Abstract
Late-onset Alzheimer’s disease is a common complex disorder of old age. Though these types of disorders can be highly heritable, they differ from single-gene (Mendelian) diseases in that their causes are often multifactorial with both genetic and environmental components. Genetic risk factors that have been firmly implicated in the cause are mutations in the amyloid precursor protein (APP), presenilin 1 (PSEN1) and presenilin 2 (PSEN2) genes, which are found in large multi-generational families with an autosomal dominant pattern of disease inheritance, the apolipoprotein E (APOE)ε4 allele and the sortilin-related receptor (SORL1) gene. Environmental factors that have been associated with late-onset Alzheimer’s disease include depressive illness, various vascular risk factors, level of education, head trauma and estrogen replacement therapy. This complexity may help explain their high prevalence from an evolutionary perspective, but the etiologic complexity makes identification of disease-related genes much more difficult. The “endophenotype” approach is an alternative method for measuring phenotypic variation that may facilitate the identification of susceptibility genes for complexly inherited traits. The usefulness of endophenotypes in genetic analyses of normal brain morphology and, in particular for Alzheimer’s disease will be reviewed as will the implications of these findings for models of disease causation. Given that the pathways from genotypes to end-stage phenotypes are circuitous at best, identifying endophenotypes more proximal to the effects of genetic variation may expedite the attempts to link genetic variants to disorders.
Late-onset Alzheimer’s disease (LOAD) is among the most frequently encountered diseases in aging societies, and its prevalence is expected to quadruple by 2047 (Brookmeyer et al., 1998). Twin studies suggest that 37% to as much as 78% of the variance in the age-at-onset of LOAD can be attributed to additive genetic effects (Meyer and Breitner, 1998). Conversely, cognitively healthy aging is also substantially influenced by genes (Gudmundsson et al., 2000). Genes increasing the risk of developing LOAD (apolipoprotein E (APOE)-ε4, sortilin-related receptor (SORL1)) (Tang et al., 1996; Rogaeva et al., 2007) have been identified and confirmed in individuals with sporadic or familial LOAD. However, they explain only a small proportion of the genetic contribution to LOAD leaving the remaining genetic risk factors to be identified. An important step towards understanding the mechanisms underlying LOAD is the identification of the genes controlling brain structure under physiological conditions. LOAD is associated with alterations in structure and function of several brain regions in particular the hippocampus and cerebral grey matter, and it is thought that these associations have a substantial genetic contribution (Posthuma et al., 2000; Thompson et al., 2001, 2002). Studies of the genetics of brain structure and function among normal individuals, which over the past decade have been extended to the entire human lifespan from childhood through extreme old age (Gogtay et al., 2004; Sowell et al., 2003; Thompson et al., 2007), have concluded that variation in brain structure and function can be expected and that pathological states represent the extremes of this variation. Cumulatively these data provide not only a backdrop for understanding the genetic influences on neuroanatomy and neurophysiology but also the basis for understanding the genetics of neurodegenerative diseases associated with changes in these brain structures including LOAD, and the concept of cognitive reserve (CR) in LOAD, a model on the reserve against brain damage that is based on the fact that there appears to be no direct relationship between the degree of LOAD pathology and degree of cognitive impairment. The few studies that explored the genetics of brain structure under physiological conditions suggest that human brain volume is genetically influenced, varies regionally within the brain, and is associated with high heritability for regional amounts of gray matter density in medial frontal cortex, Heschl’s gyrus and postcentral gyrus, and moderate to high heritability for Broca’s area, anterior cingulate, hippocampus, amygdala, gray matter of the parahippocampal gyrus and white matter of the superior occipitofrontal fasciculus (Posthuma et al., 2000; Thompson et al., 2001; Baare et al., 2001; Bartley et al., 1997; Carmelli et al., 1998; Eckert et al., 2002; Geschwind et al., 2002; Hulshoff et al., 2006; Pennington et al., 2000; Pfefferbaum et al., 2000, 2004; Reveley et al., 1984; Scamvougeras et al., 2003; Sullivan et al., 2001; Wallace et al., 2006; White et al., 2002). Furthermore, they indicate that the heritability for brain volumes, including cerebral gray and white matter, remains constant throughout life suggesting little environmental influence. However, this remains speculation and is inconsistent with imaging studies indicating alterations of brain structure in response to environmental influences (Draganski et al., 2004). The genetic influences on age-related changes in brain structure remain to be determined, and the specific genes involved in variation of brain volume are largely unknown although some candidate genes have been suggested. The continued pursuit of genetic variants associated with LOAD has been limited despite available improved analytic techniques. This may reflect the continued use of small cohorts of patients underpowered for genetic studies in this complex disease in which multiple genes with small effects each (“quantitative trait loci” (QTLs)) are likely to contribute to the various quantitative traits associated with the disease such as memory performance, amyloid/tau pathology or hippocampal atrophy. Alternatively it could reflect a failure to develop useful quantitative endophenotypes. Endophenotypes are measurable intermediate phenotypes that are generally closer to the action of the gene than affection status, and thus exhibit higher genetic signal-to-noise ratios (Gottesman, 2003). They are characteristics that are genetically correlated with disease, and can be measured in both affected and unaffected individuals. Endophenotypes often provide much greater power to localize and identify disease-related QTLs than does affection status alone (Blangero et al., 2003). Current genetic data from the study of normal brain structure and function may provide opportunities to use quantitative endophenotypic traits for understanding LOAD in light of normal brain morphology and CR.
CONCEPT OF CR
The basis for CR arose from the observation that the severity of neuropathological manifestation of LOAD does not always correlate with clinical LOAD severity (Katzman et al., 1988). The concept proposes that individuals develop CR in the presence of favorable environments such as high educational level or by genetic predisposition, or both, and that CR increases the threshold for neuropsychological responses to brain insult. It poses that those with greater brain reserve capacity can bear greater brain damage before cognitive deficit appears. Stern (2002) applied CR to any situation where there is variation in response to brain injury, suggesting that CR can be applied to individuals who are healthy as well as individuals with neurodegeneration.
Both environmental and genetic factors are likely to affect responses to injury. Gene dosage and timing will influence the response. Similarly, strength and timing of environmental factors will bring about variations among individuals. Stern (2002) proposed two forms of CR: in neural reserve, preexisting brain networks that are more efficient or have greater capacity may be less susceptible to disruption. In neural compensation, alternate networks may compensate for pathology’s disruption of preexisting networks.
Besides anatomic measures such as brain volume, head circumference, synaptic count, or dendritic branching, variables descriptive of lifetime experience are commonly used proxies for CR. These include measures of socioeconomic status, such as income or occupational or educational attainment, and several domains of neuropsychological traits including memory, general intelligence and language. Analyzing genetic contribution to CR in conjunction with neuroanatomic measures and socioeconomic measures helps clarify the concept of CR and thereby the mechanisms leading to a clinical manifestation of LOAD. To support the hypothesis of genetic contributions to CR it is in particular necessary to show that there is a differential expression of a gene(s) that influences brain morphology and cognitive function. In the following sections will discuss the genetic influences on these measures.
GENETIC INFLUENCES ON BRAIN MORPHOLOGICAL ENDOPHENOTYPES
Studies in quantitative genetics explore the decomposition of observed phenotypic variance into genetic and environmental sources by studying genetically related individuals. Heritability is the proportion of genetic variance over the total variance. Environmental variance can be further decomposed into environmental variance shared by members of a family (common environment) or non-shared variance, which is unique to a certain individual (unique environment). To determine the relative contribution of genetic, common, and unique environmental influences on variation in brain structures, the (extended) twin model is particularly powerful (Posthuma et al., 2000). In this method, heritability estimates of brain structure are usually based on data from monozygotic twin pairs (MZ) who are genetically identical (except in the rare case of a mutation) and dizygotic twin pairs (DZ) who share on average 50% of their segregating genes. If for a certain brain measure monozygotic twin pairs resemble each other more closely than DZ twin pairs, it can be inferred that variation of the brain measure is heritable. However, in addition to genetic influences, common (or shared) environmental influences may also be a contribution factor. The presence of shared environmental factors is suspected when correlations in DZ twins are >50% of the MZ correlation (Boomsma et al., 2002). A first impression of the importance of unique environmental factors can be obtained from the extent to which MZ twins do not resemble each other. To estimate contributions of additive genetic (A) effects, common (or shared) environmental (C) and unique environmental effects (E) to variation in a phenotype, structural equation modeling (SEM) is increasingly used, which is capable of explicitly testing whether genetic or environmental factors contribute to individual differences. In extended twin-studies, additional relatives of twins are included in the study design. This increases the statistical power to detect the influences of environmental influences shared by members from the same family. Later in this review we will discuss findings from (extended) twin studies on normal human brain morphology.
Brain imaging has been a useful tool to define the genetic contribution to brain structures, and the heritability of reproducible, quantitative endophenotypes (DeCarli et al., 2005). Twin studies have demonstrated substantial heritability of these endophenotypes (Carmelli et al., 2002). Although early large-scale brain-imaging research focused on young, healthy, normal adult subjects (Mazziotta et al., 2001), in the past decade normative studies of brain structure and function have been extended to the entire human lifespan, from childhood through extreme old age (Gogtay et al., 2004).
Heritability of brain volumes and structures
A total of 14 twin studies measuring brain volume have been performed. The first study that quantitatively studied brain structure in healthy MZ and DZ twin pairs using computerized axial tomography (CAT) (Table 1) (Reveley et al., 1984), found that lateral ventricle variation was mostly explained by genetic factors. Later studies using magnetic resonance imaging (MRI), found high heritability estimates of global brain measures including intracranial volume (>81%) (Baare et al., 2001; Carmelli et al., 1998; Pfefferbaum et al., 2000) and total brain volume (66%–97%) (Baare et al., 2001; Bartley et al., 1997; Pennington et al., 2000; Wright et al., 2002). The first extended twin-study that explored the genetic contributions to variation in global gray and white matter found heritability estimates of 82% for gray matter and 88% for white matter (Baare et al., 2001). The heritability of cerebral hemisphere volumes was estimated at 65% (Geschwind et al., 2002), of cerebellar volume at 88% (Posthuma et al., 2000), and of the corpus callosum between 79% and 94% (Pfefferbaum et al., 2000; Scamvougeras et al., 2003). In a study that did not include DZ twin pairs, MZ twin pair correlations were high (>0.90 for cerebellum, total brain, gray and white matter, and >0.75 for caudate nucleus, putamen, thalamus and cortical depth) when compared with a healthy, unrelated comparison group, indicating an upper limit of heritability (White et al., 2002). Brain areas that in contrast seem to be mainly under environmental control include the gyral patterning of the cortex (Bartley et al., 1997; Eckert et al., 2002), the volume of the lateral ventricles (Baare et al., 2001; Wright et al., 2002), and the volume of the hippocampus (40%) (Sullivan et al., 2001).
Table 1.
