Abstract
Histidine-aspartic acid phosphotransfer pathways are central components of prokaryotic signal transduction pathways, and are also found in many eukaryotes. Tools to study histidine kinases, however, are currently quite limited. In this paper, we present a new tool to study histidine-aspartic acid phosphotransfer pathways. We show that many histidine kinases will accept ATPγS as a substrate to form a stable thiophosphohistidine, even when they do not form stable phosphohistidines using the natural substrate ATP. An antibody that has previously been used to detect thiophosphorylated serine, threonine and tyrosine residues is shown to recognize thiophosphohistidine and thiophophoaspartic acid residues. Histidine kinase autothiophosphorylation is regulated by other protein sensor domains in the same way as autophosphorylation, and thiophosphate is transferred to downstream aspartic acid containing response regulators.
Keywords: Histidine kinase, ATPγS, thiophosphate, semisynthetic epitope, para-nitrobenzylmesylate, thiophosphohistidine, thiophosphoaspartic acid
Introduction
Histidine kinases are important components of bacterial, fungal and plant signal transduction pathways (1). In response to environmental signals, the rates of histidine kinase autophosphorylation or phosphatase activity change leading to an altered phosphorylation state of downstream response regulators. These changes lead to regulation of fundamental cellular processes such as flagellar rotation or cell growth state (1–3). At present, there are a limited number of tools available to study the activity of histidine kinases, and all use the natural substrate ATP and rely on the ability of the histidine kinase to form a stable phosphohistidine or to transfer the phosphoryl group to a downstream response regulator that is isolable (4–6). The characterization of histidine kinases remains challenging due largely to the lability of phosphohistidines. The hydrolysis rate is likely to vary widely in proteins, but studies have shown a clear difference in the stability of phosphohistidine in acidic solution compared to phosphoserine, phosphothreonine and phosphotyrosine. In the presence of 1 M HCl at 100 °C, phosphoserine and phosphothreonine are relatively stable and have half-lives of approximately 18 hours, and phosphotyrosine has a half-life of 5h (7). In contrast, 1- and 3-phosphohistidine have half-lives of 18 and 25 seconds, respectively (8). Though histidine and aspartic acid residues are likely the most common sites of phosphorylation in nature, relatively few investigations have looked at autophosphorylation with purified histidine kinases, choosing instead bioinformatic or molecular biological approaches to study the regulation of histidine kinases.
In this paper we present a new tool to study the autophosphorylation of a histidine kinase that utilizes ATPγS. We have found that 1) some histidine kinases will utilize ATPγS to form stable thiophosphohistidines even, in some cases, when the protein will not form a stable phosphohistidine, 2) the regulation of histidine kinase activity by other protein domains is similar for both ATP and ATPγS, and 3) the thiophosphate can be transferred to downstream response regulator proteins.
We demonstrate that histidine kinases tolerate S for O substitution in the catalytic cycle. This produces a product which will be stabilized against nucleophilic attack and hydrolysis relative to the corresponding phosphate. Therefore, thiophosphorylated residues in histidine kinase and response regulator proteins are more stable than their phosphophorylated analogues. This fact was recognized by those interested in studying histidine phosphorylation with mass spectrometry and where extraction of phosphorylated proteins from cells is required (9,10). Thiophosphoaspartic acid is also likely to be more stable to hydrolysis than phosphoaspartic acid given that the majority of the hydrolysis occurs through attack on the phosphate, rather than the carbonyl carbon of the thiophosphoaspartic acid ester (11).
