Abstract
Sphingosine kinase 1 (SphK1) responds to a variety of growth factor signals by increasing catalytic activity as it translocates to the plasma membrane (PM). Several studies have identified amino acids residues involved in translocation yet how SphK1 increases its catalytic activity remains to be elucidated. Herein, we report that deletion of 21 amino acids from the COOH terminus of SphK1 (1-363) results in increased catalytic activity relative to wild-type SphK1 (1-384) which is independent of the phosphorylation state of Serine 225 and PMA stimulation. Importantly, HEK293 cells stably expressing the 1-363 protein exhibit enhanced cell growth under serum-deprived cell culture conditions. Together the evidence indicates that the COOH-terminal region of SphK1 encompasses a structural element that is necessary for the increase in catalytic activity in response to PMA treatment and that its deletion renders SphK1 constitutively active with respect to PMA treatment.
Keywords: Sphingosine-1-phosphate, sphingosine kinase 1, plasma membrane, subcellular localization, PMA, protein phosphorylation
INTRODUCTION
An ever-increasing number of studies indicate that sphingolipid metabolites such as ceramide, sphingosine, ceramide-1-phosphate and sphingosine-1-phosphate (S1P) are involved in regulation of essential cell functions including cell growth, apoptosis, differentiation, migration and activation (reviewed in [1–4]). Metabolism of sphingolipids is a dynamic process. For example, hydrolysis of sphingomyelin generates ceramide which can be further deacylated by ceramidase to produce D-erythro-sphingosine. Sphingosine kinases (SphKs), of which there are two isoforms (SphK1 and SphK2), can subsequently phosphorylate D-erythro-sphingosine to form S1P which is an important first- and second messenger lipid molecule that stimulates cell growth/migration through intracellular actions and/or via binding to a cohort of cell surface receptors (S1PR1-5; [5,6]). Conversely, ceramide is a second messenger molecule closely associated with cell growth arrest and/or induction of apoptosis [7]. Hence, it has been proposed that regulation of the relative balance of ceramide and S1P levels in the cell (the ‘sphingolipid rheostat’ model) can determine whether a given cell will proliferate or undergo apoptosis. Given the important role of the sphingolipid rheostat, it stands to reason that levels of S1P are tightly regulated in the cell. In fact, numerous reports have linked deregulation of S1P generation to the development/progression of hyper-proliferative diseases such as cancer, asthma and atherosclerosis (reviewed in [8–11]).
Since the SphKs are the sole enzymes capable of generating the pro-growth sphingolipid metabolite S1P, one would expect that these enzymes are essential for cellular function. Indeed, inhibition of SphK activity with pharmacological inhibitors or molecular inhibitors (i.e. siRNA) induces apoptosis [4,9]. Additionally, the double knock-out mouse of SphK1 and SphK2 is embryonic lethal, while single knock-outs are viable, but display some defects, indicating some redundancy of function, namely S1P generation [12–15].
A number of studies demonstrate that SphK1, the more extensively studied of the two isoforms, has basal catalytic activity which is rapidly and transiently increased in response to a variety of growth factors including: EGF, PDGF, phorbol esters, TNFα and S1P [16–21]. However, unlike many protein kinases, the increase in catalytic activity of SphK1 in response to stimulation is very modest, on the order of 1.5 to 4 fold [22,23]. Similarly, SphK2 is also basally active. Although, to date, only EGF and phorbol esters have been shown to stimulate SphK2 catalytic activity [15,24]. In contrast to the pro-growth role of S1P generation, one report has indicated that SphK2 overexpression can induce apoptosis by sequestering BCL-XL [25]. The role of SphK2 in the induction of apoptosis remains unclear at this time.
Several studies have begun to elucidate the mechanism by which SphK1 activity is regulated in response to growth factor stimulation. In particular, Pitson and colleagues [26] have shown that phosphorylation of SphK1 at Serine 225 (S225), by the MAP kinases ERK1/2, enhances catalytic activity and mediates the translocation of cytoplasmic SphK1 to the plasma membrane (PM) in response to phorbol-12,13-myristic acid (PMA) or TNFα. They further demonstrated that an S225A mutation blocks cell growth under serum-deprived conditions and colony formation in soft agar [27]. Subsequent examination of SphK1 binding to phosphatidylserine containing model membranes suggested that the functional role of S225 phosphorylation may be to enhance SphK1 binding to the PM [28]. This study also demonstrated that two other SphK1 residues (T54 and N89), in conjunction with S225, mediate SphK1 binding to phosphatidylserine at the PM. We have also recently demonstrated that there are two distinct membrane locales, to which, SphK1 can translocate upon growth factor stimulation [29]. Localization to one of these locales, the PM lipid raft microdomain (PMLRM), is mediated by phosphatidylserine binding and is abrogated by mutation of S225 or T54. Blockage of localization to the PMLRM inhibits S1P formation and the survival advantage imparted by SphK1 over-expression under serum deprivation conditions [29]. Other studies indicate a role for Ca2+/Calmodulin in the relocalization of SphK1 to the PM in response to TNFα and PMA and numerous studies have implicated Protein Kinase C (PKC) in PMA stimulated SphK1 activation/translocation possibly by direct phosphorylation of SphK1 [18,30,31].
While each of these factors has been demonstrated to affect SphK1 activation/translocation, the exact mechanism by which these growth factors stimulate SphK1 catalytic activity remains unidentified. To address this issue, we have undertaken a molecular analysis of SphK1 to identify motifs important for regulation of catalytic activity in response to PMA stimulation. Herein we report that 21 amino acids in the COOH-terminal region of SphK1 are necessary for maintenance of the basally active state of SphK1. Deletion of this region results in increased catalytic activity and renders the truncation constitutively active with respect to PMA treatment (i.e. mirrors the PMA activated state of wild-type SphK1).
MATERIALS AND METHODS
Materials
D-erythro-sphingosine was purchased from Avanti Polar Lipids (Alabaster, AL). [32P] γ-ATP was purchased from MP Biomedicals (Irvine, CA). Restriction enzymes were purchased from Promega (San Luis Obispo, CA). LipofectAMINE 2000 was purchased from Invitrogen (Carlsbad, CA). Anti-His6X antibodies were from BD Biosciences. Anti-GST antibodies were from Santa Cruz. Anti-phospho S225 antibodies were from ECM Biosciences.
cDNA Cloning of Human Sphingosine Kinase 1
Human sphingosine kinase 1 was cloned as described previously [29]. For bacterial expression as an NH2-terminally tagged GST fusion protein WT human SphK1 was subcloned into the Eco R1 and Not 1 restriction sites of pGEX4T-3. SphK1 truncation mutants were generated by PCR amplification using the WT 5′ primer: (5′ CCCAGGAATTCCACCATGGATCCAGCGGGC) and 3′ primers designed to introduce stop codons into the coding region of SphK1 and were directionally cloned into pcDNA 3.1+ HISB and pGEX 4T-3 as described above. The following 3′ primers were used: 1-367, 5′ATGATCGCGGCCGCTCACTCCACGCAACC; 1-363, 5′ATGATGCGGCCGCTCAGCTGACCATCCAG; 1-315, 5′TTGATGCGGCCGCTCATTCATACTCCATATGC; 1-270, 5′ATGATGCGGCCGCTCAACTTCCCAGGTGC; 1-220, 5′ATGATGCGGCCGCTCAGGAACCCACTCTTC; 1-160, 5′ATGATGCGGCCGCTCACGTGTGCAGAGAC.
