Abstract
The Warburg effect describes a pro-oncogenic metabolism switch such that cancer cells take up more glucose than normal tissue and favor incomplete oxidation of glucose even in the presence of oxygen. To better understand how tyrosine kinase signaling, which is commonly increased in tumors, regulates the Warburg effect, we performed phosphoproteomic studies. We found that oncogenic forms of fibroblast growth factor receptor type 1 inhibit the pyruvate kinase M2 (PKM2) isoform by direct phosphorylation of PKM2 tyrosine residue 105 (Y105). This inhibits the formation of active, tetrameric PKM2 by disrupting binding of the PKM2 cofactor fructose-1,6-bisphosphate. Furthermore, we found that phosphorylation of PKM2 Y105 is common in human cancers. The presence of a PKM2 mutant in which phenylalanine is substituted for Y105 (Y105F) in cancer cells leads to decreased cell proliferation under hypoxic conditions, increased oxidative phosphorylation with reduced lactate production, and reduced tumor growth in xenografts in nude mice. Our findings suggest that tyrosine phosphorylation regulates PKM2 to provide a metabolic advantage to tumor cells, thereby promoting tumor growth.
INTRODUCTION
Cancer cells show increased aerobic glycolysis and enhanced lactate production compared to healthy cells, a phenomenon known as the Warburg effect. Furthermore, tumor tissue accumulates more glucose than does healthy tissue, because cancer cells require increased amounts of glucose as a carbon source for anabolic reactions [reviewed in (1, 2)]. Cell surface growth factor receptors, which often carry tyrosine kinase activities in their cytoplasmic domains, are overexpressed in many human cancers and are believed to play a key role in determining cell metabolism (3). Thus, we explored the hypothesis that tyrosine kinase signaling, which is commonly increased in tumors, regulates the Warburg effect and contributes to tumorigenesis and maintenance of the tumor.
Pyruvate kinase (PK), a rate-limiting enzyme during glycolysis, catalyzes the production of pyruvate and adenosine 5′-triphosphate (ATP) from phosphoenolpyruvate (PEP) and adenosine 5′-diphosphate (ADP) (4–6). Four mammalian PK isoenzymes (M1, M2, L, and R) exist, which are present in different cell types. PKM1 is a constitutively active form of PK that is found in normal adult cells. In contrast, PKM2 is found predominantly in the fetus and also in tumor cells, where the abundance of other isoforms of PK is low. PKM2 can exist in either active tetramers or inactive dimers, but in tumor cells, it predominantly occurs in dimers with low activity (4, 7–10).
Recent studies by Christofk et al. (7, 8) demonstrated that the enzymatic activity of the pyruvate kinase M2 isoform (PKM2) is inhibited by phosphotyrosine binding; moreover, these researchers found that PKM2 is crucial for aerobic glycolysis and provides a growth advantage to tumors. However, it remains unclear which tyrosine kinase pathways are physiologically responsible for this inhibition of PKM2 activity and which protein factors undergo tyrosine phosphorylation, allowing them to bind to and thereby inhibit PKM2. Furthermore, it is not clear whether PKM2 is itself tyrosine phosphorylated in cancer cells and such a physiological modification of PKM2 promotes the switch to aerobic glycolysis from oxidative phosphorylation. Here, we address all of these questions.
RESULTS
PKM2 is phosphorylated at Y105 and inhibited by FGFR1 in cancer cells
We performed a mass spectrometry (MS)–based proteomics study (11, 12) using murine hematopoietic Ba/F3 cells stably expressing ZNF198-FGFR1, a constitutively active fusion tyrosine kinase in which an N-terminal self-association motif of ZNF198 is fused to the C-terminal kinase domain of fibroblast growth factor (FGF) receptor type 1 (FGFR1). ZNF198-FGFR1 is associated with t(8;13)(p11;q12) stem cell myeloproliferative disorder (MPD) (13). Ba/F3 cells require interleukin-3 (IL-3) for cell survival and proliferation; however, constitutively active ZNF198-FGFR1 confers IL-3–independent proliferation to Ba/F3 cells (11). We identified various proteins that were tyrosine phosphorylated in Ba/F3 cells containing ZNF198-FGFR1 but not in control cells grown in the absence of IL-3. These proteins included a group of enzymes that regulate metabolism, including PKM2, lactate dehydrogenase A (LDH-A), glucose-6-phosphate dehydrogenase (G6PD), and malate dehydrogenase 2 (MDH2) (fig. S1A).
We investigated PKM2 as a possible downstream effector of FGFR1 because of its critical role in cancer cell metabolism. Figure 1A shows a schematic illustration of PKM2 and the tyrosine residues identified as phosphorylated in response to oncogenic FGFR1 signaling; these include Y83, Y105, Y148, Y175, Y370, and Y390. The MS spectrum of peptide fragments of PKM2 that contained the specified phospho-Tyr residues is shown in fig. S1B. Previous phosphoproteomic studies have shown that PKM2 tyrosine residues Y83, Y105, and Y370 are also phosphorylated in human leukemia KG-1a cells expressing FGFR1OP (FOP) 2-FGFR1, a constitutively active fusion tyrosine kinase associated with ins(12,8)(p11;p11p22) stem cell MPD (14).
Fig. 1.
PKM2 is tyrosine phosphorylated and inhibited by FGFR1 in cancer cells with oncogenic or overexpressed FGFR1. (A) Schematic representation of PKM2. The six phosphorylated tyrosine residues identified in the proteomics studies are indicated. (B) Immunoblotting (WB) of 293T cell lysates for tyrosine phosphorylation of GST-PKM2 when coexpressed with the constitutively active fusion protein 8p11 ZNF198-FGFR1 PR/TK or with FGFR1 in the presence and absence of FGFR1 ligand (bFGF). (C) FGFR1 wild type (WT) but not a kinase dead (KD) mutant inhibits PKM2 enzyme activity in 293T cells (*0.01 < P < 0.05; n.s., not significant). The error bars represent the means ± SD from three independent experiments. Relative PKM2 activity was normalized to that in control 293T cells. (D) Inhibition of FGFR1 by tyrosine kinase inhibitor TKI258 (1 μM for 2 hours) results in increased PKM2 enzyme activity in leukemia KG-1a (FOP2-FGFR1), breast cancer MDA-MB-134 (FGFR1), and lung cancer H1299 cells (FGFR1) (*0.01< P < 0.05; **P < 0.01). Relative PKM2 activity was normalized to that in control cells without TKI258 treatment.