Studies on heritability of human brain volumes
| Author | Subjects | Age in y, mean (range) | Brain region | Heritability in % (95% CI) |
|---|---|---|---|---|
| Reveley et al. (1984) | 18 MZ, 18 DZ | NA | LV | 82%–85% (NA) |
| Bartley et al. (1997) | 10 MZ, 9 DZ | MZ: 31 (19–54), DZ: 33 (18–29) | TB | 94% (NA) |
| Carmelli et al. (1998) | 74 MZ, 71 DZ | 68–79 y | IC | 91% (NA) |
| Pennington et al. (2000) | Reading disability: 25 MZ, 23 DZ; non-reading disability: 9 MZ, 9 DZ |
Reading disability: MZ: 17.1, DZ: 16.8; Non-reading disability: MZ: 19.4, DZ: 18.7 |
TB | 97% (NA) |
| Neocortex | 56% (NA) | |||
| Pfefferbaum et al. (2000) | 45 MZ, 40 DZ | MZ: 72.2, DZ: 71.4, range: 68–78 |
Subcortex | 70% (NA) |
| IC | 81% (72–90) | |||
| CC | 79% (69–89) | |||
| LV | 79% (55–100) | |||
| Posthuma et al. (2000) | See Baaré et al. (2001) | See Baaré et al. (2001) | CB | 88% (81–92) |
| Sullivan et al. (2001) | 45 MZ, 40 DZ | MZ: 72.2, DZ: 71.4, range: 68–78 |
HIP | 40% (NA) |
| Thompson et al. (2001) | 10 MZ, 10 DZ | 48.2±3.4 | Middle frontal sensomotor and anterior temporal cortices, Broca’s and Wernicke’s region (cortical thickness) |
90%–95% |
| Baaré et al. (2001) | 54 MZ, 58 DZ, 34 sibs 15 MZ, 18 DZ |
MZM: 31.2, MZF: 34.1, DZM: 30.3, DZF: 30.6, OS: 30.3, sibs: 29.0; range: 19–69 |
IC | 88%(82–92) |
| TB | 90% (85–93) | |||
| GM | 82% (73–88) | |||
| 75.7 ± y | WM | 87% (80–91) | ||
| LV | C. 59% (47–69), | |||
| Size CC | (E) 41% (31–53) | |||
| Microstructure CC (DTI) | 5:1 (NA) 3:1 (NA) |
|||
| Geschwind et al. (2002) | 72 MZ, 67 DZ | MZ: 72.3, DZ: 71.8 | Cerebral hemispheres | 65% (NA) |
| Eckert et al. (2002) | 27 MZ, 12 DZ | MZ: 6.9–16.4, DZ: 6.1–15.0 | Planum temporale asymmetry | NA |
| Wright et al. (2002) | 10 MZ, 10 DZ | MZ: 31 (19–54), DZ: 23 (18–29) | TB | 66% (17–100) |
| LV | C. 48% (0 –97), (E) 50% (32–84) |
|||
| CB | 63%, (E) 22% (NA) | |||
| Ventrolateral FR, cingulate, anterior/superior/transverse temp, retrosplenium |
58%–73% (NA) | |||
| White et al. (2002) | 12 MZ, 12 control pairs | MZ: 24.5±7.2, controls: 24.4±7.2 |
TB, GM, WM, CB | r>0.90 |
| CAU, PUT, THAL, cortical depth |
r>0.75 | |||
| Scamvougeras et al. (2003) | 14 MZ, 12 DZ | MZ: 16–41, DZ: 18–32 | CC | 94% (NA) |
| Pfefferbaum et al. (2004) | 34 MZ, 37 DZ | 4-year longitudinal follow-up T1: 68–80 y, T2: 72–84 y |
CC (T1) | 89% (NA) |
| CC (T2) | 92% (NA) | |||
| LV (T1) | 92% (NA) | |||
| LV (T2) | 88% (NA) | |||
| Wallace et al. (2006) | 90 MZ, 37 DZ | MZ: 11.9, DZ: 10.9, range: 5–19 | TB | 89% (67–92) |
| GM | 82% (50–87) | |||
| WM | 85% (56–90) | |||
| FR, TEMP, PAR | 77%–88% (50–90) | |||
| CB | 49% (13–83) | |||
| LV | 31% (0–67), (C) 24% (0 –58), (E) 45% (33–60) |
|||
| Hulshoff Pol et al. (2006) | See Baaré et al. (2001) | See Baaré et al. (2001) | WM (SOFT, CC, CST) GM, MFL, SFL, STL, CING, PARAHIP, AMYG, OCC |
69%–82% (NA) |
| 55%–85% (NA) |
It has to be pointed out that some of these studies did not correct for total cranial volume or height when measuring brain volumes. Although it is likely that the ratio of brain volume/total cranial volume is comparable among MZ twins, it remains possible that this lack of correction has led in some studies to spurious results.
The only published twin-study to date in children was consistent with previous adult studies in that additive genetic effects accounted for a substantial portion of variability in nearly all brain regions except the cerebellum (Wallace et al., 2006). While cerebellum volume was found to be mainly under influence of genes in adults (Posthuma et al., 2000; Baare et al., 2001; Wright et al., 2002), it showed an exceptionally strong susceptibility to shared (30%) and unshared or unique environmental (21%) influences in children (Wallace et al., 2006). The mechanisms underlying the strong environmental control of cerebellar volume in children remain unknown. However, the notion that the cerebellum is particularly susceptible for environmental influences is supported by the fact that it is during pediatric development the most sexually dimorphic macroscopic brain structure develops and that it is the brain structure that reaches peak volume the latest (Wallace et al., 2006). The potential for relatively strong environmental influences on cerebellar development is further consistent with its preferential susceptibility to insults such as alcohol or anoxia and its role in modulating responses to environmental stimuli. It has been postulated that the postnatal neurogenesis of cerebellar Purkinje cells may confer susceptibility to environmental insult (Welsh et al., 2002).
Three studies have more distinctively examined possible genetic effects on specific brain areas using voxel-based morphometry (Ashburner and Friston, 2001) and cortical thickness measures. In a study that constructed detailed three-dimensional maps based on a genetic continuum of similarity in grey matter in groups of unrelated subjects, DZ and MZ twins, in particular anatomical regions that include frontal and language-related cortices (i.e. sensorimotor, middle frontal, anterior temporal and Wernicke’s cortices) were found to be genetically influenced (Thompson et al., 2001). Voxel-based morphometry revealed high heritabilities of 42%–66% for temporal/parietal neocortical areas and paralimbic structures (Wright et al., 2002). Furthermore, tissue density of the left and right medial (78%, 83%) and superior frontal (76%, 80%) cortex, superior temporal cortex (80%, 77%), occipital gray matter (85%) and connecting white matter of the superior occipitofrontal fasciculus (79%, 77%) and corpus callosum (82%, 80%) were also found to be genetically determined (Hulshoff et al., 2006). In contrast, unique environmental factors influenced vast gray matter and white matter areas surrounding the lateral ventricles (up to 50%) (Hulshoff et al., 2006).
Only one study has measured the heritability estimates for changes in brain volumes over time. However, while the genetic contributions to variability in intracranial volume, corpus callosum, and lateral ventricles in healthy elderly were high (88%–92%) (Pfefferbaum et al., 2000) they did not change after 4 years’ follow-up (Pfefferbaum et al., 2004).
GENETICS OF LOAD AND LOAD ENDOPHENOTYPES
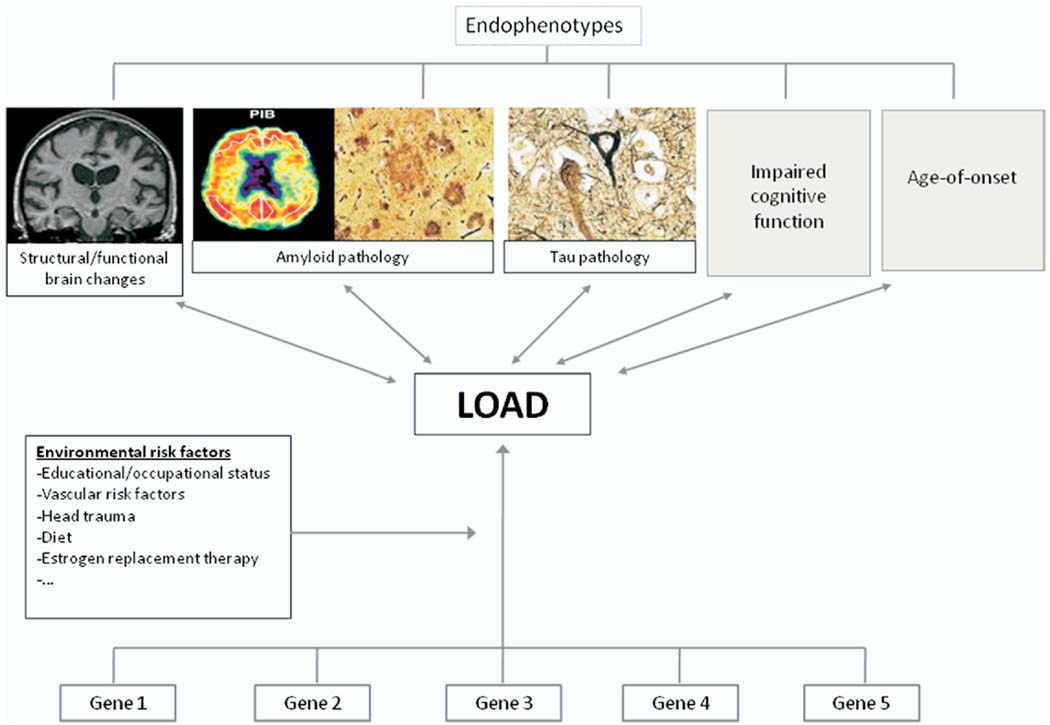
Age-at-onset of disease and cognitive test performance are the more frequently used endophenotypes in genetic studies of LOAD. Plasma amyloid β levels, a putative risk factor, have been studied rarely (Ertekin-Taner et al., 2004, 2005; Farris et al., 2004). The rationale for use of these endophenotypes is that quantitative traits provide more informative phenotypes than simply considering affection status, and thus provide more statistical power to detect small polygenic effects (Fig. 1). Genetic risk factors firmly implicated in LOAD that have been repeatedly explored in relation to age-of-onset of LOAD and cognitive function are APOE mapping to chromosome 19q13.2, and the SORL1 mapping to chromosome 11q23.3. Plasma amyloid β levels have in particular been repeatedly associated with genes mapping to chromosome 10q21–25.
Fig. 1.
Endophenotypes in the cascade between a genetic sequence variation and the Alzheimer’s disease syndrome.
APOE
APOE, a lipid-binding protein, is expressed in humans as three common isoforms coded for by three alleles, APOEε2, ε3, and ε4. The seminal finding that the APOEε4 allele is a major determinant of risk for sporadic and late-onset familial AD (Saunders et al., 1993; Corder et al., 1993, 1995; Strittmatter et al., 1993) prompted investigations of the possibility that APOEε4 may also constitute a risk factor for LOAD endophenotypes. The first reports linking APOE genotype with LOAD found a significant increase in the APOEε4 allele frequency in patients with the disease. The large body of epidemiologic data that subsequently accumulated clarified this effect by demonstrating that APOEε4 decreases the age-at-onset of LOAD in a gene dosage-dependent manner (Tang et al., 1996; Corder et al., 1993; Breitner et al., 1999; Gomez-Isla et al., 1996; Holmes et al., 1996; Hyman et al., 1996; Kurz et al., 1996; Murman et al., 1996; Poirier et al., 1993; Roses, 1997), and that APOEε4 is associated with lower cognitive performance, in particular the memory domain.
APOE and age-at-onset of LOAD
In the majority of studies, both clinical and epidemiological, age-at-onset of LOAD was strongly related to the presence of the APOE-ε4 allele (Table 2) (Tang et al., 1996; Corder et al., 1993; Breitner et al., 1999; Gomez-Isla et al., 1996; Holmes et al., 1996; Hyman et al., 1996; Kurz et al., 1996; Murman et al., 1996; Poirier et al., 1993; Roses, 1997). Taken together, these studies which include both clinical and epidemiological studies, suggest that APOEε4 may decrease the age-at-onset by as much as 7–9 years per allele. They further suggest that this effect is present across the lifespan including children and adolescents (Kurz et al., 1996; Murman et al., 1996; Gozal et al., 2007; Caselli et al., 1999; Flory et al., 2000; Liu et al., 2008; Schultz et al., 2008; Wehling et al., 2007) and across various ethnic groups although it may be stronger in Caucasians and Hispanics than African Americans (Tang et al., 1996). Mak et al. (1996) studied the APOE allele frequencies in Hong Kong elderly Chinese (65 LOAD patients and 82 age- and sex-matched controls). Both the mean and the median age-at-onset tended to be lower in subjects with one or two copies of ε4 compared to persons without ε4 allele (mean age-of-onset (SD) no ε4 vs. one ε4, one ε4 vs. two ε4s: 73.3 (8.5) vs. 72.0 (6.4) vs. 71.2 (5.0)). There was in addition a tendency for the mean and median ages at onset to be higher in subjects with ε2/ε2 or ε2/ε3 than in subjects with ε3/ε3. Although these differences only approached statistical significance (P=0.078, Z=1.419) these findings suggest that APOE also exerts its effect in Chinese populations. This notion is supported by the fact that in the same study the APOE-ε4 allele frequency was significantly higher in the AD group than in the control group (0.169 versus 0.067, P<0.01), and the fact that in Chinese the ε4 frequency is low which decreases the power to obtain statistically significant results (Hallman et al., 1991).
Table 2.
