In addition to cross-linking F-actin, Drosophila Kelch is a component of a cullin-RING ubiquitin ligase complex required for morphogenesis of ring canals during oogenesis.
Abstract
Drosophila melanogaster Kelch (KEL) is the founding member of a diverse protein family defined by a repeated sequence motif known as the KEL repeat (KREP). Several KREP proteins, including Drosophila KEL, bind filamentous actin (F-actin) and contribute to its organization. Recently, a subset of KREP proteins has been shown to function as substrate adaptor proteins for cullin-RING (really interesting new gene) ubiquitin E3 ligases. In this study, we demonstrate that association of Drosophila KEL with Cullin-3, likely in a cullin-RING ligase, is essential for the growth of Drosophila female germline ring canals. These results suggest a role for protein ubiquitylation in the remodeling of a complex F-actin cytoskeletal structure.
Introduction
The growth and development of Drosophila melanogaster oocytes relies on large intercellular bridges or ring canals that interconnect the oocyte and 15 associated nurse cells. The ring canals allow factors essential for embryonic development to move from the nurse cells to the transcriptionally inactive oocyte. Mutations that compromise ring canal formation are often female sterile, producing small eggs that cannot support embryonic development. Drosophila kelch (kel) was identified as a female-sterile mutation that produced small, cuplike eggs (Schüpbach and Wieschaus, 1991; Xue and Cooley, 1993). In kel mutants, ring canals become obstructed with F-actin and other components of the ring canal cytoskeleton, blocking transport to the oocyte (Xue and Cooley, 1993; Robinson et al., 1994; Sokol and Cooley, 1999).
The protein encoded by kel is the founding member of a large family of proteins containing KEL repeats (KREPs), which fold into a β-barrel structure similar to WD-repeat proteins (Hudson and Cooley, 2008). KREP proteins are found in nearly all eukaryotes and function in diverse cellular processes (Adams et al., 2000). Drosophila KEL has an N-terminal BTB dimerization domain and six C-terminal KREPs. Because kel mutants display disorganized ring canal F-actin, we tested whether KEL functions as a dimeric F-actin cross-linking protein and demonstrated that KEL binds and cross-links F-actin (Kelso et al., 2002).
Cullin-RING (really interesting new gene) ligases (CRLs) are ubiquitin E3 ligases responsible for ubiquitylation of specific target proteins. CRLs consist of three core components: a cullin scaffold protein, a RING domain protein that recruits ubiquitin conjugation enzymes, and a substrate adaptor protein that binds target proteins. The genomes of higher eukaryotes encode seven cullin paralogues, and each cullin associates with a distinct set of substrate-targeting proteins. CRLs assembled with Cullin-3 (CUL-3 [CRL3s]) bind to substrate adaptor proteins containing a BTB domain, and several BTB-KEL proteins have been shown to function as CRL3 substrate-targeting proteins (Petroski and Deshaies, 2005; Stogios et al., 2005).
In this paper, we demonstrate that in addition to its F-actin cross-linking function, Drosophila KEL appears to be a component of a CRL that we designate CRL3KEL, according to recent nomenclature suggestions (Bosu and Kipreos, 2008; Merlet et al., 2009). We show that CRL3KEL is required for cytoskeletal rearrangements during ring canal growth.
Results and discussion
kel mutant ovaries accumulate ring canal proteins during growth
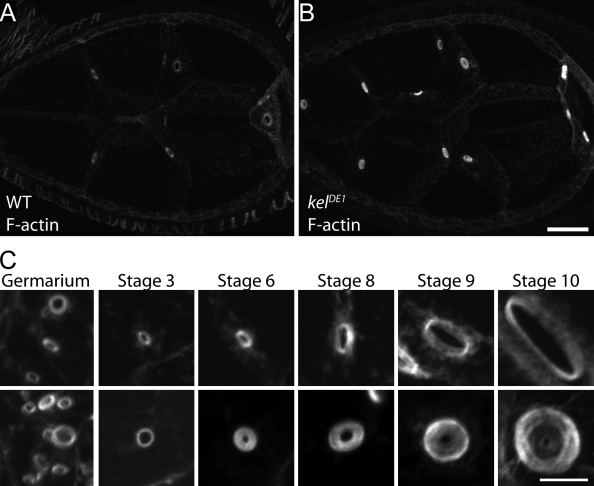
The F-actin cross-linking activity of KEL can explain the disorganization of ring canal F-actin in kel mutant egg chambers. However, we also observed an increase in the quantity of F-actin (Fig. 1, A and B), as well as the ovarian hu-li tai shao–ring canal protein (Ovhts-RC) and filamin ring canal proteins (Robinson et al., 1994; Sokol and Cooley, 1999) in kel egg chambers. These observations are not fully explained by the loss of an F-actin cross-linking protein. A potential cause of the increase in ring canal proteins was suggested by comparing the development of kel mutant ring canals with wild type over the course of oogenesis (Fig. 1 C). Consistent with previous studies (Xue and Cooley, 1993; Tilney et al., 1996), the kel ring canal phenotype first appeared at approximately stage 6 of oogenesis (for an explanation of developmental staging of oogenesis, see Spradling, 1993). From stage 6 until the end of oogenesis, the growth of the outer diameters of kel ring canals was similar to wild type; however, the inner diameter failed to expand, leaving a small lumen similar in size to ring canals of very young egg chambers. These observations suggested that KEL might function to disassemble the innermost ring canal cytoskeleton as the outer diameter grows.
