Abstract
For the first 30 years since its discovery, reversible protein acetylation has been studied and understood almost exclusively in the context of histone modification and gene transcription. With the discovery of non–histone acetylated proteins and acetylation-modifying enzymes in cellular compartments outside the nucleus, the regulatory potential of reversible acetylation has slowly been recognized in the last decade. However, the scope of protein acetylation involvement in complex biological processes remains uncertain. The recent development of new technology has enabled, for the first time, the identification and quantification of the acetylome, acetylation events at the whole-proteome level. These efforts have uncovered a stunning complexity of the acetylome that potentially rivals that of the phosphoproteome. The remarkably ubiquitous and conserved nature of protein acetylation revealed by these new studies suggests the regulatory power of this dynamic modification. The establishment of comprehensive acetylomes will change the landscape of protein acetylation, where an exciting research frontier awaits.
Protein acetylation on lysine residues (AcK) was first discovered on histones over four decades ago (1). The ensuing 30 years of intensive research established a fundamental role for reversible histone acetylation in chromatin remodeling that is crucial for gene transcription (2). The demonstration of histone acetyltransferases (HATs) and histone deacetylases (HDACs) as transcriptional coactivators and corepressors added overwhelming support to this dominant theme (3). Although histones have been the primary focus of acetylation studies, other acetylated proteins have long been known to exist (4–6). The discovery of HDAC6 and sirtuin 2 (SIR2)–related SirT3 as a microtubule-associated and mitochondrial matrix deacetylase, respectively, argued that acetylation is not exclusively reserved for nuclear histones (7–9). Although the proposition that lysine acetylation works as a versatile signaling modification has been slowly gaining support (10, 11), a critical question remains in this nascent field: Is reversible acetylation used widely as a regulatory modification, a role predominantly played by reversible phosphorylation, or is it a more specialized modification for a limited number of proteins and biological processes? A report by Choudhary et al. may have provided the answer (12).
Unlike protein phosphorylation, which can be readily identified and studied by powerful tools such as phosphate labeling in situ and sensitive phospho-specific antibodies, the reagents available for characterizing protein acetylation are much less robust. This technical difficulty has limited studies of protein acetylation mostly to a protein-by-protein basis. Although valuable, this approach is biased, rather inefficient, and does not provide a global view of protein acetylation. In 2006, Kim et al. developed a method to study protein acetylation at the whole-proteome level by using antibodies that recognize acetylated lysine (anti-AcK) to enrich for acetylated peptides, which were then identified by nano-HPLC/MS/MS (high-performance liquid chromatography–tandem mass spectrometry) analysis (13). Kim and colleagues reported about 400 lysine acetylation sites in almost 200 proteins. In addition to histones and transcriptional regulators that are known to be acetylated, acetylation was found in proteins involved in a number of cellular pathways not previously linked to acetylation, such as RNA splicing and metabolism, providing the first glimpse of the complexity of the acetylome. Furthermore, this study uncovered a surprising prevalence of acetylation in the mitochondria, where >20% of mitochondrial proteins were acetylated. It is therefore not a coincidence that SIR2-related deacetylases are residents of the mitochondrial matrix (8, 9). Although the identity of the mitochondrial acetyltransferase remains elusive, these findings suggest that mitochondrial function and metabolism are likely major regulatory targets that are controlled by reversible acetylation.
The approach taken by Kim et al. provides a snapshot of the acetylome at steady state. As we have learned from histones, acetylation is dynamic and highly regulated. Therefore, the ability to determine a quantitative “change” in specific acetylation events would provide more power to assign specific functions, an essential step to establishing a functional acetylome. The work by Choudhary and colleagues has accomplished this goal by adopting SILAC (stable-isotope labeling by amino acid in cell culture) technology and by using an LTQ Orbitrap mass spectrometer with high resolution and sensitivity (12). By labeling cellular proteomes with isotopes of different molecular weight, SILAC allows simultaneous quantification of specific acetylated peptides of mixed proteomes prepared under different experimental conditions with a reported false-discovery rate of only 0.1 to 0.3% (14).
