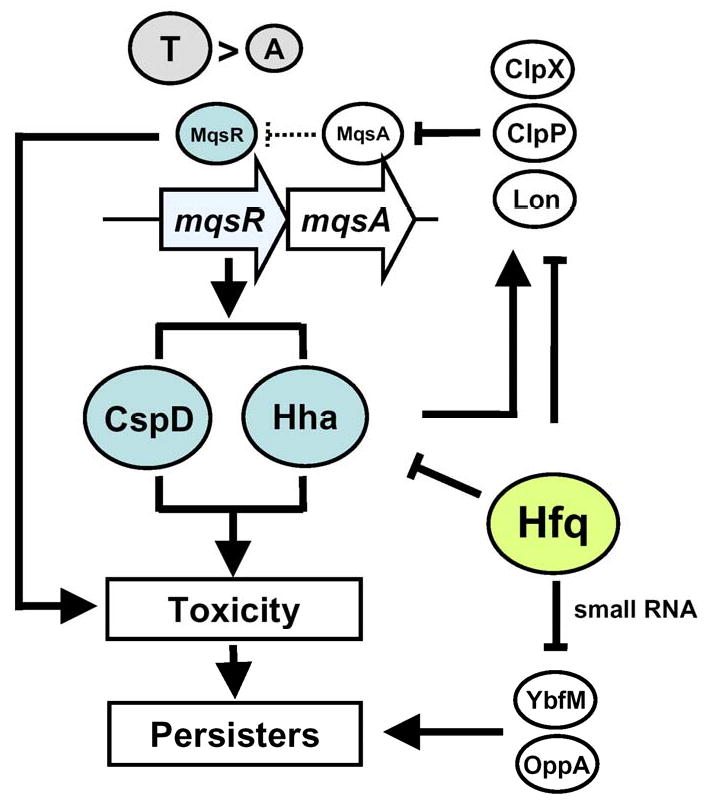
Schematic of the mechanism of persister production via MqsR. CspD and Hha are directly involved in persister formation via MqsR, and Hfq negatively regulates transport and outer membrane proteins. → indicates induction, and ⊥ indicates repression.
Hha and CspD are also related to persister cell formation with MqsR. Hha and CspD increase the production of persister cells, deletion of hha and cspD decreases persistence significantly, and deletion of hha and cspD diminish the ability of MqsR to cause persistence (Fig. 2AB). Also, hha and cspD are induced by stress and by the production of MqsR. Hha also induces genes encoding toxins in other TA pairs, including RelE (3-fold), YoeB (7-fold), and YafQ (2-fold) and induces specific proteases including Lon (4-fold), ClpP (4-fold), and ClpX (4-fold) which may, in turn, activate toxins by degrading antitoxins [18].
We also show that Hfq reduces persistence (Fig. 2A); hence, this is the first protein found to have a negative role in persistence, and this is the first report of Hfq being related to persistence. Also, the whole transcriptome analysis indicates that hfq deletion induces formation of Hha and several proteases including ClpX, ClpP, and Lon (Supplemental Table S2); induction of these genes should increase persistence, and we show ClpP and Lon increase persistence (Supplemental Fig. S1). Therefore, it appears deletion of hfq increases persistence by up-regulating hha, clpX, clpP, and lon. Also, consistent with previous results, dnaJ, dnaK, fis, hns, dksA were induced by the deletion of hfq under antibiotic conditions (Supplemental Table S2), and these genes have been identified as induced in persister cells using ofloxaxin with E. coli [35]. Also, ybfM and dppA/oppA are strongly regulated by small RNAs, micM [36] and gcvB [37], respectively, and are dependent on the RNA chaperone Hfq.
Since ybfM and oppA are induced by the deletion of hfq under persister conditions (Supplemental Table S2), we tested for their impact on persistence. YbfM is a putative outer membrane porin protein and is similar to OprD of P. aeruginosa (38% identity) [36]. The porin proteins are involved in the uptake of extracellular compounds, including antibiotics, and are related to the multi-drug resistance phenotype [38]. The link between YbfM and persistence was corroborated by a 30-fold change in survival upon deleting and overexpressing ybfM (Fig. 3). Also, OppA is a periplasmic oligopeptide-transport protein [39], and similar to YbfM, OppA also increased persistence by 12-fold upon overexpressing oppA (Fig. 3). Taken together, these results suggest that Hfq may indirectly influence persister cell formation under antibiotic stress by regulating mRNA encoding transport and outer membrane proteins including porins as well as by regulating hha, clpX, clpP, and lon.