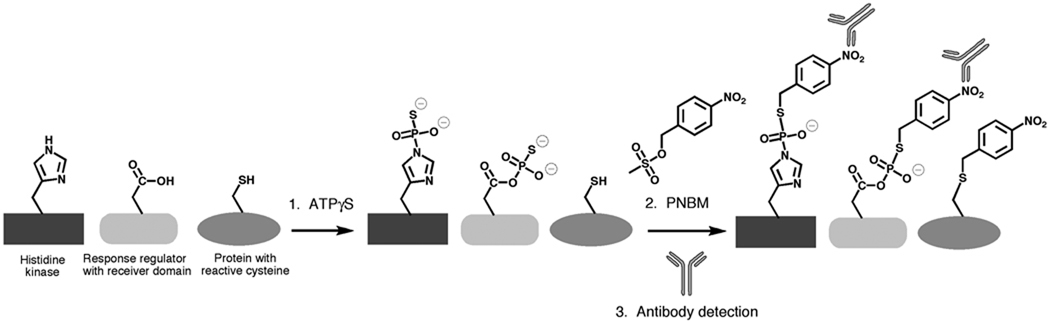
Previous work from Allen et al. has demonstrated the utility of ATPγS as a tool to study serine, threonine and tyrosine kinase activity (12,13). After reaction with ATPγS, the kinases and kinase substrates are incubated with para-nitrobenzylmesylate (PNBM), which reacts with both cysteine and with thiophosphates. An antibody specific for the alkylated thiophosphate is then used to detect the thiophosphate esters. As long as the protein of interest does not have epitopes that cross-react with the antibody, this method will work to selectively detect thiophosphorylated proteins both in vitro and in vivo. The basic method, as applied to histidine-aspartic acid phosphotransfer pathways, is outlined in Scheme 1.
Scheme 1.
A scheme outlining the method used to detect thiophosphorylated proteins. Step 1. Autophosphorylation reactions are inititated with ATPγS, and quenched with EDTA. Step 2. para-nitrobenzylmeslyate (PNBM) is added to the quenched reactions to alkylate the thiophosphorylated residues. Cross-reactivity occurs with some cysteines. Step 3. An antibody specific for the PNBM derivatized thiophosphate epitope is used to detect proteins that were thiophosphorylated in Step 1.
Materials and Methods
Histidine kinase cloning and expression
CheA and NtrB proteins were gifts from Prof. Brian Crane (Cornell University) and Prof. Sydney Kustu (UC Berkeley). S. oneidensis H-NOX associated histidine kinase (SO2145) was cloned and expressed as described previously (14). Other histidine kinase proteins (LPG2458, HCH03701, VFA0072) were cloned out of genomic DNA and ligated into pET-20b(+) expression vector (Invitrogen) cut with NcoI and XbaI restriction enzymes (NEB) and transformed into E. coli DH5α. The cytoplasmic domain of E. coli EnvZ (residues 223–450) (15) was cloned out of genomic DNA and ligated into pET-20b(+) vector cut with NdeI and NotI restriction enzymes (NEB). The receiver domain of E. coli OmpR (residues 1–128) was cloned out of genomic DNA and inserted into the Gateway entry vector pENTR/SD/D-TOPO using a TOPO cloning kit (Invitrogen). N-terminal MBP-tagged OmpR was constructed by transferring the gene into the Gateway destination vector pHMGWA (16) using LR clonase II mix (Invitrogen). Positive transformants of all constructs were screened for on LB plates containing 100 µg/mL ampicillin and the DNA sequences were confirmed by sequencing (Elim Biopharmaceuticals). With the exception of CheA and NtrB, all proteins were expressed as follows: E. coli BL21(DE3)pLysS cells containing the appropriate plasmid were grown at 37 °C to an OD600 of 0.6–0.9, induced with 10 µM IPTG, and grown for 16–18 hours at 25 °C. Cells were harvested by centrifugation at 7,000 rpm (6370 × g) for 15 minutes in an Avanti J-20 I centrifuge with a JLA 8.1 rotor (Beckman), resuspended in lysis buffer (100 mM sodium phosphate, 250 mM NaCl, 5% glycerol, , 20 mM imidazole, pH 7.9) and lysed with a high-pressure homogenizer (Avestin). Lysate was clarified by centrifugation at 42,000 rpm (200,000 × g) for 1 hour in an Optima XL-100K ultracentrifuge with a Ti-45 rotor (Beckman) prior to loading on Ni-NTA agarose resin (Qiagen). The Ni-NTA Resin was washed with lysis buffer and eluted with elution buffer (250 mM imidazole in 100 mM sodium phosphate, 250 mM NaCl, 5% glycerol, pH 7.9). Gel filtration chromatography with a S200 26/60 HiLoad Resin column (Pharmacia Biotech) connected to a Biologic HR FPLC was used for further purification. The gel filtration column was equilibrated and run in 50 mM TEA, 50 mM NaCl, 5% glycerol, pH 7.5 buffer. All proteins were >95% pure for assays, as determined by SDS-PAGE and Coomassie staining. Protein concentrations were determined by the method of Bradford and Quantitative Amino Acid Analysis.