Choudhary et al. identified over 3500 acetylation sites in ~1700 acetylated proteins. In comparison, the phosphoproteome has been estimated to comprise about 6600 phosphorylation sites in ~2200 proteins (15). Therefore, the acetylome is approaching the size of the phosphoproteome, which represents the most dominant protein modification. The total number of acetylation events will certainly be even higher because the antibody against AcK that was used in this study showed sequence preference, as revealed by the conspicuous absence of acetylated tubulin (Ac-K40). The sheer size of the acetylome points to the ubiquitous nature and potential regulatory power of reversible acetylation.
Several interesting characteristics emerged from the acetylome study. Previous analysis of the phosphoproteome, which contains tens of thousands of proteins, revealed that phosphorylation is highly conserved (16). In vertebrates, acetylation sites are as conserved as those in phosphorylated proteins, hinting at the selective pressure to maintain this protein modification. Extensive lysine acetylation is also found in Escherichia coli, further indicating an evolutionarily conserved function of acetylation (17). This conservation can also be easily appreciated by the striking finding that acetyl–coenzyme A (CoA) synthetase (ACS) activity from Salmonella to humans is regulated by the same Sir2-dependent deacetylation of a conserved lysine residue (18–20).
Unlike phosphorylation, which often clusters in unstructured regions (16), acetylation appears to be concentrated in regions with an ordered secondary structure (12), with the exception of histone tails, which are disordered by nature (21). The reason for this distribution pattern is unclear. Indeed, our knowledge of how acetylation affects protein activity remains scarce. Studies on histone acetylation show that acetylation works, at least in part, by neutralizing the positive charge on lysine (22). Acetylated lysine can also be recognized and bound by the bromodomain, an evolutionarily conserved protein module found in many chromatin-associated proteins (23). It is possible that lysine acetylation, in a manner analogous to lysine ubiquitination and specific phosphorylation, might be used to recruit and assemble protein complexes with AcK-binding activity. Whether there exists an acetyl-lysine binding activity equivalent to the bromodomain in the cytoplasm or mitochondria is an exciting possibility that remains to be determined.
The study by Choudhary et al. also revealed that many proteins undergoing acetylation are components of large macromolecular complexes. Several acetylation-modulated functional networks were identified in this study, including chromatin-remodeling complexes, RNA splicing, nuclear transport, and the actin cytoskeletal-remodeling machinery. In the actin-remodeling Arp2/3 (actin-related protein 2/3) complex, all but one of the subunits are acetylated. These findings indicate that the acetylome has high network connectivity. The unusual enrichment of acetylation suggests that multiple and possibly coordinated acetylation events might be important for the regulation of this multiprotein machinery. If AcK-binding activity does exist in such complexes, the abundance of acetylated lysine could provide a possible mechanism to regulate their assembly.
The global acetylome also showed that many acetylated proteins are involved in other posttranslational modifications. For example, protein kinases, such as those in the ATM (ataxia telangiectasia mutated) and Cdc2 families, are subject to acetylation. Furthermore, mitogen-activated protein kinase kinase (MKK6) can be acetylated by the bacterial acetyltransferase VopA, leading to its inactivation (24). These findings suggest that acetylation might have a broad regulatory role for kinase activity. In addition, several enzymes involved in other forms of lysine modification, such as ubiquitination and methylation, were also acetylated. These include the methyltransferase MLL and the demethylase JARID (Jumonji AT-rich interactive domain). Crosstalk between acetylation and other posttranslational modifications could further amplify the regulatory power of acetylation events.
