Abstract
Polyamines are important in both prokaryotic and eukaryotic translational systems. Spermine is a quaternary aliphatic amine that is cationic under physiological conditions. In this paper, we demonstrate that spermine stimulates fMet-tRNA binding to mammalian mitochondrial 55S ribosomes. The stimulatory effect of spermine is independent of the identity of the mRNA. The degree of stimulation of spermine is the same at all concentrations of mitochondrial initiation factor 2 (IF2mt) and mitochondrial initiation factor 3 (IF3mt). This observation indicates that IF2mt and IF3mt, while essential for initiation, are not the primary components of the translation initiation system affected by spermine. IF3mt dissociates 55S ribosomes detectably in the absence of spermine, but this effect is strongly inhibited in the presence of spermine. This observation indicates that the positive effect of spermine on initiation is not due to an increase in the availability of the small subunits for initiation. Spermine also promotes fMet-tRNA binding to small subunits of the mitochondrial ribosome in the presence of IF2mt. The major effect of spermine in promoting initiation complex formation thus appears to be on the interaction of fMet-tRNA with the ribosome.
Keywords: Spermine, Mitochondria, Translation, Initiation, Polyamine, Ribosome
Introduction
The initiation of protein synthesis in mitochondria has many features in common with that in prokaryotes; however, it is also characterized by several important differences. First, prokaryotic ribosomes exist as 70S monomers that are in equilibrium with 50S and 30S subunits [1]. Mammalian mitochondrial monosomes sediment at 55S and dissociate to form 28S and 39S subunits. These ribosomal subunits have small rRNAs and are highly enriched in ribosomal proteins. Second, protein synthesis in mitochondria occurs with the help of two initiation factors, instead of the three factors found in the bacterial system. In mitochondria, initiation factor 2 (IF2mt) promotes the binding of fMet-tRNA to the small subunit, and initiation factor 3 (IF3mt) stimulates initiation complex formation by facilitating the dissociation of mitochondrial 55S ribosomes [2,3]. While these two factors are also present in prokaryotes, an additional factor (initiation factor 1, IF1) exists in prokaryotes that is not present in mitochondria. IF2mt has a 37 amino acid insertion not present in bacterial IF2, which is believed to function in place of IF1 [4].
Spermine and spermidine are aliphatic polyamines that have a number of roles in both prokaryotic and eukaryotic cells. Both polyamines are cationic at physiological pH due to pK values >8 [5]. In prokaryotic cells, polyamines are known to affect protein synthesis in several ways. The structure of the Shine-Dalgarno region on the mRNA is rearranged in the presence of polyamines, which helps promote the formation of the prokaryotic initiation complex [6]. In addition to this structural effect, polyamines lower the optimal concentration of Mg2+ required for protein synthesis. Translation initiation on the minor start codon UUG, used, for example, in the translation of adenylate cyclase, is stimulated by polyamines [7]. The tertiary polyamine spermidine reduces the dissociation of 70S ribosomes, partially counteracting the action of initiation factor 3 (IF3) in the initiation of translation [8]. Spermine, a quaternary polyamine, has binding sites on both 16S and 23S rRNA, indicating that it could be important for ribosomal structure [9,10]. Spermine crosslinks near the decoding center of the small subunit, also suggesting that it could be involved in aminoacyl-tRNA binding [9].
In wheat germ, spermidine has been shown to affect translation by increasing the fidelity of protein synthesis [11]. Spermine exists in the cytosol of eukaryotic cells with concentrations in the μM range [12] and is known to be present in mitochondria although it is not synthesized there [13]. In fact, spermine plays a metabolic role in mitochondria by stimulating the activity of the pyruvate dehydrogenase complex and by activating citrate synthase in the citric acid cycle [13]. Because spermine binds to the mitochondrial membrane, the concentration of spermine inside the mitochondria is difficult to measure accurately and is, therefore, not known [14].
Spermine affects translation in mitochondria by lowering the Mg2+ requirement for the association of mitochondrial ribosomal subunits into 55S monosomes by about 1 mM [15]. Little is known about other effects that spermine may have on protein synthesis in mitochondria. Because mitochondrial mRNAs are largely leaderless, the effects of spermine on mitochondrial translation may be different from those observed with prokaryotic mRNAs, most of which contain Shine-Dalgarno sequences. In this report, we systematically examine the effects of spermine on initiation of protein synthesis in mammalian mitochondria.