Histidine kinase autophosphorylation and response regulator assays using ATPγ32P or ATPγ35S
Histidine kinases (1 or 5 µM) and response regulators (5 µM) were mixed with 500 µM ATP (Sigma-Aldrich) plus 10 µCi ATPg32P (6000 Ci/mmol, Perkin Elmer) or 500 µM ATPγS (Sigma-Aldrich) plus 10 µCi ATPγ35S (1250 Ci/mMol, Perkin Elmer) and 10 mM MgCl2 in 50 mM TEA, 50 mM NaCl, 5% glycerol pH 7.5 in 25 µL reactions. At endpoints, the reaction was quenched with 5 µL of a 6x concentrated stock of SDS-PAGE running buffer. Proteins in SDS-PAGE running buffer were not boiled because of concerns about the stability of the phosphorylated species. Proteins were separated from nucleotides on 10–20% Tris-Glycine SDS-PAGE gels (Invitrogen). Gels were exposed to a phosphorimager plate (Molecular Dynamics) for at least 16 hours and imaged using a Typhoon (Molecular Dynamics).
Histidine kinase autothiophosphorylation and response regulator assays using ATPγS
Histidine kinases (1 or 5 µM) and response regulators (5 µM or 20 µM) were mixed with 500 µM ATPγS and 10 mM MgCl2 in 50 mM TEA, 50 mM NaCl, 5% glycerol pH 7.5 in 25 µL reactions. At endpoints, the reaction was quenched with 5 µL of 500 mM EDTA and 1.5 µL of para-nitrobenzylmesylate (PNBM) was added from a 50 mM stock in 100% DMSO to give a final concentration of 1 mM PNBM and 6% DMSO. After allowing the alkylation reaction to proceed for 1.5 hrs, 6 µL of a 6x concentrated stock of SDS-PAGE gel running buffer was added. Proteins in SDS-PAGE running buffer were not boiled to be consistent with the procedure used in autophosphorylation assays. Proteins were separated from other components of the reaction mixture on 10–20% Tris-glycine SDS-PAGE gels (Invitrogen).
Thiophosphohistidine detection assays
Proteins were transferred from SDS-PAGE gels to nitrocellulose membranes (Whatman) and blocked with 5% non-fat dry milk (Carnation) in phosphate buffered saline, pH 8, containing 0.5% Tween 20 (PBST) for 1 hour at room temperature. Primary antibody specific for the alkylated thiophosphate (Epitomics, monoclonal Ab 51-8) was added at 1:5,000 in 5% non-fat dry milk in PBST and incubated with the blot overnight at 4 °C. The blot was then washed 3 times, for 10 minutes each, with PBST at 25 °C. Secondary antibody, goat anti-rabbit HRP (Pierce), was added at 1:1,000 also in 5% non-fat dry milk in PBST at 25 °C for 1 hour. The blot was again washed 3 times, for 10 minutes each, with PBST at 25 °C, and developed using SuperSignal West Femto Maximum Sensitivity Substrate (Pierce) and imaged using a Fluor-S MultiImager (Bio-Rad). Negative control reactions in which ATPγS was not added or PNBM was not added were run to confirm that there was no cross-reactivity with alkylated cysteines or other protein epitopes.