The mapping of the acetylome also has important clinical implications. Over 100 clinical trials for HDAC inhibitors (HDACIs) for cancer treatment are listed on the National Cancer Institute (NCI) Web site (http://www.cancer.gov/clinicaltrials/). These drugs are also promising therapies for neurodegenerative and inflammatory disorders (25, 26). How HDACIs achieve their therapeutic effects, however, remains poorly understood. The ability to identify “acetylation targets” of these therapeutic HDACIs could potentially provide a window into acetylation events relevant to therapeutic activity. Toward this goal, Choudhary et al. have determined the acetylome controlled by clinically approved HDAC inhibitor SAHA and by another that is under evaluation, MS-275. SAHA is an inhibitor that targets all HDACs, except the SIR2 family (27). MS-275, by contrast, is selective for class I HDACs, which are predominantly localized to the nucleus (28, 29). Surprisingly, the SAHA- and MS-275–sensitive acetylomes revealed that only 10% of all acetylation sites showed increased acetylation following treatment, indicating that these inhibitors have more specific and limited in vivo effects than previously thought. This analysis also uncovered remarkable differences in acetylation events affected by these two HDACIs. For example, Hsp90 acetylation, which inhibits Hsp90 activity (30, 31), was induced by SAHA but not MS-275, whereas the opposite effect was observed for p53 acetylation. These results clearly demonstrate that different HDACIs have distinct effects on specific acetylation events, which could contribute to their unique therapeutic activity. Accordingly, the functional acetylation map based on HDACI-specific acetylomes could help elucidate the mechanism by which a specific HDACI accomplishes its therapeutic activity, potentially pointing to better therapeutic strategies for these compounds.
The SILAC-coupled mass spectrometry technology is powerful but has limitations. For example, given the potential link between protein acetylation and metabolism, it would be interesting to quantify changes in liver acetylomes following metabolic stress, such as fasting. Because SILAC technology relies on labeling proteins with radioisotopes in paired samples, this procedure is difficult to perform in live animals. The development of label-free quantification (LFQ) mass spectrometry could provide an alternative approach to fill this need, although its accuracy is lower than that of SILAC. Using this approach, Schwer et al. determined changes in liver acetylation events in response to calorie restriction (CR), a regimen that extends life span (32). Remarkably, acetylation of ~70 mitochondrial proteins changed by at least 2.5-fold in response to CR. This finding is interesting, because CR-induced longevity requires SIR2 deacetylase (33), and there are at least two SIR2-related deacetylases in the mitochondria (8, 9, 34). This report not only provides provocative evidence linking CR and life-span regulation to the remodeling of the mitochondrial acetylome, but also demonstrates the feasibility of using LFQ-mass spectrometry to quantify acetylation events in tissues.
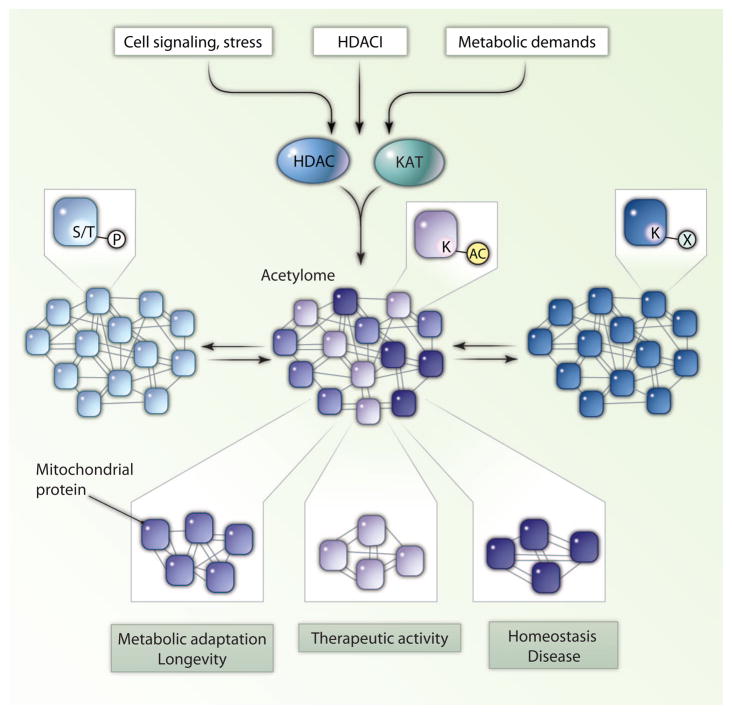
SILAC- and LFQ-based mass spectrometry holds tremendous potential to revolutionize the study of protein acetylation. For example, it will now be possible to determine acetylomic changes in defined physiological, pathological, and pharmacological contexts. The unique acetylomic signatures and comparative acetylomes could provide a rich source of information for uncovering how coordinated reversible acetylation events might control specific signaling networks important for homeostasis, disease development, and therapeutic activity (Fig. 1). A similar approach could be used to establish specific acetylomes regulated by any HDAC or HAT members through standard gain-of-function and loss-of-function approaches. Such information could provide a much more complete picture of how a specific HDAC or HAT controls complex biological processes. The adoption of these new technologies will undoubtedly expand and change the landscape of protein acetylation.