Materials and Methods
Materials
General chemicals were purchased from Sigma-Aldrich or Fisher Scientific. Bovine mitochondrial ribosomes (55S), ribosomal subunits (28S and 39S), and yeast [35S]fMet-tRNA were prepared as described [16–19]. Poly(A,U,G) was synthesized and purified as described [20]. The AUG triplet was purchased from Dharmacon. A 29-mer RNA oligonucleotide containing the 5' AUG start site (underlined) for subunit I of bovine mitochondrial cytochrome oxidase (CoI): AUGUUCAUUAACCGCUGACUAUUCUCAAC was also purchased from Dharmacon.
Cloning, expression and purification
IF2mt and IF3mt were cloned, expressed, and purified as described previously [21]. Both proteins required further purification following the Ni-NTA column. IF2mt and IF3mt were purified on DEAE-5PW and SP-5PW HPLC columns, respectively, as described [21].
Dissociation of mitochondrial 55S ribosomes by IF3mt in the presence and absence of spermine
Mitochondrial ribosomes (8 pmol, 80 nM) were incubated in the presence or absence of IF3mt (41 pmol, 410 nM) and in the presence and absence of spermine (0.1 mM) in 100 μL of gradient buffer (25 mM Tris-HCl, pH 7.6, 5 mM MgCl2, 40 mM KCl and 1 mM dithiothreitol) for 15 min at 37 °C. After incubation, reaction mixtures were placed on ice for 10 min and then layered onto cold 4.8 mL 10–30% linear sucrose gradients prepared in gradient buffer with and without 0.1 mM spermine. Gradients were centrifuged for 1 h and 45 min at 220,000 gave in the Beckman SW55 Ti rotor and fractionated as described [22].
Initiation complex formation on mitochondrial ribosomes
Stimulation of initiation complex formation by spermine was examined by measuring the increase of [35S]fMet-tRNA binding to 55S ribosomes or 28S subunits in a filter binding assay. Reaction mixtures (100 μL) were prepared as described previously [19,23] and contained the indicated amounts of IF2mt and/or IF3mt, [35S]fMet-tRNA (60 nM), 0.25 mM GTP, 1.25 mM phosphoenolpyruvate, 0.9 U pyruvate kinase, 55S ribosomes (60 nM) and, unless otherwise indicated, 10 μg poly(A,U,G). Mixtures were incubated for 10 min at 37 °C. The amount of [35S]fMet-tRNA bound to ribosomes was measured using a nitrocellulose filter binding assay [2,23].
Results and Discussion
Effect of spermine on initiation complex formation on mitochondrial 55S ribosomes
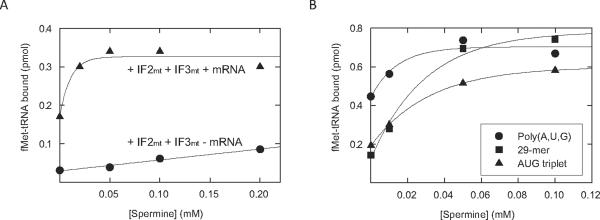
In the absence of mRNA, fMet-tRNA is still able to bind mitochondrial ribosomes to a limited extent [23]. Thus, the effect of spermine on the binding of fMet-tRNA to mitochondrial 55S ribosomes was tested in the presence and absence of mRNA (Figure 1). In the presence of both IF2mt, IF3mt, and poly(A,U,G), a two-fold increase in fMet-tRNA binding correlated with increasing concentrations of spermine up to 0.05 mM (Figure 1A). This result indicates that spermine has a positive effect on initiation. This effect could occur at one or more of several steps, including: fMet-tRNA binding, mRNA binding, the action of IF2mt, or the action of IF3mt.
Figure 1. Effect of spermine on initiation complex formation on 55S ribosomes in the presence and absence of various mRNAs.
A. [35S]fMet-tRNA binding to mitochondrial 55S particles (6 pmol, 60 nM) was tested in the presence and absence of 10 μg poly(A,U,G) mRNA and the presence of IF2mt (8 pmol, 80 nM) and IF3mt (8 pmol, 80 nM) at different concentrations of spermine. B. [35S]fMet-tRNA binding to mitochondrial 55S ribosomes was tested in the presence of saturating amounts of poly(A,U,G) (5 μg), the 29-mer cytochrome oxidase start site (2.24 μM), or the AUG triplet (6 μg, 53 μM) at different concentrations of spermine as described in Materials and Methods.