Relation between APOE genotype and LOAD endophenotypes
| Author | Subjects | Age in y, mean (range) | Endophenotype | Finding |
|---|---|---|---|---|
| Age-at-onset | ||||
| Lehtovirta et al., 1995 | 202 Finnish LOAD patients and 55 age- and sex-matched controls |
Disease onset: ε4: −/− 76±10, −/+: 77α8, 2,+/+ 71±7 |
Age-at-onset | Age-at-onset decreased from 76 to 69 as the number of ε4 alleles increased from 0 to 2 |
| Gomez-Isla et al., 1996 | 359 Patients LOAD, age and sex matched 129 controls |
LOAD group: mean age of 77.8 y; control group: mean age of 77.8 y |
Age-at-onset | Age of onset declined significantly as number of ε4 alleles increased (P<0.0001 for linear contrast ε3/ε3 to ε3ε4 to ε4/ε4) |
| Holmes et al., 1996 | 164 Patients | 60 y And older | Age-at-onset | Trend for decreasing age-at-onset of 3–4 y in carriers of the APOEε4 allele (mean age (SD): no ε4-vs ε4: 78.7 (7.9) vs. 75.5 (5.9), P=0.004)) |
| Murman et al., 1996 | 107 Normal, elderly control subjects and 123 LOAD patients |
45 y And older | Age-at-onset | Increased APOEε4 frequencies associated with onset ages of 55 and 75 y, but not at the extremes of onset ages (i.e. onset between 45 and 54 y of age and after age 75) |
| Breitner et al., 1999 | 5677 Elderly residents of Cache County, Utah |
65 y And older | Prevalence and age- at-onset |
Age-specific prevalence of LOAD reached in APOEε4 heterozygotes the maximum at age 87, in homozygotes at age 73 and in non-carriers at age 95 |
| Tang et al., 1996 | 305 LOAD patients, 485 nondemented controls |
LOAD cases: 76.4±9.1 y, controls: 72.9±6.7 y |
Relative risk of LOAD, age-at-onset |
RR for LOAD associated with APOEε4 homozygosity increased in all ethnic groups (African American relative risk [RR]=3.0; 95% confidence interval [CI]=1.5–5.9; Caucasian RR=7.3, 95% CI=2.5–21.6; and Hispanic RR=2.5, 95% CI=1.1–5.7), compared with those with APOE-epsilon 3/epsilon 3 genotypes. The risk was also increased for APOE-epsilon 4 heterozygous Caucasians (RR=2.9, 95% CI=1.7–5.1) and Hispanics (RR=1.6, 95% CI=1.1–2.3), but not for African Americans (RR=0.6, 95% CI=0.4–0.9). The age distribution of the proportion of Caucasians and Hispanics without LOAD was consistently lower for ε4 homozygous and heterozygous individuals than for those with other APOE genotypes |
| Kurz et al., 1996 | 91 Patients, 69 healthy age-matched controls |
44–95 y | Age-at-onset | Inheritance of at least one ε4 allele associated with significant reduction of age-at-onset by 7.7 y among patients 83 y or older, and a weaker relationship among patients aged 44–63 y |
| Poirier et al., 1993 | 91 Patients with LOAD and 74 controls |
Mean age (SD): 75.1 (10.3) |
Prevalence of LOAD, age-at-onset |
Significant association between ε4 and sporadic LOAD (ε4 frequency 0.380 in LOAD and 0.122 in controls, P<0.01). Age-at-onset in ε4 carriers earlier than in ε2 or ε4 carriers |
| Mak et al., 1996 | 65 LOAD patients and 82 controls |
Mean age of 76.5 y | Age-at-onset | Tendency towards lower age-at-onset in subjects with one or two copies of ε4 (mean age-of-onset (SD) −/− vs. 4/− vs. 4/4: 73.3 (8.5) vs. 72.0 (6.4) vs. 71.2 (5.0)), and higher in subjects with ε2/ε2 or ε2/ε3 than in subjects with ε3/ε3 but differences not statistically significant (P=0.078, Z=1.419) |
| do Couto et al., 1998 | 68 Patients with LOAD | Mean age (SD): 68.8 (7.9) |
Age-at-onset | Age-at-onset significantly higher in patients bearing the APOEε4 allele (ε3/ε4 and ε4/ ε4, 65.7 (7.1), n=40) compared with patients without ε4 allele (ε3/ε3, 61.6 (7.6), n=28, P<0.05) |
| Dal forno et al.,1996 | 101 LOAD subjects | Mean age: 69.6 y | Age-at-onset | Age-at-onset highest for ε4 heterozygous subjects and least for ε4 negative subjects. Heterozygous subjects declined more rapidly on MMSE and the Category Fluency Test than subjects without ε4 or ε4 homozygosity |
| Cognitive performance | ||||
| Welsh-Bomer et (l), 2008 | 507 Participants of the CCMS |
70–110 y | Cognitive performance | No association |
| Salo et al., 2001 | 46 Nondemented persons |
>85 y | Memory performance | No association |
| Murphy et al., 1997 | 86 Subjects with LOAD | Mean age of onset (SD): based on caregiver report: 65.3 (7.4); based on age when MMSE <23: 68.8 (7.0) |
Rate of decline on MMSE |
No association |
| Cosentino et al., 2008 | One incident (n=199) and two prevalent samples (n=215, n=156) of LOAD patients |
Age 65 y and older | Memory performance | Presence of an APOE ε4 allele associated with a more rapid decline in memory performance over a 7-year follow-up period |
| Wehling et al., 2007 | 70 LOAD patients | 50–75 y | Cognitive performance | APOEε4 carriers had slightly poorer performance than non-carriers on the MMSE (27.5 vs. 28.4, P=0.03) and learning trials of the CVLT (F(1,68)=5.46, P=0.022) |
| Hirono et al., 2003 | 64 LOAD patients | 60 y Or older | Memory performance | Presence of the APOEε4 allele in dose–response fashion associated with accelerated memory decline on Word Recall subtest of ADAS-Cog (mean score −/− vs. 4/− vs. 4/4: −0.2 vs. 0.4 vs. 1.0, P=0.008) |
| Mayeux et al., 2001 | 563 Healthy elderly without LOAD or questionable dementia |
65 y And older | Memory performance over 7-year follow-up |
APOEε4 allele associated with a more rapid decline in memory performance |
| Wilson et al., 2002 | 669 Participants from Religious Order Study |
65 y And older | Summary measures of episodic memory, semantic memory, working memory, perceptual speed, and visuospatial ability |
Average annual increase of 0.016 units in the ε2 subgroup and annual decreases of 0.022 units in those with ε3/3 and of 0.073 units in the ε4 subgroup |
| Lehman et al., 2006 | 2181 Elderly of the Hordaland Health Study |
70–74 y | Episodic memory | APOEε4 effect on episodic memory: OR of cognitive impairment in women 1.8 (95% CI: 1.1–2.8) for heterozygotes and 1.1 (0.3–3.7) for homozygotes; OR in men 1.1 (95% CI 0.6–2.1) for heterozygotes and 10.7 (95% CI 4.7–24) for homozygotes |
| Liu et al., 2008 | 2208 Related individuals | 50 y And older | Cognitive performance | APOEε4 significantly associated with reduced test scores for Adult Verbal Learning Test, particularly on the memory and learning subdomains |
| Bondi et al., 1995 | 52 Elderly non-demented | 59–83 y | Performance on CVLT | APOEε4 associated with poorer performance on CVLT. Six of the 14 APOEε4 subjects developed either LOAD or questionable LOAD, whereas none of the 26 non APOEε4 subjects demonstrated any cognitive decline |
| Dik et al., 2001 | 1168 Subjects from the population-based Longitudinal Aging Study Amsterdam |
62–85 y | Performance on MMSE, immediate recall and delayed recall, and the Alphabet Coding Task 15 |
APOEε4 carriers had a greater rate of cognitive decline shown by MMSE scores and slower information processing speeds after 6 y. The effects of both memory complaints and APOEε4 allele carriage were additive: subjects with both factors had a two times higher cognitive decline than did subjects without both factors |
| Hsiung et al., 2004 | 1469 Cases with cognitive impairment, 582 controls |
Control group: mean age 75.6, group with CIND: mean age 77.8, group with AD: mean age 82.7 |
Progression from normal cognition to CIND and from CIND to AD or VaD, age-at-onset of LOAD |
Possession of an APOEε4 allele associated with increased risk of LOAD developing from CIND (OR 2.6, 95% CI 1.48–4.92), and associated with decrease in the age- at-onset of LOAD |
| Petersen et al., 1995 | 66 Patients with MCI from Mayo Clinic |
Mean age: 79.8 y | Conversion from MCI to dementia |
APOEε4 strong predictor for conversion to dementia |
| Caselli et al., 1999 | 100 Nondemented individuals |
Mean age 56 y | Immediate and delayed recall |
Tests sensitive to immediate and delayed recall showed significant negative correlation with age in the APOEε4 homozygote group relative to the noncarrier group |
| Flory et al., 2000 | 220 Non-demented non- Hispanic Caucasian men and women |
Aged 24–60 (average age=46) |
Verbal learning and memory (e.g. learning a list of words and recalling them 30 min later), visual memory (e.g. reproducing a previously copied figure from memory), and attention span memory |
Performance on learning and memory tasks was significantly poorer in adults having any APOEε4 allele, relative to adults with APOEε2 or APOEε3 genotypes (P<0.01) |
| Reynolds et al., 2006 | 478 Non-demented twins from the SATSA |
50 y And older | Memory performance over 13 y |
APOEε4 associated with working and recall memory ability levels and working memory rate of change, with ε4 homozygotes exhibiting the worst performance at all ages over 13 year follow-up |
| Schultz et al., 2008 | 626 Male twins randomly drawn from the Vietnam Era Twin (VET) Registry |
In their 50s | Memory performance | ε4-Carriers: lower performance on immediate and delayed recall than non-carriers (mean (SD) comparing ε4+ vs. ε4−: immediate recall 22.19 (5.37) vs. 23.8 (6.2); delayed recall: 19.5 (5.9) vs. 20.12 (6.6)) |
In contrast to these studies, two studies found a higher age-at-onset for patients bearing the APOEε4 allele. In a study by do Couto et al. (1998) among 68 patients with LOAD, the age-at-onset of disease was significantly higher in the patients with the ε4 allele (mean onset (SD) of ε3/ε4 and ε4/ε4, 65.7 (7.1), n=40) compared with patients without the ε4 allele (mean onset (SD) ε3/ε3, 61.6 (7.6), n=28, P<0.05, two-tailed Student’s t-test). Among 101 LOAD patients (Dal Forno et al., 1996) age-at-onset was highest for the ε4-heterozygous subjects and lowest for the ε4-negative subjects. The heterozygous subjects declined more rapidly on the Mini-Mental State Examination and the Category Fluency Test than the subjects without the ε4allele or with ε4 homozygosity. The homozygous subjects declined only faster on the Physical Capacity subscale of the psychogeriatric Dependency Rating Scale. It is important to note that these two studies included relatively younger patients. It remains possible that the presence of the ε4 allele represents a particularly high risk in the older patients. The bulk of data on age-at-onset is consistent with the large body of studies showing an association between the APOEε4 allele and risk of LOAD, and suggests that the ε4 allele decreases age-at-onset of LOAD in a dose-dependent manner.
APOE and cognitive performance
Few studies, including the Cache County Study of Memory in Aging (CCMS) (Welsh-Bohmer et al., 2008), a study among 46 nondemented persons aged 85 years or over from a randomly selected group of 128 subjects in Vantaa, Finland (Salo et al., 2001), and the study by Murphy et al. (1997) observed no effect of the APOE locus on the rate of cognitive decline. It is important to note that these studies had unspecific assessment of memory (Murphy et al., 1997), had small sample sizes (Salo et al., 2001; Murphy et al., 1997) or consisted of samples prone to survival bias (Welsh-Bohmer et al., 2008) which may limit their ability to detect harmful associations. However, most studies exploring the association of APOE with cognitive performance were consistent with the studies reporting an association of the APOE genotype with LOAD or age-at-onset of LOAD, and showed a harmful effect of the APOEε4 variant with a dose–response-relationship of the effect (Table 2). In general, these studies can be divided into studies including and excluding subjects with cognitive impairment or dementia. Studies that explore the effect of APOE on cognitive performance in non-demented subjects provide the ability to draw conclusions about the effect of genetic risk factors on cognition in cognitively normal persons or the preclinical stage of the disease (Table 3).
Table 3.
Relation between SORL1 and LOAD
| Author (year) | Age | Haplotype 1 | Haplotype 2 | Other significant | ||||
|---|---|---|---|---|---|---|---|---|
| SNPs | ||||||||
| rs668387 SNP 8(1,2) |
rs689021 SNP 9 |
rs641120 SNP 10 |
rs3824968 SNP 23 |
rs2282649 SNP 24 |
rs1010159 SNP 25 |
|||
| Significant association | ||||||||
| Rogaeva et al. (2007) | Mean AAO: 70±9 −77±8 |
|||||||
| Caucasians (family dataset) |
T | T | C | |||||
| Caribbean Hispanics | C | G | C | |||||
| Caucasians (case-control datasets) |
C | G | C | T | T | C | ||
| Israeli Arabs | C | G | C | |||||
| African-Americans | ||||||||
| Lee et al. (2007)—Northern Manhattan |
Mean AAO: 79.1±5.1 −84.4±8.0 |
|||||||
| Caucasians | C | A | T | T | T | C | rs3824966 (SNP 20) | |
| Hispanics | rs12285364 (SNP 12) | |||||||
| African Americans | C | G | T | C | C | rs12285364, rs1784933 (SNP 26) |
||
| Meng et al. (2007) | Not released | |||||||
| Caucasians | + | + | + | |||||
| Lee et al. (2007)—autopsy | Mean AAO: cases: 80.5, controls: 79.9 |
|||||||
| Caucasians | C | G | C | A | T | C | ||
| Tan et al. (2007) | Mean AAO: 71.2±8.9 | |||||||
| Han Chinese | A | T | ||||||
| Seshadri et al. (2007) | Mean age: 62 +9 | |||||||
| Caucasians | + | rs1131497 (SNP 29) | ||||||
| Bettens et al. (2008) | Mean AAO: 79.0±5.2 | |||||||
| Caucasians | C | G | C | rs560573 (SNP 6), rs1614735 (SNP 27) |
||||
| Lee et al. (2008), reanalyzing data by Shibata et al. (2008) |
||||||||
| Japanese | C | T | ||||||
| Weak association | ||||||||
| Webster et al. (2007) | Age≥65 y | |||||||
| Caucasians | + | + | ||||||
| Li et al. (2008) | Mean age: 77.4±7.5 (WU), 76.4±6.1 (UK1), 76.5±5.6 (UK2) |
|||||||
| Caucasians | T | rs2070045 (SNP 19) | ||||||
| No association | ||||||||
| Li et al. (2008) | Mean AAO ≥60 | |||||||
| Caucasians | ||||||||
Studies including subjects with cognitive impairment or dementia
Cosentino et al. (2008) examined the impact of the APOEε4 variant on the rate of cognitive change in one incident (n=199) and two prevalent samples (n=215, n=156) of LOAD patients 65 years and older. The presence of at least one ε4 allele was associated with faster cognitive decline in the incident LOAD group (P=0.01). Similar results were observed for the two prevalent dementia samples when adjusting for disease severity or excluding the most impaired participants from the analyses, indicating that the APOEε4 may influence the rate of cognitive decline in both the early and late stages of LOAD. In a study by Wehling et al. (2007) which comprised 70 consecutively referred patients aged 50–75 years, APOEε4 carriers showed a slightly poorer performance than non-carriers on the MMSE (27.5 vs. 28.4, P=0.03) and learning trials of the California Verbal Learning Test (CVLT; F(1,68) = 5.46, P=0.022). Hirono et al. (2003) who explored the effect of APOE on cognition in 64 LOAD patients using the Alzheimer Disease Assessment Scale-Cognitive subscale (ADAS-Cog), observed that the presence of the ε4 allele was in a dose–response fashion associated with accelerated memory decline (mean ADAS-Cog score −/− vs. 4/− vs. 4/4: −0.2 vs. 0.4 vs. 1.0, P=0.008).