Glutathione S-transferase (GST)–tagged PKM2 was tyrosine phosphorylated in 293T cells co-transfected with plasmids encoding a constitutively active mutant form of ZNF198-FGFR1, PR/TK, in which an N-terminal proline-rich (PR) domain of ZNF198 is fused to the C-terminal FGFR1 tyrosine kinase (TK) domain (15), and in ligand-treated cells expressing FGFR1, but not in cells expressing GST-PKM2 without FGFR1 (Fig. 1B). Moreover, the presence of FGFR1 wild type, but not a kinase-dead (KD) mutant, significantly decreased the enzymatic activity of endogenous PKM2 in 293T cells (Fig. 1C). Overexpression of FGFR1 or its mutational activation has been implicated in various human solid tumors, including breast cancer, pancreatic adenocarcinoma, and malignant astrocytoma (16–19). We found that treatment with the FGFR1 inhibitor TKI258 significantly increased PKM2 enzymatic activity in human myeloid leukemia KG-1a cells harboring the FOP2-FGFR1 fusion protein (14), as well as breast cancer MDA-MB-134 cells (20, 21) and lung cancer NCI-H1299 cells overexpressing FGFR1 (22) (Fig. 1D). Together, these data suggest that FGFR1 may directly or indirectly phosphorylate and inhibit PKM2.
Mutational analysis revealed that expression of GST-PKM2 wild type or of several PKM2 mutants in which a Tyr residue was replaced with a Phe to abolish phosphorylation, including Y83F, Y148F, Y175F, Y370F, and Y390F, resulted in comparable, increased PKM2 enzyme activity compared with that in control 293T cells, whereas substitution of Y105 led to significantly greater PKM2 activation (Fig. 2A). To elucidate the role of FGFR1 in phosphorylation and inhibition of PKM2 in cancer cells, we used FGFR1-expressing human lung cancer H1299 cells to generate mouse PKM2 (mPKM2) wild type, Y105F, and Y390F “rescue” cell lines as described (7) by RNA interference–mediated stable knockdown of endogenous human PKM2 (hPKM2) and rescue expression of Flag-tagged mPKM2 variants (Fig. 2B). Consistent with the data in Fig. 2A, mPKM2 Y105F showed increased enzymatic activity in the rescue cells compared with that of wild-type and Y390F mPKM2.
Fig. 2.
FGFR1 inhibits PKM2 by phosphorylation at Y105. (A) Mutational analysis revealed that substitution of Y105 results in a significant increase in PKM2 activity in 293T cells (n.s., not significant; *P < 0.05). Relative enzyme activity was normalized to that of cells expressing GST-PKM2 WT. (B) Left: Immunoblotting (WB) shows shRNA-mediated stable knockdown of endogenous hPKM2 in H1299 cells by lentiviral transduction, and “rescue” expression of Flag-tagged mPKM2 proteins including WT, Y105F, and Y390F mutants. Right: Y105F has significantly higher catalytic activity than mPKM2 WT or Y390F in rescue H1299 cells. Relative catalytic activity was normalized to that of mPKM2 WT. (C) GST-PKM2 WT or Y105F mutant was incubated with active recombinant FGFR1 (rFGFR1) in an in vitro kinase assay. Phosphorylation at Y105 in PKM2 was detected by specific antibody p-PKM2 (Y105). (D) Immunoblotting revealed that inhibition of FGFR1 by TKI258 treatment in H1299 cells results in decreased Y105 phosphorylation of endogenous PKM2.
We also generated an antibody that specifically recognizes PKM2 phospho-Y105. This antibody detected PKM2 (but not the Y105F mutant) in 293T cells coexpressing FGFR1 wild type but not in cells coexpressing the KD mutant (fig. S2A). Moreover, in an in vitro kinase assay, recombinant FGFR1 (rFGFR1) phosphorylated purified GST-PKM2 at Y105, whereas phosphorylation of this site by rFGFR1 was not apparent in the GST-PKM2 Y105F mutant (Fig. 2C and fig. S2B). Using a pan-tyrosine phosphorylation antibody, pY99, we observed decreased total tyrosine phosphorylation of Y105F compared with PKM2 wild type in the in vitro assay (fig. S2B), suggesting that FGFR1 directly phosphorylates PKM2 at multiple sites including Y105, which may represent a major phosphorylation site of PKM2 by FGFR1. Furthermore, Y105 phosphorylation of PKM2 was apparent in human lung cancer H1299 cells overexpressing FGFR1 and leukemia KG-1a cells expressing FOP2-FGFR1; inhibition of FGFR1 and FOP2-FGFR1 by TKI258 resulted in decreased phosphorylation of PKM2 at Y105 (Fig. 2D and fig. S2C, respectively).