If spermine primarily affects fMet-tRNA binding by promoting the interaction of the mRNA with the ribosome, one might expect a smaller effect of spermine on fMet-tRNA binding in the absence of mRNA. Binding of fMet-tRNA to ribosomes was significantly lower in the absence of mRNA (Figure 1A); however, spermine stimulated this binding two-fold, as observed in the presence of mRNA, although higher concentrations of spermine were required to observe this effect. This observation suggests that spermine is affecting a step in initiation distinct from the mRNA binding step.
To further explore this conclusion, we have examined the effects of spermine on initiation complex formation with three different mRNAs (Figure 1B). These mRNAs included the AUG triplet, a 29-nucleotide RNA derived from the initiation site of bovine mitochondrial subunit I of cytochrome oxidase (CoI), and poly(A,U,G). The assays shown in Figure 1A were carried out using poly(A,U,G), a long RNA polymer that is not representative of natural mitochondrial mRNAs. Native mitochondrial messages contain few or no nucleotides 5' to the initiation codon, so the AUG triplet and the leaderless CoI mRNA were used.
In the absence of polyamines, substantial binding of fMet-tRNA to the ribosome was observed with poly(A,U,G). Spermine stimulated this binding nearly two-fold. When the AUG triplet was used in initiation complex formation (Figure 1B), less fMet-tRNA binding was observed in the absence of spermine, and this polylamine stimulated binding about three-fold, bringing it to the level observed with poly(A,U,G). To examine the effect of spermine on a natural leaderless initiation site, we used the 29-mer synthetic oligonucleotide based on the initiation site of the CoI mRNA. When this oligonucleotide was tested for initiation complex formation in the absence of spermine, it showed approximately the same level of binding as observed with the AUG triplet (Figure 1B). Again, spermine stimulated binding about three-fold.
The more substantial binding obtained in the absence of spermine with poly(A,U,G) probably reflects the ability of this long polymer to trap fMet-tRNA more effectively on the ribosome. Long mRNAs are known to bind more rapidly to the small subunit and to dissociate more slowly than shorter mRNAs [24]. Since stable initiation complex formation requires the simultaneous presence of both fMet-tRNA and mRNA on the ribosome, the more stable binding of poly(A,U,G) to the ribosome allows more initiation complex formation to be detected in the absence of spermine. Regardless, spermine stimulates fMet-tRNA binding in the absence of any mRNA (Figure 1A) and in the presence of all the mRNAs tested (Figure 1B), indicating that the effect of this polyamine on initiation is not directly on mRNA binding to the ribosome.
Effect of spermine on IF3mt-mediated stimulation of initiation complex formation
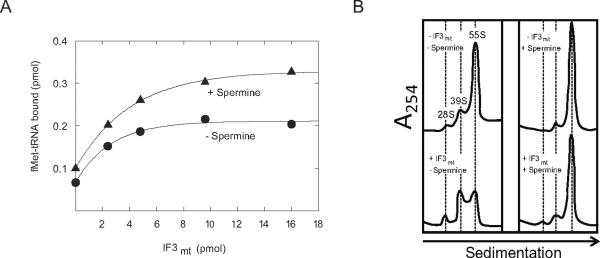
Another potential target for the effect of spermine is on the action of IF3mt. The major effect of IF3mt on initiation complex formation is to loosen the interactions between the ribosomal subunits, making the small subunit available for fMet-tRNA binding. Since spermine is expected to tighten subunit interactions, one might predict that spermine would increase the amount of IF3mt required for initiation. To test this theory, fMet-tRNA binding to mitochondrial ribosomes was tested in the presence of IF2mt and with increasing amounts of IF3mt (Figure 2A). In the presence of IF2mt and mRNA, the addition of IF3mt caused a 2-fold increase in fMet-tRNA binding in the absence of spermine (Figure 2A). This stimulation is believed to arise because IF3mt promotes the dissociation of ribosomal subunits and, therefore, enhances initiation complex formation. Surprisingly, the addition of spermine did not result in a need for a higher concentration of IF3mt to promote initiation complex formation. In the presence of spermine, IF3mt increased the amount of fMet-tRNA bound to 55S ribosomes with basically the same dose response curve as observed in the absence of spermine (Figure 2A). Spermine caused a 1.4-fold stimulation of fMet-tRNA binding independent of the amount of IF3mt added. This observation suggests that the effect of spermine is not directly on IF3mt.
