Abstract
Yeast peroxisomal NADP+-specific isocitrate dehydrogenase (IDP3) contains a canonical type I peroxisomal targeting sequence (a carboxyl-terminal Cys-Lys-Leu tripeptide), and provides the NADPH required for β-oxidation of some fatty acids in that organelle. Cytosolic yeast IDP2 carrying a PTS1 (IDP2+CKL) was only partially localized to peroxisomes, and the enzyme was able to function in lieu of either peroxisomal IDP3 or cytosolic IDP2. The analogous isocitrate dehydrogenase enzyme (IDPA) from Aspergillus nidulans, irrespective of the presence or absence of a putative PTS1, was found to exhibit patterns of dual compartmental distribution and of dual function in yeast similar to those observed for IDP2+CKL. To test a potential cellular limit on peroxisomal levels, authentic yeast IDP3, which is normally strictly peroxisomal, was over-expressed. This also resulted in dual distribution and function of the enzyme in both the cytosol and in peroxisomes, supporting the possibility of a restriction on organellar amounts of IDP.
Keywords: isocitrate dehydrogenase, NADP(H), antioxidant, β-oxidation, peroxisomes
Introduction
Because of their critical roles in energy metabolism and biosynthesis, the isocitrate dehydrogenases of multiple organisms have been analyzed with respect to structure and function. These enzymes catalyze the oxidation of isocitrate to form α-ketoglutarate with concomitant reduction of NAD(P)+. Unlike eukaryotic NAD+-dependent isocitrate dehydrogenase, a structurally complex and allosterically regulated enzyme in the tricarboxylic acid cycle, the NADP+-dependent isocitrate dehydrogenases are homodimers that are not subject to allosteric regulation [1–4].
In Saccharomyces cerevisiae, there are three homologous but genetically distinct and differentially compartmentalized NADP+-dependent isocitrate dehydrogenase isozymes: mitochondrial IDP1 [1], cytosolic IDP2 [2], and peroxisomal IDP3 [3, 4]. Although sharing >77% sequence identity, the yeast isozymes differ with respect to expression, kinetic properties, and contributions to metabolism. Yeast mitochondrial IDP1 is constitutively expressed and plays an ancillary role in cellular glutamate synthesis [5], while IDP2 is expressed during the diauxic shift, i.e. after glucose is exhausted and nonfermentable carbon sources are utilized [6, 7]. IDP2, in conjunction with glucose-6-phosphate dehydrogenase (ZWF1), the first enzyme of the pentose phosphate pathway, are the predominant cytosolic sources of NADPH for thiol-based antioxidant systems [8, 9]. Yeast peroxisomal IDP3 expression is induced by growth with fatty acid carbon sources, and provides NADPH for a specific reaction in the β-oxidation of unsaturated fatty acids containing a double bond following an even-numbered carbon [3, 4].
Our studies of the yeast IDPs have provided useful strains that allow phenotypic tests of compensation for the individual isozymes. For example, a mutant strain (idp2Δzwf1Δ) lacking the crucial cytosolic sources of NADPH exhibits lethality in medium with acetate or oleate as carbon sources due to an accumulation of deleterious by-products of oxidative metabolic pathways [8, 9]; growth under these conditions can be restored by expression of a functional IDP enzyme in the cytosol. In contrast, a strain (idp3Δ) lacking an active peroxisomal enzyme can grow with oleate as the carbon source but is unable to grow with petroselinate2, since β-oxidation of the latter but not of the former fatty acid requires peroxisomal IDP activity [3, 4]. We previously used these and other mutant strains for expression of a mammalian IDP enzyme (designated IDH1) that contains a type I peroxisomal targeting signal (PTS1, in this case a carboxyl-terminal Ala-Lys-Leu tripeptide [10]). The mammalian enzyme was found to be localized in both cytosolic and peroxisomal compartments in yeast cells and could functionally replace both IDP2 and IDP3 [11]. Dual compartmental localization of the enzyme has also been reported for the enzyme in mammalian cells [10], so comparable functions in cytosolic antioxidant systems and in peroxisomal β-oxidation can be speculated based on the studies in yeast. Interference with these functions may be relevant to the recent finding that mutations in the human enzyme have been identified in a significant number of low-grade gliomas [12, 13].
Because a single IDP enzyme in mammalian cells appears adequate to perform functions catalyzed by genetically unique IDP2 and IDP3 isozymes in yeast cells, we wished to examine, in particular, if peroxisomal localization of the normally cytosolic yeast IDP2 enzyme would permit compensation for physiological functions of the IDP3 enzyme. Another question of interest was whether the addition of a PTS1 to IDP2 would be sufficient to obtain peroxisomal localization or, alternatively, would result in the dual cytosolic and peroxisomal localization observed for the mammalian enzyme as described above [11].
Despite the significant homology among yeast IDP isozymes, they demonstrate some substantial differences in kinetic and physical properties. As previously reported [11], yeast IDP2 exhibits a narrow pH range for optimal activity with a peak at pH 7.5, and the measured pI value for the purified enzyme is 6.5. These values seem appropriate for an enzyme that functions in the cytosol, a cellular compartment with reported pH values of 7–7.2 [14, 15]. Both organellar yeast enzymes have broader pH ranges (7.5–9) for optimal activity, and both exhibit more basic pI values (8.2 for mitochondrial IDP1 and 8.9 for peroxisomal IDP3), properties more consistent for function under basic cellular conditions (pH 7.5 for the mitochondrial matrix [15] and pH 8.2 for the peroxisomal lumen [14]). Based on these physical and kinetic properties, our original prediction for current studies was that expression of IDP2 in peroxisomes might not compensate for loss of IDP3, whereas expression of IDP1 in peroxisomes might be more effective.
As a control in these studies, we examined results of expression of the IDP isozyme (IDPA) from Aspergillus nidulans, which also contains three differentially compartmentalized isozymes. These isozymes, however, are products of a single gene [16]. The mitochondrial enzyme is produced from a longer transcript of the gene that contains codons for a putative mitochondrial targeting sequence. A shorter transcript lacking the mitochondrial targeting sequence is translated to produce an enzyme that, while containing codons for a potential type I peroxisomal targeting sequence (a carboxyl-terminal Ala-Arg-Leu tripeptide), is apparently localized in both the cytosol and in peroxisomes [16]. In the current study, we wished to examine the relevance of this PTS1 sequence in dual compartmental localization and function of the same enzyme.
Experimental procedures
Yeast strains and growth conditions
All yeast mutants were derived from the parental haploid strain MMY011 (MAT ade2–1 his3–11,15 leu2–3,112 trp1–1 can 1–100), a strain previously used to examine yeast peroxisomal proteins due to its robust growth with oleate as a carbon source [17]. The idp2Δ, idp3Δ, idp2Δidp3Δ, idp2Δzwf1Δ, and idp2Δidp3Δzwf1Δ mutant strains were previously described [8, 11]. To disrupt the yeast IDP1 gene in idp3Δ or idp2Δidp3Δ mutants, a DNA fragment containing a heterologous Schizosaccharomyces pombe HIS5 gene flanked by IDP1 sequences was generated by polymerase chain reaction (PCR) and transformed into the mutant strains [18]. Yeast strains were cultivated using rich YP medium (1% yeast extract, 2% Bacto-peptone) or minimal YNB medium (0.17% yeast nitrogen base, 0.5% ammonium sulfate, pH 6.5) with appropriate supplements of 20 mg/ml to satisfy auxotrophic requirements for growth. Carbon sources were glucose or ethanol added to 2%. Oleate or petroselinate was added to 0.1% with 0.2% Tween 40 to increase solubility. Prior to growth with fatty acids, yeast strains were cultivated using rich medium plus 2% glycerol for 12 hours. Growth conditions for complementation analyses, as well as the methods for expression of tetracycline (doxycycline)-inducible proteins, were previously described [11].
Plasmid constructions
For expression of IDP2 and IDP2+CKL, DNA fragments containing both promoter and coding regions of the IDP2 gene were amplified by PCR using the genomic DNA of the parental strain as the template, and using 3′-oligonucleotides respectively lacking or containing codons for a carboxyl-terminal PTS1 (Cys-Lys-Leu). [Sequences for all oligonucletides used in these studies are presented in Supplementary Material.] The IDP1+CKL gene generated by PCR lacked the mitochondrial targeting sequence and contained codons for a Cys-Lys-Leu PTS1 tripeptide added on the 3′ end of the coding region. The IDP1+CKL gene was subcloned into pRS315 with the yeast IDP2 promoter to provide appropriate patterns of expression. For over-expression of IDP3, a DNA fragment containing the promoter and coding sequences of the IDP3 gene was generated from genomic DNA by PCR and subcloned into pRS424, a multicopy 2μ-based yeast plasmid [19].