Fig. 1.
The acetylome (K-AC) undergoes dynamic and specific changes in response to cell signaling, stress, metabolic demands, and HDAC inhibitor (HDACI) treatment, all of which can modulate activity of deacetylases (HDAC) or lysine acetyltransferases (KAT). These changes contribute to unique acetylomic signatures, represented by different shades of K-Ac–containing circles, and are integral parts of physiological and pathological responses important for homeostasis, disease development, metabolic adaptation, and therapy. For example, the mitochondrial acetylome likely plays a critical role in metabolic adaptation and longevity. The regulatory power of the acetylome is further amplified through its crosstalk with other modifications, including phosphorylation of serine or threonine residues (S/T-P) and other lysine-based modifications (K-X), where X can be ubiquitination, methylation, neddylation, or sumoylation.
The exponential growth of known cellular acetylation events, though establishing the ubiquitous nature of protein acetylation, does not in itself reveal the function of these modifications. Similar to challenges with gene array studies in early days, better computational and analytic tools will be essential to extract useful information from the large numbers of acetylation events uncovered by the ever-improving technology. One can foresee that efforts to “decode the acetylome” will become an exciting frontier in this field of research, where researchers of diverse disciplines would be brought together to make sense of the regulatory power of protein acetylation. In 2000, Tony Kouzarides asked whether acetylation was “a regulatory modification to rival phosphorylation” (35). Although the jury is still out, the explosive growth in the discovery of protein acetylation events in the last few years indicates that the answer might lie just over the horizon. Beware, the era of acetylation biology has arrived.
References and Notes
- 1.Gershey EL, Vidali G, Allfrey VG. Chemical studies of histone acetylation. The occurrence of epsilon-N-acetyllysine in the f2a1 histone. J Biol Chem. 1968;243:5018–5022. [PubMed] [Google Scholar]
- 2.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 3.Xu L, Glass CK, Rosenfeld MG. Coactivator and corepressor complexes in nuclear receptor function. Curr Opin Genet Dev. 1999;9:140–147. doi: 10.1016/S0959-437X(99)80021-5. [DOI] [PubMed] [Google Scholar]
- 4.Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 5.L’Hernault SW, Rosenbaum JL. Chlamydomonas alpha-tubulin is posttranslationally modified by acetylation on the epsilon-amino group of a lysine. Biochemistry. 1985;24:473–478. doi: 10.1021/bi00323a034. [DOI] [PubMed] [Google Scholar]
- 6.Piperno G, Fuller MT. Monoclonal antibodies specific for an acetylated form of alpha-tubulin recognize the antigen in cilia and flagella from a variety of organisms. J Cell Biol. 1985;101:2085–2094. doi: 10.1083/jcb.101.6.2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hubbert C, Guardiola A, Shao R, Kawaguchi Y, Ito A, Nixon A, Yoshida M, Wang XF, Yao TP. HDAC6 is a microtubule-associated deacetylase. Nature. 2002;417:455–458. doi: 10.1038/417455a. [DOI] [PubMed] [Google Scholar]