Site directed mutants of SphK1 were created using the QUIKCHANGE II Site-Directed Mutagenesis kit (Stratagene). Primers pairs for site-directed mutagenesis were designed according to the manufacturer’s recommendations as follows S225A, TS (5′AAGACACCTGCCGCCCCCGTTGTGGTC), BS (5′GACCACAACGGGGGCGGCAGGTGTCTT). 1-363GA3, TS (5′GGATGGTCAGCGGTGCCGCGGCATGAGCGGCCGC), BS (5′GCGGCCGCTCATGCCGCGGCACCGCTGACCATGC).
Bacterial Expression of WT Sphingosine Kinase 1 and its truncation mutants
GST-SphK1 constructs were produced in BL21-AI E. coli (Invitrogen). Overnight cultures of transformed isolates were grown with shaking at 250 rpm at 37°C in 2xYT containing 100 μg/mL ampicillin. The cultures were diluted 1:10 into fresh medium and grown at 30°C until reaching an OD600 of 0.8. The culture was then induced with 0.2% arabinose for 30 min followed by 100 mM IPTG and further incubated at 30°C for 5h. The cells were then harvested by centrifugation at 5000 xg for 20 min at 4 °C. The resulting pellet was resuspended in 1.0 mL of resuspension solution (PBS, 15 mM NaF, and 1 mM Na3VO4, 5 mg lysozyme, 1x Complete Mini EDTA-free Protease Inhibitor Tablet (Roche)). Resuspended samples were incubated at 4°C for 1 h, followed by sonication 3x each for 30 s on ice. The lysate was then clarified by centrifugation at 100,000 xg for 30 min. The supernatant was diluted to 10 mL with PBS binding buffer, filtered through 0.45 μm filters and applied to a pre-equilibrated 1 mL GST HiTrap Affinity Column (Amersham). The column was washed with at least 10 column volumes of binding buffer. GST-SphK1 fusion proteins were then eluted with 3 mL of elution buffer (binding buffer supplemented with 10 mM reduced glutathione, pH 8.0). Eluted proteins were buffer exchanged into SphK Activity Assay Buffer (SKAAB; 20 mM Tris pH 7.4, 20% Glycerol, 1 mM β-mercaptoethanol, 1 mM EDTA, 1 mM Na3VO4, 15 mM NaF, 0.5 mM 4-deoxypyridoxine) and concentrated to 1 mL on Amicon Ultra 30,000 MWCO centrifugal filters (Millipore; Bedford MA).
SphK1 Activity Assays
To assess the catalytic activity of WT SphK1 and its truncation mutants, in vitro SphK1 activity assays were performed. The formation of S-1-32P from [32P] ATP and D-erythro-sphingosine was determined, as described previously by Olivera et al. [32], using either partially affinity purified bacterially expressed GST-fusion proteins or partially affinity purified His6X-fusion proteins from stably transfected HEK293 cell lines. Briefly, protein samples were combined with 50 μM D-erythro-sphingosine, 200 μM ATP and 2 μCi [32P] ATP in a 100 μL final reaction volume of SKAAB for 30 min at 37 °C with shaking. Kinase reactions were terminated and extracted using the Bligh and Dyer method [33] with minor modifications. Briefly, 10 μL 6N HCl was added and lipids were extracted in 400 μL of Chloroform/MeOH 100:200 v/v and 125 μL Chloroform and 125 μL 1M KCl. The organic phase was removed, dried onto filter paper squares and directly transferred to scintillation tubes for liquid scintillation counting [34].
Expression of Sphingosine Kinase 1 in mammalian cells
HEK293 cells (ATCC CRL-1573) were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM; Mediatech INC) supplemented with 10% Fetal Bovine Serum (FBS) at 37°C in 5% CO2. For stable transfection, nearly confluent wells of 6 well culture dishes were transfected with 4 μg of the appropriate cDNA using LipofectAMINE 2000 (Invitrogen). Cells were selected for resistance to G418 (500 μg/mL; Gibco) and were maintained as mixed clones to avoid variability due to isolation of a single clone. To harvest SphK1 protein, the media was aspirated from confluent monolayers of the stably expressing clones; the cells were washed once in PBS, and lysed by flash freezing in liquid nitrogen.
Affinity purification of His6X-tagged SphK1 protein
For affinity purification of His6X tagged SphK1 from mammalian cells, confluent 10 cm2 plates of HEK293 cells stably expressing either WT SphK1 or the 1-363 SphK1 protein were harvested in His6X Binding Buffer (20 mM NaPO4 buffer pH 7.4 with 500 mM NaCl and an EDTA-free Protease Inhibitor Tablet (Roche)). Lysates were filtered through a 0.45 μm filter and then applied to a pre-equilibrated HisTrap affinity column (Amersham Biosciences). The column was washed with 20 mL His6X Binding buffer. Partially purified SphK1 protein was eluted in His6X Binding buffer supplemented with 500 mM Imidazole as three 1 mL fractions. The fractions were pooled, buffer exchanged into SKAAB buffer, concentrated to 1 mL on Amicon Ultra 30,000 MWCO centrifugal filters (Millipore; Bedford MA), and analyzed by Western blot analysis. Kinetic parameters were determined by varying the D-erythro-sphingosine substrate concentration in the standard in vitro SphK1 activity assay as indicated.
Subcellular fractionation of SphK1 by pool assay
Nearly confluent (~90%) 10 cm2 plates of His6X-SphK1 transfected HEK293 cells were flash frozen and resuspended in 1 mL of 1xTBS containing protease and phosphatase inhibitor cocktails (Roche and Calbiochem respectively). Post nuclear supernatants were prepared by centrifugation at 500 xg for 5 min at 4°C, and then separated into cytoplasmic and total membrane fractions by centrifugation at 100,000 xg for 30 min at 4°C. The pelleted membranes were washed once in 1xTBS, and resuspended in 1% Triton X-100. Membranes were centrifuged at 100,000 xg for 30 min at 4°C, separated into supernatant and pellet fractions. Pelleted membranes were washed once in Triton X-100 buffer, and finally resuspended in 2% SDS. To isolate the remaining membrane fraction, membranes were centrifuged at 100,000 xg for 30 min at 4°C. The supernatant was removed and the pellet fraction discarded.
PMA activation of SphK1
HEK293 cells stably expressing SphK1 constructs were seeded at 2 × 105 cells/mL into six well culture plates in DMEM containing 2% FBS. After 18 h, cells were stimulated by the addition of 300 nM PMA, or vehicle (DMSO) for 20 minutes. The cells were washed with cold PBS and snap frozen in liquid nitrogen. Thawed cells were scraped in 200 μL lysis buffer (50 mM HEPES, 50 mM MgCl2, 10 mM KCl, 10 mM NaF, 2 mM Na3VO4, 500 μM 4-deoxypyridoxine, 0.05% Triton X-100, pH 7.4) and probe sonicated for 15 s on ice. The lysates were centrifuged at 100,000 xg for 30 min at 4°C. The supernatant (cytosolic fraction) was removed and the pellet (membrane fraction) was washed with ice-cold PBS and resuspended by brief probe sonication in 150 μL lysis buffer for determination of SphK1 activity.
S1P analysis
HEK293 cells were seeded at 4×105 cells/mL in 15 cm2 culture dishes in DMEM containing 2% FBS. After 18h, 2 μM D-erythro-sphingosine (Avanti Polar Lipids) and 0.4 μCi [3H] sphingosine (ARC, St. Louis, MO) complexed with 4 mg/mL BSA in serum-free DMEM was added for 30 min prior to stimulation with 300 nM PMA or DMSO vehicle for 20 min as described previously by Olivera et al. [32]. Cells were washed in 1 mL PBS and the lipids were extracted using the Bligh and Dyer method [33] with minor modifications. Briefly, samples were acidified with 50 μL 6N HCl. Chloroform:methanol (3.75 mL, 1:2 v:v) was then added to each fraction, followed by 1.25 mL chloroform and 1.25 mL KCl. The samples were centrifuged at 2,000xg for 5 min and the organic phase containing lipids was dried down under nitrogen stream. Samples were then resuspended in 30μL chloroform and applied to a Silica Gel TLC plate (Whatman, Florham Park, NJ) and the lipids were separated using a butanol:water:acetic acid 3:1:1 v:v:v solvent system. The plates were developed in an iodine chamber and the region of the TLC plate corresponding to the Rf value (0.32) of S1P was isolated [35]. The amount of [3H] S1P present was determined by scintillation counting.