Figure 1.
kel mutant ring canals accumulate high levels of F-actin. (A and B) Comparison of the actin content of wild-type (WT; A) and kel (B) mutant egg chambers fixed, prepared, and imaged under identical conditions. (C) Representative ring canals from wild-type and kel mutant egg chambers from the indicated stages of egg chamber development labeled with fluorescent phalloidin. Bars: (A and B) 20 µm; (C) 5 µm.
Cul-3 germline clones exhibit a kel-like phenotype and altered KEL localization
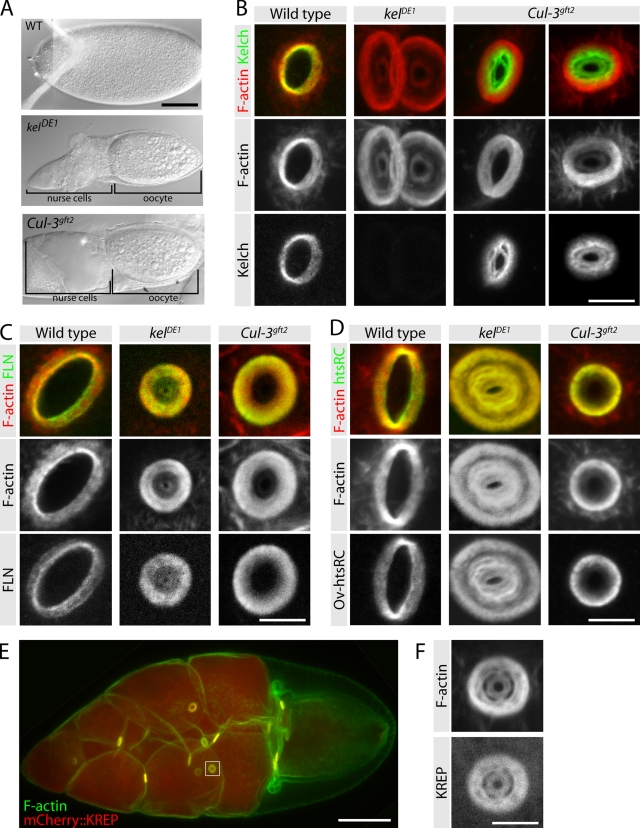
The finding that BTB-KEL proteins serve as substrate-targeting components for CRL3s prompted us to investigate whether KEL also functions as a substrate adaptor protein. If KEL and CUL-3 function together as a CRL3, Cul-3 mutants should exhibit a phenotype similar to that of kel. The Cul-3 locus is complex, producing transcripts encoding two genetically distinct protein products (designated Cul-3Soma and Cul-3Testis; Arama et al., 2007). Because the Cul-3Testis transcript is restricted to the male germline, we examined strong loss-of-function alleles affecting the broadly expressed Cul-3Soma transcripts. These Cul-3 mutations are lethal, and a previous study indicated that germline clones were infertile, producing small eggs (Mistry et al., 2004). We found that the small egg phenotype resulted from a failure to transfer cytoplasm from the nurse cells to the oocyte, which is a phenotype characteristic of ring canal mutations (Fig. 2 A). Examination of Cul-3 mutant ring canals revealed a small lumen phenotype and F-actin disorganization similar to kel (Fig. 2, B–D).
Figure 2.
Cul-3 germline clones have a kel-like ovarian ring canal phenotype. (A) DIC images of mature eggs produced by egg chambers of the indicated genotypes. (B) Distribution of F-actin and KEL on ring canals from wild-type, kel mutant, and Cul-3 germline-clone egg chambers. (C and D) Localization of NFLN (C) and Ovhts-RC (D) ring canal proteins in wild-type, kel, and Cul-3 egg chambers. (E) Expression of mCherry::KREP driven by maternal tubulin-Gal4::VP16 results in strong, uniform germline expression and produces a fully penetrant dominant-negative kel-like phenotype. The box indicates the region shown at higher magnification in F. (F) High-magnification image of mCherry::KREP (bottom) and F-actin (top) in a ring canal from an egg chamber overexpressing mCherry::KREP. Bars: (A) 100 µm; (B–D and F) 5 µm; (E) 50 µm.
The F-actin at ring canals is present with several other proteins that we collectively refer to as the ring canal cytoskeleton. Assembly of the ring canal cytoskeleton depends on filamin, which is among the first proteins to localize to ring canals and is required for all other known ring canal components to localize (Robinson et al., 1997; Sokol and Cooley, 1999). Another constituent of the ring canal cytoskeleton is the novel Ovhts-RC protein, which is essential for F-actin recruitment (Petrella et al., 2007). We found that both filamin and Ovhts-RC colocalized with F-actin in the ring canal lumens of Cul-3 mutant egg chambers, much as they do in kel mutants (Fig. 2, C and D). The Cul-3 F-actin phenotype was less severe than the kel phenotype, perhaps because the Cul-3 alleles we used are strong loss-of-function alleles rather than nulls (Mistry et al., 2004). However, another possibility is that the F-actin cross-linking activity of KEL is independent of CUL-3.
KEL localization in Cul-3 mutants was dramatically affected. Instead of colocalizing with the ring canal F-actin, KEL accumulated at high levels in the ring canal lumen where F-actin was relatively sparse (Fig. 2 B). The substrate-targeting proteins of CRLs can be subject to cullin-dependent autocatalytic degradation as part of the CRL assembly–disassembly cycle (Bosu and Kipreos, 2008; Merlet et al., 2009). We speculate that in the absence of CUL-3, its substrate adaptor protein KEL does not turn over, resulting in its aberrant accumulation in the ring canal lumen; we investigate this idea further below (see Fig. 4). The similarity of the kel and Cul-3 phenotypes, together with the altered localization of KEL in Cul-3 mutant egg chambers suggested that KEL and CUL-3 are functioning as a CRL at ring canals.