To express A. nidulans IDPA isozymes in yeast, the coding region for the cytosolic/peroxisomal protein (IDPA) was generated by PCR using a cDNA template [16] and subcloned into pCM252 (EUROSCARF [20]), a centromere-based plasmid containing a tetracycline-inducible promoter. This plasmid (pCM-IDPA) lacked the mitochondrial targeting sequence and could express both cytosolic and peroxisomal forms of IDPA. For expression of only cytosolic IDPA, PCR was used to construct a form of the IDPA coding region (IDPA−ARL) lacking codons for the putative carboxyl-terminal PTS1, generating plasmid pCM-IDPA−ARL. Plasmids were transformed initially into a yeast mutant (idp1Δidp2Δidp3Δ) with disruptions in all IDP chromosomal loci to assess amounts of doxycycline needed to produce levels of IDPA and IDPA−ARL comparable to those of endogenous IDP2 in the parental yeast strain. The plasmids were then transformed into various yeast mutants to assess compartmental localization and the extent of physiological complementation for yeast IDP isozymes.
Subcellular fractionation and immunoblot analyses
The parental strain and mutant transformants were transferred from minimal selective medium for cultivation in rich YP medium with oleate or petroselinate as carbon sources. When culture A600nm values reached ~2, organellar pellets enriched for peroxisomes and mitochondria, and cytosolic supernatant fractions were prepared from cellular homogenates by differential centrifugation [21]. Enzyme assays were performed as previously described [11], and protein concentrations were determined using the Bradford dye binding assay [22].
Protein samples (5–10 μg) of cellular homogenates and of cytosolic or organellar fractions were electrophoresed using 10% polyacrylamide/sodium dodecyl sulfate gels [23]. Immunoblot analyses were conducted using an antiserum that reacts with both yeast IDP1 and IDP2 [7], as well as with IDPA [16]. Other antisera used to determine cellular fractionation efficiency were specific for yeast enzymes including IDP3 [3] and peroxisomal MDH3 [24]. An antiserum for β-actin (Abcam) was used to confirm that cytosolic contamination of organellar fractions was minimal. Immunoreactivity was detected using the enhanced chemiluminescent method, and densitometry was conducted using ImageJ software. To quantify levels of IDP or IDPA enzymes in cellular and organellar fractions, samples of known concentrations of the enzymes purified by affinity chromatography, as previously described [11] or as detailed below, were co-electrophoresed with cellular protein samples for immunoblots and densitometric comparison.
Expression, purification, and kinetic analyses of histidine-tagged form of IDPA
For expression in Escherichia coli and subsequent affinity purification, the coding region for the IDPA protein with six adjacent histidine codons added onto 3′ end of the gene was constructed using PCR and subcloned into plasmid pET17b (Novagen). The plasmid was transformed into E. coli strain BL21(DE3) (Novagen), and expression was induced by addition of isopropyl β--D-thiogalactopyranoside to 1 mM when culture A600nm values reached 0.7; incubation was continued for 6 h at 30°C. IDPA was purified using Ni2+ –nitrilotriacetic acid (NTA) affinity chromatography (QIAGEN), resulting in yields of 10 mg enzyme/liter.
Enzyme assays were conducted at A340nm as previously described [11]. pH optima with respect to isocitrate were determined using saturation kinetic curves conducted at various pH values with assays containing buffers of either 50 mM Tris-HCl (for pH values ranging from 6 to 9) or 50 mM MES-NaOH (for pH 5.5). Assays contained 3 mM MgCl2, 0.25 mM NADP+, and concentrations of D-isocitrate ranging from 0 to 0.7 mM. pH optima with respect to NADP+ were determined using similar saturation kinetic curves conducted with 1 mM D-isocitrate and concentrations of NADP+ ranging from 0 to 0.4 mM. Protein concentrations for purified enzymes were determined using molar extinction coefficients calculated as described by Pace et al. [25]. Isoelectric focusing was performed to determine pI values for purified proteins using commercial gels (pH 3–10, Invitrogen).
Results
Expression and tests for complementation by a peroxisomal form of yeast IDP2
To obtain peroxisomal localization of the normally cytosolic yeast IDP2 enzyme, PCR was used to generate an IDP2+CKL gene containing the IDP2 promoter and coding region, with additional codons added onto the 3′-end of the coding region for a Cys-Lys-Leu tripeptide. This tripeptide is the PTS1 sequence for peroxisomal yeast IDP3, and has been shown to be necessary for organellar localization of that isozyme [3, 4]. Aligned amino and carboxyl terminal sequences for proteins in this study are shown in Fig. 1. The authentic IDP2 gene was also generated for use as a control in transformations. The IDP2 and IDP2+CKL genes were subcloned into pRS315, a centromere-based plasmid for single-copy expression in yeast [19], and the plasmids used for transformation of various mutant strains. Following growth of transformants with rich YP medium containing fatty acid carbon sources to induce peroxisome proliferation [17, 26], cell pellets were fractionated to obtain samples enriched in cytosolic or organellar (mitochondria plus peroxisomes) proteins. Immunoblot analysis was conducted using an antiserum that recognizes IDP1 and IDP2 [7].
Fig. 1.
Aligned amino and carboxyl terminal sequences for IDP enzymes. The amino terminus of yeast IDP1 was determined by amino acid sequence analysis [1]. Amino termini for yeast IDP2 and IDP3 [2–3] and for the non-mitochondrial form of IDPA [16] were assigned based on nucleotide sequences, the location of likely translation initiation methionine residues, and alignments with IDP1 [2–3].
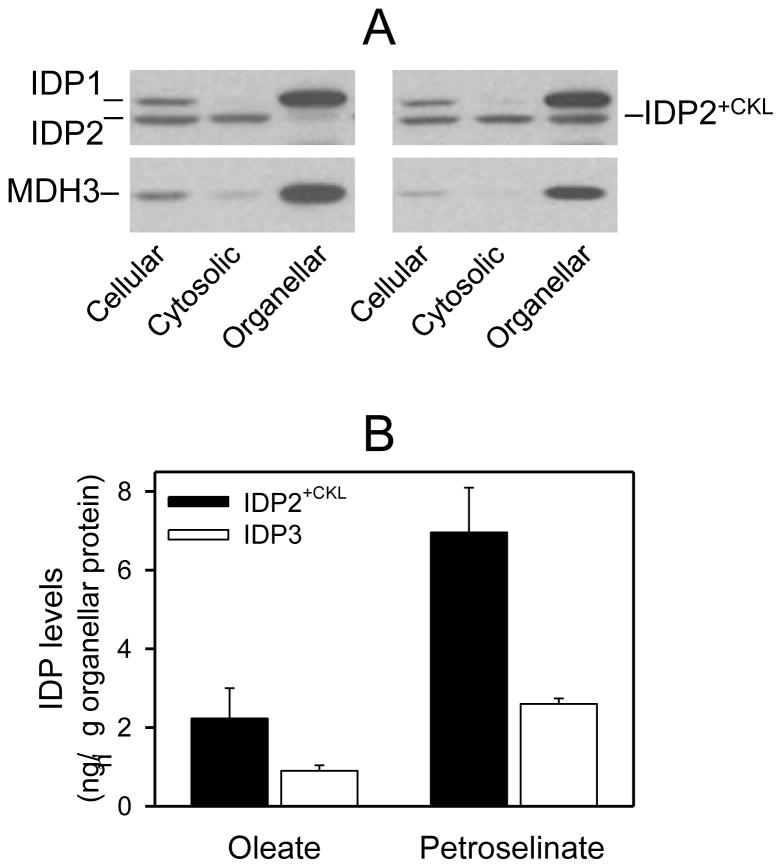
As illustrated in Fig. 2A for an idp2Δ transformant expressing the authentic plasmid-borne IDP2 gene (left panels) and grown using YP petroselinate medium, cellular levels of IDP1 and IDP2 were approximately equivalent (as they are in parental cells grown under this condition [11]). IDP2 was primarily located in the cytosolic fraction, whereas endogenous mitochondrial IDP1 was located in the organellar fraction. For an idp2Δ transformant expressing IDP2+CKL (right panels), cellular levels of IDP1 and IDP2 were approximately equivalent, but IDP2+CKL was partitioned between cytosolic and peroxisomal compartments. An antiserum for peroxisomal MDH3 [24] was used in these experiments as a control to assess the degree of peroxisomal rupture during cellular fractionation resulting in detection of some MDH3 in the cytosolic fraction (Fig. 2A).
Fig. 2.