Cell Growth Assays
HEK293 cells stably transfected with the specific SphK1 constructs were seeded into six-well culture dishes (2.5 × 104 cells/well) in DMEM containing 10% FBS. After 24 h, the media was changed to DMEM containing 0.5% FBS. Viable cells, as determined by Trypan blue exclusion, were counted using a hemocytometer over a period of 4 days.
Statistical Analysis
Data is presented as mean ± standard deviation for the number of experiments indicated in the figure legend. Unpaired Student’s t tests were employed for SphK1 activity assays, S1P analysis and cell growth assays. Differences were considered significant at p values <0.05. Kinetic analysis included non-linear regression analysis by the least-squares method. Student’s t test was employed on the best-fit kinetic curves. Differences were considered significant at p values <0.05.
RESULTS
Kinase activity of recombinant WT SphK1 and its truncated mutants produced in E. coli
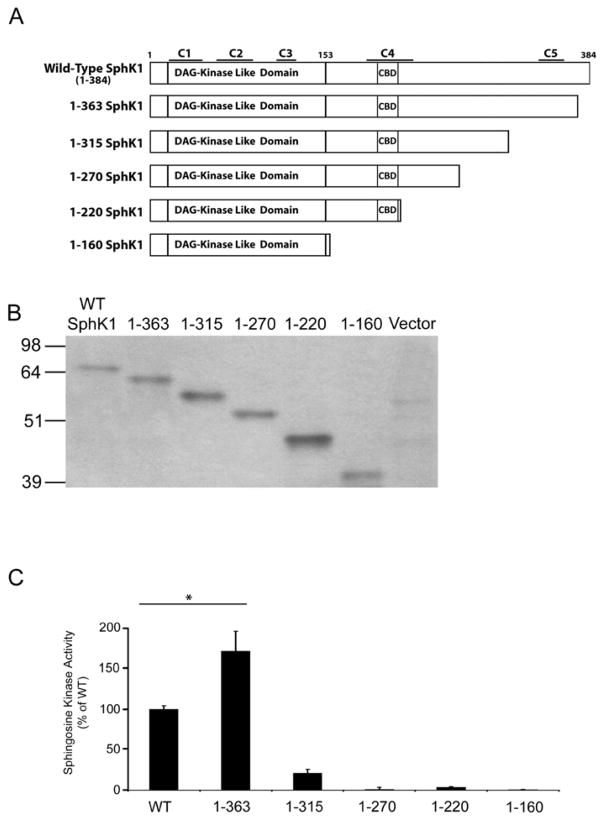
To initiate our molecular analysis of SphK1, we sought to define the minimal structural elements of SphK1 necessary for catalytic activity and identify potential regulatory regions within SphK1. Early studies identified a region of SphK1, encompassing amino acids 16–153, with homology to the DAG kinase catalytic domain [36]. Therefore, we designed a series of truncation mutants that retain this region of SphK1 and systematically deleted specific structural elements from the COOH-terminus of SphK1. Using the PROF secondary structure determination program [37] we identified 5 potential sites for truncation of SphK1 (Figure 1A). Full-length WT SphK1 and the 5 truncated SphK1 mutants were produced as GST (~26 KDa)-tagged fusion proteins in E. coli and partially affinity purified over GST HiTrap Affinity columns (Figure 1B). We next determined the catalytic activity of the SphK1 truncated mutants, using the standard in vitro SphK1 activity assay (Figure 1C). Surprisingly, the GST1-363 mutant possessed significantly (p<0.005) higher levels of kinase activity (~1.6 fold increase) than that observed for WT GST-SphK1. In contrast, the GST1-315 mutant retained minimal kinase activity (~20% of WT GST-SphK1) while further truncation of amino acids from the COOH-terminus abrogated kinase activity indicating that amino acids beyond residue 153 are required for SphK1 kinase activity. Since E. coli bacteria do not produce post-translationally modified (i.e. phosphorylation) recombinant proteins, the enhanced catalytic activity of the 1-363 mutant appears to be independent of the post-translational modification state of the protein.
Figure 1. Truncation mutagenesis of bacterially generated SphK1.
(A) WT-SphK1 was systematically truncated from the COOH terminus to identify the minimal catalytically active fragment of SphK1. The positions of the 5 conserved sphingolipid kinase domains (C1-C5), the calmodulin binding domain (CBD) and the region of homology to the diacylglycerol (DAG) kinase domain are annotated. (B) SphK1 truncations were partially purified by GST-affinity chromatography and visualized by Western blot analysis with anti-GST antibodies. (C) Sphingosine kinase activity was examined in triplicate using equal amounts of partially purified GST-tagged protein determined by densitometric analysis using Image J. Relative activity levels for each mutant are expressed as the percentage of WT GST-SphK1 activity. (n=5, * p<0.005)
Kinase activity of recombinant WT SphK1 and truncated mutants produced in HEK293 cells
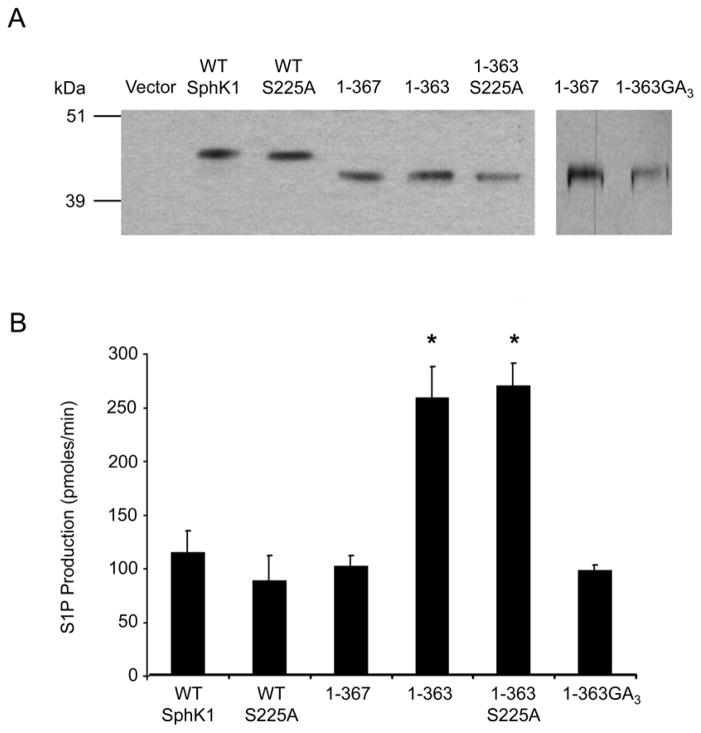
To ascertain whether post-translational modification further enhances or abrogates the increased catalytic activity of the 1-363 mutant, we generated epitope tagged mammalian expression constructs of WT His6X-SphK1 and the His6X-1-363 mutant and stably expressed them in HEK293 cells. As a control, we also generated a stable line expressing a His6X-1-367 mutant, which reportedly maintains wild-type levels of SphK1 catalytic activity in HEK293T cells [38]. His-tagged SphK1 proteins were isolated, partially affinity purified from HEK293 cell cultures and analyzed by Western blot analysis (Figure 2A). The catalytic activity of WT His6X-SphK1, and the His6X-1-367 and His6X-1-363 mutants were determined. Consistent with the results of the E. coli produced WT GST-SphK1 and GST-1-363 SphK1, the His6X-1-363 mutant demonstrated a significant increase (~2.2 fold, p<0.001) in kinase activity relative to WT His6X-SphK1 (Figure 2B). The His6X-1-367 mutant possessed catalytic activity comparable to WT His6X-SphK1, as previously reported [38].