Studies excluding subjects with cognitive impairment or dementia
Most studies exploring these associations among non-demented subjects yielded consistent results, indicating that APOE also exerts its effect in cognitively normal subjects or preclinical stages of the disease. In a study by Mayeux et al. (2001) presence of an APOEε4 allele was in 563 non-demented elderly associated with a more rapid decline in a composite score of memory performance over a 7-year follow-up period. Among 669 participants of the Religious Order Study (Wilson et al., 2002), possession of one or more ε4 alleles was over an 8-year follow-up associated with faster decline in episodic memory compared to the ε3/3 genotype, while possession of one or more APOEε2 alleles was associated with reduced decline. The rate of change in episodic memory was an average annual increase of 0.016 U in the ε2 subgroup and annual decreases of 0.022 U in those with ε3/3 and of 0.073 U in the ε4 subgroup. In 2181 elderly participants (aged 70–74 years) from the Hordaland Health Study the APOEε4 allele was in a dose-dependent fashion also associated with lower episodic memory performance. The strongest effect was seen in homozygous men (OR 10.7; 95% CI 4.7–24.0) (Lehmann et al., 2006). In a Dutch sample of 2208 related individuals, the ε4 variant was associated with reduced test scores for the Adult Verbal Learning Test, and within this test strongest for the memory and learning subdomains (Liu et al., 2008). Bondi et al. (1995) explored the effect of APOE on cognition in 52 non-demented elderly using the CVLT. Consistent with the studies described above, APOEε4 carriers demonstrated significantly poorer mean performances than non-carriers. Six of the 14 APOEε4 carriers who completed annual follow-up evaluations developed either LOAD or questionable LOAD, whereas none of the 26 non-carriers demonstrated any cognitive decline.
The longitudinal population-based Longitudinal Aging Study Amsterdam (Dik et al., 2001) explored to what extent subjective memory complaints and APOEε4 allele carriage interact in their prediction of future cognitive decline. In this study of 1168 elderly subjects, APOEε4 carriers had after a six year follow-up a greater rate of cognitive decline measured by MMSE scores and slower information processing speeds. This effect appeared to be additive with the effect of memory complaints: subjects with both factors showed a two times higher cognitive decline than did subjects without memory complaints and ε4 allele.
In the Canadian Study of Health and Aging (Hsiung et al., 2004) and a consecutive sample of 66 patients from the Mayo Clinic Alzheimer’s Disease Center/Alzheimer’s Disease Patient Registry who met criteria for a diagnosis of a mild cognitive impairment (MCI) and who had at least one clinical reevaluation (Petersen et al., 1995), possession of an APOEε4allele increased the risk of conversion from cognitive impairment no dementia (CIND) or MCI to LOAD. In the Canadian Study of Health and Aging the presence of the APOEε4 allele was also associated with a decrease in the age-at-onset of LOAD (Hsiung et al., 2004).
In two cross-sectional studies in younger subjects (average ages 46 and 56) (Caselli et al., 1999; Flory et al., 2000) the APOEε4 allele was relative to the noncarrier group associated with significantly poorer performance on learning and memory tasks and immediate and delayed recall, suggesting that age-related memory decline occurs earlier in cognitively healthy APOEε4 carriers than in non-carriers, and precedes clinically detectable LOAD.
Finally, these findings could also be replicated by twin studies. In a longitudinal study over 13 years (Reynolds et al., 2006) among 478 twins from the Swedish Adoption/Twin Study of Aging (SATSA), the APOEε4 variant was in a dose-dependent fashion at all ages associated with worse working and recall memory, and rate of change in working memory. In a second longitudinal twin study among 626 twins in their 50s (Schultz et al., 2008) ε4-carriers showed significantly lower performance on immediate and delayed recall than non-carriers (mean (SD) comparing ε4+ vs. ε4−: immediate recall 22.19 (5.37) vs. 23.8 (6.2); delayed recall: 19.5 (5.9) vs. 20.12 (6.6)), supporting the genetic contribution of APOE to LOAD.
It has been proposed that statistical linear models to examine cognitive status are less sensitive since the rate of cognitive decline will change continually if it follows a nonlinear model (Plassman and Breitner, 1996). On the other hand, nonlinear and mixed effects statistical models can estimate the change of decline over time, and also model the variability in the age for individuals with same degree of disease severity. This approach would increase the sensitivity of the test and therefore might increase the statistical power to detect possible associations. Martins et al., 2005 reported a larger rate of decline estimates associated with APOE genotypes when using nonlinear models.
SORL1
Sorting mechanisms that cause APP and the β- and γ-secretases to colocalize in the same membranous compartment play important roles in the regulation of Aβ production, the main putative culprit in LOAD. SORL1 is involved in trafficking of APP from the cell surface to the Golgi–endoplasmic reticulum complex. Under-expression of SORL1 leads to over-expression of Aβ and an increased risk of AD (Rogaeva et al., 2007). Most studies exploring the effect of SORL1 on cognitive impairment or dementia used LOAD as a dichotomized trait in the analyses. Rogaeva and colleagues (2007) first reported the allelic and haplotypic associations between LOAD and variants in SORL1. Subsequently several studies supported the initial finding by showing that genetic variants in SORL1 contribute toward LOAD (Bettens et al., 2008; Lee et al., 2007, 2008; Meng et al., 2007; Seshadri et al., 2007; Tan et al., 2007). The original study included four different ethnic groups, ranging from North American and European Caucasians, Caribbean Hispanics, African-Americans, and Israeli-Arabs. With this investigation on over 6000 subjects, two different sets of haplotypes were identified: (1) SNPs in the 5′ end of the gene (SNP 8–10, 120873131 bp-120886175 bp) among Caribbean Hispanics (family study), Caucasians (case-control study), and Israeli-Arabs (case-control study); and (2) SNPs in the 3′ end of the gene (SNP 22–25, 120962172 bp-120988611 bp) among multiple Caucasian samples (family and case-control studies) and African-Americans (family study). Haplotype analysis strengthened statistical support further. However, as observed in many common diseases, these candidate SNPs confer a modestly elevated risk of LOAD, ranging from an odds ratio of 1.4–2.2, and the allelic association was not uniform across datasets. The authors strengthened their allelic association findings by cell biology findings which showed that suppression of SORL1 led to elevation of amyloid β levels. Two subsequent studies by the same group broadly supported one or both haplotypes or some variations of the two: haplotype C-G/C/at SNPs 8–10, or haplotype T-T-C at SNPs 23–25, or both. Lee and colleagues (2007) showed that the same set of SNPs at SNPs 23–25 were associated with LOAD in Caucasians residing in northern Manhattan. They then confirmed the allelic and haplotypic associations in autopsyconfirmed cases of Caucasian ethnicity for haplotype at SNPs 8–10 and haplotype at SNPs 23–25 (Lee et al., 2008).
Seven other groups examined the relation between SORL1 and LOAD or LOAD endophenotypes in different populations (Bettens et al., 2008; Seshadri et al., 2007; Tan et al., 2007; Li H et al., 2008; Li M et al., 2008; Webster et al., 2008). Three replication studies supported the initial findings, while the remaining three showed either negative or weak results. Three clearly positive studies included one by Tan et al. (2007) and the other by Seshadri et al. (2007). Bettens and colleagues (2008) directly replicated SNPs 8 through 10 and showed support for SNPs 25–27 in 550 Belgians with LOAD and 637 unaffected individuals. Tan et al. (2007) examined 223 cases and 263 controls from a Han Chinese population to show that haplotype G/C/A at SNP 19-22-23 was associated with LOAD (OR=1.35; 1.04–1.74), but none of the haplotypes in SNP 8 to SNP 10 were associated. Webster et al. (2008) and Li M et al. (2008) reported weak associations.
Li et al. (2008) and Shibata et al. (2008) reported no associations between SORl1 and LOAD. However, in the latter study the negative results were based on genotypic association analyses only. When Lee et al. (2008) reanalyzed the data of this study using allelic association tests, SNPs 8 and 24 were significantly associated with LOAD supporting the association in both the 3′ and 5′ regions of SORL1. Using the Framingham community-based family samples, Seshadri et al. (2007) extended the existing studies using cognitive performance as an endophenotype. The authors reported that SORL1 was significantly associated with abstract reasoning ability as measured by the Similarity test (P=3.2×10−6). However, they did not observe an association with memory. A possible explanation for this discrepancy may be that this sample consisted of 705 related persons, which can lead to limited power to uncover associations as compared to larger samples that include unrelated subjects.
Although discrepancies between studies exploring the effect of genetic risk factors on cognitive performance might be attributed to various reasons (i.e. recruitment strategies, LOAD diagnosis criteria, the degree of cognitive decline, and age-at-onset of the LOAD patients), among the most substantial differences are methodological approaches, different test battery used to assess cognition and the statistical methods applied for the analysis of the data.
Other genes
Several genes, in particular genes mapping to chromosome 10q21–25, have been reported to influence amyloid β levels in LOAD. In a study by Ertekin-Taner et al. (2005) amyloid β42 levels were related to a missense C/T polymorphism in exon 6 of the in the urokinase-type plasminogen activator (PLAU) gene at chromosome 10q24. In a second study by the same group genetic variants in a haplotype block spanning the insulin degrading enzyme (IDE) mapping to 10q23–25 were significantly associated with plasma amyloid β42 levels (Ertekin-Taner et al., 2004). The latter finding is consistent with a study by Farris et al. (2004) demonstrating that partial loss-of-function mutations in IDE, that induce diabetes, also impair degradation of amyloid β protein. Plau (Ertekin-Taner et al., 2005) and IDE (Ertekin-Taner et al., 2004) were also associated with an increased risk of LOAD, supporting the usefulness of amyloid β levels as a LOAD endophenotype. Additional genes and putative loci have been reported, but independent replication remains inconsistent. There is little concordance between case-control and family-based studies (Bertram et al., 2007; Grupe et al., 2007; Holmans et al., 2005; Liu et al., 2007; Myers et al., 2002) suggesting that clinical and genetic heterogeneity influences the outcome of these analyses. LRP6, a coreceptor for Wnt signaling, has been associated with late-onset AD and confirmed in a case-control analysis (De Ferrari et al., 2007). GAB2 modifies the risk of late-onset AD in APOEε4 carriers and is associated with hyperphosphorylation of tau protein (Reiman et al., 2007). The P86L polymorphism in CALHM1 encodes an essential component of a previously uncharacterized cerebral Ca2+ channel that controls Aβ level increases and has been putatively associated with late-onset AD (Dreses-Werringloer et al., 2008). Additional genes that have been reported but remain to be confirmed include CHRNB2, A2M, CTNNA3, GSTO1, GSTO2 and GAPD (Blacker et al., 1998; Giedraitis et al., 2006; Li et al., 2004; Ozturk et al., 2005). There also continues to be support for linkage to late-onset AD at 6p, 9q, 10q and 12p and 19q (Bertram et al., 2005; Blacker et al., 2003; Farrer et al., 2003; Hahs et al., 2006; Lee et al., 2006; Pericak-Vance et al., 2000; Rademakers et al., 2005; Scott et al., 2003; Li et al., 2006; Sweet et al., 2003; Wijsman et al., 2004), but specific causative loci could not yet be identified. In contrast, mutations in APP, PSEN1 and PSEN2 genes are associated with early-onset Alzheimer’s disease (onset ≤60 years) with an autosomal pattern of inheritance.