Figure 4.
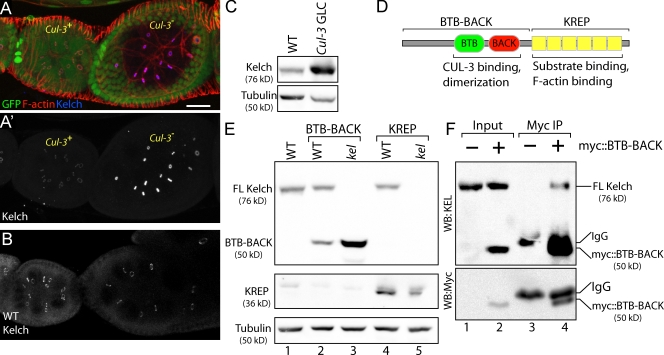
KEL accumulates in Cul-3− cells. (A and A′) The Cul-3 mutant egg chamber (marked by loss of GFP in A) exhibits highly elevated levels of KEL protein at ring canals (A′). (B) KEL levels in wild-type (WT) egg chambers of similar stage to those shown in A. (C) Western blots of ovarian lysates prepared from wild-type ovaries or ovaries containing Cul-3 germline clones (GLC). α-Tubulin serves as a loading control. (D) Drawing indicates the domain organization of KEL. The top brackets indicate N- and C-terminal regions included in BTB-BACK and KREP transgenes. (E) The N-terminal half of KEL is stabilized in kel mutant ovaries. Myc::BTB-BACK and Myc::KREP were expressed in the germlines of either wild-type or kel flies. (F) Full-length KEL is present in immunoprecipitates of Myc::BTB-BACK. Immunoprecipitations (IP) of either wild-type (−) or transgenic ovaries expressing Myc::BTB-BACK (+) were probed with antibodies to either KEL or Myc as indicated. Myc::BTB-BACK is specifically precipitated from the transgenic ovaries and migrates slightly faster than the IgG heavy chain (lane 4 on α-Myc immunoblot). The longer exposure required to see endogenous KEL in the top blot prevented the Myc::BTB-BACK and IgG bands from being resolved. FL, full length; WB, Western blot. Bar, 20 µm.
Overexpression of the KREP domain phenocopies kel
Previously, we expressed the KREP domain in the germline and found that it localized specifically to ring canals in kel mutant ovaries (Robinson and Cooley, 1997). This indicated that the KREP domain bound a ring canal–specific protein in addition to F-actin because abundant F-actin is present throughout egg chambers, whereas KREP localization was ring canal specific. If KEL is a substrate adaptor protein for a CRL, the ring canal localization of KREP could result from it binding its substrate. Our earlier experiments involved low-level expression of the KREP domain. Because the KREP domain cannot interact with CUL-3, we reasoned that expressing KREP at higher levels might exert a dominant-negative effect by sequestering substrate. To test this, we generated otherwise wild-type flies expressing mCherry-tagged KREP at high levels and observed a kel-like phenotype with ring canals mostly occluded (Fig. 2, E and F). This result is consistent with a substrate-binding function for the KREP domain.
CUL-3 is a KEL-dependent ring canal component
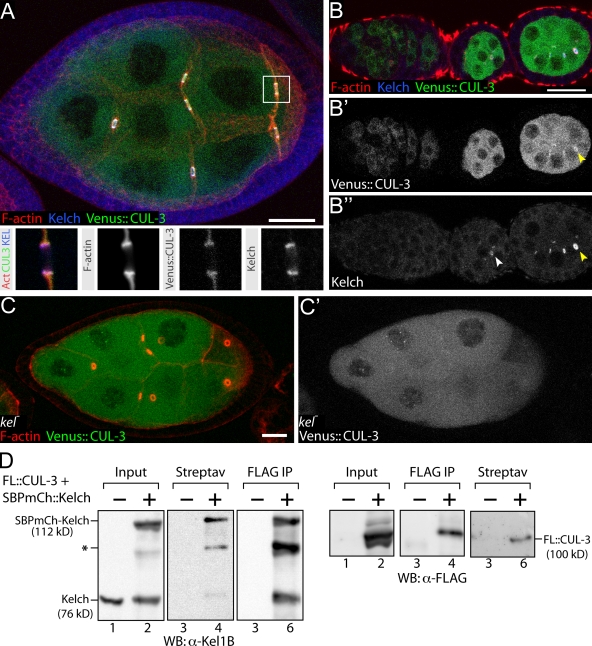
To determine whether KEL and CUL-3 function together at ring canals, we examined the localization of CUL-3 using flies that express CUL-3 fused to the Venus YFP variant (Nagai et al., 2002) for localization experiments. We used the MARCM (mosaic analysis with a repressible cell marker) clonal technique (Lee and Luo, 1999) to generate Cul-3 mutant egg chambers marked by expression of Venus::CUL-3 so that Venus::CUL-3 was the only source of CUL-3. Ring canal morphology in these egg chambers was rescued, indicating that the Venus::CUL-3 fusion protein was functional (Fig. 3 A). Venus::CUL-3 expressed in germ cells was distributed uniformly throughout the cytoplasm and was highly enriched at ring canals (Fig. 3 A). KEL localizes to ring canals beginning in egg chamber stages 1–3 (Xue and Cooley, 1993), well after filamin, Ovhts-RC, and F-actin localize to ring canals. When we drove expression of Venus::CUL-3 from the earliest stages of oogenesis, Venus::CUL-3 localized to ring canals between stages 2 and 3, similar to KEL (Fig. 3, B–B″). Furthermore, CUL-3 failed to localize to ring canals in kel mutant egg chambers (Fig. 3, C and C′), indicating that KEL is required for CUL-3 ring canal localization.