Y105 phosphorylation disrupts formation of active tetrameric PKM2 by releasing cofactor fructose-1,6-bisphosphate
To gain mechanistic insight into the role of Y105 phosphorylation in PKM2 regulation, we determined whether a phospho-Y105 peptide based on the PKM2 sequence surrounding Y105 could inhibit PKM2. We incubated recombinant PKM2 (rPKM2) preincubated with fructose-1,6-bisphosphate (FBP) with identical amounts (final concentration, 1.5 μM) of a phospho-Y105 peptide (SDPILpYRPVAV) or a non–phospho-Y105 peptide (SDPILYRPVAV) and followed this by dialysis and analysis of PKM2 enzymatic activity (23). Mock treatment without peptide and treatment with a phospho-Y390 peptide (AEAAIpYHLQLF) were included as negative controls. As shown in Fig. 3A, FBP treatment resulted in a ~65% increase in PKM2 activity compared with the mock treatment. This increase was abolished by the phospho-Y105 peptide, whereas the non–phospho-Y105 and phospho-Y390 peptides did not affect FBP-dependent activation of rPKM2. Formation of PKM2 tetramers is induced by binding of its cofactor FBP (6), and cross-linking revealed that incubation of PKM2 and FBP with phospho-Y105 peptide led to a marked decrease in formation of tetrameric, active PKM2 (Fig. 3B), an observation that correlates with the reduced PKM2 activity (Fig. 3A).
Fig. 3.
Y105 phosphorylation disrupts formation of active, tetrameric PKM2 by releasing cofactor FBP. (A) Incubation of a pY105 phosphopeptide attenuates the catalytic activity of FBP-loaded recombinant PKM2 (rPKM2). Relative PKM2 activity was normalized to rPKM2 without preincubation with FBP. The error bars represent the means ± SD from three independent experiments. (B) Incubation of phospho-Y105 peptide leads to reduced formation of tetrameric, active PKM2. rPKM2 (Myc-tagged) preincubated with FBP was incubated with each peptide, followed by 0.025% glutaraldehyde cross-linking (+) before SDS–polyacrylamide gel electrophoresis and Western blot analysis. Parallel samples without cross-linking treatment (−) were included as loading controls. The numbers represent the relative intensity of the specific bands of PKM2 tetramers, which are normalized to the value of control sample without treatment of peptide. (C) Phospho-Y105 peptide incubation results in release of FBP from PKM2. [14C]FBP was incubated with rPKM2 and the unbound FBP was dialysed away. [14C]FBP-soaked rPKM2 was incubated with the indicated peptides followed by dialysis to remove the unbound FBP peptides. Retained [14C]FBP on PKM2 was measured with a scintillation counter. (D) Inhibition of FGFR1 by TKI258 results in increased PKM2 enzyme activity in rescue H1299 cells expressing mPKM2 WT or Y390F mutant, but not in cells expressing mPKM2 Y105F and K433E mutants (*0.01 < P < 0.05; n.s., not significant). Relative PKM2 activity was normalized to WT cells without TKI258 treatment. (E) Phosphorylation of PKM2 at Y105 is detected by immunoblotting in rescue cells expressing mPKM2 WT, Y390F, or K433E mutants, but not in cells expressing the Y105F mutant.
PKM2 activity is inhibited after phosphotyrosine binding through the release of FBP from the PKM2 allosteric pocket (8). We hypothesized that, in an active PKM2 tetramer, one PKM2 molecule, when Y105 phosphorylated, might act as the unidentified, “PKM2 binding partner” that provides the inhibitory phosphotyrosine motif that releases FBP from other sister molecules in the same tetramer in an “intermolecular” manner. We thus examined the effect of phospho-Y105 peptide binding on FBP-bound rPKM2. Exposure of PKM2 to the phospho-Y105 peptide resulted in a significant decrease in the amount of [14C]FBP bound to rPKM2 (Fig. 3C). PKM2 K433 is essential for phosphotyrosine binding; a PKM2 K433E mutant is phosphotyrosine binding–deficient and resistant to inhibition mediated by tyrosine kinase signaling (8). Consistent with this, both mPKM2 K433E and Y105F mutants are constitutively active and were resistant to FGFR1-dependent inhibition in the rescue H1299 cells (Fig. 3D), even though FGFR1 phosphorylated K433E at Y105 (Fig. 3E). Together, these results suggest that inhibition of PKM2 by FGFR1 is predominantly mediated through phosphorylation at Y105, which likely involves K433-dependent phosphotyrosine binding, release of cofactor FBP, and disruption of active PKM2 tetramers.
PKM2 is specifically phosphorylated at Y105 in various cancer cell lines
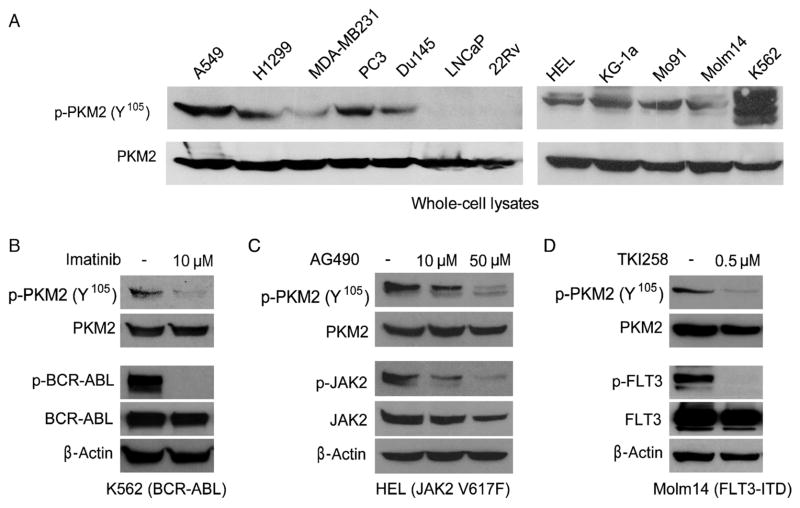
We found that PKM2 was phosphorylated at Y105 in various human solid tumor cell lines, including A549 and H1299 lung cancer cells, MDA-MB231 breast cancer cells, and PC3 and Du145 prostate cancer cells, but not in LNCaP and 22Rv prostate cancer cells (Fig. 4A). Moreover, we found that PKM2 is Y105-phosphorylated in several hematopoietic cancer cell lines associated with various constitutively activated tyrosine kinase mutants. These include HEL [which contains the Janus kinase 2 (JAK2) Val617Phe mutant], KG-1a (which contains FOP2-FGFR1 fusion tyrosine kinase), Mo91 [which contains est variant 6 (ETV6)–neurotrophic tyrosine kinase, receptor, type 3 (NTRK3) fusion tyrosine kinase], Molm14 [which contains fms-related tyrosine kinase 3 (FLT3)–internal tandem duplication (ITD) mutant], and K562 [which contains breakpoint cluster region (BCR)–ABL fusion tyrosine kinase] (Fig. 4A). We observed that inhibiting FGFR1 decreased PKM2 Y105 phosphorylation in lung cancer H1299 cells and leukemia KG-1a cells (Fig. 2D and fig. S2C, respectively). Furthermore, experiments using different tyrosine kinase inhibitors [imatinib to inhibit BCR-ABL (Fig. 4B), AG490 to inhibit JAK2 (Fig. 4C), and TKI258 to inhibit FLT3 (Fig. 4D)] revealed that BCR-ABL, JAK2, and FLT3-ITD are responsible for phosphorylation of PKM2 at Y105 in the pertinent human cancer cell lines. We also found that ABL, JAK2, and FLT3 directly phosphorylated PKM2 in the in vitro kinase assays using recombinant proteins (fig. S3).