Expression and localization of IDP2+CKL. [A] Yeast idp2Δidp3Δ transformants expressing plasmid-borne copies of the IDP2 gene (left panels) or the IDP2+CKL gene (right panels) were grown using YP oleate medium to A600nm = 2. Cellular, cytosolic, and organellar protein extracts (5 μg each) prepared from transformants were electrophoresed for immunoblot analysis using an antiserum for IDP1/IDP2 [7] or an antiserum for peroxisomal MDH3 [24]. [B] Quantitative results are show for densitometric analyses of multiple immunoblots (similar to those shown in [A]) used to visualize organellar isozyme levels. Organellar levels of IDP3 in the parental strain and of IDP2+CKL in transformed mutant strains (idp2Δidp3Δ, idp2Δidp3Δzwf1Δ, and idp2Δzwf1Δ) used for complementation analyses were determined for strains grown in YP oleate or YP petroselinate medium. Organellar levels were quantified relative to samples of purified IDP2 and IDP3 as described under Experimental Procedures.
Multiple similar immunoblots of cellular fractions from various transformant strains expressing IDP2 or IDP2+CKL and grown with either oleate or petroselinate as the carbon source were used for densitometric analyses. Values were compared with those for known concentrations of co-electrophoresed samples of purified IDP isozymes. These analyses showed that cellular levels (as well as activities) of IDP2+CKL were similar to those for authentic IDP2. IDP2 was located almost entirely in the cytosolic fraction, whereas 11% of total cellular IDP2+CKL was contained in the organellar fraction in transformants grown with oleate as the carbon source. The percentage of organellar IDP2+CKL increased to 32% of the total cellular level with petroselinate as the carbon source. Thus, the addition of a PTS1 signal to the carboxyl-terminus of IDP2 provides partial peroxisomal localization.
Similarly quantitative analyses of immunoblots demonstrated that, even though only a portion of the cellular IDP2+CKL in transformants was organellar, the actual levels of organellar IDP2+CKL exceeded those of the authentic peroxisomal IDP3 isozyme in control strains (Fig. 2B). Peroxisomal levels of both IDP2+CKL and IDP3 were three-fold higher in cells grown with petroselinate as the carbon source than those observed with oleate. However, organellar levels of IDP2+CKL were elevated two- to three-fold relative to those of IDP3 under both growth conditions. This suggests that organellar levels of the peroxisomal isozyme are increased by growth with petroselinate, a carbon source which requires a functional IDP isozyme for β-oxidation. These results, along with other similar results discussed below, also suggest a limit to the amount of IDP enzymes in peroxisomes, perhaps explaining the incomplete organellar localization of IDP2+CKL.
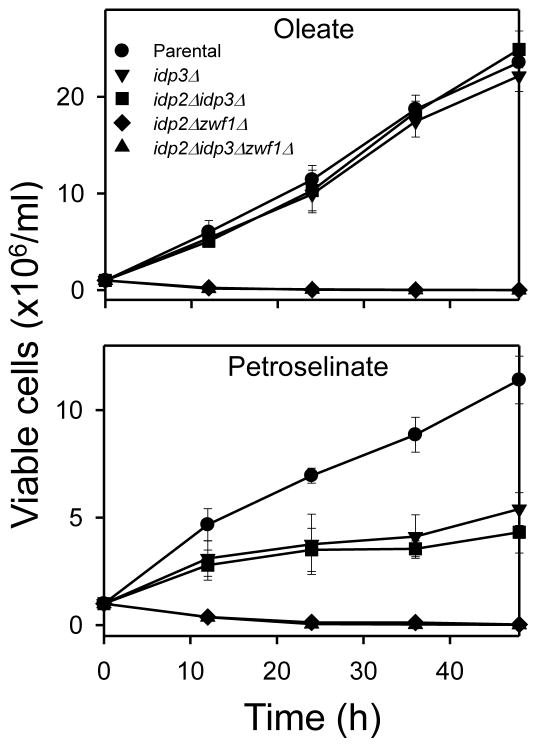
To assess the degree of physiological compensation of organellar IDP2+CKL for endogenous isozymes (and for other complementation experiments described below), we used a panel of yeast gene disruption strains with defined growth phenotypes when oleate or petroselinate is used as the carbon source. As shown in Fig. 3, test strains lacking IDP3 (idp3Δ[▼] or idp2Δidp3Δ[■]) grew as well as the parental strain [●] with oleate as the carbon source, but these mutant strains exhibited only one doubling of viable cell numbers and little additional growth with petroselinate as the carbon source. These strains, if transformed and expressing a competent peroxisomal IDP enzyme, would be expected to regain the ability to grow with petroselinate as the carbon source. Another strain lacking IDP2 and ZWF1 (idp2Δzwf1Δ [◆]) rapidly lost viability when grown with either fatty acid carbon source and provides a test for restoration of growth by expression of a functional cytosolic IDP enzyme. A similar strain (idp2Δidp3Δzwf1Δ[▲]) provides a test for restoration of growth by an enzyme that can compensate for functions of both cytosolic and peroxisomal IDP enzymes.
Fig. 3.
Growth of parental and mutant test strains. The indicated strains were grown in rich YP medium with oleate or petroselinate as the carbon source. Viable cell numbers were determined by plating 10-fold dilutions of the cultures onto YP glucose plates at the indicated times. Numbers represent three independent measurements (± S.D.).
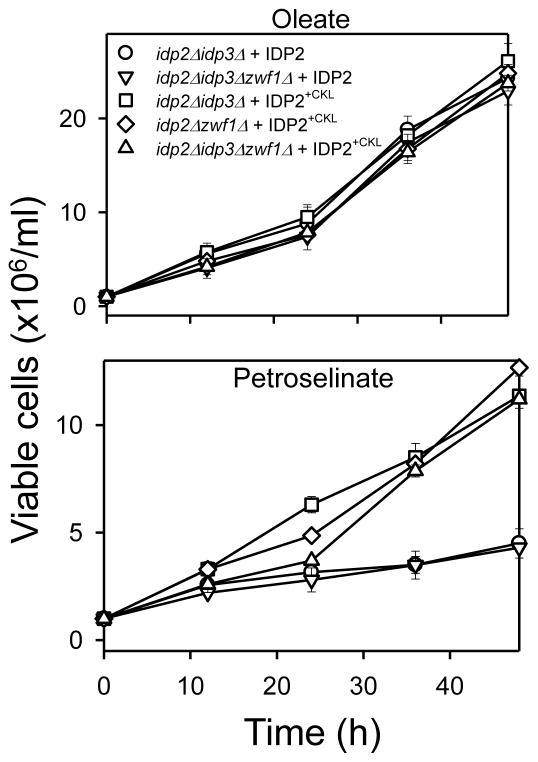
As shown in Fig. 4, expression of the authentic IDP2 enzyme in test disruption strains did not compensate for the absence of IDP3 in the idp2Δidp3Δ[○] or the idp2Δidp3Δzwf1Δ[▽] strain, since these transformants were unable to grow with petroselinate as the carbon source. However, expression of IDP2 in the latter strain did prevent loss of viability under this condition, indicating complementation for cytosolic IDP2 as expected. In contrast, expression of IDP2+CKL provided complementation for both IDP2 and IDP3, permitting growth of mutants lacking either or both isozymes (idp2Δidp3Δ [□], idp2Δzwf1Δ [◇], and idp2Δidp3Δzwf1Δ [△]) using medium containing oleate or petroselinate as the carbon source. This indicates that the dual compartmental localization of IDP2+CKL provides functional compensation in both cellular compartments.
Fig. 4.
Growth complementation tests for IDP2+CKL. The indicated transformant strains (control strains expressing IDP2 and test strains expressing IDP2+CKL) were grown in rich YP medium with oleate or petroselinate as the carbon source. Viable cell numbers were determined by plating dilutions of the cultures onto YP glucose plates at the indicated times. Numbers represent three independent measurements (± S.D.).