- 8.Onyango P, Celic I, McCaffery JM, Boeke JD, Feinberg AP. SIRT3, a human SIR2 homologue, is an NAD-dependent deacetylase localized to mitochondria. Proc Natl Acad Sci USA. 2002;99:13653–13658. doi: 10.1073/pnas.222538099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schwer B, North BJ, Frye RA, Ott M, Verdin E. The human silent information regulator (Sir)2 homologue hSIRT3 is a mitochondrial nicotinamide adenine dinucleotide-dependent deacetylase. J Cell Biol. 2002;158:647–657. doi: 10.1083/jcb.200205057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cohen T, Yao TP. AcK-knowledge reversible acetylation. Sci STKE. 2004;2004:pe42. doi: 10.1126/stke.2452004pe42. [DOI] [PubMed] [Google Scholar]
- 11.Yang XJ, Seto E. Lysine acetylation: Codified crosstalk with other posttranslational modifications. Mol Cell. 2008;31:449–461. doi: 10.1016/j.molcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 13.Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, Cheng T, Kho Y, Xiao H, Xiao L, Grishin NV, White M, Yang XJ, Zhao Y. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol Cell. 2006;23:607–618. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- 14.Mann M. Functional and quantitative proteomics using SILAC. Nat Rev Mol Cell Biol. 2006;7:952–958. doi: 10.1038/nrm2067. [DOI] [PubMed] [Google Scholar]
- 15.Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 16.Gnad F, Ren S, Cox J, Olsen JV, Macek B, Oroshi M, Mann M. PHOSIDA (phosphorylation site database): Management, structural and evolutionary investigation, and prediction of phosphosites. Genome Biol. 2007;8(13):R250–R250. doi: 10.1186/gb-2007-8-11-r250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang J, Sprung R, Pei J, Tan X, Kim S, Zhu H, Liu CF, Grishin NV, Zhao Y. Lysine acetylation is a highly abundant and evolutionarily conserved modification in Escherichia coli. Mol Cell Proteomics. 2009;8:215–225. doi: 10.1074/mcp.M800187-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hallows WC, Lee S, Denu JM. Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proc Natl Acad Sci USA. 2006;103:10230–10235. doi: 10.1073/pnas.0604392103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schwer B, Bunkenborg J, Verdin RO, Andersen JS, Verdin E. Reversible lysine acetylation controls the activity of the mitochondrial enzyme acetyl-CoA synthetase 2. Proc Natl Acad Sci USA. 2006;103:10224–10229. doi: 10.1073/pnas.0603968103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Starai VJ, Celic I, Cole RN, Boeke JD, Escalante-Semerena JC. Sir2-dependent activation of acetyl-CoA synthetase by deacetylation of active lysine. Science. 2002;298:2390–2392. doi: 10.1126/science.1077650. [DOI] [PubMed] [Google Scholar]
- 21.Hansen JC, Lu X, Ross ED, Woody RW. Intrinsic protein disorder, amino acid composition, and histone terminal domains. J Biol Chem. 2006;281:1853–1856. doi: 10.1074/jbc.R500022200. [DOI] [PubMed] [Google Scholar]
- 22.Ren Q, Gorovsky MA. Histone H2A.Z acetylation modulates an essential charge patch. Mol Cell. 2001;7:1329–1335. doi: 10.1016/s1097-2765(01)00269-6. [DOI] [PubMed] [Google Scholar]
- 23.Mujtaba S, He Y, Zeng L, Yan S, Plotnikova O, Sachchidanand R, Sanchez NJ, Zeleznik-Le Z, Ronai MM. Zhou, Structural mechanism of the bromodomain of the coactivator CBP in p53 transcriptional activation. Mol Cell. 2004;13:251–263. doi: 10.1016/s1097-2765(03)00528-8. [DOI] [PubMed] [Google Scholar]