DISCUSSION
The work reviewed above indicates that there are various measures that are useful endophenotypes associated with genetic liability for both normal brain aging, CR and LOAD. Among the endophenotypes showing the strongest evidence of heritability, linkage and/or association with normal brain aging are the medial frontal cortex, Heschl’s gyrus and postcentral gyrus, Broca’s area, anterior cingulate, gray matter of the parahippocampal gyrus and white matter of the superior occipitofrontal fasciculus. The high heritability for these endophenotypes seems to be present throughout life and seems to be functionally relevant. In contrast, the heritability of hippocampus volume seems to be modest. This finding is in the study of cognitive aging and dementia of particular interest as the hippocampus is central to the formation of new memories and memory consolidation, the process for converting short-term memory into stored or long-term memory (Wittenberg and Tsien, 2002). In particular atrophic changes in the hippocampus play a major role in the memory impairment observed at early stages of LOAD (deToledo-Morrell et al., 2004; Stoub et al., 2006). Further support for an environmental rather than genetic control of hippocampus volume comes from a twin study of Ammon’s horn sclerosis (Jackson et al., 1990) in which only the twin of MZ pairs who had experienced prolonged, childhood febrile seizures developed sclerosis. This discordance in hippocampal response to trauma suggests susceptibility of this structure to environmental events. Mammalian studies reporting neurogenesis of the hippocampal dentate gyrus in adult animals even into senescence (Eriksson et al., 1998; Gould et al., 1999) suggest that the relatively stable size of the hippocampus throughout adulthood (Gallagher et al., 1996; Harding et al., 1998) may reflect a lifelong relative maintenance of volume (Cameron and McKay, 1999; Kempermann et al., 1998) through mechanisms such as neurogenesis and synaptogenesis with rich environmental stimulation (Gould et al., 1999) even when genetically compromised (Rampon et al., 2000; Rampon and Tsien, 2000). This speculation must, however, be tempered by the relatively small number of new neurons generated in confined regions of the hippocampus (Kornack and Rakic, 1999) and the lack of evidence that volume necessarily reflects cell number. In any case, if neurogenesis could be adequately and functionally amplified, it may carry new promise for conditions affecting the hippocampus, such as LOAD, providing that eradication of the disease process permitted reinstatement of neurogenesis, synaptogenesis, and accompanying hippocampal functions.
Taken together these studies of the genetics of brain structure and function among normal individuals suggest that variation in brain structure and function can be expected and that pathological states represent the extremes of this variation. They further indicate that the morphological characteristics of each brain structure represent differential vulnerability to environmental influences and may also be phenotypical expressions of different sets of genes, which may operate on morphology at different times throughout development and aging.
As a consequence, these data provide a valuable background for understanding the genetics of neurodegenerative diseases associated with changes in brain structures including LOAD and the mechanisms underlying CR preventing individuals developing cognitive impairment.
Useful endophenotypes in LOAD are besides structural brain measures, age-at-onset of LOAD, neuropsychological test measures such as memory performance, and amyloid β levels. Using these quantitative traits provides a more informative phenotype than simply considering affection status and thus increases statistical power.
Although encouraging, this also raises some additional questions and challenges. First, are the genes mediating each endophenotype involved in CR, abnormal brain aging and cognition at least partially distinct from each other? This is a key assumption of the endophenotype approach, yet empiric proof of this remains to be determined. A substantial degree of overlap appears likely for a number of the known genes associated with early and late-onset AD, at least including the amyloid precursor protein (APP), presenilin 1 (PSEN1), presenilin 2 (PSEN2), APOE and SORL1, given that these genes are involved in either the production or processing of the β amyloid peptide. Nevertheless, each gene has a unique role in this cascade and it thus seems likely these loci will differ in their magnitudes of influence across the brain systems affected in this disorder. How do these genes (along with others that remain to be identified) coalesce in influencing liability to overt expression of LOAD? Are their effects additive or interactive? The answers to these questions depend on large-scale studies of genetically at-risk samples with and without environmental exposures and the use of sophisticated statistical modeling algorithms that can powerfully probe the resulting datasets for evidence of gene–gene and gene–environment interactions. Finally, are these endophenotypes and associated genes unique to cognitive impairment in LOAD, or are they shared by other diseases such as dementia with Lewy bodies, Parkinson’s disease or depression? Lewy body inclusions and Lewy neurites, the key pathological hallmarks of dementia with Lewy bodies and Parkinson’s disease, are a frequent coexistent pathologic change observed in autopsy-confirmed LOAD.
The questions posed above raise considerable challenges for investigators attempting to unravel the genetic complexity of LOAD. Nevertheless, we have entered a new era in which conjoint advances in molecular genetics and dissection of the dementing phenotype are enabling rapid progress with multiple gene discoveries. These discoveries validate the dissection of this disorder into its more discretely determined subcomponents in order to elucidate the mechanisms underlying cognitive impairment in the elderly.
Despite their utility in the context of etiological research on LOAD, endophenotypes have not proven to have great utility in the clinical distinction of dementing disorders. As described above, the different forms of dementia show substantial clinical and pathological overlap, and likely do not reflect completely separate underlying pathologies or genetic causes but rather a continuous spectrum of disease. Therefore, although they more realistically reflect variation in the underlying causes of illness, the use of endophenotypic assessments in diagnostic or treatment contexts is difficult.
CONCLUSION
In conclusion, given that the pathways from genotypes to end-stage phenotypes are circuitous at best, discernment of endophenotypes more proximal to the effects of genetic variation can improve statistical power and thereby be a powerful tool in the identification of genes linked to complex disorders. They can help us understand how environmental and genetic factors interact to influence disease susceptibility and expression, and can help identify targets for the development of new treatment and prevention strategies.
Acknowledgments
This work was supported by federal grants from the National Institute on Aging of the National Institutes of Health (P01AG07232, R37AG15473, P50 AG08702) and by grants from the Alzheimer Association, the Blanchette Hooker Rockefeller Fund, the Robertson Gift from the Banbury Fund and the Merrill Lynch Foundation.
Abbreviations
- ADAS-Cog
Alzheimer Disease Assessment Scale-Cognitive subscale
- APOE
apolipoprotein E
- CCMS
Cache County Study of Memory in Aging
- CIND
cognitive impairment no dementia
- CR
cognitive reserve
- CVLT
California Verbal Learning Test
- DZ
dizygotic
- IDE
insulin degrading enzyme
- LOAD
late-onset Alzheimer’s disease
- MCI
mild cognitive impairment
- MZ
monozygotic
- QTL
quantitative trait loci
- SATSA
Swedish Adoption/Twin Study of Aging
- SORL1
sortilin-related receptor
REFERENCES
- Ashburner J, Friston KJ. Why voxel-based morphometry should be used. Neuroimage. 2001;14(6):1238–1243. doi: 10.1006/nimg.2001.0961. [DOI] [PubMed] [Google Scholar]
- Baare WF, Hulshoff Pol HE, Boomsma DI, Posthuma D, de Geus EJ, Schnack HG, van Haren NE, van Oel CJ, Kahn RS. Quantitative genetic modeling of variation in human brain morphology. Cereb Cortex. 2001;11(9):816–824. doi: 10.1093/cercor/11.9.816. [DOI] [PubMed] [Google Scholar]
- Bartley AJ, Jones DW, Weinberger DR. Genetic variability of human brain size and cortical gyral patterns. Brain. 1997;120(Pt 2):257–269. doi: 10.1093/brain/120.2.257. [DOI] [PubMed] [Google Scholar]
- Bertram L, Hiltunen M, Parkinson M, Ingelsson M, Lange C, Ramasamy K, Mullin K, Menon R, Sampson AJ, Hsiao MY, Elliott KJ, Velicelebi G, Moscarillo T, Hyman BT, Wagner SL, Becker KD, Blacker D, Tanzi RE. Family-based association between Alzheimer’s disease and variants in UBQLN1. N Engl J Med. 2005;352(9):884–894. doi: 10.1056/NEJMoa042765. [DOI] [PubMed] [Google Scholar]
- Bertram L, McQueen MB, Mullin K, Blacker D, Tanzi RE. Systematic meta-analyses of Alzheimer disease genetic association studies: the AlzGene database. Nat Genet. 2007 Jan;39(1):17–23. doi: 10.1038/ng1934. [DOI] [PubMed] [Google Scholar]
- Bettens K, Brouwers N, Engelborghs S, De Deyn PP, Van Broeckhoven C, Sleegers K. SORL1 is genetically associated with increased risk for late-onset Alzheimer’s disease in the Belgian population. Hum Mutat. 2008;29(5):769–770. doi: 10.1002/humu.20725. [DOI] [PubMed] [Google Scholar]
- Blacker D, Wilcox MA, Laird NM, Rodes L, Horvath SM, Go RC, Perry R, Watson B, Jr, Bassett SS, McInnis MG, Albert MS, Hyman BT, Tanzi RE. Alpha-2 macroglobulin is genetically associated with Alzheimer disease. Nat Genet. 1998;19(4):357–360. doi: 10.1038/1243. [DOI] [PubMed] [Google Scholar]
- Blacker D, Bertram L, Saunders AJ, Moscarillo TJ, Albert MS, Wiener H, Perry RT, Collins JS, Harrell LE, Go RC, Mahoney A, Beaty T, Fallin MD, Avramopoulos D, Chase GA, Folstein MF, McInnis MG, Bassett SS, Doheny KJ, Pugh EW, Tanzi RE. Results of a high-resolution genome screen of 437 Alzheimer’s disease families. Hum Mol Genet. 2003;12(1):23–32. doi: 10.1093/hmg/ddg007. [DOI] [PubMed] [Google Scholar]
- Blangero J, Williams JT, Almasy L. Novel family-based approaches to genetic risk in thrombosis. J Thromb Haemost. 2003;1(7):1391–1397. doi: 10.1046/j.1538-7836.2003.00310.x. [DOI] [PubMed] [Google Scholar]
- Bondi MW, Salmon DP, Monsch AU, Galasko D, Butters N, Klauber MR, Thal LJ, Saitoh T. Episodic memory changes are associated with the APOE-epsilon 4 allele in nondemented older adults. Neurology. 1995;45(12):2203–2206. doi: 10.1212/wnl.45.12.2203. [DOI] [PubMed] [Google Scholar]
- Boomsma D, Busjahn A, Peltonen L. Classical twin studies and beyond. Nat Rev. 2002;3(11):872–882. doi: 10.1038/nrg932. [DOI] [PubMed] [Google Scholar]
- Breitner JC, Wyse BW, Anthony JC, Welsh-Bohmer KA, Steffens DC, Norton MC, Tschanz JT, Plassman BL, Meyer MR, Skoog I, Khachaturian A. APOE-epsilon4 count predicts age when prevalence of AD increases, then declines: the Cache County study. Neurology. 1999;53(2):321–331. doi: 10.1212/wnl.53.2.321. [DOI] [PubMed] [Google Scholar]