Expression and tests for complementation by a peroxisomal form of yeast IDP1
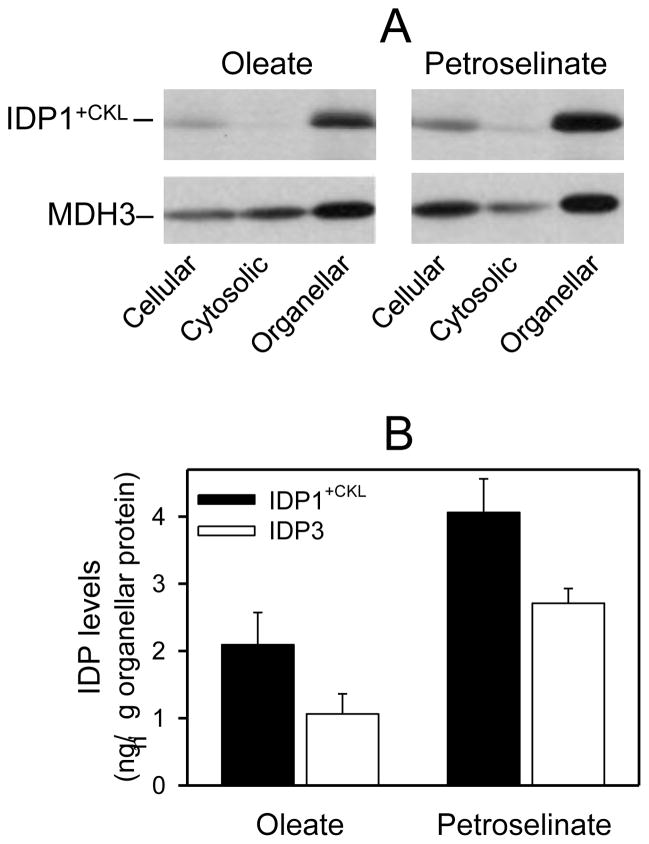
We have previously shown that expression of an IDP1 gene lacking its mitochondrial targeting pre-sequence results in cytosolic localization of IDP1 [27]. To obtain peroxisomal localization of IDP1, PCR was used to generate a form of this truncated IDP1 gene containing codons for the PTS1 Cys-Lys-Leu tripeptide at the 3′-end of the coding region. Also, to ensure appropriate expression of the enzyme during growth on non-fermentable carbon sources, the altered IDP1+CKL gene was linked to the IDP2 promoter. This construct was subcloned into pRS315, and the plasmid used for transformation of test strains lacking IDP1 and/or other IDP isozymes. Immunoblot analyses of cellular fractions obtained from these transformants indicated that essentially all of the IDP1+CKL protein was associated with organellar fractions, as illustrated for an idp1Δidp2Δidp3Δ transformant in Fig. 5A. Densitometry, conducted as described above, was used to quantify IDP1+CKL and marker protein levels in cellular and organellar fractions. As shown in Fig. 5B, organellar levels of IDP1+CKL in yeast transformants exceeded those of authentic IDP3 in control strains expressing the endogenous peroxisomal enzyme, by ~two-fold in cells grown with oleate and by ~50% in cells grown with petroselinate as the carbon source. Expression of both IDP1+CKL and IDP3 were ~two-fold higher in organellar fractions of cells grown with petroselinate versus oleate as the carbon source, again suggesting an increase in peroxisomal levels under conditions requiring peroxisomal IDP activity for efficient growth.
Fig. 5.
Organellar localization of IDP1+CKL. [A] Yeast idp1Δidp2Δidp3Δ transformants expressing plasmid-borne copy of the IDP1+CKL gene were grown using YP oleate or petroselinate medium as indicated to A600nm = 2. Cellular, cytosolic, and organellar protein extracts (5 μg each) prepared from transformants were electrophoresed for immunoblot analysis using an antiserum for IDP1/IDP2 [7] or an antiserum for peroxisomal MDH3 [24]. [B] Quantitative results are shown for densitometric analyses of multiple immunoblots (similar to those shown in [A]) used to visualize organellar levels of IDP3 in the parental strain and of IDP1+CKL in transformed mutant strains (idp1Δ, Δidp3, and idp1Δidp3Δ) used for complementation analyses. Organellar levels were quantified relative to samples of purified IDP1 and IDP3 as described under Experimental Procedures.
Since the IDP1+CKL enzyme was localized entirely in the organellar pellet, we assessed the level of physiological compensation using yeast gene disruption strains lacking IDP3. As shown in Fig. 6, expression and organellar localization of IDP1+CKL was not adequate to compensate for loss of IDP3, since idp3Δ [▽] and idp1Δidp3Δ [□] transformants exhibited growth patterns similar to those observed for the untransformed strains in medium with petroselinate as the carbon source (see Fig. 3). [A control transformant strain [◦] containing IDP3 but lacking IDP1 grew with petroselinate as the carbon source, indicating that expression of IDP1+CKL does not interfere with other peroxisomal functions.] Since immunochemical levels of IDP1+CKL in organellar fractions of these transformants was substantial and even higher than levels of IDP3 in organellar pellets from control strains (Fig. 5), we assayed cellular and organellar levels of IDP activity. Activity due to expression of IDP1+CKL was only ~5% of that due to similar levels of endogenous IDP3 and, in fact, activity due to IDP1+CKL was much lower than the activity measured for a similar immunochemical level of endogenous mitochondrial IDP1 in the parental strain. Thus, one possibility, as discussed below in the context of functional and structural comparisons, is that IDP1+CKL may be unable to adopt an active conformation in the peroxisomal matrix.
Fig. 6.
Growth complementation tests for IDP1+CKL. The indicated transformant strains expressing IDP1+CKL were grown in rich YP medium with oleate or petroselinate as the carbon source. Viable cell numbers were determined by plating 10-fold dilutions of the cultures onto YP glucose plates at the indicated times. Numbers represent three independent measurements (± S.D.).
Results of these studies suggest that yeast IDP2+CKL can function in lieu of IDP3 when the normally cytosolic enzyme is partially localized to peroxisomes. Thus, the neutral pH optimum for catalysis and the neutral pI value of IDP2 do not prevent function in the basic pH environment of the peroxisomal matrix, contrary to our original prediction. In contrast, the normally mitochondrial IDP1 enzyme, despite its basic pH optimum for catalysis and its basic pI value, is apparently inactive when localized to peroxisomes. This suggests that the environment of the mitochondrial matrix may be important for catalytic activity and/or that factors in that compartment are important for maturation of the active IDP1 homodimer.
Kinetic and physical properties of A. nidulans IDP
Because of the differences we found in the ability of yeast IDP2 and IDP1 isozymes to function in peroxisomes, we wished to compare properties of A. nidulans IDP (IDPA) with those of the yeast enzymes. As described above, the same gene in A. nidulans is expressed to produce mitochondrial, peroxisomal, and cytosolic isozymes in fungal cells [16]. The mitochondrial isozyme is generated from a longer transcript containing a putative mitochondrial targeting signal, whereas the peroxisomal and cytosolic enzymes are generated from a shorter transcript of the same gene.
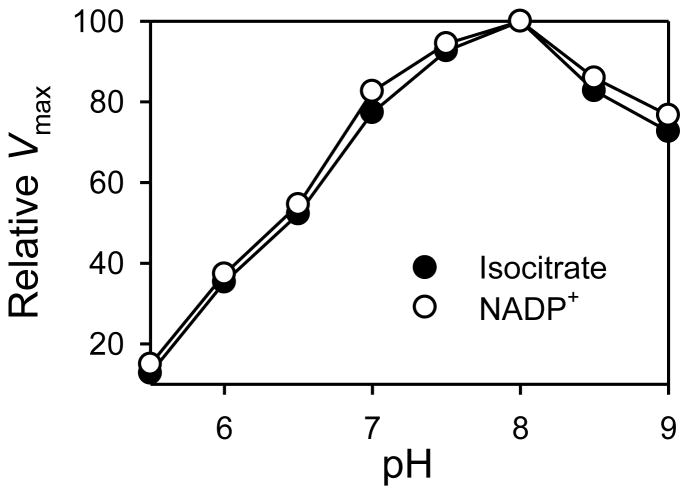
To analyze kinetic and physical properties of IDPA, the shorter coding region was expressed in E. coli and affinity purified using Ni2+-NTA chromatography. To determine the optimal pH for catalysis, apparent Vmax values were determined with respect to either isocitrate or NADP+ using saturation kinetic analyses conducted at various pH values ranging from 5.5–9.0. As shown in Fig. 7, IDPA exhibited similar broad optimal pH ranges with both substrate and cofactor, with peaks at pH 8.0 but with at least 80% of the peak value observed from pH 7.0–9.0. Thus, optimal pH values for catalysis by IDPA overlap those of all the yeast isozymes, but the broad optimum in the basic pH ranges is more similar to those exhibited by yeast mitochondrial IDP1 and peroxisomal IDP3 [11]. The experimentally determined pI value for IDPA was 7.4. This value is intermediate between that measured for yeast cytosolic IDP2 (6.5) and those measured for organellar IDP1 (8.2) or IDP3 (8.9) isozymes [11].
Fig. 7.
pH optima for the IDPA enzyme. Saturation velocity curves with respect to substrate (D-isocitrate) or cofactor (NADP+) were determined for the purified IDPA enzyme at each indicated pH value. Apparent Vmax values were expressed relative to the maximum value (109 ± 1.5 units/mg) obtained at pH 8.0.
With respect to kinetic parameters summarized in Table 1, IDPA was found to be unique but shared some characteristics with each of the yeast isozymes. The apparent Vmax value for IDPA measured at the optimum pH of 8.0 was found to be similar to that for yeast IDP2. The apparent Km value for IDPA with respect to isocitrate was similar to that for IDP1, whereas the apparent Km value for IDPA with respect to NADP+ was similar to that for IDP3. The general similarities in kinetic and physical properties for IDPA and the various yeast isozymes suggest that IDPA should be functionally competent to substitute for the differentially compartmentalized yeast IDPs.