Figure 3.
Venus::CUL-3 localizes to ring canals and binds KEL. (A) Venus::CUL-3 fluorescence is distributed throughout the cytoplasm and is enriched on ring canals with KEL and F-actin. Insets show individual channels for a single focal plane of boxed ring canal in panel A and are 10 × 10 µm. (B–B′) Venus::CUL-3 is first observed on ring canals at approximately stage 3 of oogenesis (yellow arrowheads mark Venus::CUL-3 and KEL on a ring canal). In B′, KEL is first visible in stage 2 and is marked by a white arrowhead. (C and C′) Venus::CUL-3 does not localize to kel mutant ring canals. (D) Copurification experiments from either wild-type (−) ovaries or ovaries expressing Flag::CUL-3 and SBP-mCherry::KEL driven by maternal-αtub67C Gal4::VP16 (+). Streptav indicates SBP purification with streptavidin resin; Flag IP indicates immunoprecipitation with Flag antibodies. Protein species are labeled. The asterisk indicates either a proteolytic product of SBP-mCherry::KEL or cross reactivity with overexpressed Flag::CUL-3 (FL::CUL-3). WB, Western blot. Bars, 20 µm.
To test whether KEL and CUL-3 are in a molecular complex, we generated epitope- or affinity-tagged forms of KEL and CUL-3 for copurification experiments. When both streptavidin-binding peptide (SBP)–mCherry::KEL and Flag::CUL-3 were expressed in ovaries, each protein copurified with the other (Fig. 3 D). Together, these results indicate that like other substrate adaptors for CUL-3–based E3 ligases, KEL is present in a complex with CUL-3, and this complex localizes to ring canals in a KEL-dependent manner.
KEL levels are regulated by its association with CUL-3
Substrate adaptor proteins in CRLs are often subject to autoubiquitylation and degradation. This phenomenon has been observed for several CRL3 substrate adaptors (Pintard et al., 2003; Zhang et al., 2005), in which steady-state levels of substrate adaptor proteins increase as CUL-3 function is reduced. We examined Cul-3 mutant clones to directly compare KEL levels in similarly staged mutant and wild-type egg chambers. KEL was present on ring canals at highly elevated levels in Cul-3 mutants compared with wild type (Fig. 4, A and A′; Fig. 4 B shows the normal levels of KEL in similarly staged egg chambers). The KEL antibody used for immunofluorescence was raised against the BTB domain, the region which mediates association with CUL-3. Therefore, it was possible that the increase in fluorescence intensity we observed was caused by greater antigen accessibility of KEL in Cul-3 mutant cells. To test this, we compared KEL levels in lysates from wild-type and Cul-3 mutant ovaries and found that KEL levels were elevated approximately fourfold in Cul-3 mutant extracts (Fig. 4 C). The Cul-3 extracts were prepared from tissue enriched for Cul-3 mutant germline cells but that also contained wild-type tissue (see Materials and methods); therefore, this measurement underestimates the extent of KEL accumulation in Cul-3 mutant cells.
To identify regions of KEL important for its degradation, we examined the steady-state levels of the N- and C-terminal halves of KEL. When expressed in wild-type egg chambers, the N-terminal BTB-BACK domain (Fig. 4 D) was present at low levels (Fig. 4 E, lane 2). However, the BTB-BACK domain accumulated to levels approximately fourfold greater than wild type when expressed in a kel mutant background (Fig. 4 E, lane 3). In contrast, levels of the C-terminal KREP domain were similar in wild-type and kel mutant ovaries (Fig. 4 E, lanes 4 and 5). Therefore, the BTB-BACK domain is subject to turnover, but turnover requires the presence of full-length KEL. To investigate this idea further, we looked for direct evidence that the BTB-BACK protein associates with full-length KEL in ovaries by immunoprecipitating Myc-tagged BTB-BACK (Fig. 4 F). Endogenous full-length KEL was present in Myc immunoprecipitates from ovaries expressing Myc::BTB-BACK (Fig. 4 F, lanes 3 and 4), supporting the idea that Myc::BTB-BACK instability is caused by its association with KEL.
The disassembly of the ring canal cytoskeleton is regulated by KEL
The major defect in kel mutant ring canals is a failure to expand the ring canal lumen as the outer rim grows. In light of our finding that KEL functions with CUL-3, we hypothesize that a ring canal component is ubiquitylated and that this facilitates the disassembly of the ring canal cytoskeleton from the inner portion of the ring canal during ring canal expansion. This model predicts that the accumulated ring canal cytoskeleton in kel mutants results from reduced disassembly of the ring canal cytoskeleton and further suggests that turnover of ring canal proteins should be reduced in kel mutants. To test this, we performed an in vivo pulse–chase experiment using GFP-tagged Ovhts-RC to mark the ring canal cytoskeleton.