Results and Discussion
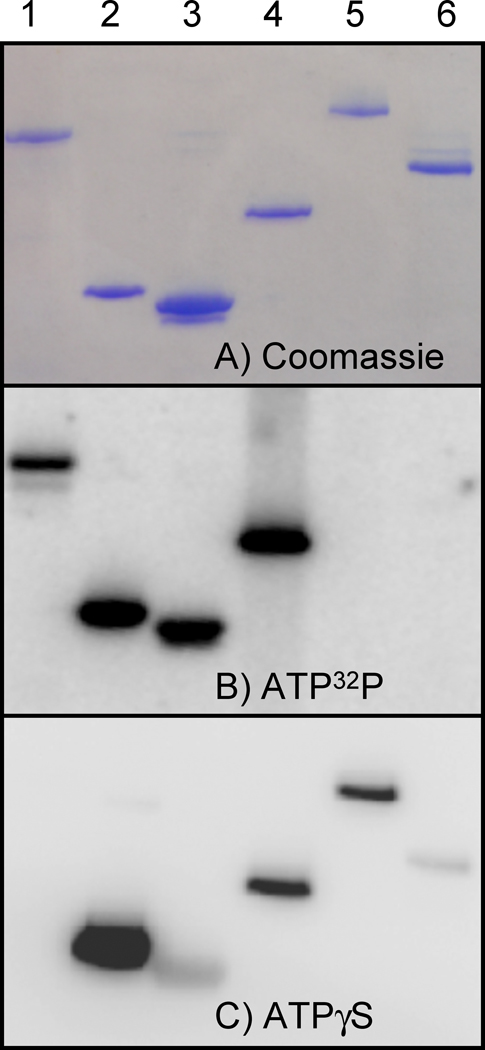
The panel of histidine kinases chosen for this study was largely based on previous work to study the regulation of histidine kinases by adjacent Heme-Nitric Oxide OXygen sensor (H-NOX) proteins. In that work, we demonstrated that the H-NOX from Shewanella oneidensis regulates the activity of a histidine kinase found in the same operon (14). Unfortunately, attempts to show similar regulation in proteins from other organisms have proven more difficult. Figure 1 illustrates the fact that many histidine kinases that do not form a stable phosphohistidine will form a stable thiophosphohistidine. For comparison, the well studied histidine kinases Thermotoga maritima CheA and Escherichia coli NtrB are shown in lanes 1 and 2, respectively. Lanes 3–6 show the H-NOX associated histidine kinases from Shewanella oneidensis, Legionella pneumophila, Hahella chejuensis, and Vibrio fischeri. Of particular note are the histidine kinases from Vibrio fischeri and Hahella chejuensis. Neither of these proteins form a stable phosphohistidine with ATP, but both form stable thiophosphohistidines with ATPγS. In the case of the Hahella kinase, though no stable autophosphorylated protein could be detected, the thiophosphate was stable overnight at 25 °C (Supplemental Fig. 2). Also of interest is the CheA protein from Thermotoga maritima. It does not accept ATPγS as a substrate (Figure 1 and Supplemental Figure 1). Given that the conditions used for autophosphorylation and autothiophosphorylation reactions are identical except for the nucleotide used, the stability of the thiophosphate is likely what allows for detection in cases where the phosphorylated protein cannot be detected. In some of the histidine kinases, such as the H-NOX associated kinase from Shewanella oneidensis, the intensity of the signal in the ATP32P gel is different from the signal in the ATPγS Western blot (Figure 1). This can be explained by either a difference in the ability of the protein to accept ATPγS rather than ATP or by differences in the degree of PNBM alkylation or antibody recognition of the alkylated thiophosphate in the protein. In the case of the Shewanella kinase, the differences in Figure 1 are likely explained by differences in alkylation or recognition of the epitope, as the protein was exceptionally active with ATPγ35S when compared with other the proteins in the panel, but much less active in the ATPγS antibody assay (Supplemental Figure 1).
Figure 1.