Table 1.
Kinetic properties of yeast and fungal enzymes.
| S. cerevisiaea |
A. nidulans |
|||
|---|---|---|---|---|
| IDP1 | IDP2 | IDP3 | IDPA | |
| Vmax (units/mg) | 65 (±17) | 126 (±25) | 48 (±1) | 109 (±1) |
| Km isocitrate (μM) | 30 (±0) | 220 (±30) | 15 (±1) | 30 (±1) |
| Km NADP+ (μM) | 30 (±0) | 20 (±0) | 10 (±2) | 9 (±1) |
Data previously presented in [11].
Expression in yeast and tests for complementation by IDPA
To examine complementation for yeast IDP2 and/or IDP3, we designed a construct containing the shorter IDPA coding region (i.e. lacking the mitochondrial targeting sequence). We also designed another construct containing this IDPA coding region but lacking the PTS1 Ala-Arg-Leu tripeptide. Although the latter form of IDPA (IDPA−ARL) is not expressed in A. nidulans [16], we wished to test the relevance of the PTS1 sequence to peroxisomal localization and to try to restrict localization of IDPA to the cytosol. These constructs were transferred to pCM252, a single-copy vector containing a tetracycline (doxycycline)-inducible promoter. We previously used this induction system to obtain desired expression levels of another heterologous protein in yeast [11].
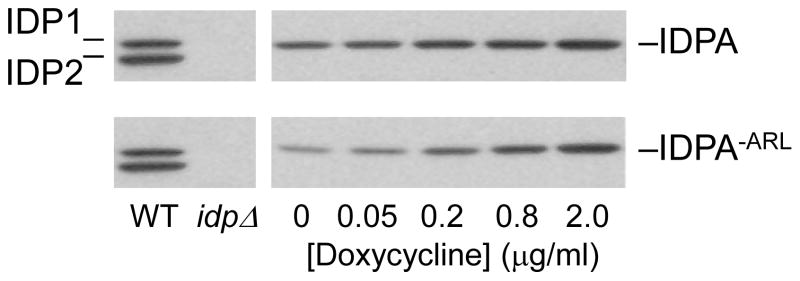
To assess levels of doxycycline needed to obtain levels of IDPA enzymes approximating those of the yeast IDPs, the pRS315 plasmids for expression of IDPA were used for transformation of a yeast strain (idp1Δidp2Δidp3Δ) lacking all endogenous IDP isozymes. Expression was monitored using immunoblots of protein extracts taken from transformants cultivated using medium with oleate as the carbon source and containing various concentrations of doxycycline (Fig. 8). Immunoblots were conducted using the yeast IDP antiserum, which we and others [16] have found to react well with IDPA. For both IDPA and IDPA−ARL, doxycycline concentrations of 2 μg/ml produced immunochemical levels of expression approximately equivalent to those of IDP2 in the parental strain. Similar levels of cellular IDP activity (0.3–0.4 units/mg cellular protein) were observed using enzyme assays with cellular extracts obtained after growth of the transformants with this concentration of doxycycline. Therefore, we used this concentration in subsequent complementation tests.
Fig. 8.
Expression of IDPA and IDPA−ARL enzymes in yeast. Immunoblot analysis of cellular protein extracts (5 μg each) was conducted to examine expression of IDPA and IDPA−ARL enzymes in an idp1Δidp2Δidp3Δ yeast strain (idpΔ). Expression was induced by growth of cultures in YP oleate medium for 24 h with the indicated concentrations of doxycycline. The IDPA enzymes were detected using the antiserum for yeast IDP1/IDP2 [7], and levels were compared with levels of IDP1 and IDP2 in the parental strain (WT).
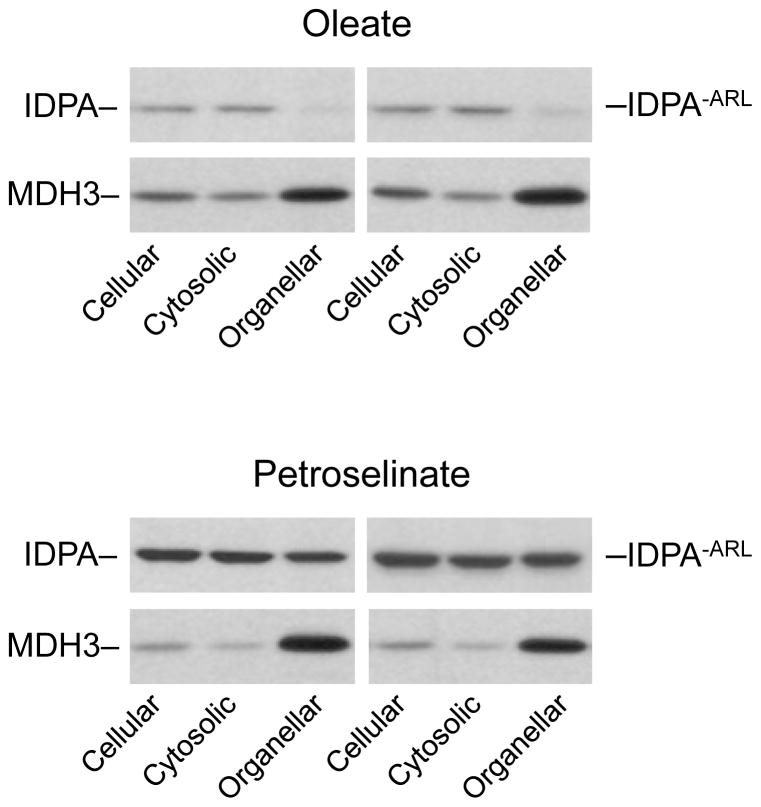
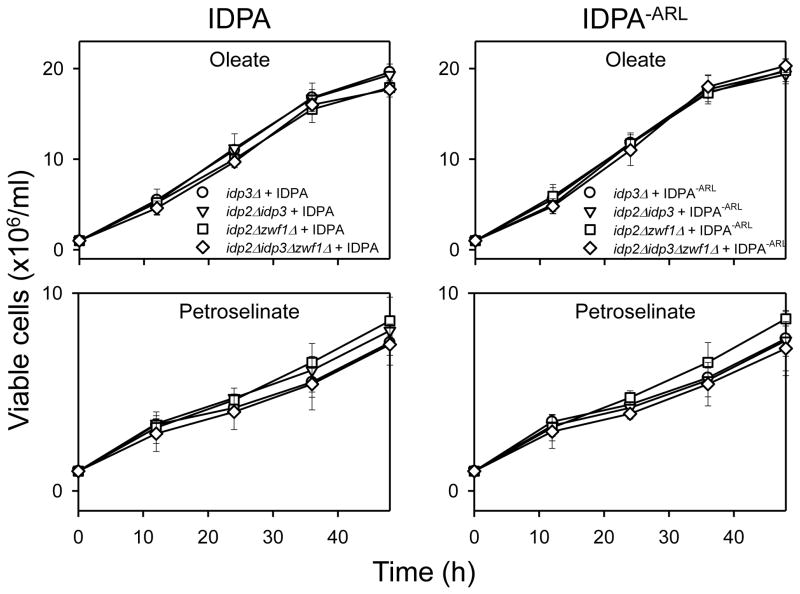
The compartmental distributions of IDPA and of IDPA−ARL enzymes were examined following growth of idp1Δidp2Δidp3Δ transformants expressing these enzymes in medium with oleate or petroselinate as the carbon source. With oleate as the carbon source (Fig. 9), overall cellular levels of both IDPA and IDPA−ARL were relatively low and, interestingly, both enzymes were primarily localized to the cytosolic compartment with only ~10% of the total cellular levels of both enzymes associated with organellar fractions. Cellular levels of both IDPA and IDPA−ARL were elevated, ~9-fold and ~5-fold, respectively, in cells grown with petroselinate versus oleate as the carbon source, and organellar levels of both enzymes represented ~20% of the total cellular levels. Thus, it appears that the carboxyl-terminal Ala-Arg-Leu tripeptide, which is present in IDPA but absent in IDPA−ARL, is irrelevant to the extent of peroxisomal localization of the protein. Also, since both enzymes were expressed using the same concentrations of doxycycline, the type of fatty acid used for growth apparently affects the stability of the IDPA enzymes in yeast.
Fig. 9.
Organellar localization of IDPA and IDPA−ARL enzymes. Cellular, cytosolic, and organellar protein extracts were prepared from idp1Δidp2Δidp3Δ transformants expressing IDPA (left panels) or IDPA−ARL (right panels). The transformant strains were cultivated to A600nm ≅ 2 using YP oleate or YP petroselinate medium supplemented with 2 μg/ml doxycycline. Protein extracts (5 μg each) were electrophoresed for immunoblot analysis using the antiserum for IDP1/IDP2 [7] or an antiserum for peroxisomal MDH3 [24].