- Brookmeyer R, Gray S, Kawas C. Projections of Alzheimer’s disease in the United States and the public health impact of delaying disease onset. Am J Public Health. 1998;88(9):1337–1342. doi: 10.2105/ajph.88.9.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron HA, McKay RD. Restoring production of hippocampal neurons in old age. Nat Neurosci. 1999;2(10):894–897. doi: 10.1038/13197. [DOI] [PubMed] [Google Scholar]
- Carmelli D, DeCarli C, Swan GE, Jack LM, Reed T, Wolf PA, Miller BL. Evidence for genetic variance in white matter hyperintensity volume in normal elderly male twins. Stroke. 1998;29(6):1177–1181. doi: 10.1161/01.str.29.6.1177. [DOI] [PubMed] [Google Scholar]
- Carmelli D, Swan GE, DeCarli C, Reed T. Quantitative genetic modeling of regional brain volumes and cognitive performance in older male twins. Biol Psychol. 2002;61(1–2):139–155. doi: 10.1016/s0301-0511(02)00056-x. [DOI] [PubMed] [Google Scholar]
- Caselli RJ, Graff-Radford NR, Reiman EM, Weaver A, Osborne D, Lucas J, Uecker A, Thibodeau SN. Preclinical memory decline in cognitively normal apolipoprotein E-epsilon4 homozygotes. Neurology. 1999;53(1):201–207. doi: 10.1212/wnl.53.1.201. [DOI] [PubMed] [Google Scholar]
- Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Jr, Roses AD, Pericak-Vance MA, Small GW, Haines JL. The apolipoprotein E E4 allele and sex-specific risk of Alzheimer’s disease. JAMA. 1995;273(5):373–374. [PubMed] [Google Scholar]
- Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, Roses AD, Haines JL, Pericak-Vance MA. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science. 1993;261(5123):921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- Cosentino S, Scarmeas N, Helzner E, Glymour MM, Brandt J, Albert M, Blacker D, Stern Y. APOE epsilon 4 allele predicts faster cognitive decline in mild Alzheimer’s disease. Neurology. 2008;70(19 Pt 2):1842–1849. doi: 10.1212/01.wnl.0000304038.37421.cc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dal Forno G, Rasmusson DX, Brandt J, Carson KA, Brookmeyer R, Troncoso J, Kawas CH. Apolipoprotein E genotype and rate of decline in probable Alzheimer’s disease. Arch Neurol. 1996;53(4):345–350. doi: 10.1001/archneur.1996.00550040085017. [DOI] [PubMed] [Google Scholar]
- DeCarli C, Massaro J, Harvey D, Hald J, Tullberg M, Au R, Beiser A, D’Agostino R, Wolf PA. Measures of brain morphology and infarction in the Framingham Heart Study: establishing what is normal. Neurobiol Aging. 2005;26(4):491–510. doi: 10.1016/j.neurobiolaging.2004.05.004. [DOI] [PubMed] [Google Scholar]
- De Ferrari GV, Papassotiropoulos A, Biechele T, Wavrant De-Vrieze F, Avila ME, Major MB, Myers A, Saez K, Henriquez JP, Zhao A, Wollmer MA, Nitsch RM, Hock C, Morris CM, Hardy J, Moon RT. Common genetic variation within the low-density lipoprotein receptor-related protein 6 and late-onset Alzheimer’s disease. Proc Natl Acad Sci U S A. 2007;104(22):9434–9439. doi: 10.1073/pnas.0603523104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- deToledo-Morrell L, Stoub TR, Bulgakova M, Wilson RS, Bennett DA, Leurgans S, Wuu J, Turner DA. MRI-derived entorhinal volume is a good predictor of conversion from MCI to AD. Neurobiol Aging. 2004;25(9):1197–1203. doi: 10.1016/j.neurobiolaging.2003.12.007. [DOI] [PubMed] [Google Scholar]
- Dik MG, Jonker C, Comijs HC, Bouter LM, Twisk JW, van Kamp GJ, Deeg DJ. Memory complaints and APOE-epsilon4 accelerate cognitive decline in cognitively normal elderly. Neurology. 2001;57(12):2217–2222. doi: 10.1212/wnl.57.12.2217. [DOI] [PubMed] [Google Scholar]
- do Couto FS, de Mendonca A, Garcia C, Rocha L, Lechner MC. Age of onset in patients with Alzheimer’s disease with different apoE genotypes. J Neurol Neurosurg Psychiatry. 1998;64(6):817. doi: 10.1136/jnnp.64.6.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draganski B, Gaser C, Busch V, Schuierer G, Bogdahn U, May A. Neuroplasticity: changes in grey matter induced by training. Nature. 2004;427(6972):311–312. doi: 10.1038/427311a. [DOI] [PubMed] [Google Scholar]
- Dreses-Werringloer U, Lambert JC, Vingtdeux V, Zhao H, Vais H, Siebert A, Jain A, Koppel J, Rovelet-Lecrux A, Hannequin D, Pasquier F, Galimberti D, Scarpini E, Mann D, Lendon C, Campion D, Amouyel P, Davies P, Foskett JK, Campagne F, Marambaud P. A polymorphism in CALHM1 influences Ca2+ homeostasis, Abeta levels, and Alzheimer’s disease risk. Cell. 2008;133(7):1149–1161. doi: 10.1016/j.cell.2008.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert MA, Leonard CM, Molloy EA, Blumenthal J, Zijdenbos A, Giedd JN. The epigenesis of planum temporale asymmetry in twins. Cereb Cortex. 2002;12(7):749–755. doi: 10.1093/cercor/12.7.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson PS, Perfilieva E, Bjork-Eriksson T, Alborn AM, Nordborg C, Peterson DA, Gage FH. Neurogenesis in the adult human hippocampus. Nat Med. 1998;4(11):1313–1317. doi: 10.1038/3305. [DOI] [PubMed] [Google Scholar]
- Ertekin-Taner N, Allen M, Fadale D, Scanlin L, Younkin L, Petersen RC, Graff-Radford N, Younkin SG. Genetic variants in a haplotype block spanning IDE are significantly associated with plasma Abeta42 levels and risk for Alzheimer’s disease. Hum Mutat. 2004;23(4):334–342. doi: 10.1002/humu.20016. [DOI] [PubMed] [Google Scholar]
- Ertekin-Taner N, Ronald J, Feuk L, Prince J, Tucker M, Younkin L, Hella M, Jain S, Hackett A, Scanlin L, Kelly J, Kihiko-Ehman M, Neltner M, Hersh L, Kindy M, Markesbery W, Hutton M, de Andrade M, Petersen RC, Graff-Radford N, Estus S, Brookes AJ, Younkin SG. Elevated amyloid beta protein (Abeta42) and late onset Alzheimer’s disease are associated with single nucleotide polymorphisms in the urokinase-type plasminogen activator gene. Hum Mol Genet. 2005;14(3):447–460. doi: 10.1093/hmg/ddi041. [DOI] [PubMed] [Google Scholar]
- Farrer LA, Bowirrat A, Friedland RP, Waraska K, Korczyn AD, Baldwin CT. Identification of multiple loci for Alzheimer disease in a consanguineous Israeli-Arab community. Hum Mol Genet. 2003;12(4):415–422. doi: 10.1093/hmg/ddg037. [DOI] [PubMed] [Google Scholar]
- Farris W, Mansourian S, Leissring MA, Eckman EA, Bertram L, Eckman CB, Tanzi RE, Selkoe DJ. Partial loss-of-function mutations in insulin-degrading enzyme that induce diabetes also impair degradation of amyloid beta-protein. Am J Pathol. 2004;164(4):1425–1434. doi: 10.1016/s0002-9440(10)63229-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flory JD, Manuck SB, Ferrell RE, Ryan CM, Muldoon MF. Memory performance and the apolipoprotein E polymorphism in a community sample of middle-aged adults. Am J Med Genet. 2000;96(6):707–711. doi: 10.1002/1096-8628(20001204)96:6<707::aid-ajmg1>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- Gallagher M, Landfield PW, McEwen B, Meaney MJ, Rapp PR, Sapolsky R, West MJ. Hippocampal neurodegeneration in aging. Science. 1996;274(5287):484–485. doi: 10.1126/science.274.5287.484. [DOI] [PubMed] [Google Scholar]
- Geschwind DH, Miller BL, DeCarli C, Carmelli D. Heritability of lobar brain volumes in twins supports genetic models of cerebral laterality and handedness. Proc Natl Acad Sci U S A. 2002;99(5):3176–3181. doi: 10.1073/pnas.052494999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giedraitis V, Hedlund M, Skoglund L, Blom E, Ingvast S, Brundin R, Lannfelt L, Glaser A New. Alzheimer’s disease locus on chromosome 8. J Med Genet. 2006;43(12):931–935. doi: 10.1136/jmg.2006.043000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogtay N, Giedd JN, Lusk L, Hayashi KM, Greenstein D, Vaituzis AC, Nugent TF, III, Herman DH, Clasen LS, Toga AW, Rapoport JL, Thompson PM. Dynamic mapping of human cortical development during childhood through early adulthood. Proc Natl Acad Sci U S A. 2004;101(21):8174–8179. doi: 10.1073/pnas.0402680101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Isla T, West HL, Rebeck GW, Harr SD, Growdon JH, Locascio JJ, Perls TT, Lipsitz LA, Hyman BT. Clinical and pathological correlates of apolipoprotein E epsilon 4 in Alzheimer’s disease. Ann Neurol. 1996;39(1):62–70. doi: 10.1002/ana.410390110. [DOI] [PubMed] [Google Scholar]
- Gottesman GTD., II The endophenotype concept in psychiatry: etymology and strategic intentions. Am J Psychiatry. 2003;160(4):636–645. doi: 10.1176/appi.ajp.160.4.636. [DOI] [PubMed] [Google Scholar]
- Gould E, Beylin A, Tanapat P, Reeves A, Shors TJ. Learning enhances adult neurogenesis in the hippocampal formation. Nat Neurosci. 1999;2(3):260–265. doi: 10.1038/6365. [DOI] [PubMed] [Google Scholar]
- Gozal D, Capdevila OS, Kheirandish-Gozal L, Crabtree VM. APOE epsilon 4 allele, cognitive dysfunction, and obstructive sleep apnea in children. Neurology. 2007;69(3):243–249. doi: 10.1212/01.wnl.0000265818.88703.83. [DOI] [PubMed] [Google Scholar]
- Grupe A, Abraham R, Li Y, Rowland C, Hollingworth P, Morgan A, Jehu L, Segurado R, Stone D, Schadt E, Karnoub M, Nowotny P, Tacey K, Catanese J, Sninsky J, Brayne C, Rubinsztein D, Gill M, Lawlor B, Lovestone S, Holmans P, O’Donovan M, Morris JC, Thal L, Goate A, Owen MJ, Williams J. Evidence for novel susceptibility genes for late-onset Alzheimer’s disease from a genome-wide association study of putative functional variants. Hum Mol Genet. 2007;16(8):865–873. doi: 10.1093/hmg/ddm031. [DOI] [PubMed] [Google Scholar]
- Gudmundsson H, Gudbjartsson DF, Frigge M, Gulcher JR, Stefansson K. Inheritance of human longevity in Iceland. Eur J Hum Genet. 2000;8(10):743–749. doi: 10.1038/sj.ejhg.5200527. [DOI] [PubMed] [Google Scholar]
- Hahs DW, McCauley JL, Crunk AE, McFarland LL, Gaskell PC, Jiang L, Slifer SH, Vance JM, Scott WK, Welsh-Bohmer KA, Johnson SR, Jackson CE, Pericak-Vance MA, Haines JL. A genome-wide linkage analysis of dementia in the Amish. Am J Med Genet B Neuropsychiatr Genet. 2006;141(2):160–166. doi: 10.1002/ajmg.b.30257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallman DM, Boerwinkle E, Saha N, Sandholzer C, Menzel HJ, Csazar A, Utermann G. The apolipoprotein E polymorphism: a comparison of allele frequencies and effects in nine populations. Am J Hum Genet. 1991;49(2):338–349. [PMC free article] [PubMed] [Google Scholar]
- Harding AJ, Halliday GM, Kril JJ. Variation in hippocampal neuron number with age and brain volume. Cereb Cortex. 1998;8(8):710–718. doi: 10.1093/cercor/8.8.710. [DOI] [PubMed] [Google Scholar]
- Hirono N, Hashimoto M, Yasuda M, Kazui H, Mori E. Accelerated memory decline in Alzheimer’s disease with apolipoprotein epsilon4 allele. J Neuropsychiatr Clin Neurosci. 2003;15(3):354–358. doi: 10.1176/jnp.15.3.354. [DOI] [PubMed] [Google Scholar]
- Holmans P, Hamshere M, Hollingworth P, Rice F, Tunstall N, Jones S, Moore P, Wavrant DeVrieze F, Myers A, Crook R, Compton D, Marshall H, Meyer D, Shears S, Booth J, Ramic D, Williams N, Norton N, Abraham R, Kehoe P, Williams H, Rudrasingham V, O’Donovan M, Jones L, Hardy J, Goate A, Lovestone S, Owen M, Williams J. Genome screen for loci influencing age at onset and rate of decline in late onset Alzheimer’s disease. Am J Med Genet B Neuropsychiatr Genet. 2005;135(1):24–32. doi: 10.1002/ajmg.b.30114. [DOI] [PubMed] [Google Scholar]