Fig. 4.
PKM2 is specifically phosphorylated at Y105 in various cancer cell lines. (A) Immunoblotting detects Y105 phosphorylation of PKM2 in diverse lung cancer (A549, H1299), breast cancer (MDA-MB231), prostate cancer (PC3, Du145), and leukemia (HEL, KG-1a, Mo91, Molm14, K562) cell lines, but not in two prostate cancer cell lines, LNCaP and 22Rv. Immunoblotting shows that targeting BCR-ABL by imatinib in K562 cells (B), JAK2 by AG490 in HEL cells (C), and FLT3 by TKI258 in Molm14 cells (D) decreases phosphorylation of PKM2 Y105.
Presence of the PKM2 Y105F mutant in cancer cells leads to decreased proliferation under hypoxic conditions, increased oxidative phosphorylation, and reduced tumor growth
We used the H1299 rescue cell lines to elucidate the role of PKM2 Y105 phosphorylation in cancer cell metabolism and tumor growth. Under normoxic conditions, cells rescued with any of the mPKM2 variants showed a comparable rate of proliferation that was greater than that of parental cells, in which endogenous hPKM2 was stably knocked down. However, cells rescued with mPKM2 Y105F showed a significantly slower proliferation rate under hypoxic conditions than did cells rescued with mPKM2 wild type or mPKM2 Y390F (Fig. 5A and fig. S4). The mPKM2 Y105F rescue cells also had a higher rate of oxygen consumption than did cells rescued with mPKM2 wild type (Fig. 5B). Moreover, under normoxia, a significant decrease in lactate production was apparent in the Y105F rescue cells compared with that in mPKM2 wild type and Y390F rescue cells (Fig. 5C). In addition, treatment with oligomycin, a specific inhibitor of mitochondrial ATP synthase, led to a significant decrease in the proliferation rate, oxygen consumption rate, and intracellular ATP concentration of Y105F rescue cells compared to those in cells rescued with mPKM2 wild type (Fig. 5D, left, middle, and right, respectively). Together, these data suggest that rescue cells with a form of PKM2 that is catalytically more active (Y105F) rely more on oxidative phosphorylation for cell proliferation than do cells with PKM2 wild type or the Y390F mutant.
Fig. 5.
Expression of PKM2 Y105F mutant in H1299 cells leads to decreased proliferation under hypoxic conditions, increased oxidative phosphorylation and reduced tumor growth. (A) Rescue expression of mPKM2 Y105F in H1299 cells results in reduced cell proliferation under hypoxic conditions (1%) but not at normal oxygen tension (normoxic; 17% oxygen) compared with cells expressing PKM2 WT or Y390F mutant. Cell proliferation was determined by the increase in cell number 96 hours after seeding compared to that at seeding for each cell line (T = 0). Error bars represent the means ± SD from three independent experiments. (B) Y105F rescue H1299 cells have a higher rate of oxygen consumption than do cells with mPKM2 WT. (C) Y105F rescue H1299 cells show significantly reduced lactate production under normoxia. (D) Proliferation (left), oxygen consumption rate (middle), and intracellular ATP concentration (right) of Y105F rescue H1299 cells are significantly decreased relative to cells expressing PKM2 WT or the Y390F mutant when treated with oligomycin. (E) Upper left: Tumors (indicated by red arrows) in representative nude mice injected with mPKM2 WT H1299 cells on the left flank and mPKM2 Y105F H1299 cells on the right flank. Lower left: Dissected tumors from the depicted nude mice. Right: expression of Flag-tagged mPKM2 WT and Y105F detected by immunoblotting in injected rescue cells (upper) and tumor lysates (lower). Phosphorylation of PKM2 at Y105 was detected in tumors formed by cells expressing mPKM2 WT, but not in tumors derived from Y105F-expressing cells. (F) H1299 cells expressing PKM2 Y105F show significantly reduced tumor formation in xenograft nude mice (P value was determined by the paired Student’s t test).
We performed xenograft experiments in which we injected nude mice with mPKM2 wild type and Y105F rescue H1299 cells. The mice were injected with 10 million cells (PKM2 wild type rescue cells on the left flank and Y105F cells on the right flank; n = 6) and monitored for tumor growth over a 6-week period. The masses of tumors derived from Y105F rescue cells were significantly reduced compared to those of tumors formed by mPKM2 wild type rescue cells (Fig. 5, E and F); indeed, Y105F rescue cells failed to form a tumor in one mouse (#874). These results demonstrate that the presence of PKM2 Y105F in cancer cells results in attenuated tumor growth in vivo, suggesting that inhibitory phosphorylation at Y105 of PKM2 confers a proliferative advantage.