Figure 2. Effect of spermine on IF3mt-induced 55S dissociation.
A. [35S]fMet-tRNA binding to mitochondrial 55S particles (8 pmol, 80 nM) was tested at different concentrations of IF3mt with and without spermine (0.1 mM) using 10 μg poly(A,U,G) as the mRNA at a fixed concentration of IF2mt (8 pmol, 80 nM). B. Fractionation profiles of mitochondrial 55S ribosomes after centrifugation on 10–30% sucrose gradients in 5 mM Mg2+. Mitochondrial 55S ribosomes (8 pmol, 80 nM) were incubated at 5 mM MgCl2 with or without spermine (0.1 mM) in the absence or presence of IF3mt (41 pmol, 410 nM) at 37 °C for 15 min. The samples were then placed on ice for 10 min and subsequently layered on a 10–30% sucrose density gradient (gradients were prepared with and without spermine, respectively, corresponding to the reaction conditions used) and analyzed as described in Materials and Methods.
In prokaryotes, spermine has been shown to change the conformation of the 30S ribosomal subunit to favor subunit association [9]. It also reduces the electrostatic repulsion between the subunits by masking the charges on the rRNA [25]. In both prokaryotes and mitochondria, spermine has been shown to tighten the interaction between the ribosomal subunits [15,26]. The effect of spermine on the ability of IF3mt to promote subunit dissociation was tested directly using sucrose density gradient centrifugation (Figure 3B). In the absence of both IF3mt and spermine, mitochondrial ribosomes existed primarily as 55S particles at 5 mM Mg2+, although a small amount of dissociation was observed (Figure 3B). The addition of spermine led to a stronger interaction between the ribosomal subunits, and very few subunits were observed in its presence. In the absence of spermine, IF3mt was able to promote the dissociation of ribosomal subunits. However, in the presence of spermine, IF3mt had almost no effect on the dissociation of 55S ribosomes. Thus, spermine appears to play a role in tightening the interactions between the mitochondrial 28S and 39S subunits. It is possible that, in the presence of spermine, IF3mt loosens mitochondrial subunit contacts enough to allow initiation complex formation but not enough to stably dissociate the ribosomal subunits. This observation argues that the effect of spermine is not on the action of IF3mt in promoting ribosomal subunit dissociation.
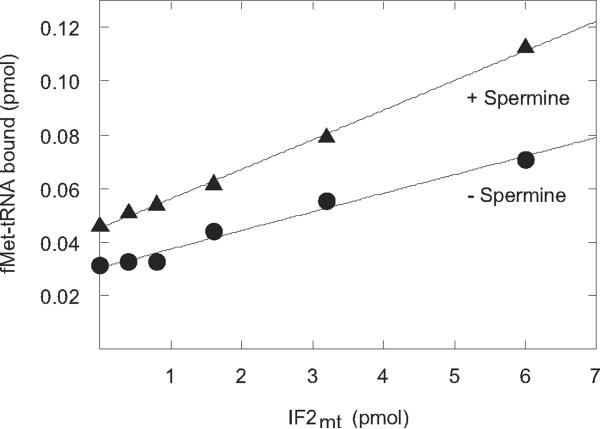
Figure 3. Effect of IF2mt on initiation complex formation in the presence and absence of spermine.
[35S]fMet-tRNA binding to mitochondrial 55S particles (6 pmol, 60 nM) was tested with and without spermine (0.1 mM) the indicated concentrations of IF2mt.
Effect of spermine on the action of IF2mt in fMet-tRNA binding to 55S ribosomes
IF2mt is known to stabilize the binding of fMet-tRNA to the P-site of the ribosome and, as expected, the amount of fMet-tRNA bound to the ribosome was quite low in the absence of IF2mt (Figure 3). However, even in the absence of IF2mt, spermine enhanced the binding of fMet-tRNA to the ribosome. This observation is in agreement with the data above, suggesting that the effect of spermine is directly on fMet-tRNA binding to the P-site. The presence of spermine did not significantly alter the stimulatory effect of IF2mt. As the IF2mt concentration was increased, considerable stimulation of fMet-tRNA binding was observed as expected. However, the degree of stimulation of fMet-tRNA binding to the ribosome caused by spermine (1.6-fold) was constant at all concentrations of IF2mt. This observation suggests that spermine does not directly affect the action of IF2mt.