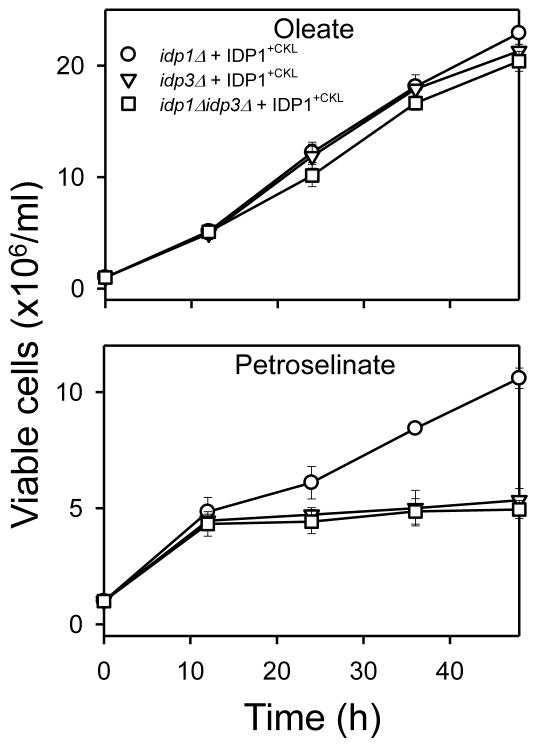
To examine the extent that IDPA and IDPA−ARL can functionally replace yeast IDP2 and IDP3, mutant strains described above (Fig. 3) were used for transformation and complementation analyses. As shown in Fig. 10, expression of either IDPA or IDPA−ARL was able to compensate for function of IDP2 in idp2Δzwf1Δ (□) and idp2Δidp3Δzwf1Δ (◇) transformants, preventing the loss of viability of the untransformed strains grown using medium with oleate or petroselinate as the carbon source. In addition, substantial growth with petroselinate as the carbon source was observed for idp3Δ (○) and idp2Δidp3Δ (▽) transformants expressing either IDPA or IDPA−ARL, suggesting that both enzymes can also compensate for the authentic yeast IDP3 enzyme. The similarity in complementation patterns obtained with IDPA or IDPA−ARL is consistent with the similarity in cellular distribution patterns for these enzymes (Fig. 9).
Fig. 10.
Growth complementation tests for IDPA and IDPA−ARL. The indicated transformant strains expressing IDPA (left panels) or IDPA−ARL (right panels) were grown in rich YP medium with oleate or petroselinate as the carbon source. Viable cell numbers were determined by plating dilutions of the cultures onto YP glucose plates at the indicated times. Numbers represent three independent measurements (± S.D.).
In addition to complementation for yeast IDP2 and IDP3 enzymes, we also found that a form of IDPA localized to mitochondria can compensate for loss of IDP1 in S. cerevisiae mutants. These results are presented in Supplemental Information
Over-expression of IDP3
As described above and as summarized in Table 2, expression of yeast IDP2+CKL, of IDPA, or of IDPA−ARL produced patterns of dual compartmentalization in peroxisomes and in the cytosol similar to those obtained following expression of the mammalian NADP+-specific IDH1 enzyme in yeast cells, as previously reported [11]. Possible explanations for this dual localization include (a) the presence of some sort of cytosolic ‘retention’ signal in these proteins and/or (b) the existence of some restriction on the amount of an IDP enzyme that can be imported into peroxisomes. The latter would produce, by default, cytosolic localization of some of the expressed enzymes. To distinguish between these possibilities, we over-expressed the yeast IDP3 isozyme. This enzyme, at normal levels of expression, is localized in the peroxisomal matrix, as previously established using both cellular fractionation methods [3, 4] and immunoelectron microscopy [4] and, thus, would lack a cytosolic retention signal.
Table 2.
Summary of cellular distribution of functional peroxisomal IDPs.
| Enzyme | Peroxisomal levelsa (ng/μg protein) |
||
|---|---|---|---|
| Oleate (% cellular) | Petroselinate (% cellular) | Difference | |
| IDP3b | 1.1 ± 0.3 (100) | 2.7 ± 0.2 (100) | 2.5-fold |
| IDP2+CKL | 2.2 ± 0.7 (11) | 7.0 ± 1.1 (32) | 3.2-fold |
| IDPA | 0.2 ± 0 (10) | 3.6 ± 0.8 (19) | 18-fold |
| IDPA−AKL | 0.5 ± 0.1 (11) | 4.6 ± 0.5 (22) | 9.2-fold |
| 2μ IDP3 | 9.0 ± 1.4 (56) | 9.0 ± 1.3 (56) | none |
Numbers represent three or four independent measurements (±S.D.). The percentage of total cellular levels represented by peroxisomal levels was estimated from immunoblots using co-electrophoresed samples of purified proteins to determine ng IDP enzyme/μg in cellular protein samples and ng IDP enzyme/μg in peroxisomal protein samples. Volumes of cellular and peroxisomal fractions were used to calculate total protein content.
Levels of the authentic IDP3 enzyme in peroxisomes were determined using immunoblots of cellular fractions from the parental strain. In this strain, IDP3 was detected only in the organellar fraction.
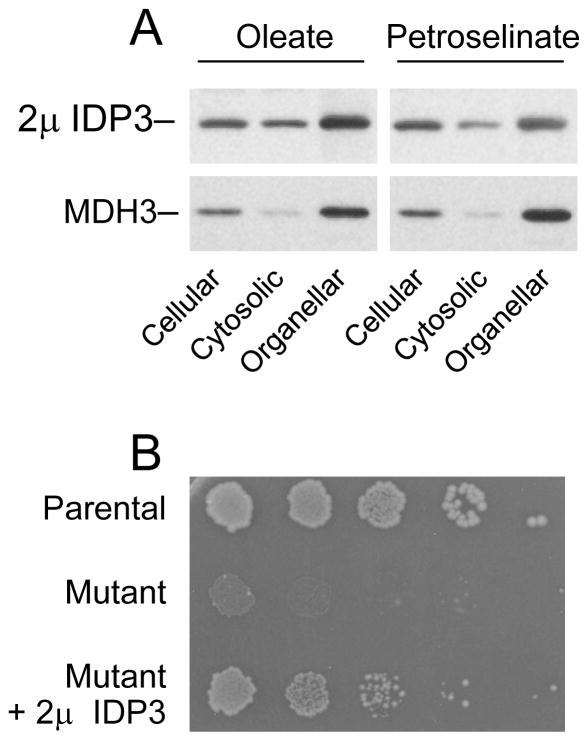
For over-expression of IDP3, we tested both the doxycycline-inducible system described above and the use of a multicopy 2μ yeast plasmid. We found that the latter, which contained the IDP3 gene under control of its authentic promoter, resulted in more reproducible and higher levels of cellular expression (five- to six-fold above those of authentic IDP3 in the parental strain). For analysis of cellular compartmentalization, the 2μ IDP3 plasmid was transformed into the idp1Δidp2Δidp3Δ yeast mutant lacking all endogenous IDP isozymes. Transformants were cultivated in medium with oleate or petroselinate as the carbon source prior to cellular fractionation. As shown in Fig. 11A and as summarized in Table 2, cellular expression of IDP3 using the 2μ plasmid resulted in a substantial increase in peroxisomal levels of the enzyme with both fatty acids (~nine-fold with oleate and ~three-fold with petroselinate relative to peroxisomal levels of the authentic enzyme in parental cells). However, a substantial fraction of the total expressed enzyme (~44% with both carbon sources) was localized in the soluble cytosolic fraction.
Fig. 11.
Expression and localization of 2μ IDP3. [A] Cellular, cytosolic, and organellar protein extracts were prepared from an idp1Δidp2Δidp3Δ transformant expressing IDP3 using a 2μ plasmid and grown using YP oleate medium (left panels) or YP petroselinate medium (right panels). Protein samples (5 μg each) were electrophoresed for immunoblot analysis using an antiserum for IDP3 [3] or an antiserum for peroxisomal MDH3 [24]. [B] Complementation for IDP2 by 2μ IDP3 was assessed by plating dilutions of cultures of the parental strain, the idp2Δzwf1Δ mutant, and of the idp2Δzwf1Δ transformant expressing 2μ IDP3 onto YP oleate plates. Colony sizes and numbers were assessed after 6 days of growth at 30°C.