Figure 2. Examination of the 364–367 region and the effect of ERK1/2 phosphorylation on the catalytic activity of the 1–363 mutant.
(A) Truncation and site-directed mutants of SphK1 were stably transfected into HEK293 cells, partially affinity purified and the relative expression levels of the SphK1 proteins were determined by Western blot analysis and densitometric analysis using Image J. Representative Western blots of the normalized protein expression are shown. The 1-363GA3 protein is from a separate blot with 1–367 as a loading control. (B) In vitro SphK1 activity assays were performed in triplicate using equal quantities of partially affinity purified protein determined by densitometric analysis. SphK1 activity is expressed as pmoles of S1P formed per min. (n=4, * p<0.001)
Because previous studies have demonstrated that phosphorylation at Serine 225 (S225) by ERK1/2 increases catalytic activity of SphK1 in response to PMA treatment [27], we next wanted to determine whether phosphorylation of SphK1 at S225 is necessary for the observed increase in catalytic activity for the His6X-1-363 mutant. S225 was mutated to Alanine (S225A) in WT His6X-SphK1 and the His6X-1-363 mutant, the proteins were partially affinity purified (Figure 2A), and the effect of the S225A mutation on kinase activity was determined. The His6X-S225A mutant possessed levels of catalytic activity comparable to WT His6X-SphK1 (Figure 2B) as has been previously demonstrated [26]. The enhanced kinase activity of the His6X-1-363 mutant, relative to WT His6X-SphK1, is preserved in the His6X-1-363-S225A mutant (Figure 2B, p<0.001). This observation is consistent with the bacterially produced GST1-363 mutant, which was not phosphorylated at S225, indicating that the increased catalytic activity of the 1-363 mutant is independent of S225 phosphorylation.
The enhancement of catalytic activity observed for the 1-363 mutant appears to result from deletion of amino acid residues 364-367, as the 1-367 mutant retains wild-type levels of catalytic activity. This increase in SphK1 activity could be due to the removal of a structural element that maintains SphK1 in a basally active state. One possible mechanism by which these residues maintain SphK1 basal activity would be through polar interactions mediated by glutamate 367. To examine whether the observed difference in catalytic activity between the His6X-1-363 and His6X-1-367 mutants is due to polar interactions mediated by glutamate 367 (E367), we mutated E367 to alanine and arginine. These mutations did not affect the catalytic activity of the His6X-1-367 mutant (data not shown) indicating that polar interactions are not necessary for maintenance of basal SphK1 activity.
A second possible explanation for the observed increase in catalytic activity of the His6X-1-363 mutant is a change in the overall structure of SphK1 due to the deletion. We next examined this possibility by mutating the residues 364GCVE367 to 364GAAA367 (1-363GA3). Mutation of these residues resulted in kinase activity levels comparable to the His6X-1-367 mutant (Figure 2A and B). Together these data indicate deletion of the region between residue 367 and 363 is sufficient to induce an increase in SphK1 catalytic activity. These results further suggest that the specific sequence of the 364GCVE367 tetrapeptide is not important for maintenance of the basally active state of SphK1.
Kinetic analysis of the 1-363 truncated mutant protein expressed in HEK293 cells
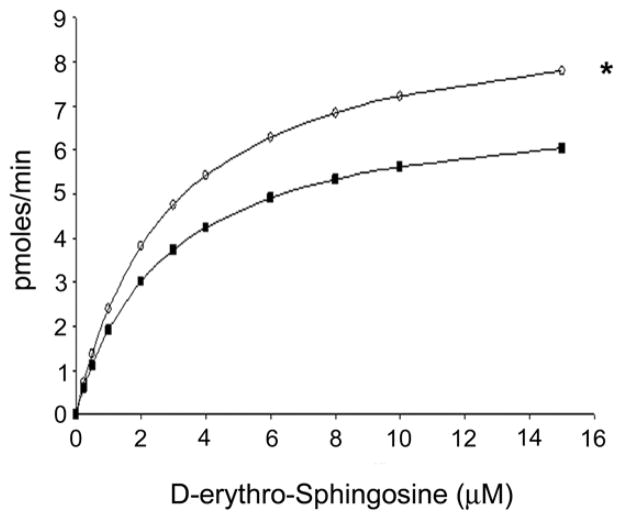
To determine whether the enhanced activity of the 1-363 mutant results from alterations in the sphingosine substrate binding capacity or from enhancement of the catalytic efficiency, we next examined the kinetic parameters of WT His6X-SphK1 and the His6X-1-363 mutant. To accomplish this, the in vitro SphK1 kinase activities of the partially affinity purified WT His6X-SphK1 and His6X-1-363 mutant proteins were measured using increasing concentrations of D-erythro-sphingosine as a substrate (Figure 3). The kinetic parameters for WT His6X-SphK1 and the His6X-1-363 mutant were examined and the KM and Vmax for both proteins determined using the nonlinear regression analysis tool ANEMONA [39]. Both the WT His6X-SphK1 and the His6X-1-363 mutant display typical Michaelis-Menten kinetics (Figure 3). The KM toward D-erythro-sphingosine and the Vmax for WT His6X-SphK1 were determined to be 2.75 μM and 7.15 pmoles/min respectively. There was slight decrease in the KM for D-erythro-sphingosine of the His6X-1-363 mutant compared to WT His6X-SphK1 (2.47 μM) indicating that deletion of 21 amino acids from COOH-terminus likely does not alter D-erythro-sphingosine substrate binding. There was, however, a significant increase in the Vmax (9.17 pmoles/min) for the His6X-1-363 mutant (p<0.05) correlating to a 1.43 fold increase in the substrate specificity constant indicating that the His6X-1-363 mutant is more catalytically active than WT His6X-SphK1.
Figure 3. Kinetic analysis of SphK1 and the 1–363 mutant.