Effect of spermine on fMet-tRNA binding to 28S subunits
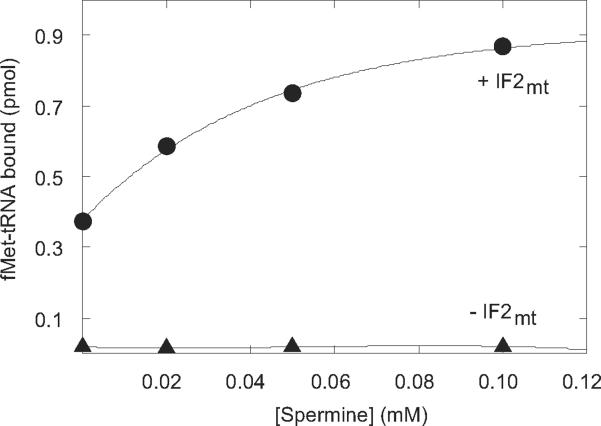
The data above demonstrate that the effect of spermine is on fMet-tRNA binding to the ribosome; however, the question arises as to whether spermine affects fMet-tRNA interactions on the small or large ribosomal subunit. If fMet-tRNA binding to the small subunit is being affected, the same increase in fMet-tRNA binding should be observed on 28S subunits that was seen on 55S ribosomes. In the presence of increasing amounts of spermine, an increase in fMet-tRNA binding was observed in the presence of IF2mt (Figure 4). IF3mt was not required in this assay, since no dissociation of ribosomal subunits was needed. The stimulatory effect of spermine was absolutely dependent on IF2mt, reflecting the lower stability of fMet-tRNA bound to the small subunit alone. However, the same two-fold stimulation of fMet-tRNA binding by spermine was observed on both 28S subunits and 55S ribosomes. Thus, the full effect of spermine was seen on 28S subunits and can be attributed to a direct stimulation of fMet-tRNA binding to the P-site of the small ribosomal subunit.
Figure 4. Effect of spermine on initiation complex formation on 28S subunits in the presence and absence of initiation factors.
[35S]fMet-tRNA binding to mitochondrial 28S subunits (4 pmol, 40 nM) was tested using 10 μg poly(A,U,G) as the mRNA in the presence and absence of IF2mt (14 pmol, 140 nM) at different concentrations of spermine.
Effect of polyamines on fMet-tRNA binding to prokaryotic and mitochondrial ribosomes
Polyamines have been shown to be important for prokaryotic translation initiation, and the current work demonstrates that spermine is also important in the mitochondrial translation system. Because spermine was shown to crosslink to the decoding center of the prokaryotic ribosome, it was assumed that the effect of spermine could be on fMet-tRNA binding to the small ribosomal subunit. However, it was unclear whether the effect of spermine was primarily on the interaction of the tRNA with the rRNA or the proteins of the small subunit. The crystal structure of the Thermus thermophilus ribosome demonstrates that the P-site bound tRNA has several important contacts with the small subunit of the ribosome that could be affected by the presence of spermine [27]. Ten of those contacts are with rRNA nucleotides on the small subunit, and these ten residues are present in the truncated mitochondrial 12S rRNA. Spermine could enhance these interactions by shielding the repulsion between the backbones of the tRNA and the rRNA. The tRNA in the P-site of the prokaryotic ribosome only interacts with one small subunit protein (S13), which is not present in mitochondrial ribosomes. However, additional protein contacts are made between the mitochondrial small subunit and the P-site tRNA [18]. An interaction occurs between the T-loop of the tRNA bound at the P-site and the P-site finger protein, a structure unique to the mitochondrial system [18]. A further interaction is observed between the CCA-arm of the tRNA and an unknown small subunit protein. These protein interactions, unique to the mitochondrial system, could also be enhanced by spermine.
Conclusion
Initiation of translation in mitochondria is stimulated by the presence of spermine, a polyamine that occurs naturally in mitochondria. The stimulatory effect of spermine does not appear to depend on the mRNA used, the action of IF3mt, or the action of IF2mt. The data suggest that the binding of fMet-tRNA to the P-site of mitochondrial 55S ribosomes is directly stabilized by spermine. This effect could be due to the alteration of the conformations of the ribosomal subunits by spermine or to a direct stabilization of the contacts between fMet-tRNA and the ribosome. The observation that the full effect of spermine is observed on 28S subunits indicates that this polyamine is primarily enhancing the interaction of the fMet-tRNA with the small subunit. Future work on translation in mitochondria should carefully consider the spermine concentration used.