- Holmes C, Levy R, McLoughlin DM, Powell JF, Lovestone S. Apolipoprotein E: non-cognitive symptoms and cognitive decline in late onset Alzheimer’s disease. J Neurol Neurosurg Psychiatry. 1996;61(6):580–583. doi: 10.1136/jnnp.61.6.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiung GY, Sadovnick AD, Feldman H. Apolipoprotein E epsilon4 genotype as a risk factor for cognitive decline and dementia: data from the Canadian Study of Health and Aging. CMAJ. 2004;171(8):863–867. doi: 10.1503/cmaj.1031789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulshoff Pol HE, Schnack HG, Posthuma D, Mandl RC, Baare WF, van Oel C, van Haren NE, Collins DL, Evans AC, Amunts K, Burgel U, Zilles K, de Geus E, Boomsma DI, Kahn RS. Genetic contributions to human brain morphology and intelligence. J Neurosci. 2006;26(40):10235–10242. doi: 10.1523/JNEUROSCI.1312-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman BT, Gomez-Isla T, Rebeck GW, Briggs M, Chung H, West HL, Greenberg S, Mui S, Nichols S, Wallace R, Growdon JH. Epidemiological, clinical, and neuropathological study of apolipoprotein E genotype in Alzheimer’s disease. Ann N Y Acad Sci. 1996;802:1–5. doi: 10.1111/j.1749-6632.1996.tb32592.x. [DOI] [PubMed] [Google Scholar]
- Jackson GD, Berkovic SF, Tress BM, Kalnins RM, Fabinyi GC, Bladin PF. Hippocampal sclerosis can be reliably detected by magnetic resonance imaging. Neurology. 1990;40(12):1869–1875. doi: 10.1212/wnl.40.12.1869. [DOI] [PubMed] [Google Scholar]
- Katzman R, Terry R, DeTeresa R, Brown T, Davies P, Fuld P, Renbing X, Peck A. Clinical, pathological, and neurochemical changes in dementia: a subgroup with preserved mental status and numerous neocortical plaques. Ann Neurol. 1988;23(2):138–144. doi: 10.1002/ana.410230206. [DOI] [PubMed] [Google Scholar]
- Kempermann G, Kuhn HG, Gage FH. Experience-induced neurogenesis in the senescent dentate gyrus. J Neurosci. 1998;18(9):3206–3212. doi: 10.1523/JNEUROSCI.18-09-03206.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornack DR, Rakic P. Continuation of neurogenesis in the hippocampus of the adult macaque monkey. Proc Natl Acad Sci U S A. 1999;96(10):5768–5773. doi: 10.1073/pnas.96.10.5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurz A, Altland K, Lautenschlager N, Zimmer R, Busch R, Gerundt I, Lauter H, Muller U. Apolipoprotein E type 4 allele and Alzheimer’s disease: effect on age at onset and relative risk in different age groups. J Neurol. 1996;243(6):452–456. doi: 10.1007/BF00900498. [DOI] [PubMed] [Google Scholar]
- Lee JH, Cheng R, Santana V, Williamson J, Lantigua R, Medrano M, Arriaga A, Stern Y, Tycko B, Rogaeva E, Wakutani Y, Kawarai T, St George-Hyslop P, Mayeux R. Expanded genomewide scan implicates a novel locus at 3q28 among Caribbean Hispanics with familial Alzheimer disease. Arch Neurol. 2006;63(11):1591–1598. doi: 10.1001/archneur.63.11.1591. [DOI] [PubMed] [Google Scholar]
- Lee JH, Cheng R, Honig LS, Vonsattel JP, Clark L, Mayeux R. Association between genetic variants in SORL1 and autopsy-confirmed Alzheimer’s disease. Neurology. 2008;70(11):887–889. doi: 10.1212/01.wnl.0000280581.39755.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Cheng R, Schupf N, Manly J, Lantigua R, Stern Y, Rogaeva E, Wakutani Y, Farrer L, St George-Hyslop P, Mayeux R. The association between genetic variants in SORL1 and Alzheimer’s disease in an urban, multiethnic, community-based cohort. Arch Neurol. 2007;64(4):501–506. doi: 10.1001/archneur.64.4.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Shibata N, Cheng R, Mayeux R. Possible association between SORL1 and Alzheimer Disease? Reanalysing the data of Shibata et al. Dement Geriatr Cogn Disord. 2008;26(5):482. doi: 10.1159/000167792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann DJ, Refsum H, Nurk E, Warden DR, Tell GS, Vollset SE, Engedal K, Nygaard HA, Smith AD. Apolipoprotein E epsilon4 and impaired episodic memory in community-dwelling elderly people: a marked sex difference. The Hordaland Health Study. J Neurol Neurosurg Psychiatry. 2006;77(8):902–908. doi: 10.1136/jnnp.2005.077818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehtovirta M, Helisalmi S, Mannermaa A, Soininen H, Koivisto K, Ryynanen M, Riekkinen P., Sr Apolipoprotein E polymorphism and Alzheimer’s disease in eastern Finland. Neurosci Lett. 1995;185(1):13–15. doi: 10.1016/0304-3940(94)11213-3. [DOI] [PubMed] [Google Scholar]
- Li H, Wetten S, Li L, St Jean PL, Upmanyu R, Surh L, Hosford D, Barnes MR, Briley JD, Borrie M, Coletta N, Delisle R, Dhalla D, Ehm MG, Feldman HH, Fornazzari L, Gauthier S, Goodgame N, Guzman D, Hammond S, Hollingworth P, Hsiung GY, Johnson J, Kelly DD, Keren R, Kertesz A, King KS, Lovestone S, Loy-English I, Matthews PM, Owen MJ, Plumpton M, Pryse-Phillips W, Prinjha RK, Richardson JC, Saunders A, Slater AJ, St George-Hyslop PH, Stinnett SW, Swartz JE, Taylor RL, Wherrett J, Williams J, Yarnall DP, Gibson RA, Irizarry MC, Middleton LT, Roses AD. Candidate single-nucleotide polymorphisms from a genomewide association study of Alzheimer’s disease. Arch Neurol. 2008;65(1):45–53. doi: 10.1001/archneurol.2007.3. [DOI] [PubMed] [Google Scholar]
- Li M, Atmaca-Sonmez P, Othman M, Branham KE, Khanna R, Wade MS, Li Y, Liang L, Zareparsi S, Swaroop A, Abecasis GR. CFH haplotypes without the Y402H coding variant show strong association with susceptibility to age-related macular degeneration. Nat Genet. 2006;38(9):1049–1054. doi: 10.1038/ng1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Rowland C, Catanese J, Morris J, Lovestone S, O’Donovan MC, Goate A, Owen M, Williams J, Grupe A. SORL1 variants and risk of late-onset Alzheimer’s disease. Neurobiol Dis. 2008;29(2):293–296. doi: 10.1016/j.nbd.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Nowotny P, Holmans P, Smemo S, Kauwe JS, Hinrichs AL, Tacey K, Doil L, van Luchene R, Garcia V, Rowland C, Schrodi S, Leong D, Gogic G, Chan J, Cravchik A, Ross D, Lau K, Kwok S, Chang SY, Catanese J, Sninsky J, White TJ, Hardy J, Powell J, Lovestone S, Morris JC, Thal L, Owen M, Williams J, Goate A, Grupe A. Association of late-onset Alzheimer’s disease with genetic variation in multiple members of the GAPD gene family. Proc Natl Acad Sci U S A. 2004;101(44):15688–15693. doi: 10.1073/pnas.0403535101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Arias-Vasquez A, Sleegers K, Aulchenko YS, Kayser M, Sanchez-Juan P, Feng BJ, Bertoli-Avella AM, van Swieten J, Axenovich TI, Heutink P, van Broeckhoven C, Oostra BA, van Duijn CM. A genomewide screen for late-onset Alzheimer disease in a genetically isolated Dutch population. Am J Hum Genet. 2007;81(1):17–31. doi: 10.1086/518720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Pardo LM, Schuur M, Sanchez-Juan P, Isaacs A, Sleegers K, de Koning I, Zorkoltseva IV, Axenovich TI, Witteman JC, Janssens AC, van Swieten JC, Aulchenko YS, Oostra BA, van Duijn CM. The apolipoprotein E gene and its age-specific effects on cognitive function. Neurobiol Aging. 2008 doi: 10.1016/j.neurobiolaging.2008.09.015. in press. [DOI] [PubMed] [Google Scholar]
- Mak YT, Chiu H, Woo J, Kay R, Chan YS, Hui E, Sze KH, Lum C, Kwok T, Pang CP. Apolipoprotein E genotype and Alzheimer’s disease in Hong Kong elderly Chinese. Neurology. 1996;46(1):146–149. doi: 10.1212/wnl.46.1.146. [DOI] [PubMed] [Google Scholar]
- Martins CA, Oulhaj A, de Jager CA, Williams JH. APOE alleles predict the rate of cognitive decline in Alzheimer’s disease: a nonlinear model. Neurology. 2005;65(12):1888–1893. doi: 10.1212/01.wnl.0000188871.74093.12. [DOI] [PubMed] [Google Scholar]
- Mayeux R, Small SA, Tang M, Tycko B, Stern Y. Memory performance in healthy elderly without Alzheimer’s disease: effects of time and apolipoprotein-E. Neurobiol Aging. 2001;22(4):683–689. doi: 10.1016/s0197-4580(01)00223-8. [DOI] [PubMed] [Google Scholar]
- Mazziotta J, Toga A, Evans A, Fox P, Lancaster J, Zilles K, Woods R, Paus T, Simpson G, Pike B, Holmes C, Collins L, Thompson P, MacDonald D, Iacoboni M, Schormann T, Amunts K, Palomero-Gallagher N, Geyer S, Parsons L, Narr K, Kabani N, Le Goualher G, Boomsma D, Cannon T, Kawashima R, Mazoyer B. A probabilistic atlas and reference system for the human brain: International Consortium for Brain Mapping (ICBM) Philos Trans R Soc Lond. 2001;356(1412):1293–1322. doi: 10.1098/rstb.2001.0915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Y, Lee JH, Cheng R, St George-Hyslop P, Mayeux R, Farrer LA. Association between SORL1 and Alzheimer’s disease in a genome-wide study. Neuroreport. 2007;18(17):1761–1764. doi: 10.1097/WNR.0b013e3282f13e7a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer JM, Breitner JC. Multiple threshold model for the onset of Alzheimer’s disease in the NAS-NRC twin panel. Am J Med Genet. 1998;81(1):92–97. doi: 10.1002/(sici)1096-8628(19980207)81:1<92::aid-ajmg16>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- Murman DL, Foster NL, Kilgore SP, McDonagh CA, Fink JK. Apolipoprotein E and Alzheimer’s disease: strength of association is related to age at onset. Dementia. 1996;7(5):251–255. doi: 10.1159/000106888. [DOI] [PubMed] [Google Scholar]
- Murphy GM, Jr, Taylor J, Kraemer HC, Yesavage J, Tinklenberg JR. No association between apolipoprotein E epsilon 4 allele and rate of decline in Alzheimer’s disease. Am J Psychiatry. 1997;154(5):603–608. doi: 10.1176/ajp.154.5.603. [DOI] [PubMed] [Google Scholar]
- Myers A, Wavrant De-Vrieze F, Holmans P, Hamshere M, Crook R, Compton D, Marshall H, Meyer D, Shears S, Booth J, Ramic D, Knowles H, Morris JC, Williams N, Norton N, Abraham R, Kehoe P, Williams H, Rudrasingham V, Rice F, Giles P, Tunstall N, Jones L, Lovestone S, Williams J, Owen MJ, Hardy J, Goate A. Full genome screen for Alzheimer disease: stage II analysis. Am J Med Genet. 2002;114(2):235–244. doi: 10.1002/ajmg.10183. [DOI] [PubMed] [Google Scholar]
- Ozturk A, Desai PP, Minster RL, Dekosky ST, Kamboh MI. Three SNPs in the GSTO1, GSTO2 and PRSS11 genes on chromosome 10 are not associated with age-at-onset of Alzheimer’s disease. Neurobiol Aging. 2005;26(8):1161–1165. doi: 10.1016/j.neurobiolaging.2004.11.001. [DOI] [PubMed] [Google Scholar]
- Pennington BF, Filipek PA, Lefly D, Chhabildas N, Kennedy DN, Simon JH, Filley CM, Galaburda A, DeFries JC. A twin MRI study of size variations in human brain. J Cogn Neurosci. 2000;12(1):223–232. doi: 10.1162/089892900561850. [DOI] [PubMed] [Google Scholar]
- Pericak-Vance MA, Grubber J, Bailey LR, Hedges D, West S, Santoro L, Kemmerer B, Hall JL, Saunders AM, Roses AD, Small GW, Scott WK, Conneally PM, Vance JM, Haines JL. Identification of novel genes in late-onset Alzheimer’s disease. Exp Gerontol. 2000;35(9–10):1343–1352. doi: 10.1016/s0531-5565(00)00196-0. [DOI] [PubMed] [Google Scholar]
- Petersen RC, Smith GE, Ivnik RJ, Tangalos EG, Schaid DJ, Thibodeau SN, Kokmen E, Waring SC, Kurland LT. Apolipoprotein E status as a predictor of the development of Alzheimer’s disease in memory-impaired individuals. JAMA. 1995;273(16):1274–1278. [PubMed] [Google Scholar]
- Pfefferbaum A, Sullivan EV, Carmelli D. Morphological changes in aging brain structures are differentially affected by time-linked environmental influences despite strong genetic stability. Neurobiol Aging. 2004;25(2):175–183. doi: 10.1016/s0197-4580(03)00045-9. [DOI] [PubMed] [Google Scholar]
- Pfefferbaum A, Sullivan EV, Swan GE, Carmelli D. Brain structure in men remains highly heritable in the seventh and eighth decades of life. Neurobiol Aging. 2000;21(1):63–74. doi: 10.1016/s0197-4580(00)00086-5. [DOI] [PubMed] [Google Scholar]