DISCUSSION
Our finding that direct phosphorylation at Y105 inhibits PKM2 activity provides new insight into the molecular mechanism underlying tyrosine kinase–dependent regulation of tumor cell metabolism. We identified PKM2 as a direct substrate of the oncogenic tyrosine kinase FGFR1, which phosphorylates PKM2 at Y105. Consistent with these findings, our colleagues at Cell Signaling Technologies (CST) have found in phosphoproteomics-based studies that Y105 of PKM2 is phosphorylated in human cancer cell lines established from different malignancies, including leukemias associated with the oncogenic tyrosine kinases BCR-ABL and FLT3, and solid tumors such as ovarian cancer, glial tumor, lung cancer, and stomach cancer (24). Therefore, our finding that phosphorylation of Y105 inhibits PKM2 activity may represent a common, short-term molecular mechanism underlying the Warburg effect in both leukemias and solid tumors, in addition to the long-term changes believed to be regulated by transcription factors, including hypoxia-inducible factor 1 and Myc. However, the mechanism by which lactate production is increased in cancer cells harboring phospho-PKM2 with low activity is unknown.
It has been argued that the stoichiometry of tyrosine phosphorylation of glycolytic enzymes, including pyruvate kinase, is too low to affect their catalytic activity (8). Indeed, only a small fraction of PKM2 is phosphorylated in FOP2-FGFR1–expressing KG-1a cells, which could not be visualized in isoelectric focusing (IEF) experiments (fig. S5A). However, our “intermolecular”, or transprotein, FBP-release model suggests that a single PKM2 molecule, when phosphorylated at Y105, can directly and transiently mediate FBP release from many PKM2 molecules, as proposed by Christofk et al. (8). This would allow a small amount of phosphorylated PKM2-Y105 to convert substantial amounts of PKM2 to the low-activity FBP-unbound state. However, the stoichiometry of PKM2 tyrosine phosphorylation may vary in different cellular contexts. For example, our IEF experiment showed that FGFR1 wild type causes a stoichoimetric shift (>90%) of PKM2 to a more phosphorylated form in 293T cells, compared with cells expressing the FGFR1 KD control (fig. S5B). Such high stoichiometry could potentially allow Y105 phosphorylation to inhibit PKM2 in an “intramolecular manner,” in which Y105 phosphorylation causes a conformational alteration within the same molecule of PKM2 to affect K433-dependent FBP binding. Pyruvate kinase transmits regulatory signals across large distances within a single PKM2 molecule, and the intersubunit interfaces are important for allosteric signal transmission between the binding sites of the PKM2 substrate PEP and cofactor FBP (25, 26). Y105 is located on the interface between the A and C domains of PKM2, ~17 Å distal from FBP (fig. S5C). Because long-range allosteric regulation in PKM2 is possible, phosphorylation of Y105 could potentially transmit an allosteric signal to the FBP binding site within the same PKM2 molecule, leading to decreased FBP binding. We hypothesize that such an allosteric signal could contribute to FBP release in PKM2 molecules that are Y105-phosphorylated and act in concert with the “intermolecular” model that may represent the predominant mechanism for phospho-Y105–dependent inhibition of PKM2.
Christofk et al. (8) proposed that binding of tyrosine-phosphorylated proteins inhibits PKM2 by inducing the release of FBP. We found that FGFR1 binds to PKM2 in a tyrosine phosphorylation–dependent manner; however, FGFR1 still binds to PKM2 K433E and Y105F mutants (fig. S6), and both mutants are catalytically active and resistant to FGFR1-dependent inhibition (Fig. 3D). This suggests that Y105 phosphorylation is the predominant mechanism underlying FGFR1-dependent inhibition of PKM2 through K433, and it is unlikely that the binding of FGFR1 to PKM2 affects PKM2 activity directly. Such an interaction may contribute to inhibition of PKM2 indirectly, because it may be required for FGFR1 to phosphorylate Y105.
Our finding that cancer cells expressing the active mPKM2 Y105F mutant are more dependent on oxidative phosphorylation for cell metabolism and proliferation than cells with WT mPKM2 is consistent with previous observations, made by Christofk et al., when they replaced endogenous hPKM2 with mouse PKM1 (mPKM1) in H1299 cells (8). Most noticeably, both the PKM2 Y105F mutant and PKM1 are catalytically more active than PKM2 and are resistant to tyrosine kinase–dependent inhibition. These studies suggest that the physiological phosphorylation and dephosphorylation kinetics at Y105 of PKM2 may regulate the switch between aerobic glycolysis and oxidative phosphorylation, perhaps by balancing the ratio between the active and inactive forms of PKM2. Moreover, because either knockdown of PKM2 (8) or replacement of PKM2 with the catalytically more active Y105F mutant (Fig. 5) or PKM1 (8) effectively attenuates cancer cell proliferation in vitro and in vivo, PKM2 may serve as an interesting therapeutic target in cancer treatment, such that either inhibition or activation of PKM2 may affect cancer cell metabolism and cause tumor regression.
MATERIALS AND METHODS
Proteomics studies
Phosphopeptides were prepared with the PhosphoScan Kit (Cell Signaling Technology). In brief, 2 × 108 to 3 × 108 Ba/F3 cells (~20 to 40 mg of total protein) and cells that stably express distinct ZNF198-FGFR1 variants were treated with IL-3 and serum withdrawal for 4 hours before preparation of cell lysates as described (12). Protein extracts from whole-cell lysates were trypsin-digested. Tyrosine-phosphorylated peptides were enriched by immunoaffinity purification (IAP) with antibody against phosphotyrosine (P-Tyr-100) and analyzed by liquid chromatography coupled with MS. Tandem mass spectra were collected in a data-dependent manner with an LTQ ion trap mass spectrometer (ThermoFinnigan).