Acknowledgments
Funds for this project, but no other contribution, were provided by NIH grant GM32734 to LLS.
Abbreviations
- (IF3)
Initiation factor 3
- (IF2)
initiation factor 2
- (IF3mt)
mitochondrial initiation factor 3
- (IF2mt)
mitochondrial initiation factor 2
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Spremulli LL, Coursey A, Navratil T, Hunter SE. Initiation and elongation factors in mammalian mitochondrial protein synthesis. In: Moldave K, editor. Prog. Nuc. Acid Res. Mol. Biol. Elsevier; 2004. pp. 211–261. [DOI] [PubMed] [Google Scholar]
- [2].Koc EC, Spremulli LL. Identification of mammalian mitochondrial translational initiation factor 3 and examination of its role in initiation complex formation with natural mRNAs. J. Biol. Chem. 2002;277:35541–35549. doi: 10.1074/jbc.M202498200. [DOI] [PubMed] [Google Scholar]
- [3].Liao H-X, Spremulli LL. Initiation of protein synthesis in animal mitochondria: Purification and characterization of translational initiation factor 2. J. Biol. Chem. 1991;266:20714–20719. [PubMed] [Google Scholar]
- [4].Gaur R, Grasso D, Datta PP, Krishna PDV, Das G, Spencer A, Agrawal RK, Spremulli L, Varshney U. A single mammalian mitochondrial translation initiation factor functionally replaces two bacterial factors. Mol. Cell. 2008;29:180–190. doi: 10.1016/j.molcel.2007.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Takeda Y, Samejima K, Nagano K, Watanabe M, Sugeta H, Kyogoku Y. Determination of protonation sites in thermospermine and in some other polyamines by 15N and 13C nuclear magnetic resonance spectroscopy. Eur. J. Biochem. 1983;130:383–389. doi: 10.1111/j.1432-1033.1983.tb07164.x. [DOI] [PubMed] [Google Scholar]
- [6].Yoshida M, Meksuriyen D, Kashiwagi K, Kawai G, Igarashi K. Polyamine stimulation of the synthesis of oligopeptide-binding protein (OppA). Involvement of a structural change of the Shine-Dalgarno sequence and the initiation codon AUG in OppA mRNA. J. Biol. Chem. 1999;274:22723–22728. doi: 10.1074/jbc.274.32.22723. [DOI] [PubMed] [Google Scholar]
- [7].Higashi K, Terui Y, Suganami A, Tamura Y, Nishimura K, Kashiwagi K, Igarashi K. Selective structural change by spermidine in the bulged-out region of double-stranded RNA and its effect on RNA function. J. Biol. Chem. 2008;283:32989–32994. doi: 10.1074/jbc.M806027200. [DOI] [PubMed] [Google Scholar]
- [8].Umekage S, Ueda T. Spermidine inhibits transient and stable ribosome subunit dissociation. FEBS Lett. 2006;580:1222–1226. doi: 10.1016/j.febslet.2006.01.033. [DOI] [PubMed] [Google Scholar]
- [9].Amarantos I, Zarkadis IK, Kalpaxis DL. The identification of spermine binding sites in 16S rRNA allows interpretation of the spermine effect on ribosomal 30S subunit functions. Nuc. Acids Res. 2002;30:2832–2843. doi: 10.1093/nar/gkf404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Xaplanteri MA, Petropoulos AD, Dinos GP, Kalpaxis DL. Localization of spermine binding sites in 23S rRNA by photoaffinity labeling: parsing the spermine contribution to ribosomal 50S subunit functions. Nuc. Acids Res. 2005;33:2792–2805. doi: 10.1093/nar/gki557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Igarashi K, Kashiwagi K, Aoki R, Kojima M, Hirose S. Comparative studies on the increase by polyamines of fidelity of protein synthesis in Escherichia coli and wheat germ cell-free systems. Biochem. Biophys. Res. Commun. 1979;91:440–448. doi: 10.1016/0006-291x(79)91541-9. [DOI] [PubMed] [Google Scholar]
- [12].Igarashi K, Kashiwagi K. Polyamines: Mysterious modulators of cellular functions. Biochem. Biophys. Res. Commun. 2000;271:559–564. doi: 10.1006/bbrc.2000.2601. [DOI] [PubMed] [Google Scholar]