The kinetic parameters of WT His6X-SphK1 (black squares) and the His6X-1-363 truncation (open circles) were determined by performing in vitro SphK1 activity assays in the presence of various concentrations of D-erythro-sphingosine. Kinase activity was expressed as pmoles of product formed per min and the Michaelis-Menten plots of the data were obtained by non-linear regression analysis using the program ANEMONA. (n=3; * p <0.05)
Subcellular localization is unaffected in the 1-363 mutant
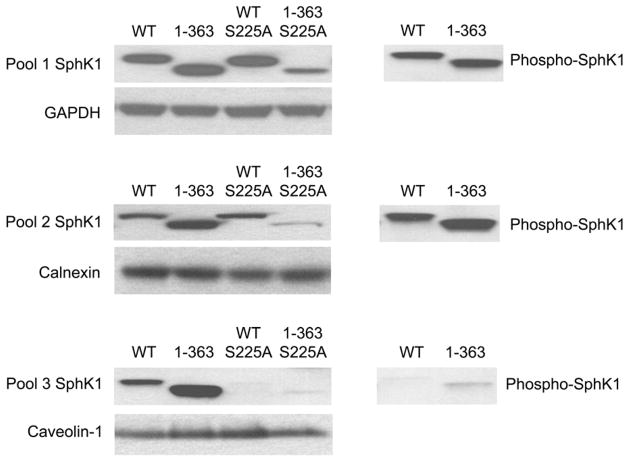
We have previously demonstrated that localization of SphK1 to the PMLRM mediates the ability of SphK1 to overcome serum deprivation induced growth inhibition [29]. SphK1 localizes to two distinct membrane subfractions, a membrane associated fraction of SphK1 that can be dissociated from the membrane pellet by high salt (1M NaCl), high pH (500 mM Na2CO3 pH 11.5) and 1% Triton X-100 and a second fraction, resistant to these treatments, localized to the PMLRM [29]. Hence, we next examined the subcellular localization of the 1-363 truncation mutant to determine whether removal of the COOH terminal 21 amino acids affected the intracellular localization of SphK1 by performing a “pool assay” employing 1% Triton X-100, as previously described [29]. Whole cell lysates of HEK293 cells stably expressing SphK1 proteins were divided into the three intracellular “pools” including cytosolic (Pool 1), membrane associated fractions (Pool 2) and PMLRM fractions (Pool 3) and the localization of SphK1 and loading control/marker proteins for each fraction were examined. As shown in Figure 4, both WT SphK1 and the 1-363 mutant proteins were present in all three fractions, whereas the S225A mutants of WT SphK1 is not localized to the PMLRM as previously reported [29]. Interestingly, the 1-363 mutant is present at higher levels in the PMLRM fraction indicating that removal of the COOH terminal 21 amino acids of SphK1 enhances PM localization in addition to enhancing SphK1 catalytic activity. However, this enhanced localization is blocked by the S225A mutation in the 1-363S225A mutant. Taken together, the data suggests that the COOH terminal 21 amino acids of SphK1 play a role in the maintenance of basal activity and subcellular localization and that their removal leads to enhanced catalytic activity and enhanced PMLRM localization.
Figure 4. Subcellular localization of SphK1, the 1–363 mutant and their S225A derivatives.
HEK293 whole cell lysates expressing WT His6X-SphK1, His6X-1-363 and the S225A mutants were separated into cytosolic, and Triton X-100 soluble and Triton X-100 insoluble membrane fractions. 10 μg of total protein from each fraction was separated by SDS-PAGE and Western blot analyses using anti-His6X antibodies and anti-phospho-S225 antibodies were performed. Loading controls for Pool 1 (GAPDH), Pool 2 (calnexin) and Pool 3 (caveolin-1) fractions were included as indicated.
Recent studies have indicated the protein phosphatase 2A (PP2A) dephosphorylates SphK1 at S225 [40]. Importantly, the binding site for PP2A was localized to the COOH terminal 21 amino acids of SphK1 which are removed in the 1-363 mutant. Since phosphorylation of S225 has been implicated in the relocalization of SphK1 to the PM [27], we expected that deletion of 21 amino acids from the COOH terminus would generate a constitutively phosphorylated S225 in the His6X-1-363 mutant. Using anti-phospho S225 antibodies, phosphorylated WT SphK1 and His6X-1-363 mutant proteins were detected in Pool 1 and Pool 2, however, very little phospho-SphK1 protein was detected in the PMLRM fraction (Pool 3; Figure 4). The lack of phospho-SphK1 protein in Pool 3 indicates that phosphorylation of S225 is not required for PMLRM localization.
The response of the 1-363 SphK1 mutant to PMA treatment in HEK293 cells
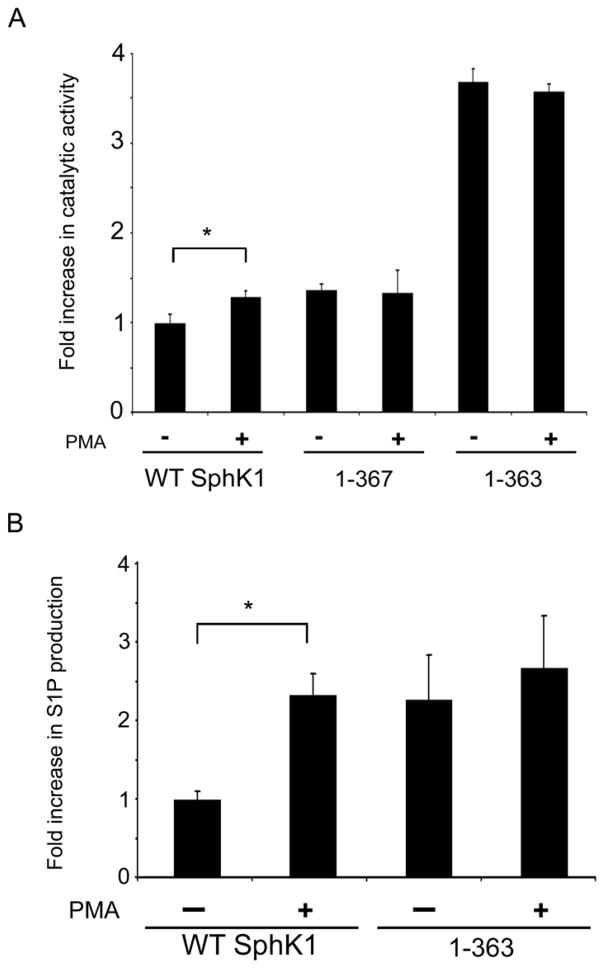
Previous studies show that SphK1 is translocated to the PM and activated in response to PMA [18,21]. To assess whether the COOH-terminal region of SphK1 is important for the regulation of SphK1 catalytic activity in response to PMA treatment, we examined the kinase activity of WT His6X-SphK1, and the His6X-1-367 and His6X-1-363 mutants in HEK293 cells. As shown in Figure 5A, we observed a statistically significant increase (p<0.05) in the catalytic activity of WT His6X-SphK1 in response to PMA treatment. In contrast, we did not observe an increase in catalytic activity for the either the His6X-1-367 or the His6X-1-363 mutants upon PMA treatment. This lack of response to PMA treatment indicates that the COOH-terminal region is necessary for the response of SphK1 to PMA treatment.
Figure 5. Effects of PMA stimulation on 1–363 mutant catalytic activity and S1P production.
(A) HEK293 cells stably expressing His6X-SphK1, the His6X-1-367 and His6X-1-363 truncation mutants were cultured overnight in DMEM containing 2% FBS and subsequently treated with vehicle (DMSO) or 300 nM PMA for 20 minutes. Cells were lysed and total membrane fractions were prepared. In vitro SphK1 activity assays were performed in triplicate on 10 μg of each total membrane fraction. The response of WT His6X-SphK1 and the truncations to PMA treatment was adjusted to account for the contribution of endogenous SphK1 activity. (n=3 in triplicates; * p<0.05) (B) HEK293 cells stably expressing WT His6X-SphK1 and the His6X-1-363 truncation were cultured in triplicate overnight in DMEM containing 2% FBS and subsequently prelabeled with 2 μM D-erythro-sphingosine containing 0.4 μCi of [3H] D-erythro-sphingosine delivered as a 4 mg/ml BSA conjugate prior to treatment with vehicle (DMSO) or 300 nM PMA for 20 minutes. After treatment, lipids were extracted and were separated by TLC with appropriate standards. (n=3 in triplicates; * p<0.01)
To provide further evidence for the enhancement of the catalytic activity of the His6X-1-363 mutant, we next examined the production of S1P after PMA stimulation in HEK293 cells. We preloaded HEK293 cells stably expressing WT His6X-SphK1 and the His6X-1-363 mutant with [3H] D-erythro-sphingosine. After 20 min of PMA treatment, we examined the amounts of [3H] S1P formed using thin layer chromatography. As shown in Figure 5B, HEK293 cells stably expressing the WT His6X-SphK1 significantly (p<0.01) increased the production of S1P in response to PMA stimulation. Importantly, while the cells stably expressing the His6X-1-363 mutant displayed little, if any, change in S1P levels after PMA treatment, we observed similar levels of S1P production in His6X-1-363 expressing cells compared to the PMA activated state of WT His6X-SphK1 expressing cells. This indicates that deletion of 21 amino acids from the COOH terminus of SphK1 mimics the PMA activated state of WT SphK1 and further supports the hypothesis that the COOH terminus of SphK1 is involved in the regulation of SphK1 catalytic activity in response to PMA treatment.