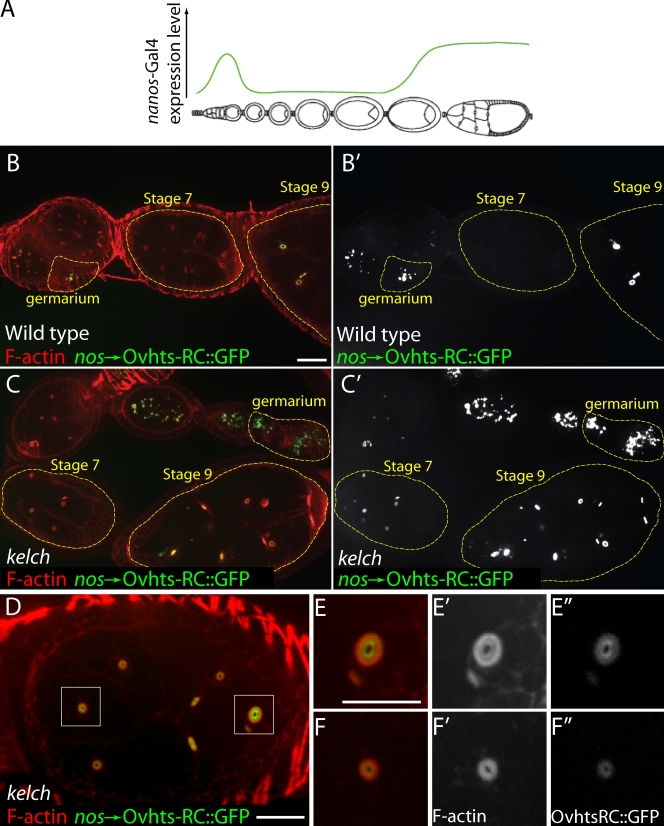
Driving expression of Ovhts-RC::GFP using the nanos promoter (nos-Gal4::VP16; Van Doren et al., 1998) results in two periods of expression in the developing germline: first in the germarium and a second, sustained phase beginning around stage 9 of oogenesis (Fig. 5, A and B; Petrella et al., 2007). After the pulse of expression in the germarium, Ovhts-RC::GFP was not detected during stages 3–8 in wild-type ovarioles (Fig. 5, B and B′). Interestingly, in kel mutants, Ovhts-RC::GFP was apparent in the germarium but did not disappear from stage 3–8 egg chambers (Fig. 5, C and C′), indicating that the turnover of the ring canal cytoskeleton was impaired in kel mutants.
Figure 5.
Pulsed expression of the Ovhts-RC ring canal protein in kel mutants reveals a defect in ring canal cytoskeleton disassembly. (A) Diagram indicating the temporal expression profile of the nanos-Gal4::VP16 driver in green. A strong pulse of expression in the germarium is followed by little or no expression during the midstages of oogenesis. High levels of expression resume at approximately stage 9. (B and B′) In a wild-type background, Ovhts-RC::GFP is present at high levels on ring canals in the germarium and in early stage egg chambers, but there is no detectable expression in the stage 7 egg chamber (highlighted in yellow dotted outlines). (C and C′) In kel ovaries, Ovhts-RC::GFP is present at high levels in the germarium and persists into midstages (yellow dotted outlines). (D) Stage 7 kel egg chamber expressing Ovhts-RC::GFP. Two ring canals (boxed areas) are shown at higher magnification in E–E″ and F–F″. In both ring canals, the F-actin distribution displays the kel phenotype. Ovhts-RC::GFP fluorescence is highest at the innermost portion of the ring canal and gradually decreases toward the outer part of the ring. Bars: (B–D) 20 µm; (E–F″) 10 µm.
When ring canals from stage 6–8 kel egg chambers were examined more closely, a bright ring of Ovhts-RC::GFP was visible near the center of mutant ring canals, and the GFP fluorescence gradually diminished toward the outer portion of the ring canal cytoskeleton (Fig. 5, D–F). Given the temporal expression pattern of nos-Gal4::VP16, the bright inner ring of Ovhts-RC::GFP in kel mutants likely resulted from Ovhts-RC::GFP added to ring canals in the germarium when Ovhts-RC::GFP levels were high. Therefore, the graded diminution of Ovhts-RC::GFP toward the outer part of the ring resulted from decreasing levels of nanos-driven Ovhts-RC::GFP during ring canal growth. The overall distribution of Ovhts-RC in kel mutants was more uniform (Fig. 2 D), indicating that the graded Ovhts-RC::GFP accumulation is specific to the nanos-driven Ovhts-RC::GFP.
Further work will be required to determine the identity of the CRL3KEL substrates, as well as the consequence of CRL3KEL-mediated ubiquitylation. One candidate substrate is actin because we previously demonstrated that the KREP domain binds F-actin (Kelso et al., 2002). However, we were unable to detect evidence of ubiquitylated actin in ovarian lysates (unpublished data). Furthermore, our previous characterization of the KREP domain suggests that it binds a ring canal–specific protein in addition to actin (Robinson and Cooley, 1997). Interestingly, another class of CRL3 that uses BACURD (BTB-containing adaptor for CUL-3–mediated RhoA degradation) BTB proteins as substrate adaptor proteins has recently been shown to regulate the F-actin cytoskeleton by targeting RhoA for degradation (Chen et al., 2009). If the dominant-negative effect of high-level KREP expression results from a KREP–substrate complex, we may be able to identify the substrate by identifying proteins that copurify with KREP.