A panel of six purified histidine kinases were tested using radioactive ATP32P and the ATPγS antibody assay. A) Coomassie stained gel of the panel of purified histidine kinases. B) Phosphorimage of radioactive gel showing the panel of histidine kinases tested using ATP32P. C) Western blot showing the panel of histidine kinases tested using the ATPγS antibody assay. Lane 1: T. maritima CheA, 2: E. coli NtrB and lanes 3–6: H-NOX associated kinases from S. oneidensis, L. pneumophila, H. chejuensis, and V. fischeri respectively. The molecular weights of the proteins are as follows: T. maritima CheA: 75.5 kDa, E. coli NtrB: 23.6 kDa, SO2144: 34.9 kDa, LPG2458: 49.2 kDa, HCH03701: 87.4 kDa, VFA0072: 65.3 kDa
We did not determine which isomers of the thiophosphohistidine were detected in the proteins we tested. However, given that the antibody was initially raised to detect thiophosphate esters of hydroxy amino acids (11, 12), the epitope recognized by the antibody is unlikely to include any part of the amino acid. This should permit detection of either 1-thiophosphohistidine or 3-thiophosphohistidine.
It should also be noted that the Vibrio fischeri and Hahella chejuensis histidine kinases are hybrid kinases, and have a receiver domain which may contribute to the rapid hydrolysis rate. In addition, the aspartic acid residues in the receiver domains of the hybrid kinases may be thiophosphorylated and, therefore, contribute to the overall signal detected for these proteins in the assays. From our experiments, we conclude that because 1) the reactivity of the antibody towards the alkylated thiophosphate may vary in the context of different proteins, and 2) the rate of ATPγS turnover may vary for different proteins, care should be taken when comparing the relative autothiophoshorylation rates between proteins using this assay.
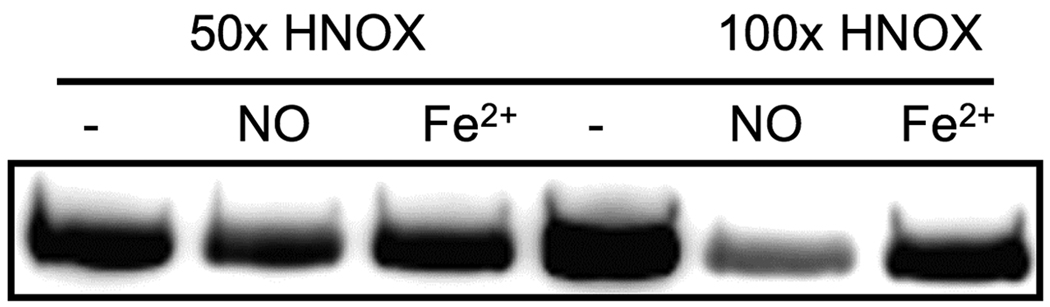
In our previous work with H-NOX/histidine kinase pairs, we found that the H-NOX domain inhibits the autophosphorylation of the histidine kinase in the NO ligated state, but not in the reduced, unligated state (14) and H.K. Carlson (unpublished work). This was further tested for other bacterial H-NOX proteins, and a similar regulation was found for the H-NOX-kinase pair from Hahella chejuensis (Figure 2). At 100-fold excess of the H-NOX, the reduced, unligated state of the protein inhibited the autothiophosphorylation rate of the Hahella chejunsensis histidine kinase to a much lesser extent than the NO ligated state (Figure 2). Autophosphorylation of the Hahella kinase in the presence or absence of the Hahella H-NOX protein using ATP was not observed (Figure 1). This is a clear example of the utility of the ATPγS method in studying the regulatory mechanism of challenging, novel phosphotransfer pathways.
Figure 2.
The autophosphorylation activity of the H-NOX-associated kinase from Hahella chejuensis is inhibited by the addition of H-NOX in the Fe2+-NO ligation state, but to a lesser extent by the Fe2+ unligated state. Kinase (1 µM) was incubated for 1.5 hours with 500 µM ATPγS and 2.5 mM MgCl2 in the presence or absence of an excess of H-NOX in either the Fe2+-NO or the Fe2+ unligated states. The inhibition of autophosphorylation activity by the NO bound H-NOX for the Hahella kinase is similar to that seen in other H-NOX-associated kinases that are stably phosphorylated upon incubation with radioactive ATP32P.