- [13].Toninello A, Salvi M, Mondovi B. Interaction of biologically active amines with mitochondria and their role in the mitochondrial-mediated pathway of apoptosis. Curr. Med. Chem. 2004;11:2349–2374. doi: 10.2174/0929867043364559. [DOI] [PubMed] [Google Scholar]
- [14].Tassani V, Campagnolo M, Toninello A, Siliprandi D. The contribution of endogenous polyamines to the permeability transition of rat liver mitochondria. Biochem. Biophys. Res. Commun. 1996;226:850–854. doi: 10.1006/bbrc.1996.1439. [DOI] [PubMed] [Google Scholar]
- [15].Spremulli LL, Kraus B. Bovine mitochondrial ribosomes: Effect of cations and heterologous dissociation factors on subunit interactions. Biochem. Biophys. Res. Commun. 1987;147:1077–1081. doi: 10.1016/s0006-291x(87)80180-8. [DOI] [PubMed] [Google Scholar]
- [16].Ma J, Spremulli LL. Expression, purification and mechanistic studies of bovine mitochondrial translational initiation factor 2. J. Biol. Chem. 1996;271:5805–5811. doi: 10.1074/jbc.271.10.5805. [DOI] [PubMed] [Google Scholar]
- [17].Matthews DE, Hessler RA, Denslow ND, Edwards JS, O'Brien TW. Protein composition of the bovine mitochondrial ribosome. J. Biol. Chem. 1982;257:8788–8794. [PubMed] [Google Scholar]
- [18].Sharma MR, Koc EC, Datta PP, Booth TM, Spremulli LL, Agrawal RK. Structure of the mammalian mitochondrial ribosome reveals an expanded functional role for its component proteins. Cell. 2003;115:97–108. doi: 10.1016/s0092-8674(03)00762-1. [DOI] [PubMed] [Google Scholar]
- [19].Graves M, Spremulli LL. Activity of Euglena gracilis chloroplast ribosomes with prokaryotic and eukaryotic initiation factors. Arch. Biochem. Biophys. 1983;222:192–199. doi: 10.1016/0003-9861(83)90516-7. [DOI] [PubMed] [Google Scholar]
- [20].Christian B, Spremulli L. Evidence for an active role of IF3mt in initiation of translation in mammalian mitochondria. Biochem. 2009;48:3269–3278. doi: 10.1021/bi8023493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Grasso DG, Christian BE, Spencer AC, Spremulli LL. Over-expression and purification of mitochondrial translational initiation factor 2 and initiation factor 3. Meth. Enz. Translation Initiation: Reconstituted Systems and Biophysical Methods. 2007:59–78. doi: 10.1016/S0076-6879(07)30004-9. [DOI] [PubMed] [Google Scholar]
- [22].Haque ME, Grasso D, Spremulli LL. The interaction of mammalian mitochondrial translational initiation factor 3 with ribosomes: evolution of terminal extensions in IF3mt. Nuc. Acids Res. 2008;36:589–597. doi: 10.1093/nar/gkm1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Bhargava K, Spremulli LL. Role of the N- and C-terminal extensions on the activity of mammalian mitochondrial translational initiation factor 3. Nuc. Acids Res. 2005;33:7011–7018. doi: 10.1093/nar/gki1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Farwell M, Schirawski J, Hager P, Spremulli LL. Analysis of the interaction between bovine mitochondrial 28S ribosomal subunits and mRNA. Biochim. Biophys. Acta. 1996;1309:122–130. doi: 10.1016/s0167-4781(96)00118-2. [DOI] [PubMed] [Google Scholar]
- [25].Favaudon V, Pochon F. Magnesium dependence of the association kinetics of Escherichia coli ribosomal subunits. Biochem. 1976;15:3903–3912. doi: 10.1021/bi00663a001. [DOI] [PubMed] [Google Scholar]
- [26].Zitomer RS, Flaks JG. Magnesium dependence and equilibrium of the Escherichia coli ribosomal subunit association. J. Mol. Biol. 1972;71:263–279. doi: 10.1016/0022-2836(72)90350-6. [DOI] [PubMed] [Google Scholar]
- [27].Korostelev A, Trakhanov S, Laurberg M, Noller HF. Crystal structure of a 70S ribosome-tRNA complex reveals functional interactions and rearrangements. Cell. 2006;126:1065–1077. doi: 10.1016/j.cell.2006.08.032. [DOI] [PubMed] [Google Scholar]