CRL-directed ubiquitylation most often leads to proteasome-mediated degradation (Petroski and Deshaies, 2005); therefore, we favor a model in which CRL3KEL functions by targeting a ring canal protein for destruction. However, we note that we did not observe ring canal defects when we attempted to inhibit proteasome function by genetic or pharmacological means (unpublished data). Thus, we cannot rule out the possibility that CRL3KEL mediates ring canal disassembly through a nonproteasomal mechanism.
Our results provide further confirmation for the proposal that BTB-KEL proteins function as substrate adaptors for CRLs assembled with CUL-3. In the case of KEL, we previously demonstrated that it has an independent function as an F-actin cross-linking protein (Kelso et al., 2002). In light of this, it is interesting to note that the Caenorhabditis elegans MEL-26, a BTB protein substrate adaptor for a CRL3 which targets a microtubule-severing protein, also has a CUL-3–independent function regulating the F-actin cytoskeleton (Luke-Glaser et al., 2005). We speculate that KEL molecules alternate between cross-linking F-actin and functioning in CRL3. Disruption of the normal cycle of interactions by genetically removing CUL-3 results in aberrant accumulation of KEL in a dead-end location in the ring canal lumen. Understanding how CUL-3–dependent and –independent functions are balanced will be an interesting avenue for future research.
Materials and methods
Genetics
Cul-3 chromosomes used were: Cul-3gft2, FRT2L-40A and Cul-3gft1, FRT2L-40A (provided by J. Skeath, University of Washington, St. Louis, MO); both Cul-3 alleles are strong hypomorphs (Mistry et al., 2004). The kel allele used was kelDE1, a protein null (Xue and Cooley, 1993). Constructs expressing the N- and C-terminal halves of KEL under the control of the germline-specific otu promoter were previously described (Robinson and Cooley, 1997). To be consistent with current literature, we refer to the ORF1-R construct described in Robinson and Cooley (1997) as BTB-BACK. To generate negatively marked Cul-3 mutant clones, y, w, hsFLP3F; Cul-3gft2, FRT2L-40A flies were crossed to y, w, hsFLP3F; GFP33, GFP38, FRT2L-40A flies, and their progeny were heat shocked for 1 h at 38°C during larval development. To generate germline clones for the analysis of KEL protein levels, y, w, hsFLP3F; Cul-3gft2, FRT2L-40A females were crossed to P[ovoD], FRT2L-40A/CyO males, and progeny were heat shocked as larvae. The presence of the ovoD mutation ensured that all late-stage egg chambers in progeny females were Cul-3− (Chou and Perrimon, 1996); however, some younger germline cysts and most associated somatic cells were likely Cul-3+. To localize Venus::CUL-3 in egg chambers, we generated Cul-3gft2 MARCM clones (Lee and Luo, 1999) using the Venus::CUL-3 transgene as both the clonal marker and source of Venus::CUL-3. Flies of genotype P[Venus::CUL-3]272.17/hsFLP12; Cul-3gft2, FRT2L-40A/P[tub-Gal80] FRT2L-40A; p[tub-Gal4] were heat shocked one time as mixed second- to third-stage larvae for 1 h at 38°C. To express mCherry::KREP, mCherry::KEL, and Flag::CUL-3 during oogenesis, we used the P[mat-αtub-GAL4::VP16]V37 line (#7063; Bloomington Stock Center).
Molecular biology
Cul-3 sequence encoding the 773-aa CUL-3 product encoded by Cul-3Soma transcripts (equivalent to CUL-3–PC in FlyBase release 5.22; Arama et al., 2007) was PCR amplified from Berkeley Drosophila Genome Project cDNA clone LD03316 and recombined into the Gateway Donor vector pDONR201. The Venus::CUL-3 fusion protein construct was generated by recombining the Cul-3 entry clone with the pPVW vector from the Drosophila Gateway collection (Drosophila Genomics Resource Center). To make the KEL full-length and KREP expression constructs, DNA encoding KEL and the KREP domain (as defined in Robinson and Cooley, 1997) were PCR amplified from cDNA clones and recombined into pDONR201. After sequence verification, the ORFs were recombined into pTAPmCherryW-attB, a UASp phiC31 integration vector constructed in our laboratory that adds an N-terminal tag consisting of the SBP affinity tag (Keefe et al., 2001) and mCherry (Shaner et al., 2004). P element transgenic lines were made using standard procedures (Spradling, 1986), and phiC31 integration constructs were injected by Duke University Model System Genomics into the AttP2 landing site (Groth et al., 2004).
Western blots and immunoprecipitation
For immunoprecipitations, ovaries from ∼100 flies were homogenized with 10 strokes in a glass homogenizer (Duall; Kontes Glass Co.) with a Teflon pestle in lysis buffer (50 mM Hepes, pH 7.5, 150 mM NaCl, 5 mM EGTA, 0.5% Triton X-100, 10% glycerol supplemented with 1× complete protease inhibitor mix [Roche]). Protein concentrations were determined using the Bio-Rad Laboratories protein assay, and 2 mg of protein extract was incubated with ∼2 µg anti–Myc 9E10 antibody (Roche) or 2 µg anti–Flag M2 (Sigma-Aldrich). Antibody–antigen complexes were precipitated on protein A/G agarose (Santa Cruz Biotechnology, Inc.), washed four times in lysis buffer, and eluted by boiling in SDS-PAGE loading buffer. SBP pull-downs of SBP-mCherry::KEL were performed as described for immunoprecipitations, except SBP-mCherry::KEL was precipitated with 30 µl streptavidin UltraLink resin (Thermo Fisher Scientific). Precipitated proteins were separated by SDS-PAGE and analyzed by Western blotting. Antibodies used for Western blots were: α–KEL 1B (Developmental Studies Hybridoma Bank; Xue and Cooley, 1993), 1:00 α–Myc 9E10 (Santa Cruz Biotechnology, Inc.), and 1:300 α–tubulin E7 (Developmental Studies Hybridoma Bank). Western blots were developed with the SuperSignal West Dura ECL kit (Thermo Fisher Scientific), and luminescence was captured either on a charge-coupled device (Alpha Innotech) or Hyperfilm ECL (GE Healthcare).