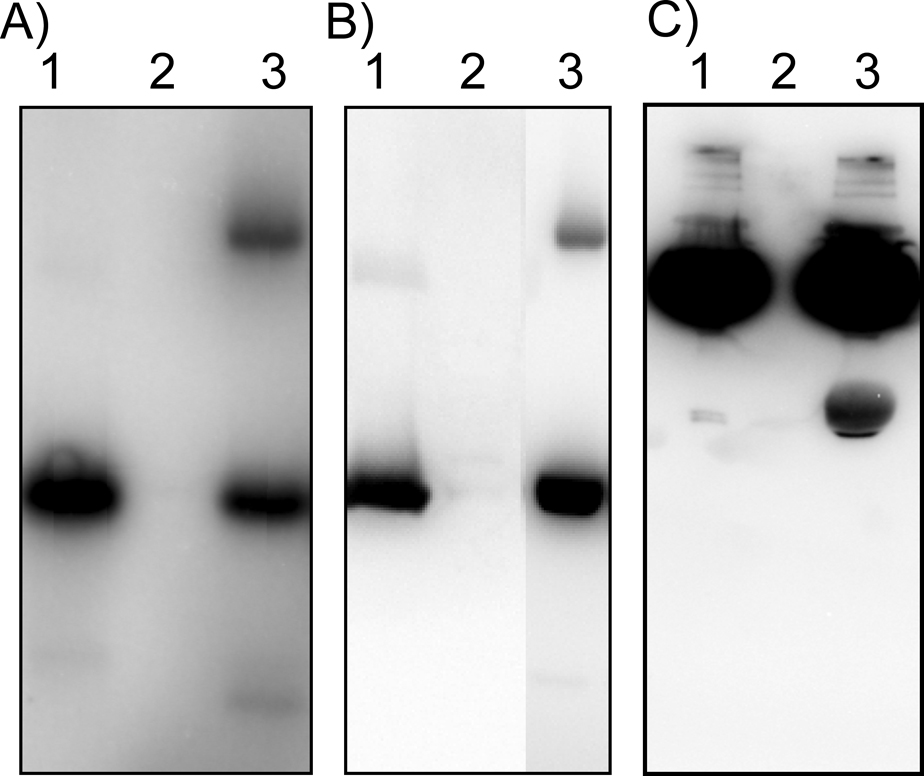
In contrast to many serine, threonine and tyrosine kinases, histidine kinases autophosphorylate prior to transferring the phosphate to a response regulator. This poses some problems for the ATPγS method to study phosphotransfer. The thiophosphohistidine is expected to be more stable to hydrolysis and might also display sluggish phosphotransfer kinetics. However, the transfer of thiophosphate from the histidine kinase EnvZ to its cognate response regulator OmpR was observed, as well as the transfer of thiophosphate from the H-NOX-associated kinase from Hahella chejuensis to the response regulator in the same predicted operon (Figure 3). Transfer from EnvZ to OmpR was detectable at equal concentrations (5 µM), but higher concentrations of the Hahella kinase (20 µM) were needed to observe transfer to its response regulator. A method to selectively detect the downstream partner of a histidine kinase would be broadly useful in the discovery of prokaryotic phosphotransfer pathways. Present methods for determining the cognate response regulator for a histidine kinase require purification of all response regulators from an organism and determining the kinetic preference for phosphotransfer (17).
Figure 3.
Thiophosphate transfer from histidine kinases to response regulators. A) reaction with ATP32P showing phosphate transfer from the histidine kinase EnvZ to MBP-tagged OmpR. Lane 1: EnvZ, 2: MBP-OmpR alone, 3: MBP-OmpR + EnvZ [5 µM EnvZ, 5 µM MBP-OmpR]. B) reaction using ATPγS and the antibody assay showing thiophosphate transfer from the histidine kinase EnvZ to MBP-tagged OmpR. Lane 1: EnvZ, 2: MBP-OmpR alone, 3: MBP-OmpR + EnvZ [5 µM EnvZ, 5 µM MBP-OmpR]. C) thiophosphate transfer from the H-NOX associated histidine kinase in Hahella chejuensis to the response regulator found in the same predicted operon. Lane 1: Hahella histidine kinase, 2: Hahella response regulator alone, 3: Hahella histidine kinase + response regulator [20 µM Hahella histidine kinase, 5 µM Hahella response regulator].