The 1-363 mutant displays enhanced survival under serum deprivation conditions
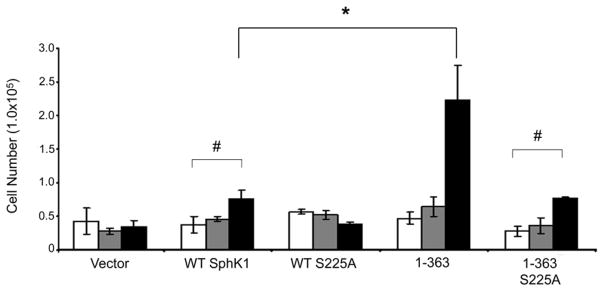
Localization of SphK1 to the PMLRM is required for the ability of SphK1 to overcome serum deprivation induced growth inhibition [29]. We, therefore, reasoned that the enhanced catalytic activity, enhanced S1P production and enhanced PMLRM localization of the His6X-1-363 mutant should directly enhance survival of HEK293 cells. To test this, we performed cell growth assays using HEK293 cell lines stably expressing vector only, WT His6X-SphK1, His6X-1-363, and their S225A derivatives. For cell growth analysis, HEK293 cell lines were grown in the presence of DMEM containing either 10% FBS or 0.5% FBS for the days indicated. All HEK293 cells expressing WT SphK1 and its mutants grew in the presence of 10% FBS to an approximately equal extent (data not shown). However, when grown under serum deprivation conditions (0.5% FBS), vector transfected cells did not increase in number over the days indicated. In contrast, WT SphK1 over-expressing cells increased in number as expected which is consistent with previous studies (p<0.05) [27,29]. Interestingly, HEK293 cells expressing the His6X-1-363 mutant significantly increased in number from day 0 to day 4 (p<0.01 Figure 6), to an extent greater than WT SphK1 expressing cells. Additionally, consistent with previous reports, the S225A mutation abrogated the enhanced growth effects of WT SphK1 over-expression. Similarly, the S225A mutation in the His6X-1-363S225A mutant also abrogated the enhanced growth effect observed for the His6X-1-363 mutant. Taken together, these data indicate that the increased activity, S1P production, and enhanced PMLRM localization of the His6X-1-363 mutant work in concert to enhance survival/growth.
Figure 6. Growth of SphK1 and the truncation and site-directed mutants under normal and reduced serum conditions.
HEK293 cells stably expressing vector only, WT His6X-SphK1, His6X-1-363, His6X-S225A and His6X-1-363-S225A constructs were plated on day 0, in triplicate, and were grown in culture medium containing 0.5% FBS for 4 days. On days 1 (white bars), 2 (gray bars) and 4 (black bars) cells were stained with Trypan Blue. Cells that excluded Trypan Blue were counted with a hemocytometer. (n=3 in triplicates; # p<0.05, * p<0.01)
DISCUSSION
In this study, we sought to identify regions of SphK1 that are necessary for minimal catalytic activity as well as the regulation of catalytic activity. To achieve these goals we initially employed a bacterial expression system wherein we produced five SphK1 fusion proteins (GST-SphK1) that were truncated from the COOH-terminus based on secondary structure analyses. All of these truncated mutants retain a motif that shares homology with the DAG kinase catalytic domain encompassing amino acid residues 16-153 of WT-SphK1 [36]. For this reason, we anticipated that each of the truncation mutants of SphK1 would be catalytically active. However, the only mutant that retained >20% of WT catalytic activity was the 1-363 truncation mutant.
In fact, the GST-1-363 mutant displayed catalytic activity that was significantly higher than WT GST-SphK1. It is of note that this observed increase in activity occurs in the absence of post-translational modifications since the GST-1-363 mutant is produced in E. coli bacteria. It is also interesting that the observed increase in catalytic activity of the GST-1-363 mutant (~1.6 fold) closely resembles the increase in SphK1 activity in response to growth factor treatment reported by several groups (1.5–4 fold; [22,23]. We further confirmed this observation in HEK293 mammalian cells by comparing the catalytic activity of the His6X-1-363 mutant to the His6X-1-367 mutant that was previously reported to possess wild-type catalytic activity [38]. We also demonstrate that truncation of SphK1 beyond amino acid 363 from the COOH-terminus abrogated SphK1 catalytic activity in bacterial cells indicating that the region between amino acids 315 and 363 represents the COOH-terminal boundary of the catalytic domain of SphK1.
Detailed examination of the secondary structure predictions of this region using PROF [37] revealed three β sheets (β sheets 12 – 14). The conserved C5 domain (337FAVDGEL343) is present within β sheet 13 [41]. To further refine the minimal catalytically active fragment of SphK1 we have tested COOH terminal truncation mutants of SphK1 encompassing residues 1–334 and 1–347, in HEK293 cells. These truncations were also minimally catalytically inactive (data not shown) indicating that the catalytically active fragment of SphK1 must include the region between 347 and 363 amino acids and likely encompasses all of β Sheet 14 (residues 350–362). Consistent with our observations, a previous report has noted that truncation of SphK1 to residue 344 (1–344) also eliminated catalytic activity [38].
We have also employed truncation mutagenesis of the NH2-terminal region of SphK1 and have found that deletion of only the first 10 amino acids of SphK1 is tolerated (data not shown). Taken together with our current findings, we believe that the region of SphK1 from amino acid 154 to amino acid 363 is essential for catalytic activity and may participate in sphingolipid substrate binding. Consistent with this hypothesis, Yokota et al. [42] has identified Asp 177 within the conserved C4 region, as part of the substrate binding cleft of mouse SphK1. These findings further imply that SphK1 may not contain distinct catalytic and regulatory regions but is instead a compact kinase domain with short NH2- and COOH- terminal extensions that are dispensable for catalytic activity.
Our mutagenesis experiments identified 364GCVE367 as residues necessary for the maintenance of basal catalytic activity for SphK1. Mutation of these residues to Alanine (His6X-1-363GA3) maintained the wild-type basally active state whereas their removal led to increased catalytic activity. A possible explanation for this difference would be that backbone hydrogen bonds made by these four amino acid residues stabilize the interaction of β sheet 14 with another region of SphK1. Deletion of these four amino acids may destabilize this interaction allowing SphK1 to undergo a conformational shift to an “open” or “active” conformation that results in higher catalytic activity.
A second possible explanation would be that the COOH-terminal region of SphK1 acts as an active site “occluding loop” (i.e. a pseudo-substrate binding motif), as seen in PKC [43]. In the case of SphK1 this loop would partially block the active site, allowing basal kinase activity, and would shift out of the active site upon post-translational modification of SphK1. This model is contingent upon the four amino acids 364–367 being sufficient to block the active site (i.e. the His6X-1-367 mutant maintains basal activity whereas the His6X-1-363 mutant has higher activity because its active site is unblocked). The fact that no change in substrate binding relative to WT His6X-SphK1 was observed for the His6X-1-363 mutant in our kinetic studies argues against the idea of an active site “occluding loop”. The kinetic studies of the mammalian His6X-1-363 mutant instead indicate that the increase in catalytic activity is due to a change in the Kcat for the enzyme. This relatively small change in the Kcat for the enzyme is consistent with the observed change in Kcat for the constitutively active G113A SphK1 mutant generated by Pitson et al. [44].