To assess the functionality of the cytosolically localized IDP3 in cells over-expressing the enzyme, we transformed an idp2Δzwf1Δ mutant with the 2μ IDP3 plasmid and analyzed growth of resulting transformants using medium with oleate as the carbon source. As shown in Fig. 11B, using cells diluted onto YP oleate plates, the parental strain grew well, but the idp2Δzwf1Δ mutant failed to grow under these conditions. In contrast, a transformant of the mutant strain over-expressing IDP3 grew well on YP oleate plates, although colony sizes were somewhat reduced relative to those of the parental strain. These results suggest that over-expression of IDP3 does result in dual compartmental localization, and that the partial cytosolic localization of IDP3 permits compensation for antioxidant functions normally contributed by IDP2.
Discussion
One interesting unifying observation of these and previous studies [11] is that cytosolic forms of IDP containing a carboxyl-terminal PTS1 sequence exhibit dual localization in peroxisomes and in the cytosol of yeast cells. This was not unexpected for the non-mitochondrial mammalian enzyme (IDH1) or for IDPA from A. nidulans, since these enzymes also exhibit similar patterns of dual compartmental localization in native host cells [10, 16]. We have now extended these results to show that IDP2, normally found only in the cytosol, is partially localized to peroxisomes when a PTS1 tripeptide is added to the carboxyl terminus. We previously speculated that the non-mitochondrial mammalian enzyme might contain a cytosolic retention signal that prevents complete peroxisomal localization [11]. However, in yeast transformants grown with petroselinate as the carbon source, we have found that the amounts of IDP2+CKL, of IDPA (or IDPA−ARL), and of the mammalian enzyme imported into peroxisomes are quite similar (1.5-fold to 3-fold in excess of endogenous IDP3 in control cells), whereas cytosolic levels as a percentage of the total cellular level of the enzymes vary from ~33% for mammalian IDH1 [11] to a range of 68%–80% for IDP2CKL and IDPA enzymes (Table 2). Thus, there appears to be a mechanism for limiting the amount of IDP enzyme in peroxisomes. This possibility was supported by the finding that over-expression of yeast peroxisomal IDP3 produced an ~3-fold increase in peroxisomal levels in cells grown with petroselinate as well as localization of ~44% of the total expressed cellular protein in the soluble cytosolic fraction. Collectively, these results suggest a limit in peroxisomal levels of IDP of ~3-fold that of authentic IDP3. Since no changes in cellular distribution were observed for other peroxisomal proteins (e.g. MDH3), this limit appears to apply to cells expressing substantial amounts of an IDP enzyme.
We have also found that growth with petroselinate relative to growth with oleate as the carbon source produces an increase in levels of peroxisomal IDP. This is presumably due to some unknown cellular mechanism to ensure a peroxisomal source of the NADPH needed for β-oxidation of the former fatty acid which contains a double bond following an even-numbered carbon [28]. In parental cells, cellular levels of endogenous IDP3, which is solely peroxisomal [3, 4], were elevated by 2.5-fold with growth using petroselinate relative to levels observed with oleate as the carbon source. Both cellular and peroxisomal levels were also increased by growth with petroselinate of yeast transformants expressing IDPA. However, while cellular levels of yeast IDP2+CKL were similar following growth with oleate or petroselinate, the proportion of the enzyme localized in peroxisomes was increased ~3-fold with petroselinate as the carbon source. Thus, increased peroxisomal localization with petroselinate can occur in the absence of increased cellular expression.
Despite the relatively high degree of sequence conservation among the various IDP enzymes in these studies, we have observed substantial differences related to the importance of PTS1 sequences for peroxisomal import in yeast. For authentic yeast IDP3 expressed at normal cellular levels, the Cys-Lys-Leu carboxyl-terminal tripeptide is both necessary and sufficient for peroxisomal import [3, 4]. Similarly for IDP1, addition of a Cys-Lys-Leu carboxyl-terminal tripeptide to a cytosolic form of the isozyme lacking its mitochondrial targeting sequence [27] appears to result in complete peroxisomal localization, although the enzyme is essentially inactive. In contrast, as described above, the presence of a PTS1 sequence in mammalian IDH1 or in yeast IDP2 produces dual compartmental localization in peroxisomes and in the cytosol. We have previously shown that removal of the PTS1 sequence from mammalian IDH1 results in solely cytosolic localization [11] and, clearly, the absence of such a sequence in authentic yeast IDP2 also produces only cytosolic localization. Thus the PTS1 sequences in these cases are necessary but are not sufficient for complete peroxisomal localization.
For the A. nidulans IDPA enzyme, the presence or absence of its carboxyl-terminal Ala-Arg-Leu tripeptide, which fits the classic definition for a PTS1 sequence [11, 29, 30], appears to have no impact on peroxisomal/cytosolic distribution patterns of the enzyme expressed in yeast. Localization of several other proteins has been shown to be mediated by an alternative PTS2 peroxisomal targeting sequence, a nonapeptide generally near the amino terminus with a consensus sequence of (R/K)(L/V/I)X5(H/Q)(L/A/F) where X is any amino acid [31, 32]. There is a weak match (KVKNPVVEL) with the PTS2 consensus sequence near the amino-terminus of IDPA. However, yeast IDP1 has a similar sequence (KVKQPVVEL) in an analogous position, and removal of the mitochondrial targeting sequence produces cytosolic but not peroxisomal localization of IDP1 [27]. IDPA is not unique, in that other peroxisomal proteins that lack a typical PTS1 or that contain a redundant PTS1 are still efficiently imported (reviewed in [33]). In some cases, conformational epitopes formed by interaction with a cofactor, or by interaction with another peroxisomal protein, have been invoked as factors critical for import. In any event, the PTS1 sequence in IDPA, which was previously assumed to be a peroxisomal targeting sequence [16], is clearly not necessary nor is it sufficient for complete peroxisomal localization.
In terms of the evolution of compartmentalized IDP enzymes in various organisms, sequence analyses [34] suggest that the cytosolic and mitochondrial enzymes originated from independent gene duplication events in animals and in yeast. The cytosolic mammalian IDH presumably gained a PTS1 sequence that facilitates partial peroxisomal localization. An additional duplication event would need to be invoked for evolution of genes for specific peroxisomal isozymes like IDP3 in yeast. Based on functional similarities discussed below, we assume this event involved duplication of the IDP2 gene and subsequent acquisition of a PTS1 sequence. For plants, which contain genetically distinct IDP isozymes in the cytosol, in mitochondria, in peroxisomes, and in plastids (reviewed in [35]), yet another duplication event would be necessary.
Among evolutionary pressures that might have influenced the origin of independent loci for differentially compartmentalized IDPs in yeast and plants is the importance in these organisms for provision of α-ketoglutarate to support nitrogen assimilation in the form of glutamate. As reviewed by Hodges [36], there is evidence that the cytosolic plant IDP isozyme may play a key role in this process. We have also proposed that yeast cytosolic IDP2 plays a key role in shuttling isocitrate and α-ketoglutarate among different cellular compartments and may even have a unique capacity among the yeast isozymes for bidirectional enzymatic function in vivo [5]. Interestingly, the most remarkable kinetic characteristic of yeast IDP2, a relatively low apparent affinity for isocitrate (~seven-fold lower than that of IDP1, Table 1), is shared by cytosolic enzymes in plants (with apparent affinities for isocitrate ranging from four- to eight-fold lower than that for a mitochondrial isozyme [35]). This suggests that some common evolutionary pressure has led to similar kinetic properties for these cytosolic enzymes, a process that might be facilitated by independent genetic loci for cytosolic and peroxisomal enzymes.
In terms of interchangeable function, heterologous mammalian IDH1 functions well in both peroxisomal and cytosolic cellular compartments in yeast cells [11]. A. nidulans IDPA can also compensate for both IDP2 and IDP3, and for mitochondrial IDP1 (Supplemental Information). For the yeast isozymes, we found that IDP2+CKL can concomitantly replace cytosolic IDP2 and peroxisomal IDP3. The latter function indicates that the physical properties of IDP2 (narrow and neutral pH optimum, and slightly acidic pI value) do not reduce catalytic capacity in the basic peroxisomal lumen as we had initially predicted. In fact, in current and previous studies [27], we have found that yeast mitochondrial IDP1 is the least adaptable in terms of function in other cellular compartments. In a previous study [27], we found that even elevated expression of the cytosolic form of IDP1 provided minimal compensation for IDP2 function. Essentially the same result was obtained in the current study, since immunochemical levels of the peroxisomal form of IDP1+CKL were found to exceed those of IDP3 in parental cells, but the normally mitochondrial enzyme was not active and did not complement idp3Δ mutants. We therefore speculate that the dimeric IDP1 enzyme may not properly fold or assemble into a catalytically active form in cellular compartments other than the mitochondrial matrix, where necessary chaperones and/or other factors may be located.