Immunofluorescence and microscopy
Ovaries were dissected in ionically matched adult Drosophila saline buffer (Singleton and Woodruff, 1994) and fixed in 6% heptane-saturated formaldehyde, followed by washing and incubations in PBS supplemented with 0.3% Triton X-100 and 0.5% BSA. Primary antibodies and dilutions used were: 1:1 mouse monoclonal α–KEL 1B (Xue and Cooley, 1993), 1:500 rat polyclonal α-NFLN (N-terminal filamin antigen; Sokol and Cooley, 2003), and 1:5 mouse monoclonal α–HtsRC 7C (Robinson et al., 1994). Fluorescent labels used were Alexa Fluor 488, 568, or 633 conjugated to phalloidin, goat anti–mouse IgG, or goat anti–rat IgG (Invitrogen). Samples were viewed on either a confocal microscope (LSM 510; Carl Zeiss, Inc.) or an inverted microscope (Axiovert 200; Carl Zeiss, Inc.) equipped with a confocal imaging system (CARV II; BD) and a camera (CoolSNAP HQ2; Photometrics). One of three Carl Zeiss, Inc. objectives was used: 25× NA 0.8 Neofluar or 40× or 63× NA 1.2 W C-Apochromat. Images were edited using IPLab/iVision (BD), ImageJ (National Institutes of Health), LSM software (Carl Zeiss, Inc.), and/or Photoshop CS4 (Adobe). In some cases, a despeckle filter was uniformly applied to images to reduce nonbiological noise. Figures were assembled with Illustrator CS2 (Adobe).
Acknowledgments
We are grateful to Jim Skeath for providing reagents and to members of the Cooley laboratory for comments on the manuscript. We received cDNA and Gateway vectors from the Drosophila Genomics Resource Center, and monoclonal antibodies were obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the National Institute of Child Health and Human Development and maintained by the University of Iowa.
This work was supported by National Institutes of Health grant GM052702 to L. Cooley.
Footnotes
Abbreviations used in this paper:
- CRL
- cullin-RING ligase
- CRL3
- CRL assembled with CUL-3
- CUL-3
- Cullin-3
- KEL
- Kelch
- KREP
- KEL repeat
- Ovhts-RC
- ovarian hu-li tai shao–ring canal protein
- SBP
- streptavidin-binding peptide
References
- Adams J., Kelso R., Cooley L. 2000. The kelch repeat superfamily of proteins: propellers of cell function. Trends Cell Biol. 10:17–24 10.1016/S0962-8924(99)01673-6 [DOI] [PubMed] [Google Scholar]
- Arama E., Bader M., Rieckhof G.E., Steller H. 2007. A ubiquitin ligase complex regulates caspase activation during sperm differentiation in Drosophila. PLoS Biol. 5:e251 10.1371/journal.pbio.0050251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosu D.R., Kipreos E.T. 2008. Cullin-RING ubiquitin ligases: global regulation and activation cycles. Cell Div. 3:7 10.1186/1747-1028-3-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Yang Z., Meng M., Zhao Y., Dong N., Yan H., Liu L., Ding M., Peng H.B., Shao F. 2009. Cullin mediates degradation of RhoA through evolutionarily conserved BTB adaptors to control actin cytoskeleton structure and cell movement. Mol. Cell. 35:841–855 10.1016/j.molcel.2009.09.004 [DOI] [PubMed] [Google Scholar]
- Chou T.B., Perrimon N. 1996. The autosomal FLP-DFS technique for generating germline mosaics in Drosophila melanogaster. Genetics. 144:1673–1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth A.C., Fish M., Nusse R., Calos M.P. 2004. Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics. 166:1775–1782 10.1534/genetics.166.4.1775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson A.M., Cooley L. 2008. Phylogenetic, structural and functional relationships between WD- and Kelch-repeat proteins. In The coronin family of proteins. Subcellular Biochemistry, vol. 48 Clemen C.S., Eichinger L., Rybakin V., Landes Bioscience, Austin, TX: 6–19 [DOI] [PubMed] [Google Scholar]
- Keefe A.D., Wilson D.S., Seelig B., Szostak J.W. 2001. One-step purification of recombinant proteins using a nanomolar-affinity streptavidin-binding peptide, the SBP-Tag. Protein Expr. Purif. 23:440–446 10.1006/prep.2001.1515 [DOI] [PubMed] [Google Scholar]
- Kelso R.J., Hudson A.M., Cooley L. 2002. Drosophila Kelch regulates actin organization via Src64-dependent tyrosine phosphorylation. J. Cell Biol. 156:703–713 10.1083/jcb.200110063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T., Luo L. 1999. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 22:451–461 10.1016/S0896-6273(00)80701-1 [DOI] [PubMed] [Google Scholar]
- Luke-Glaser S., Pintard L., Lu C., Mains P.E., Peter M. 2005. The BTB protein MEL-26 promotes cytokinesis in C. elegans by a CUL-3-independent mechanism. Curr. Biol. 15:1605–1615 10.1016/j.cub.2005.07.068 [DOI] [PubMed] [Google Scholar]