A recent report demonstrates that S225 of SphK1 is a substrate for dephosphorylation by PP2A and that the region of SphK1 from 368–384 contributes to the association of SphK1 with PP2A [40]. This region of SphK1 deleted in the His6X-1-367 and His6X-1-363 truncation mutants. Therefore we examined the phosphorylation state of S225 in WT His6X-SphK1 and in the His6X-1-363 mutant. It was expected that deletion of this region of SphK1 would increase the level of S225 phosphorylation for the His6X-1-363 mutant relative to WT His6X-SphK1. However, in our system, we observed that that S225 phosphorylation occurs to an equal extent in both WT His6X-SphK1 and the His6X-1-363 mutant. Phosphorylated SphK1 protein was observed in Pool 1 (cytosol), and Pool 2 (membrane associated fraction). Strikingly, very little phosphorylated protein was detected in the PMLRM (Pool 3).
This observation seems at odds with the data indicating that the S225A abrogates localization of SphK1 to the PMLRM fraction, however, several possible explanations for this observation can be proposed. The first explanation would be that our recent report was the first to examine the role of S225 in targeting to the PMLRM [29]. Earlier reports that did examine the phosphorylation status of S225 examined only the Triton X-100 soluble (Pool 2) SphK1 membrane fraction [26,27]. Our results are consistent with the observation that S225 is phosphorylated in the Triton X-100 soluble membrane fraction, indicating that the lack of S225 phosphorylation may be a characteristic unique to the PMLRM localized fraction of SphK1. Indeed, a previous study demonstrated that WT-SphK1 targeted to the PMLRM via the Lck-myristoylation motif [45] was hypo-phosphorylated at S225 relative to a non-myristoylated WT-SphK1 protein [27].
Secondly, it is possible that a Serine residue at position 225, regardless of phosphorylation state, is required for localization to the PMLRM and that the S225A mutation disrupts the ability of SphK1 to interact with phosphatidylserine. Previous studies of SphK1 binding to PM mimetic model membranes demonstrated that an insect cell produced phospho-mimetic mutant of SphK1 (S225E) and WT SphKl (a fraction of which was phosphorylated at S225) displayed similar levels of binding to PM model micelles [28]. On the other hand, the S225A mutant had much lower levels of PM mimetic binding than WT SphK1. The binding of WT SphK1 that was not phosphorylated at S225 (i.e. bacterially produced SphK1 or de-phosphorylated insect cell SphK1) was not measured, so it is not known at this time if phosphorylation of S225 or the mere presence of a Serine residue at position 225 is required for PMLRM binding.
Our data also demonstrate that the COOH-terminus of SphK1 (367-384) is required for stimulation of SphK1 catalytic activity in response to PMA as neither the His6X-1-367 nor the His6X-1-363 mutant displayed increased catalytic activity after PMA treatment. This indicates that the COOH terminus of SphK1 contains regulatory elements necessary for SphK1 activation in response to PMA. PMA, a well characterized direct activator of PKC and stimulator of SphK1 activity [18], could exert its effects on SphK1 activity by stimulating PKC to directly phosphorylate SphK1. Consistent with this, Shu et al. demonstrated that PKCε phosphorylates and stimulates SphK1 catalytic activity under in vitro assay conditions [30]. Our sequence analyses, identified four potential PKC phosphorylation sites within SphK1 (T54, S181, T205 and S371). S371, which resides in the COOH terminal region deleted in the 1-363 mutant, is therefore an interesting candidate for post-translational modification in response to PMA treatment. Whether the activation of SphK1 in response to PMA treatment is mediated by phosphorylation of S371 is currently under investigation in our laboratory.
Lastly, the observed survival advantage of the His6X-1-363 mutant relative to WT His6X-SphK1, under serum deprivation conditions, is likely due to the increased catalytic activity at the PMLRM and higher intracellular levels of S1P generation of the His6X-1-363 mutant. Consistent with this observation, several studies have reported that membrane association and generation of S1P are absolute requirements for the growth of SphK1 over-expressing cells under serum deprived conditions [27,29] [38]. Further supporting this view, enforced targeting of a S225A mutant of SphK1 to the PM, by attaching an Lck-myristoylation motif to S225A, restored the survival of the myr-S225A SphK1 expressing cells under reduced serum conditions [27].
CONCLUSIONS
In conclusion, truncation of 21 amino acids from the COOH-terminus of SphK1 increases catalytic activity, localization to the PMLRM, enhances SphK1 over-expression mediated cell growth under serum deprivation conditions and renders SphK1 non-responsive to PMA. These findings indicate that the COOH terminal region of SphK1 is required for the stimulatory/relocalization effects of PMA treatment. Other reports have previously demonstrated that this region of SphK1 elicits similar effects on SphK1 activity/localization in response to TNFα [46]. Additionally, this region of SphK1 has been shown to mediate the binding of PP2A, a S/T protein phosphatase, implicated in attenuation of SphK1 signaling [40]. Taken together, these data indicate the vital importance of the COOH terminal region of SphK1 in the regulation of its catalytic activity and subcellular localization. The manner in which the COOH-terminal region of SphK1 regulates SphK1 activity/localization will become clear once the 3-D structure of SphK1 is determined. In the meantime, the 1-363 truncated mutant of SphK1 can serve as a tool for generation of high levels of intracellular S1P without the need for PMA treatment.
Acknowledgments
This study was funded by NIH grant CA 91155 and the Jake Gittlen Research Foundation.