We have speculated, based on complementation analyses of yeast mutants, that the mammalian IDP enzyme (IDH1) may function in mammalian cells to support both antioxidant functions in the cytosol and β-oxidation of some fatty acids in peroxisomes. It is known that cellular levels of IDH1 vary considerably in a tissue-specific fashion in mammals [37], and we have found that patterns of cytosolic and peroxisomal localization vary substantially in different mammalian cell lines (Lu and McAlister-Henn, unpublished data). However, little is known about mechanisms for, or influences on, these different patterns of cellular compartmentalization. Since specific mutations of IDH1 have recently been found to be associated with many high grade and secondary human glioblastomas [12, 13, 38], interference with either or both compartmentalized cellular functions may be highly deleterious. Thus, this is an area important for future investigation.
Supplementary Material
Acknowledgments
This word was supported by National Institutes of Health Grant AG017477. We thank Dr. Michael Hynes for providing the IDPA cDNA and Dr. Ralf Erdmann for yeast IDP3 antiserum. We also thank Dr. Karyl Minard and Sondra Anderson for critical comments on the manuscript.
Abbreviations used
- IDP
NADP+-specific isocitrate dehydrogenase
- IDP1
yeast mitochondrial IDP
- IDP2
yeast cytosolic IDP
- IDP3
yeast peroxisomal IDP
- IDPA
Aspergillus nidulans IDP
- IDH1
mammalian non-mitochondrial IDP
- MDH3
yeast peroxisomal malate dehydrogenase
- IDP1+CKL
yeast IDP1 with a PTS1
- IDP2+CKL
yeast IDP2 with a PTS1
- IDPA−ARL
A. nidulans IDPA lacking a PTS1
- mitoIDPA
A. nidulans IDPA with a yeast mitochondrial targeting sequence
- PCR
polymerase chain reaction
- NTA
nitrilotriacetic acid
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Haselbeck RJ, McAlister-Henn L. J Biol Chem. 1991;266:2339–2345. [PubMed] [Google Scholar]
- 2.Loftus TM, Hall LV, Anderson SL, McAlister-Henn L. Biochemistry. 1994;33:9661–9667. doi: 10.1021/bi00198a035. [DOI] [PubMed] [Google Scholar]
- 3.Henke B, Girzalsky W, Berteaux-Lecellier V, Erdmann R. J Biol Chem. 1998;273:3702–3711. doi: 10.1074/jbc.273.6.3702. [DOI] [PubMed] [Google Scholar]
- 4.van Roermund CW, Hettema EH, Kal AJ, van den Berg M, Tabak HF, Wanders RJ. Embo J. 1998;17:677–687. doi: 10.1093/emboj/17.3.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Contreras-Shannon V, Lin AP, McCammon MT, McAlister-Henn L. J Biol Chem. 2005;280:4469–4475. doi: 10.1074/jbc.M410140200. [DOI] [PubMed] [Google Scholar]
- 6.DeRisi JL, Iyer VR, Brown PO. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 7.Haselbeck RJ, McAlister-Henn L. J Biol Chem. 1993;268:12116–12122. [PubMed] [Google Scholar]
- 8.Minard KI, McAlister-Henn L. J Biol Chem. 1999;274:3402–3406. doi: 10.1074/jbc.274.6.3402. [DOI] [PubMed] [Google Scholar]
- 9.Minard KI, McAlister-Henn L. Free Radic Biol Med. 2001;31:832–843. doi: 10.1016/s0891-5849(01)00666-9. [DOI] [PubMed] [Google Scholar]
- 10.Geisbrecht BV, Gould SJ. J Biol Chem. 1999;274:30527–30533. doi: 10.1074/jbc.274.43.30527. [DOI] [PubMed] [Google Scholar]
- 11.Lu Q, Minard KI, McAlister-Henn L. Arch Biochem Biophys. 2008;472:17–25. doi: 10.1016/j.abb.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Balss J, Meyer J, Mueller W, Korshunov A, Hartmann C, von Deimling A. Acta Neuropathol. 2008;116:597–602. doi: 10.1007/s00401-008-0455-2. [DOI] [PubMed] [Google Scholar]
- 13.Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, Kos I, Batinic-Haberle I, Jones S, Riggins GJ, Friedman H, Friedman A, Reardon D, Herndon J, Kinzler KW, Velculescu VE, Vogelstein B, Bigner DD. N Engl J Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Roermund CW, de Jong M, IJL, van Marle J, Dansen TB, Wanders RJ, Waterham HR. J Cell Sci. 2004;117:4231–4237. doi: 10.1242/jcs.01305. [DOI] [PubMed] [Google Scholar]
- 15.Orij R, Postmus J, Ter Beek A, Brul S, Smits GJ. Microbiology. 2009;155:268–278. doi: 10.1099/mic.0.022038-0. [DOI] [PubMed] [Google Scholar]
- 16.Szewczyk E, Andrianopoulos A, Davis MA, Hynes MJ. J Biol Chem. 2001;276:37722–37729. doi: 10.1074/jbc.M105645200. [DOI] [PubMed] [Google Scholar]
- 17.McCammon MT, Veenhuis M, Trapp SB, Goodman JM. J Bacteriol. 1990;172:5816–5827. doi: 10.1128/jb.172.10.5816-5827.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gueldener U, Heinisch J, Koehler GJ, Voss D, Hegemann JH. Nucleic Acids Res. 2002;30:e23. doi: 10.1093/nar/30.6.e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sikorski RS, Hieter P. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Belli G, Gari E, Piedrafita L, Aldea M, Herrero E. Nucleic Acids Res. 1998;26:942–947. doi: 10.1093/nar/26.4.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Daum G, Bohni PC, Schatz G. J Biol Chem. 1982;257:13028–13033. [PubMed] [Google Scholar]
- 22.Bradford MM. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 23.Laemmli UK, Favre M. J Mol Biol. 1973;80:575–599. doi: 10.1016/0022-2836(73)90198-8. [DOI] [PubMed] [Google Scholar]
- 24.Steffan JS, McAlister-Henn L. J Biol Chem. 1992;267:24708–24715. [PubMed] [Google Scholar]
- 25.Pace CN, Vajdos F, Fee L, Grimsley G, Gray T. Protein Sci. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Veenhuis M, Mateblowski M, Kunau WH, Harder W. Yeast. 1987;3:77–84. doi: 10.1002/yea.320030204. [DOI] [PubMed] [Google Scholar]
- 27.Contreras-Shannon V, McAlister-Henn L. Arch Biochem Biophys. 2004;423:235–246. doi: 10.1016/j.abb.2003.12.038. [DOI] [PubMed] [Google Scholar]
- 28.Kunau WH, Dommes V, Schulz H. Prog Lipid Res. 1995;34:267–342. doi: 10.1016/0163-7827(95)00011-9. [DOI] [PubMed] [Google Scholar]
- 29.Subramani S. J Biol Chem. 1996;271:32483–32486. doi: 10.1074/jbc.271.51.32483. [DOI] [PubMed] [Google Scholar]
- 30.Gould SJ, Keller GA, Schneider M, Howell SH, Garrard LJ, Goodman JM, Distel B, Tabak H, Subramani S. Embo J. 1990;9:85–90. doi: 10.1002/j.1460-2075.1990.tb08083.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schliebs W, Kunau WH. Biochim Biophys Acta. 2006;1763:1605–1612. doi: 10.1016/j.bbamcr.2006.08.051. [DOI] [PubMed] [Google Scholar]
- 32.Lazarow PB. Biochim Biophys Acta. 2006;1763:1599–1604. doi: 10.1016/j.bbamcr.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 33.van der Klei IJ, Veenhuis M. Biochim Biophys Acta. 2006;1763:1794–1800. doi: 10.1016/j.bbamcr.2006.08.013. [DOI] [PubMed] [Google Scholar]
- 34.Nekrutenko A, Hillis DM, Patton JC, Bradley RD, Baker RJ. Mol Biol Evol. 1998;15:1674–1684. doi: 10.1093/oxfordjournals.molbev.a025894. [DOI] [PubMed] [Google Scholar]
- 35.Galvez S, Lancien M, Hodges M. Trends Plant Sci. 1999;4:484–490. doi: 10.1016/s1360-1385(99)01500-9. [DOI] [PubMed] [Google Scholar]
- 36.Hodges M. J Exp Bot. 2002;53:905–916. doi: 10.1093/jexbot/53.370.905. [DOI] [PubMed] [Google Scholar]
- 37.Jennings GT, Sechi S, Stevenson PM, Tuckey RC, Parmelee D, McAlister-Henn L. J Biol Chem. 1994;269:23128–23134. [PubMed] [Google Scholar]
- 38.Watanabe T, Nobusawa S, Kleihues P, Ohgaki H. Am J Pathol. 2009;174:1149–1153. doi: 10.2353/ajpath.2009.080958. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.