Reagents
Tyrosine kinase inhibitor (TKI258) was provided by Novartis Pharma. Short hairpin RNA (shRNA) constructs for PKM2 knockdown were purchased from Open Biosystems. The nonphospho- and phosphopeptides were synthesized by American Peptide Company. Murine PKM2 was Flag-tagged by polymerase chain reaction and subcloned into pLHCX retroviral vector. PKM2 variants were subcloned into pDEST27 and pET100 vectors (Invitrogen) for GST-tagged PKM2 expression in mammalian cells and histidine (His)-tagged PKM2 expression in bacterial cells, respectively. Mutations Y83F, Y105F, Y148F, Y175F, Y370F, and Y390F were introduced into PKM2 with QuikChange-XL site-directed mutagenesis kit (Stratagene).
Cell culture
H1299, A549, MDA-MB-134, MDA-MB231, HEL, KG-1a, Mo91, Molm14, and K562 cells were cultured in RPMI 1640 medium with 10% fetal bovine serum (FBS). 293T and GP2-293 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) with 10% FBS. LNCaP and 22Rv cells were cultured in RPMI 1640 medium with 10% FBS, 1 mM sodium pyruvate, and 10 mM Hepes. PC3 cells were cultured in F12 Kaighn’s medium with 5% FBS. Du145 cells were cultured in minimum essential medium with 5% FBS, NaHCO3 (1.5 g/liter), 0.1 mM nonessential amino acids, and 1 mM sodium pyruvate. In the cell proliferation assay, 5 × 104 cells were seeded in a six-well plate and cultured at 37°C in normoxia (5% CO2 and 95% air). Twenty-four hours after seeding, cells used in hypoxia experiments were incubated at 37°C in a sealed hypoxia chamber filled with 1% O2, 5% CO2, and 94% N2. Cells used for oligomycin treatment were incubated at 37°C under normoxic condition. To generate the PKM2 rescue H1299 cell lines, Flag-tagged mouse PKM2 wild type, Y105F, and Y390F were cloned into the retroviral vector pLHCX (Clontech). The constructs were cotransfected with pAmpho cassette vector (Clontech) into GP2-293 cells. Retrovirus was harvested 48 hours after transfection. H1299 cells were infected with harvested retro-virus and were selected by hygromycin (300 μg/ml) for 2 weeks. For lentiviral infection to knock down endogenous hPKM2, shRNA construct was obtained from Open Biosystems. The sequence of shRNA used for knockdown is as follows: 5′-CCGGGCTGTGGCTCTAGACACTAA-ACTCGAGTTTAGTGTCTAGAGCCACAGCTTTTTG-3′. The shRNA construct was cotransfected with two packaging plasmids (pCMV-VSV-G and pCMV-dR8.2 dvpr) into 293T cells. Lentivirus was harvested 48 hours after transfection. H1299 cells stably expressing Flag-tagged PKM2 variants were infected with harvested lentivirus and were selected by puromycin (2 μg/ml) for 1 week.
Antibodies
Antibodies against phospho-Tyr (pY99) and against FGFR1, c-ABL, and FLT-3 were from Santa Cruz Biotechnology; antibodies against PKM2 and JAK2 were from Cell Signaling Technology; antibodies against GST, Flag, and β-actin and Flag M2 beads were from Sigma. Specific antibody against phospho-PKM2 (Tyr105) was generated by Cell Signaling Technology.
Purification of recombinant PKM2 proteins
Hexahistidine-tagged PKM2 proteins were purified by sonication of BL21 (DE3)pLysS cells obtained from 250 ml of culture with IPTG (isopropyl-β-D-thiogalactopyranoside) induction for 4 hours. Cell lysates were resolved by centrifugation and loaded onto a Ni-NTA column in 20 mM imidazole. After washing twice, the protein was eluted with 250 mM imidazole. Proteins were desalted on a PD-10 column and the purification efficiency was examined by Coomassie staining and Western blotting.
PKM2 enzyme assay
Pyruvate kinase activity was measured by an LDH-coupled enzyme assay (27). The assay was carried out with 1 μg of cell lysates or 20 ng of recombinant PKM2 with an enzyme buffer [50 mM tris-HCl, 100 mM KCl, 5 mM MgCl2, 1 mM ADP, 0.5 mM PEP, 0.2 mM NADH (reduced form of NAD+), and 8 U of LDH]. The decrease in absorbance at 340 nm from the oxidation of NADH was measured as pyruvate kinase activity by a spectrophotometer. For the peptide competition assay, 1 μM recombinant His-PKM2 was incubated with or without 10 μM FBP (Sigma) for 30 min at room temperature in a dialysis buffer containing 50 mM tris-HCl (pH 7.5), 100 mM KCl, 5 mM MgCl2, and 5 % glycerol. After incubation, samples were dialysed against 2 liters of the dialysis buffer with 10,000 molecular weight cutoff (MWCO) dialysis cassettes (Pierce) for 7 hours. FBP-loaded samples were divided into four samples and incubated with each peptide (mock, pY390, Y105, and pY105) at the final concentration of 1.5 μM for 30 min at room temperature, and each sample was subjected to the PKM2 enzyme assay as described above.
[14C]FBP dialysis assay
Recombinant His-tagged PKM2 (1 μM) was incubated with 10 μM [14C]FBP (MP Biomedicals) for 30 min at room temperature in a dialysis buffer containing 50 mM tris-HCl (pH 7.5), 100 mM KCl, 5 mM MgCl2, and 5 % glycerol. After incubation, samples were dialysed against 2 liters of dialysis buffer with 10,000 MWCO dialysis cassettes (Pierce) for 7 hours. The dialysed samples were divided into four samples and incubated with each peptide (mock, pY390, Y105, and pY105) at the final concentration of 1.5 μM for 30 min at room temperature, and each sample was redialysed against 2 liters of the dialysis buffer with 10,000 MWCO dialysis cassettes for 7 hours. After redialysis, samples were recovered and the amount of [14C]FBP was measured by scintillation counting.