ABBREVIATIONS
- SphK1
Sphingosine Kinase 1
- Sph
Sphingosine
- S1P
Sphingosine-1-phosphate
- PM
plasma membrane
- PMLRM
plasma membrane lipid raft microdomain
- PMA
phorbol-12,13-myristic acid
- PKC
protein kinase C
- S225
Serine 225
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Argraves KM, Obeid LM, Hannun YA. Adv Exp Med Biol. 2002;507:439–44. doi: 10.1007/978-1-4615-0193-0_68. [DOI] [PubMed] [Google Scholar]
- 2.Cuvillier O. Biochim Biophys Acta. 2002;1585:153–62. doi: 10.1016/s1388-1981(02)00336-0. [DOI] [PubMed] [Google Scholar]
- 3.Maceyka M, Payne SG, Milstien S, Spiegel S. Biochim Biophys Acta. 2002;1585:193–201. doi: 10.1016/s1388-1981(02)00341-4. [DOI] [PubMed] [Google Scholar]
- 4.Hait NC, Oskeritzian CA, Paugh SW, Milstien S, Spiegel S. Biochim Biophys Acta. 2006;1758:2016–26. doi: 10.1016/j.bbamem.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 5.Meyer zu Heringdorf D, Jakobs KH. Biochim Biophys Acta. 2007;1768:923–40. doi: 10.1016/j.bbamem.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 6.Takabe K, Paugh SW, Milstien S, Spiegel S. Pharmacol Rev. 2008;60:181–95. doi: 10.1124/pr.107.07113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Blitterswijk WJ, van der Luit AH, Veldman RJ, Verheij M, Borst J. Biochem J. 2003;369:199–211. doi: 10.1042/BJ20021528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Billich A, Baumruker T. Subcell Biochem. 2008;49:487–522. doi: 10.1007/978-1-4020-8831-5_19. [DOI] [PubMed] [Google Scholar]
- 9.Vadas M, Xia P, McCaughan G, Gamble J. Biochim Biophys Acta. 2008;1781:442–7. doi: 10.1016/j.bbalip.2008.06.007. [DOI] [PubMed] [Google Scholar]
- 10.Shida D, Takabe K, Kapitonov D, Milstien S, Spiegel S. Curr Drug Targets. 2008;9:662–73. doi: 10.2174/138945008785132402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oskeritzian CA, Milstien S, Spiegel S. Pharmacol Ther. 2007;115:390–9. doi: 10.1016/j.pharmthera.2007.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Allende ML, et al. J Biol Chem. 2004;279:52487–92. doi: 10.1074/jbc.M406512200. [DOI] [PubMed] [Google Scholar]
- 13.Zemann B, et al. Blood. 2006;107:1454–8. doi: 10.1182/blood-2005-07-2628. [DOI] [PubMed] [Google Scholar]
- 14.Mizugishi K, Yamashita T, Olivera A, Miller GF, Spiegel S, Proia RL. Mol Cell Biol. 2005;25:11113–21. doi: 10.1128/MCB.25.24.11113-11121.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hait NC, Sarkar S, Le Stunff H, Mikami A, Maceyka M, Milstien S, Spiegel S. J Biol Chem. 2005;280:29462–9. doi: 10.1074/jbc.M502922200. [DOI] [PubMed] [Google Scholar]
- 16.Olivera A, Spiegel S. Nature. 1993;365:557–60. doi: 10.1038/365557a0. [DOI] [PubMed] [Google Scholar]
- 17.Mazurek N, Megidish T, Hakomori S, Igarashi Y. Biochem Biophys Res Commun. 1994;198:1–9. doi: 10.1006/bbrc.1994.1001. [DOI] [PubMed] [Google Scholar]
- 18.Johnson KR, Becker KP, Facchinetti MM, Hannun YA, Obeid LM. J Biol Chem. 2002;277:35257–62. doi: 10.1074/jbc.M203033200. [DOI] [PubMed] [Google Scholar]
- 19.Xia P, et al. Proc Natl Acad Sci U S A. 1998;95:14196–201. doi: 10.1073/pnas.95.24.14196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meyer zu Heringdorf D, Lass H, Kuchar I, Lipinski M, Alemany R, Rumenapp U, Jakobs KH. Eur J Pharmacol. 2001;414:145–54. doi: 10.1016/s0014-2999(01)00789-0. [DOI] [PubMed] [Google Scholar]
- 21.Sarkar S, Maceyka M, Hait NC, Paugh SW, Sankala H, Milstien S, Spiegel S. FEBS Lett. 2005;579:5313–7. doi: 10.1016/j.febslet.2005.08.055. [DOI] [PubMed] [Google Scholar]
- 22.Rius RA, Edsall LC, Spiegel S. FEBS Lett. 1997;417:173–6. doi: 10.1016/s0014-5793(97)01277-5. [DOI] [PubMed] [Google Scholar]
- 23.Yamanaka M, et al. J Biol Chem. 2004;279:53994–4001. doi: 10.1074/jbc.M410144200. [DOI] [PubMed] [Google Scholar]
- 24.Hait NC, Bellamy A, Milstien S, Kordula T, Spiegel S. J Biol Chem. 2007;282:12058–65. doi: 10.1074/jbc.M609559200. [DOI] [PubMed] [Google Scholar]
- 25.Liu H, et al. J Biol Chem. 2003;278:40330–6. doi: 10.1074/jbc.M304455200. [DOI] [PubMed] [Google Scholar]
- 26.Pitson SM, Moretti PA, Zebol JR, Lynn HE, Xia P, Vadas MA, Wattenberg BW. Embo J. 2003;22:5491–500. doi: 10.1093/emboj/cdg540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pitson SM, Xia P, Leclercq TM, Moretti PA, Zebol JR, Lynn HE, Wattenberg BW, Vadas MA. J Exp Med. 2005;201:49–54. doi: 10.1084/jem.20040559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stahelin RV, Hwang JH, Kim JH, Park ZY, Johnson KR, Obeid LM, Cho W. J Biol Chem. 2005;280:43030–8. doi: 10.1074/jbc.M507574200. [DOI] [PubMed] [Google Scholar]
- 29.Hengst JA, Francy-Guilford JM, Fox TE, Wang X, Conroy EJ, Yun JK. Arch Biochem Biophys. 2009 doi: 10.1016/j.abb.2009.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shu X, Wu W, Mosteller RD, Broek D. Mol Cell Biol. 2002;22:7758–68. doi: 10.1128/MCB.22.22.7758-7768.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sutherland CM, Moretti PA, Hewitt NM, Bagley CJ, Vadas MA, Pitson SM. J Biol Chem. 2006;281:11693–701. doi: 10.1074/jbc.M601042200. [DOI] [PubMed] [Google Scholar]
- 32.Olivera A, Rosenthal J, Spiegel S. Anal Biochem. 1994;223:306–12. doi: 10.1006/abio.1994.1589. [DOI] [PubMed] [Google Scholar]
- 33.Bligh EG, Dyer WJ. Can J Biochem Physiol. 1959;37:911–7. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 34.Itagaki K, Yun JK, Hengst JA, Yatani A, Hauser CJ, Spolarics Z, Deitch EA. J Pharmacol Exp Ther. 2007 doi: 10.1124/jpet.107.121210. [DOI] [PubMed] [Google Scholar]
- 35.Kralik SF, Du X, Patel C, Walsh JP. Anal Biochem. 2001;294:190–3. doi: 10.1006/abio.2001.5166. [DOI] [PubMed] [Google Scholar]
- 36.Pitson SM, D’Andrea RJ, Vandeleur L, Moretti PA, Xia P, Gamble JR, Vadas MA, Wattenberg BW. Biochem J. 2000;350(Pt 2):429–41. [PMC free article] [PubMed] [Google Scholar]
- 37.Ouali M, King RD. Protein Sci. 2000;9:1162–76. doi: 10.1110/ps.9.6.1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pitson SM, et al. J Biol Chem. 2002;277:49545–53. doi: 10.1074/jbc.M206687200. [DOI] [PubMed] [Google Scholar]
- 39.Hernandez A, Ruiz MT. Bioinformatics. 1998;14:227–8. doi: 10.1093/bioinformatics/14.2.227. [DOI] [PubMed] [Google Scholar]
- 40.Barr RK, Lynn HE, Moretti PA, Khew-Goodall Y, Pitson SM. J Biol Chem. 2008;283:34994–5002. doi: 10.1074/jbc.M804658200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nava VE, et al. FEBS Lett. 2000;473:81–4. doi: 10.1016/s0014-5793(00)01510-6. [DOI] [PubMed] [Google Scholar]
- 42.Yokota S, Taniguchi Y, Kihara A, Mitsutake S, Igarashi Y. FEBS Lett. 2004;578:106–10. doi: 10.1016/j.febslet.2004.10.081. [DOI] [PubMed] [Google Scholar]
- 43.House C, Kemp BE. Science. 1987;238:1726–8. doi: 10.1126/science.3686012. [DOI] [PubMed] [Google Scholar]
- 44.Pitson SM, Moretti PA, Zebol JR, Vadas MA, D’Andrea RJ, Wattenberg BW. FEBS Lett. 2001;509:169–73. doi: 10.1016/s0014-5793(01)03162-3. [DOI] [PubMed] [Google Scholar]
- 45.Zlatkine P, Mehul B, Magee AI. J Cell Sci. 1997;110 (Pt 5):673–9. doi: 10.1242/jcs.110.5.673. [DOI] [PubMed] [Google Scholar]
- 46.Xia P, et al. J Biol Chem. 2002;277:7996–8003. doi: 10.1074/jbc.M111423200. [DOI] [PubMed] [Google Scholar]