- Merlet J., Burger J., Gomes J.E., Pintard L. 2009. Regulation of cullin-RING E3 ubiquitin-ligases by neddylation and dimerization. Cell. Mol. Life Sci. 66:1924–1938 10.1007/s00018-009-8712-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mistry H., Wilson B.A., Roberts I.J., O’Kane C.J., Skeath J.B. 2004. Cullin-3 regulates pattern formation, external sensory organ development and cell survival during Drosophila development. Mech. Dev. 121:1495–1507 10.1016/j.mod.2004.07.007 [DOI] [PubMed] [Google Scholar]
- Nagai T., Ibata K., Park E.S., Kubota M., Mikoshiba K., Miyawaki A. 2002. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 20:87–90 10.1038/nbt0102-87 [DOI] [PubMed] [Google Scholar]
- Petrella L.N., Smith-Leiker T., Cooley L. 2007. The Ovhts polyprotein is cleaved to produce fusome and ring canal proteins required for Drosophila oogenesis. Development. 134:703–712 10.1242/dev.02766 [DOI] [PubMed] [Google Scholar]
- Petroski M.D., Deshaies R.J. 2005. Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 6:9–20 10.1038/nrm1547 [DOI] [PubMed] [Google Scholar]
- Pintard L., Willis J.H., Willems A., Johnson J.L., Srayko M., Kurz T., Glaser S., Mains P.E., Tyers M., Bowerman B., Peter M. 2003. The BTB protein MEL-26 is a substrate-specific adaptor of the CUL-3 ubiquitin-ligase. Nature. 425:311–316 10.1038/nature01959 [DOI] [PubMed] [Google Scholar]
- Robinson D.N., Cooley L. 1997. Drosophila kelch is an oligomeric ring canal actin organizer. J. Cell Biol. 138:799–810 10.1083/jcb.138.4.799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson D.N., Cant K., Cooley L. 1994. Morphogenesis of Drosophila ovarian ring canals. Development. 120:2015–2025 [DOI] [PubMed] [Google Scholar]
- Robinson D.N., Smith-Leiker T.A., Sokol N.S., Hudson A.M., Cooley L. 1997. Formation of the Drosophila ovarian ring canal inner rim depends on cheerio. Genetics. 145:1063–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schüpbach T., Wieschaus E. 1991. Female sterile mutations on the second chromosome of Drosophila melanogaster. II. Mutations blocking oogenesis or altering egg morphology. Genetics. 129:1119–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner N.C., Campbell R.E., Steinbach P.A., Giepmans B.N., Palmer A.E., Tsien R.Y. 2004. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 22:1567–1572 10.1038/nbt1037 [DOI] [PubMed] [Google Scholar]
- Singleton K., Woodruff R.I. 1994. The osmolarity of adult Drosophila hemolymph and its effect on oocyte-nurse cell electrical polarity. Dev. Biol. 161:154–167 10.1006/dbio.1994.1017 [DOI] [PubMed] [Google Scholar]
- Sokol N.S., Cooley L. 1999. Drosophila filamin encoded by the cheerio locus is a component of ovarian ring canals. Curr. Biol. 9:1221–1230 10.1016/S0960-9822(99)80502-8 [DOI] [PubMed] [Google Scholar]
- Sokol N.S., Cooley L. 2003. Drosophila filamin is required for follicle cell motility during oogenesis. Dev. Biol. 260:260–272 10.1016/S0012-1606(03)00248-3 [DOI] [PubMed] [Google Scholar]
- Spradling A. 1986. P-element mediated transformation. In Drosophila: A Practical Approach. Roberts D.B., editor IRL Press, Washington, DC: 175–197 [Google Scholar]
- Spradling A. 1993. Developmental genetics of oogenesis. In The Development of Drosophila melanogaster, vol. 1 Bate M., Arias A.M., Cold Spring Harbor Laboratory Press, Plainview, NY: 1–70 [Google Scholar]
- Stogios P.J., Downs G.S., Jauhal J.J., Nandra S.K., Privé G.G. 2005. Sequence and structural analysis of BTB domain proteins. Genome Biol. 6:R82 10.1186/gb-2005-6-10-r82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilney L.G., Tilney M.S., Guild G.M. 1996. Formation of actin filament bundles in the ring canals of developing Drosophila follicles. J. Cell Biol. 133:61–74 10.1083/jcb.133.1.61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Doren M., Williamson A.L., Lehmann R. 1998. Regulation of zygotic gene expression in Drosophila primordial germ cells. Curr. Biol. 8:243–246 10.1016/S0960-9822(98)70091-0 [DOI] [PubMed] [Google Scholar]
- Xue F., Cooley L. 1993. kelch encodes a component of intercellular bridges in Drosophila egg chambers. Cell. 72:681–693 10.1016/0092-8674(93)90397-9 [DOI] [PubMed] [Google Scholar]
- Zhang D.D., Lo S.C., Sun Z., Habib G.M., Lieberman M.W., Hannink M. 2005. Ubiquitination of Keap1, a BTB-Kelch substrate adaptor protein for Cul3, targets Keap1 for degradation by a proteasome-independent pathway. J. Biol. Chem. 280:30091–30099 10.1074/jbc.M501279200 [DOI] [PubMed] [Google Scholar]