In vitro kinase assay
GST-PKM2 construct was transfected into 293T cells with Lipofectamine 2000 (Invitrogen). Cells were lysed 24 hours after transfection, and GST-PKM2 was pulled down by Glutathione Sepharose 4B beads (Amersham Bioscience), followed by treatment of 50 U of YOP phosphatase (New England Biolab) at 30°C for 1 hour in bovine serum albumin (1 mg/ml) and 1 × YOP reaction buffer containing 50 mM tris (pH 7.0), 100 mM NaCl, 2 mM Na2EDTA, and 5 mM dithiothreitol (DTT). The beads were then washed with PBS and subjected to FGFR1 kinase assay according to manufacturer’s protocol (Invitrogen). In brief, the YOP-treated beads were incubated with 100 ng of recombinant FGFR1 for 30 min at room temperature in FGFR1 kinase buffer [10 mM Hepes (pH 7.5), 150 mM NaCl, 5 mM DTT, 0.01 % Triton X-100, 10 mM MnCl2, and 200 μM ATP]. The samples were electrophoresed on 10% SDS–acrylamide gel, transferred onto a nitro-cellulose membrane, and then detected with antibody against phosphotyrosine (pY99) and specific antibody against phospho-PKM2 (Y105).
Lactate production, oxygen consumption, and intracellular ATP assays
Cellular lactate production was measured under normoxia with a fluorescence-based lactate assay kit (MBL). Phenol red–free RPMI medium without FBS was added to a six-well plate of subconfluent cells and incubated for 1 hour at 37°C. After incubation, 1 μl of medium from each well was assessed with the lactate assay kit. Cell numbers were counted by a microscope (×40). The oxygen consumption assay was performed as described previously (8). Intracellular ATP concentration was measured by an ATP bioluminescent somatic cell assay kit (Sigma).
Xenograft studies
Nude mice (nu/nu, male, 6 to 8 weeks old, Charles River Laboratories) were subcutaneously injected with 10 × 106 H1299 cells stably expressing mPKM2 wild type and Y105F mutant in conjunction with stable knockdown of endogenous hPKM2 on the left and right flanks, respectively. Tumor formation was assessed every 2 to 3 days. Tumor growth was recorded by measuring two perpendicular diameters of the tumors over a 6-week time course with the formula 4π/3 × (width/2)2 × (length/2). The tumors were harvested and weighed at the experimental endpoint, and the masses of tumors (grams) derived from cells expressing mPKM2 wild type or Y105F mutant in both flanks of each mouse were compared. Statistical analyses were done in comparison to the control group with a paired Student’s t test.
Supplementary Material
REFERENCES AND NOTES
- 1.Brahimi-Horn MC, Chiche J, Pouyssegur J. Hypoxia signalling controls metabolic demand. Curr Opin Cell Biol. 2007;19:223–229. doi: 10.1016/j.ceb.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 2.Kroemer G, Pouyssegur J. Tumor cell metabolism: Cancer’s Achilles’ heel. Cancer Cell. 2008;13:472–482. doi: 10.1016/j.ccr.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 3.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 4.Hathurusinghe HR, Goonetilleke KS, Siriwardena AK. Current status of tumor M2 pyruvate kinase (tumor M2-PK) as a biomarker of gastrointestinal malignancy. Ann Surg Oncol. 2007;14:2714–2720. doi: 10.1245/s10434-007-9481-x. [DOI] [PubMed] [Google Scholar]
- 5.Mazurek S, Boschek CB, Hugo F, Eigenbrodt E. Pyruvate kinase type M2 and its role in tumor growth and spreading. Semin Cancer Biol. 2005;15:300–308. doi: 10.1016/j.semcancer.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 6.Mazurek S, Grimm H, Boschek CB, Vaupel P, Eigenbrodt E. Pyruvate kinase type M2: A crossroad in the tumor metabolome. Br J Nutr. 2002;87(Suppl 1):S23–S29. [PubMed] [Google Scholar]
- 7.Christofk HR, Vander Heiden MG, Harris MH, Ramanathan A, Gerszten RE, Wei R, Fleming MD, Schreiber SL, Cantley LC. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452:230–233. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- 8.Christofk HR, Vander Heiden MG, Wu N, Asara JM, Cantley LC. Pyruvate kinase M2 is a phosphotyrosine-binding protein. Nature. 2008;452:181–186. doi: 10.1038/nature06667. [DOI] [PubMed] [Google Scholar]
- 9.Guminska M, Ignacak J, Kedryna T, Stachurska MB. Tumor-specific pyruvate kinase isoenzyme M2 involved in biochemical strategy of energy generation in neoplastic cells. Acta Biochim Pol. 1997;44:711–724. [PubMed] [Google Scholar]
- 10.Kumar Y, Tapuria N, Kirmani N, Davidson BR. Tumour M2-pyruvate kinase: A gastrointestinal cancer marker. Eur J Gastroenterol Hepatol. 2007;19:265–276. doi: 10.1097/MEG.0b013e3280102f78. [DOI] [PubMed] [Google Scholar]