- 24.Trosky JE, Li Y, Mukherjee S, Keitany G, Ball H, Orth K. VopA inhibits ATP binding by acetylating the catalytic loop of MAPK kinases. J Biol Chem. 2007;282:34299–34305. doi: 10.1074/jbc.M706970200. [DOI] [PubMed] [Google Scholar]
- 25.Halili MA, Andrews MR, Sweet MJ, Fairlie DP. Histone deacetylase inhibitors in inflammatory disease. Curr Top Med Chem. 2009;9:309–319. doi: 10.2174/156802609788085250. [DOI] [PubMed] [Google Scholar]
- 26.Kazantsev AG, Thompson LM. Therapeutic application of histone deacetylase inhibitors for central nervous system disorders. Nat Rev Drug Discov. 2008;7:854–868. doi: 10.1038/nrd2681. [DOI] [PubMed] [Google Scholar]
- 27.Marks PA, Xu WS. Histone deacetylase inhibitors: Potential in cancer therapy. J Cell Biochem. 2009;107:600–608. doi: 10.1002/jcb.22185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu E, Dul E, Sung CM, Chen Z, Kirkpatrick R, Zhang GF, Johanson K, Liu R, Lago A, Hofmann G, Macarron R, de los Frailes M, Perez P, Krawiec J, Winkler J, Jaye M. Identification of novel isoform-selective inhibitors within class I histone deacetylases. J Pharmacol Exp Ther. 2003;307:720–728. doi: 10.1124/jpet.103.055541. [DOI] [PubMed] [Google Scholar]
- 29.Saito A, Yamashita T, Mariko Y, Nosaka Y, Tsuchiya K, Ando T, Suzuki T, Tsuruo T, Nakanishi O. A synthetic inhibitor of histone deacetylase, MS-27-275, with marked in vivo antitumor activity against human tumors. Proc Natl Acad Sci USA. 1999;96:4592–4597. doi: 10.1073/pnas.96.8.4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kovacs JJ, Murphy PJ, Gaillard S, Zhao X, Wu JT, Nicchitta CV, Yoshida M, Toft DO, Pratt WB, Yao TP. HDAC6 regulates Hsp90 acetylation and chaperone-dependent activation of glucocorticoid receptor. Mol Cell. 2005;18:601–607. doi: 10.1016/j.molcel.2005.04.021. [DOI] [PubMed] [Google Scholar]
- 31.Bali P, Pranpat M, Bradner J, Balasis M, Fiskus W, Guo F, Rocha K, Kumaraswamy S, Boyapalle S, Atadja P, Seto E, Bhalla K. Inhibition of histone deacetylase 6 acetylates and disrupts the chaperone function of heat shock protein 90: A novel basis for antileukemia activity of histone deacetylase inhibitors. J Biol Chem. 2005;280:26729–26734. doi: 10.1074/jbc.C500186200. [DOI] [PubMed] [Google Scholar]
- 32.Schwer B, Eckersdorff M, Li Y, Silva JC, Fermin D, Kurtev MV, Giallourakis C, Comb MJ, Alt FW, Lombard DB. Calorie restriction alters mitochondrial protein acetylation. Aging Cell. 2009;8:604–606. doi: 10.1111/j.1474-9726.2009.00503.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin SJ, Defossez PA, Guarente L. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science. 2000;289:2126–2128. doi: 10.1126/science.289.5487.2126. [DOI] [PubMed] [Google Scholar]
- 34.Nakagawa T, Lomb DJ, Haigis MC, Guarente L. SIRT5 Deacetylates carbamoyl phosphate synthetase 1 and regulates the urea cycle. Cell. 2009;137:560–570. doi: 10.1016/j.cell.2009.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kouzarides T. Acetylation: A regulatory modification to rival phosphorylation? EMBO J. 2000;19:1176–1179. doi: 10.1093/emboj/19.6.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.We thank Y.-M. Zhao, B. Harvat, and T. Cohen for helpful comments. This work is supported by NIH (grants NS054022 and AR055613).