Supplementary Material
Acknowledgments
This work was supported by the NIH (R01 EB003872) and the ARO (W911NF-06-1-0408). T. W. is the T. Michael O'Connor II Endowed Chair. We are grateful for the Keio and ASKA strains provided by the Genome Analysis Project in Japan.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lewis K. Persister cells, dormancy and infectious disease. Nat Rev Micro. 2007;5:48–56. doi: 10.1038/nrmicro1557. [DOI] [PubMed] [Google Scholar]
- 2.Spoering AL, Lewis K. Biofilms and planktonic cells of Pseudomonas aeruginosa have similar resistance to killing by antimicrobials. J Bacteriol. 2001;183:6746–6751. doi: 10.1128/JB.183.23.6746-6751.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.LaFleur MD, Kumamoto CA, Lewis K. Candida albicans biofilms produce antifungal-tolerant persister cells. Antimicrob Agents Chemother. 2006;50:3839–3846. doi: 10.1128/AAC.00684-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gerdes K, Christensen SK, Løbner-Olesen A. Prokaryotic toxin-antitoxin stress response loci. Nat Rev Microbiol. 2005;3:371–382. doi: 10.1038/nrmicro1147. [DOI] [PubMed] [Google Scholar]
- 5.Engelberg-Kulka H, Amitai S, Kolodkin-Gal I, Hazan R. Bacterial programmed cell death and multicellular behavior in bacteria. PLoS Genet. 2006;2:e135. doi: 10.1371/journal.pgen.0020135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown JM, Shaw KJ. A novel family of Escherichia coli toxin-antitoxin gene pairs. J Bacteriol. 2003;185:6600–6608. doi: 10.1128/JB.185.22.6600-6608.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yamaguchi Y, Park JH, Inouye M. MqsR, a crucial regulator for quorum sensing and biofilm formation, is a GCU-specific mRNA interferase in Escherichia coli. J Biol Chem. 2009;284:28746–28753. doi: 10.1074/jbc.M109.032904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang XS, García-Contreras R, Wood TK. Escherichia coli transcription factor YncC (McbR) regulates colanic acid and biofilm formation by repressing expression of periplasmic protein YbiM (McbA) ISME J. 2008;2:615–631. doi: 10.1038/ismej.2008.24. [DOI] [PubMed] [Google Scholar]
- 9.Brown BL, Grigoriu S, Kim Y, Arruda J, Davenport A, Wood TK, Peti W, Page R. Three dimensional structure of the MqsR:MqsA complex: a novel TA pair with a toxin homologous to RelE and an antitoxin with unique properties. PLoS Pathog. 2009 doi: 10.1371/journal.ppat.1000706. In revision. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim Y, Wang X, Zhang XS, Page R, Peti W, Wood TK. Escherichia coli Toxin/Antitoxin Pair MqsR/MqsA Regulate Toxin CspD. Environ Microbiol. 2009 doi: 10.1111/j.1462-2920.2009.02147.x. In revision. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aizenman E, Engelberg-Kulka H, Glaser G. An Escherichia coli chromosomal “addiction module” regulated by 3′,5′-bispyrophosphate: A model for programmed bacterial cell death. Proc Natl Acad Sci USA. 1996;93:6059–6063. doi: 10.1073/pnas.93.12.6059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gotfredsen M, Gerdes K. The Escherichia coli relBE genes belong to a new toxin-antitoxin gene family. Mol Microbiol. 1998;29:1065–1076. doi: 10.1046/j.1365-2958.1998.00993.x. [DOI] [PubMed] [Google Scholar]
- 13.Masuda Y, Miyakawa K, Nishimura Y, Ohtsubo E. chpA and chpB, Escherichia coli chromosomal homologs of the pem locus responsible for stable maintenance of plasmid R100. J Bacteriol. 1993;175:6850–6856. doi: 10.1128/jb.175.21.6850-6856.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cherny I, Gazit E. The YefM antitoxin defines a family of natively unfolded proteins: Implications as a novel antibacterial target. J Biol Chem. 2004;279:8252–8261. doi: 10.1074/jbc.M308263200. [DOI] [PubMed] [Google Scholar]
- 15.Motiejyūnaitė R, Armalytė JM, Sužiedėlienė E. Escherichia coli dinJ-yafQ genes act as a toxin-antitoxin module. FEMS Microbiol Lett. 2007;268:112–119. doi: 10.1111/j.1574-6968.2006.00563.x. [DOI] [PubMed] [Google Scholar]
- 16.Black DS, Kelly AJ, Mardis MJ, Moyed HS. Structure and organization of hip, an operon that affects lethality due to inhibition of peptidoglycan or DNA synthesis. J Bacteriol. 1991;176:4081–4091. doi: 10.1128/jb.173.18.5732-5739.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim Y, Wang X, Qun M, Zhang XS, Wood TK. Toxin-antitoxin systems in Escherichia coli influence biofilm formation through YjgK (TabA) and fimbriae. J Bacteriol. 2008;191:1258–1267. doi: 10.1128/JB.01465-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.García-Contreras R, Zhang XS, Kim Y, Wood TK. Protein translation and cell death: the role of rare tRNAs in biofilm formation and in activating dormant phage killer genes, PLoS One. 2008:e2394. doi: 10.1371/journal.pone.0002394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keren I, Shah D, Spoering A, Kaldalu N, Lewis K. Specialized persister cells and the mechanism of multidrug tolerance in Escherichia coli. J Bacteriol. 2004;186:8172–8180. doi: 10.1128/JB.186.24.8172-8180.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shah D, Zhang Z, Khodursky A, Kaldalu N, Kurg K, Lewis K. Persisters: a distinct physiological state of E. coli. BMC Microbiol. 2006;6:53. doi: 10.1186/1471-2180-6-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ren D, Bedzyk LA, Thomas SM, Ye RW, Wood TK. Gene expression in Escherichia coli biofilms. Appl Microbiol Biotechnol. 2004;64:515–524. doi: 10.1007/s00253-003-1517-y. [DOI] [PubMed] [Google Scholar]