- 11.Chen J, Deangelo DJ, Kutok JL, Williams IR, Lee BH, Wadleigh M, Duclos N, Cohen S, Adelsperger J, Okabe R, Coburn A, Galinsky I, Huntly B, Cohen PS, Meyer T, Fabbro D, Roesel J, Banerji L, Griffin JD, Xiao S, Fletcher JA, Stone RM, Gilliland DG. PKC412 inhibits the zinc finger 198-fibroblast growth factor receptor 1 fusion tyrosine kinase and is active in treatment of stem cell myeloproliferative disorder. Proc Natl Acad Sci USA. 2004;101:14479–14484. doi: 10.1073/pnas.0404438101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rush J, Moritz A, Lee KA, Guo A, Goss VL, Spek EJ, Zhang H, Zha XM, Polakiewicz RD, Comb MJ. Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nat Biotechnol. 2005;23:94–101. doi: 10.1038/nbt1046. [DOI] [PubMed] [Google Scholar]
- 13.Xiao S, Nalabolu SR, Aster JC, Ma J, Abruzzo L, Jaffe ES, Stone R, Weissman SM, Hudson TJ, Fletcher JA. FGFR1 is fused with a novel zinc-finger gene, ZNF198, in the t(8;13) leukaemia/lymphoma syndrome. Nat Genet. 1998;18:84–87. doi: 10.1038/ng0198-84. [DOI] [PubMed] [Google Scholar]
- 14.Gu TL, Goss VL, Reeves C, Popova L, Nardone J, Macneill J, Walters DK, Wang Y, Rush J, Comb MJ, Druker BJ, Polakiewicz RD. Phosphotyrosine profiling identifies the KG-1 cell line as a model for the study of FGFR1 fusions in acute myeloid leukemia. Blood. 2006;108:4202–4204. doi: 10.1182/blood-2006-06-026666. [DOI] [PubMed] [Google Scholar]
- 15.Xiao S, McCarthy JG, Aster JC, Fletcher JA. ZNF198-FGFR1 transforming activity depends on a novel proline-rich ZNF198 oligomerization domain. Blood. 2000;96:699–704. [PubMed] [Google Scholar]
- 16.Kobrin MS, Yamanaka Y, Friess H, Lopez ME, Korc M. Aberrant expression of type I fibroblast growth factor receptor in human pancreatic adenocarcinomas. Cancer Res. 1993;53:4741–4744. [PubMed] [Google Scholar]
- 17.Luqmani YA, Graham M, Coombes RC. Expression of basic fibroblast growth factor, FGFR1 and FGFR2 in normal and malignant human breast, and comparison with other normal tissues. Br J Cancer. 1992;66:273–280. doi: 10.1038/bjc.1992.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Penault-Llorca F, Bertucci F, Adelaide J, Parc P, Coulier F, Jacquemier J, Birnbaum D, deLapeyriere O. Expression of FGF and FGF receptor genes in human breast cancer. Int J Cancer. 1995;61:170–176. doi: 10.1002/ijc.2910610205. [DOI] [PubMed] [Google Scholar]
- 19.Morrison RS, Yamaguchi F, Bruner JM, Tang M, McKeehan W, Berger MS. Fibroblast growth factor receptor gene expression and immunoreactivity are elevated in human glioblastoma multiforme. Cancer Res. 1994;54:2794–2799. [PubMed] [Google Scholar]
- 20.Pole JC, Courtay-Cahen C, Garcia MJ, Blood KA, Cooke SL, Alsop AE, Tse DM, Caldas C, Edwards PA. High-resolution analysis of chromosome rearrangements on 8p in breast, colon and pancreatic cancer reveals a complex pattern of loss, gain and translocation. Oncogene. 2006;25:5693–5706. doi: 10.1038/sj.onc.1209570. [DOI] [PubMed] [Google Scholar]
- 21.Reis-Filho JS, Simpson PT, Turner NC, Lambros MB, Jones C, Mackay A, Grigoriadis A, Sarrio D, Savage K, Dexter T, Iravani M, Fenwick K, Weber B, Hardisson D, Schmitt FC, Palacios J, Lakhani SR, Ashworth A. FGFR1 emerges as a potential therapeutic target for lobular breast carcinomas. Clin Cancer Res. 2006;12:6652–6662. doi: 10.1158/1078-0432.CCR-06-1164. [DOI] [PubMed] [Google Scholar]
- 22.Marek L, Ware KE, Fritzsche A, Hercule P, Helton WR, Smith JE, McDermott LA, Coldren CD, Nemenoff RA, Merrick DT, Helfrich BA, Bunn PA, Jr, Heasley LE. Fibroblast growth factor (FGF) and FGF receptor-mediated autocrine signaling in non-small-cell lung cancer cells. Mol Pharmacol. 2009;75:196–207. doi: 10.1124/mol.108.049544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abbreviations for the amino acids are as follows: A, Ala; D, Asp; E, Glu; F, Phe; H, His; I, Ile; L, Leu; P, Pro; Q, Gln; R, Arg; S, Ser; V, Val; and Y, Tyr.
- 24.http://www.phosphosite.org/siteAction.do?id=10858
- 25.Mattevi A, Bolognesi M, Valentini G. The allosteric regulation of pyruvate kinase. FEBS Lett. 1996;389:15–19. doi: 10.1016/0014-5793(96)00462-0. [DOI] [PubMed] [Google Scholar]
- 26.Mattevi A, Rizzi M, Bolognesi M. New structures of allosteric proteins revealing remarkable conformational changes. Curr Opin Struct Biol. 1996;6:824–829. doi: 10.1016/s0959-440x(96)80013-3. [DOI] [PubMed] [Google Scholar]
- 27.Shimada N, Shinagawa T, Ishii S. Modulation of M2-type pyruvate kinase activity by the cytoplasmic PML tumor suppressor protein. Genes Cells. 2008;13:245–254. doi: 10.1111/j.1365-2443.2008.01165.x. [DOI] [PubMed] [Google Scholar]
- 28.We gratefully acknowledge the critical reading of the manuscript by B. J. P. Huntly. Plasmid encoding murine PKM2 was provided by H. Fujii at New York University. This work was supported in part by NIH grant CA120272 (J.C.), the American Cancer Society (J.C.), and the Multiple Myeloma Research Foundation (J.C. and S.L.). S.K. is a special fellow of the Leukemia and Lymphoma Society. J.C. is a Georgia Cancer Coalition Distinguished Cancer Scholar and an American Cancer Society Basic Research Scholar.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.