Conclusion
The assay method described in this paper is likely to be broadly useful for studying the activity of histidine kinases—both as an alternative to radioactive ATP, and as a means to probe the activity of histidine kinases that do not accept ATP to form stable phospohistidines. Unlike phosphospecific antibody methods used in eukaryotic kinase studies, where each antibody exhibits specificity for the specific phosphorylated residue based on the neighboring amino acids, the thiophosphate-PNBM specific antibody is context independent and can be used for detection of the phosphate modification in any protein context. Further work is necessary to determine the utility of this assay for characterizing new phosphotransfer pathways. However, the transfer of thiophosphate from EnvZ to OmpR and between the Hahella kinase and response regulator are very promising results. Further work will also focus on using this technique for the preparation of thiophosphorylated proteins as an alternative to BeF3 for structural studies or for mass spectrometry.
Supplementary Material
Supplemental Figure 1. All kinases that react with the ATPγS antibody are also reactive with ATPγ35S. The CheA from Thermotoga maritima is not reactive. Lane 1: Thermotoga maritima CheA, 2: E. coli NtrB, and lanes 3–5: H-NOX associated kinases from H. chejuensis, S. oneidensis and V. fischeri respectively. L. pneumophila kinase is poorly expressed and was was included in this assay.
Supplemental Figure 2. Stability of thiophosphohistidine in the H-NOX associated kinase from Hahella chejuensis. Pre-thiophosphorylated kinase (5 µM), separated from ATPγS by three 100-fold dilution/concentration cycles in a Vivaspin 500 10 K MWCO spin concentrator (Sartorius Stedim Biotech) was allowed to hydrolyze overnight in buffer with 10 mM MgCl2 but lacking ATPγS. Timepoints were analyzed using the ATPγS antibody assay. This histidine kinase forms a stable phosphohistidine, and the thiophosphohistidine is only hydrolyzed to a small degree over 20 hours.
Acknowledgements
The authors are very grateful to Doug Mitchell a former member of the Marletta lab who suggested that we try this approach. In addition, we would like to thank members of the Marletta and Shokat labs for stimulating discussion and advice.
Funding
This work was supported by NIH grants GM070671 to MAM and EB001987 to KMS. Thanks to Epitomics for generously providing antibody 51-8 for these studies.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annu Rev Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 2.West AH, Stock AM. Histidine kinases and response regulator proteins in two-component signaling systems. Trends Biochem Sci. 2001;26:369–376. doi: 10.1016/s0968-0004(01)01852-7. [DOI] [PubMed] [Google Scholar]
- 3.Porter SL, Armitage JP. Chemotaxis in Rhodobacter sphaeroides requires an atypical histidine protein kinase. J Biol Chem. 2004;279:54573–54580. doi: 10.1074/jbc.M408855200. [DOI] [PubMed] [Google Scholar]
- 4.Kinoshita E, Kinoshita-Kikuta E, Takiyama K, Koike T. Phosphate-binding tag, a new tool to visualize phosphorylated proteins. Mol Cell Proteomics. 2006;5:749–757. doi: 10.1074/mcp.T500024-MCP200. [DOI] [PubMed] [Google Scholar]
- 5.Yamada S, Nakamura H, Kinoshita E, Kinoshita-Kikuta E, Koike T, Shiro Y. Separation of a phosphorylated histidine protein using phosphate affinity polyacrylamide gel electrophoresis. Anal Biochem. 2007;360:160–162. doi: 10.1016/j.ab.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 6.Barbieri CM, Stock AM. Universally applicable methods for monitoring response regulator aspartate phosphorylation both in vitro and in vivo using Phos-tag-based reagents. Anal Biochem. 2008;376:73–82. doi: 10.1016/j.ab.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duclos B, Marcandier S, Cozzone AJ. Chemical properties and separation of phosphoamino acids by thin-layer chromatography and/or electrophoresis. Methods Enzymol. 1991;201:10–21. doi: 10.1016/0076-6879(91)01004-l. [DOI] [PubMed] [Google Scholar]