- Plassman BL, Breitner JC. Apolipoprotein E and cognitive decline in Alzheimer’s disease. Neurology. 1996;47(2):317–320. doi: 10.1212/wnl.47.2.317. [DOI] [PubMed] [Google Scholar]
- Poirier J, Davignon J, Bouthillier D, Kogan S, Bertrand P, Gauthier S. Apolipoprotein E polymorphism and Alzheimer’s disease. Lancet. 1993;342(8873):697–699. doi: 10.1016/0140-6736(93)91705-q. [DOI] [PubMed] [Google Scholar]
- Posthuma D, de Geus EJ, Neale MC, Hulshoff Pol HE, Baare WEC, Kahn RS, Boomsma D. Multivariate genetic analysis of brain structure in an extended twin design. Behav Genet. 2000;30(4):311–319. doi: 10.1023/a:1026501501434. [DOI] [PubMed] [Google Scholar]
- Rademakers R, Cruts M, Sleegers K, Dermaut B, Theuns J, Aulchenko Y, Weckx S, De Pooter T, Van den Broeck M, Corsmit E, De Rijk P, Del-Favero J, van Swieten J, van Duijn CM, Van Broeckhoven C. Linkage and association studies identify a novel locus for Alzheimer disease at 7q36 in a Dutch population-based sample. Am J Hum Genet. 2005;77(4):643–652. doi: 10.1086/491749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampon C, Tang YP, Goodhouse J, Shimizu E, Kyin M, Tsien JZ. Enrichment induces structural changes and recovery from nonspatial memory deficits in CA1 NMDAR1-knockout mice. Nat Neurosci. 2000;3(3):238–244. doi: 10.1038/72945. [DOI] [PubMed] [Google Scholar]
- Rampon C, Tsien JZ. Genetic analysis of learning behavior-induced structural plasticity. Hippocampus. 2000;10(5):605–609. doi: 10.1002/1098-1063(2000)10:5<605::AID-HIPO11>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Reiman EM, Webster JA, Myers AJ, Hardy J, Dunckley T, Zismann VL, Joshipura KD, Pearson JV, Hu-Lince D, Huentelman MJ, Craig DW, Coon KD, Liang WS, Herbert RH, Beach T, Rohrer KC, Zhao AS, Leung D, Bryden L, Marlowe L, Kaleem M, Mastroeni D, Grover A, Heward CB, Ravid R, Rogers J, Hutton ML, Melquist S, Petersen RC, Alexander GE, Caselli RJ, Kukull W, Papassotiropoulos A, Stephan DA. GAB2 alleles modify Alzheimer’s risk in APOE epsilon4 carriers. Neuron. 2007;54(5):713–720. doi: 10.1016/j.neuron.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reveley AM, Reveley MA, Chitkara B, Clifford C. The genetic basis of cerebral ventricular volume. Psychiatry Res. 1984;13(3):261–266. doi: 10.1016/0165-1781(84)90041-6. [DOI] [PubMed] [Google Scholar]
- Reynolds CA, Prince JA, Feuk L, Brookes AJ, Gatz M, Pedersen NL. Longitudinal memory performance during normal aging: twin association models of APOE and other Alzheimer candidate genes. Behav Genet. 2006;36(2):185–194. doi: 10.1007/s10519-005-9027-6. [DOI] [PubMed] [Google Scholar]
- Rogaeva E, Meng Y, Lee JH, Gu Y, Kawarai T, Zou F, Katayama T, Baldwin CT, Cheng R, Hasegawa H, Chen F, Shibata N, Lunetta KL, Pardossi-Piquard R, Bohm C, Wakutani Y, Cupples LA, Cuenco KT, Green RC, Pinessi L, Rainero I, Sorbi S, Bruni A, Duara R, Friedland RP, Inzelberg R, Hampe W, Bujo H, Song YQ, Andersen OM, Willnow TE, Graff-Radford N, Petersen RC, Dickson D, Der SD, Fraser PE, Schmitt-Ulms G, Younkin S, Mayeux R, Farrer LA, St George-Hyslop P. The neuronal sortilin-related receptor SORL1 is genetically associated with Alzheimer’s disease. Nat Genet. 2007;39(2):168–177. doi: 10.1038/ng1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roses AD. Alzheimer’s disease: the genetics of risk. Hosp Pract. 1997;32(7):51–55. doi: 10.1080/21548331.1997.11443525. 58–63, 67-59. [DOI] [PubMed] [Google Scholar]
- Salo A, Ylikoski R, Verkkoniemi A, Polvikoski T, Juva K, Rastas S, Kontula K, Kainulainen K, Niinisto L, Notkola IL, Sulkava R. Does apolipoprotein E influence learning and memory in the non-demented oldest old? Int Psychogeriatr. 2001;13(4):451–459. doi: 10.1017/s1041610201007864. [DOI] [PubMed] [Google Scholar]
- Saunders AM, Strittmatter WJ, Schmechel D, St George-Hyslop PH, Pericak-Vance MA, Joo SH, Rosi BL, Gusella JF, Crapper-MacLachlan DR, Alberts MJ, et al. Association of apolipoprotein E allele epsilon 4 with late-onset familial and sporadic Alzheimer’s disease. Neurology. 1993;43(8):1467–1472. doi: 10.1212/wnl.43.8.1467. [DOI] [PubMed] [Google Scholar]
- Scamvougeras A, Kigar DL, Jones D, Weinberger DR, Witelson SF. Size of the human corpus callosum is genetically determined: an MRI study in mono and dizygotic twins. Neurosci Lett. 2003;338(2):91–94. doi: 10.1016/s0304-3940(02)01333-2. [DOI] [PubMed] [Google Scholar]
- Schultz MR, Lyons MJ, Franz CE, Grant MD, Boake C, Jacobson KC, Xian H, Schellenberg GD, Eisen SA, Kremen WS. Apolipoprotein E genotype and memory in the sixth decade of life. Neurology. 2008;70(19 Pt 2):1771–1777. doi: 10.1212/01.wnl.0000286941.74372.cc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott WK, Hauser ER, Schmechel DE, Welsh-Bohmer KA, Small GW, Roses AD, Saunders AM, Gilbert JR, Vance JM, Haines JL, Pericak-Vance MA. Ordered-subsets linkage analysis detects novel Alzheimer disease loci on chromosomes 2q34 and 15q22. Am J Hum Genet. 2003;73(5):1041–1051. doi: 10.1086/379083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seshadri S, DeStefano AL, Au R, Massaro JM, Beiser AS, Kelly-Hayes M, Kase CS, D’Agostino RB, Sen Decarli C, Atwood LD, Wolf PA. Genetic correlates of brain aging on MRI and cognitive test measures: a genome-wide association and linkage analysis in the Framingham study. BMC Med Genet 8. 2007 Suppl 1:S15. doi: 10.1186/1471-2350-8-S1-S15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata N, Ohnuma T, Baba H, Higashi S, Nishioka K, Arai H. Genetic association between SORL1 polymorphisms and Alzheimer’s disease in a Japanese population. Dement Geriatr Cogn Disord. 2008;26(2):161–164. doi: 10.1159/000149821. [DOI] [PubMed] [Google Scholar]
- Sowell ER, Peterson BS, Thompson PM, Welcome SE, Henkenius AL, Toga AW. Mapping cortical change across the human life span. Nat Neurosci. 2003;6(3):309–315. doi: 10.1038/nn1008. [DOI] [PubMed] [Google Scholar]
- Stern Y. What is cognitive reserve? Theory and research application of the reserve concept. J Int Neuropsychol Soc. 2002;8(3):448–460. [PubMed] [Google Scholar]
- Stoub TR, de Toledo-Morrell L, Stebbins GT, Leurgans S, Bennett DA, Shah RC. Hippocampal disconnection contributes to memory dysfunction in individuals at risk for Alzheimer’s disease. Proc Natl Acad Sci U S A. 2006;103(26):10041–10045. doi: 10.1073/pnas.0603414103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strittmatter WJ, Saunders AM, Schmechel D, Pericak-Vance M, Enghild J, Salvesen GS, Roses AD. Apolipoprotein E: high-avidity binding to beta-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer’s disease. Proc Natl Acad Sci U S A. 1993;90(5):1977–1981. doi: 10.1073/pnas.90.5.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan EV, Pfefferbaum A, Swan GE, Carmelli D. Heritability of hippocampal size in elderly twin men: equivalent influence from genes and environment. Hippocampus. 2001;11(6):754–762. doi: 10.1002/hipo.1091. [DOI] [PubMed] [Google Scholar]
- Sweet RA, Nimgaonkar VL, Devlin B, Jeste DV. Psychotic symptoms in Alzheimer disease: evidence for a distinct phenotype. Mol Psychiatry. 2003;8(4):383–392. doi: 10.1038/sj.mp.4001262. [DOI] [PubMed] [Google Scholar]
- Tan EK, Lee J, Chen CP, Teo YY, Zhao Y, Lee WL. SORL1 haplotypes modulate risk of Alzheimer’s disease in Chinese. Neurobiol Aging. 2007 doi: 10.1016/j.neurobiolaging.2007.10.013. in press. [DOI] [PubMed] [Google Scholar]
- Tang MX, Maestre G, Tsai WY, Liu XH, Feng L, Chung WY, Chun M, Schofield P, Stern Y, Tycko B, Mayeux R. Relative risk of Alzheimer’s disease and age-at-onset distributions, based on APOE genotypes among elderly African Americans, Caucasians, and Hispanics in New York City. Am J Hum Genet. 1996;58(3):574–584. [PMC free article] [PubMed] [Google Scholar]
- Thompson P, Cannon TD, Toga AW. Mapping genetic influences on human brain structure. Ann Med. 2002;34(7–8):523–536. doi: 10.1080/078538902321117733. [DOI] [PubMed] [Google Scholar]
- Thompson PM, Cannon TD, Narr KL, van Erp T, Poutanen VP, Huttunen M, Lonnqvist J, Standertskjold-Nordenstam CG, Kaprio J, Khaledy M, Dail R, Zoumalan CI, Toga AW. Genetic influences on brain structure. Nat Neurosci. 2001;4(12):1253–1258. doi: 10.1038/nn758. [DOI] [PubMed] [Google Scholar]
- Thompson PM, Hayashi KM, Dutton RA, Chiang MC, Leow AD, Sowell ER, De Zubicaray G, Becker JT, Lopez OL, Aizenstein HJ, Toga AW. Tracking Alzheimer’s disease. Ann N Y Acad Sci. 2007;1097:183–214. doi: 10.1196/annals.1379.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace GL, Eric Schmitt J, Lenroot R, Viding E, Ordaz S, Rosenthal MA, Molloy EA, Clasen LS, Kendler KS, Neale MC, Giedd JN. A pediatric twin study of brain morphometry. J Child Psychol Psychiatry. 2006;47(10):987–993. doi: 10.1111/j.1469-7610.2006.01676.x. [DOI] [PubMed] [Google Scholar]
- Webster JA, Myers AJ, Pearson JV, Craig DW, Hu-Lince D, Coon KD, Zismann VL, Beach T, Leung D, Bryden L, Halperin RF, Marlowe L, Kaleem M, Huentelman MJ, Joshipura K, Walker D, Heward CB, Ravid R, Rogers J, Papassotiropoulos A, Hardy J, Reiman EM, Stephan DA. Sorl1 as an Alzheimer’s disease predisposition gene? Neurodegener Dis. 2008;5(2):60–64. doi: 10.1159/000110789. [DOI] [PubMed] [Google Scholar]
- Wehling E, Lundervold AJ, Standnes B, Gjerstad L, Reinvang I. APOE status and its association to learning and memory performance in middle aged and older Norwegians seeking assessment for memory deficits. Behav Brain Funct. 2007;3:57. doi: 10.1186/1744-9081-3-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh JP, Yuen G, Placantonakis DG, Vu TQ, Haiss F, O’Hearn E, Molliver ME, Aicher SA. Why do Purkinje cells die so easily after global brain ischemia? Aldolase C, EAAT4, and the cerebellar contribution to posthypoxic myoclonus. Adv Neurol. 2002;89:331–359. [PubMed] [Google Scholar]
- Welsh-Bohmer KA, Ostbye T, Sanders L, Pieper CF, Hayden KM, Tschanz JT, Norton MC. Neuropsychological performance in advanced age: influences of demographic factors and apolipoprotein E: findings from the Cache County Memory Study. Clin Neuropsychol. 2008;23:77–99. doi: 10.1080/13854040801894730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White T, Andreasen NC, Nopoulos P. Brain volumes and surface morphology in monozygotic twins. Cereb Cortex. 2002 May;12(5):486–493. doi: 10.1093/cercor/12.5.486. [DOI] [PubMed] [Google Scholar]
- Wijsman EM, Daw EW, Yu CE, Payami H, Steinbart EJ, Nochlin D, Conlon EM, Bird TD, Schellenberg GD. Evidence for a novel late-onset Alzheimer disease locus on chromosome 19p13.2. Am J Hum Genet. 2004;75(3):398–409. doi: 10.1086/423393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RS, Bienias JL, Berry-Kravis E, Evans DA, Bennett DA. The apolipoprotein E epsilon 2 allele and decline in episodic memory. J Neurol Neurosurg Psychiatry. 2002;73(6):672–677. doi: 10.1136/jnnp.73.6.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittenberg GM, Tsien JZ. An emerging molecular and cellular framework for memory processing by the hippocampus. Trends Neurosci. 2002;25(10):501–505. doi: 10.1016/s0166-2236(02)02231-2. [DOI] [PubMed] [Google Scholar]
- Wright IC, Sham P, Murray RM, Weinberger DR, Bullmore ET. Genetic contributions to regional variability in human brain structure: methods and preliminary results. Neuroimage. 2002;17(1):256–271. doi: 10.1006/nimg.2002.1163. [DOI] [PubMed] [Google Scholar]