- 22.González Barrios AF, Zuo R, Hashimoto Y, Yang L, Bentley WE, Wood TK. Autoinducer 2 controls biofilm formation in Escherichia coli through a novel motility quorum-sensing regulator (MqsR, B3022) J Bacteriol. 2006;188:305–316. doi: 10.1128/JB.188.1.305-316.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2:0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kitagawa M, Ara T, Arifuzzaman M, Ioka-Nakamichi T, Inamoto E, Toyonaga H, Mori H. Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): unique resources for biological research. DNA Res. 2005;12:291–299. doi: 10.1093/dnares/dsi012. [DOI] [PubMed] [Google Scholar]
- 25.Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. 2nd. Cold Spring Harbor Laboratory Press; N.Y: 1989. [Google Scholar]
- 26.Canada KA, Iwashita S, Shim H, Wood TK. Directed evolution of toluene ortho-monooxygenase for enhanced 1-naphthol synthesis and chlorinated ethene degradation. J Bacteriol. 2002;184:344–349. doi: 10.1128/JB.184.2.344-349.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vázquez-Laslop N, Lee H, Neyfakh AA. Increased persistence in Escherichia coli caused by controlled expression of toxins or other unrelated proteins. J Bacteriol. 2006;188:3494–3497. doi: 10.1128/JB.188.10.3494-3497.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Donegan K, Matyac C, Seidler R, Porteous A. Evaluation of methods for sampling, recovery, and enumeration of bacteria applied to the phylloplane. Appl Environ Microbiol. 1991;57:51–56. doi: 10.1128/aem.57.1.51-56.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ren D, Bedzyk LA, Ye RW, Thomas SM, Wood TK. Differential gene expression shows natural brominated furanones interfere with the autoinducer-2 bacterial signaling system of Escherichia coli. Biotechnol Bioeng. 2004;88:630–642. doi: 10.1002/bit.20259. [DOI] [PubMed] [Google Scholar]
- 30.Wang X, Kim Y, Wood TK. Control and benefits of CP4-57 prophage excision in Escherichia coli biofilms. ISME J. 2009;3:1164–1179. doi: 10.1038/ismej.2009.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guisbert E, Rhodius VA, Ahuja N, Witkin E, Gross CA. Hfq modulates the σE-mediated envelope stress response and the σ32-mediated cytoplasmic stress response in Escherichia coli. J Bacteriol. 2007;189:1963–1973. doi: 10.1128/JB.01243-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pellicer MT, Badía J, Aguilar J, Baldomà L. glc locus of Escherichia coli: characterization of genes encoding the subunits of glycolate oxidase and the glc regulator protein. J Bacteriol. 1996;178:2051–2059. doi: 10.1128/jb.178.7.2051-2059.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spoering AL, Vulić M, Lewis K. GlpD and PlsB participate in persister cell formation in Escherichia coli. J Bacteriol. 2006;188:5136–5144. doi: 10.1128/JB.00369-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Magnuson RD. Hypothetical functions of toxin-antitoxin systems. J Bacteriol. 2007;189:6089–6092. doi: 10.1128/JB.00958-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hansen S, Lewis K, Vulić M. Role of global regulators and nucleotide metabolism in antibiotic tolerance in Escherichia coli. Antimicrob Agents Chemother. 2008;52:2718–2726. doi: 10.1128/AAC.00144-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rasmussen AA, Johansen J, Nielsen JS, Overgaard M, Kallipolitis B, Valentin-Hansen P. A conserved small RNA promotes silencing of the outer membrane protein YbfM. Mol Microbiol. 2009;72:566–577. doi: 10.1111/j.1365-2958.2009.06688.x. [DOI] [PubMed] [Google Scholar]
- 37.Pulvermacher SC, Stauffer LT, Stauffer GV. Role of the Escherichia coli Hfq protein in GcvB regulation of oppA and dppA mRNAs. Microbiology. 2009;155:115–123. doi: 10.1099/mic.0.023432-0. [DOI] [PubMed] [Google Scholar]
- 38.Pagès JM, James CE, Winterhalter M. The porin and the permeating antibiotic: a selective diffusion barrier in Gram-negative bacteria. Nat Rev Microbiol. 2008;6:893–903. doi: 10.1038/nrmicro1994. [DOI] [PubMed] [Google Scholar]
- 39.Hogarth BG, Higgins CF. Genetic organization of the oligopeptide permease (opp) locus of Salmonella typhimurium and Escherichia coli. J Bacteriol. 1983;153:1548–1551. doi: 10.1128/jb.153.3.1548-1551.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