- 8.Hultquist DE. The preparation and characterization of phosphorylated derivatives of histidine. Biochim Biophys Acta. 1968;153:329–340. doi: 10.1016/0005-2728(68)90078-9. [DOI] [PubMed] [Google Scholar]
- 9.Besant PG, Attwood PV. Detection and analysis of protein histidine phosphorylation. Mol Cell Biochem. 2009;329:93–106. doi: 10.1007/s11010-009-0117-2. [DOI] [PubMed] [Google Scholar]
- 10.Lasker M, Bui CD, Besant PG, Sugawara K, Thai P, Medzihradszky G, Turck CW. Protein histidine phosphorylation: increased stability of thiophosphohistidine. Protein Sci. 1999;8:2177–2185. doi: 10.1110/ps.8.10.2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wolanin PM, Webre DJ, Stock JB. Mechanism of Phosphatase Activity in the Chemotaxis Response Regulator CheY. Biochemistry. 2003;42:14075–14082. doi: 10.1021/bi034883t. [DOI] [PubMed] [Google Scholar]
- 12.Allen JJ, Li M, Brinkworth CS, Paulson JL, Wang D, Hubner A, Chou W-H, Davis RJ, Burlingame AL, Messing RO, Katayama CD, Hedrick SM, Shokat KM. A semisynthetic epitope for kinase substrates. Nature Meth. 2007;4:511–516. doi: 10.1038/nmeth1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Allen JJ, Lazerwith SE, Shokat KM. Bio-orthogonal Affinity Purification of Direct Kinase Substrates. J. Am. Chem. Soc. 2005;127:5288–5289. doi: 10.1021/ja050727t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Price MS, Chao LY, Marletta MA. Shewanella oneidensis MR-1 H-NOX regulation of a histidine kinase by nitric oxide. Biochemistry. 2007;46:13677–13683. doi: 10.1021/bi7019035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoshida T, Phadtare S, Inouye M. Functional and structural characterization of EnvZ, an osmosensing histidine kinase of E. coli. Methods Enzymol. 2007;423:184–202. doi: 10.1016/S0076-6879(07)23008-3. [DOI] [PubMed] [Google Scholar]
- 16.Busso D, Delagoutte-Busso B, Moras D. Construction of a set Gateway-based destination vectors for high-throughput cloning and expression screening in Escherichia coli. Analytical Biochemistry. 2005;343:313–321. doi: 10.1016/j.ab.2005.05.015. [DOI] [PubMed] [Google Scholar]
- 17.Laub MT, Biondi EG, Skerker JM. Phosphotransfer profiling: systematic mapping of two-component signal transduction pathways and phosphorelays. Methods Enzymol. 2007;423:531–548. doi: 10.1016/S0076-6879(07)23026-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. All kinases that react with the ATPγS antibody are also reactive with ATPγ35S. The CheA from Thermotoga maritima is not reactive. Lane 1: Thermotoga maritima CheA, 2: E. coli NtrB, and lanes 3–5: H-NOX associated kinases from H. chejuensis, S. oneidensis and V. fischeri respectively. L. pneumophila kinase is poorly expressed and was was included in this assay.
Supplemental Figure 2. Stability of thiophosphohistidine in the H-NOX associated kinase from Hahella chejuensis. Pre-thiophosphorylated kinase (5 µM), separated from ATPγS by three 100-fold dilution/concentration cycles in a Vivaspin 500 10 K MWCO spin concentrator (Sartorius Stedim Biotech) was allowed to hydrolyze overnight in buffer with 10 mM MgCl2 but lacking ATPγS. Timepoints were analyzed using the ATPγS antibody assay. This histidine kinase forms a stable phosphohistidine, and the thiophosphohistidine is only hydrolyzed to a small degree over 20 hours.