A combination of yeast genetics, synthetic genetic array analysis, and high-throughput screening reveals that sumoylation of Mcm21p promotes disassembly of the mitotic spindle.
Abstract
We describe the application of a novel screening approach that combines automated yeast genetics, synthetic genetic array (SGA) analysis, and a high-content screening (HCS) system to examine mitotic spindle morphogenesis. We measured numerous spindle and cellular morphological parameters in thousands of single mutants and corresponding sensitized double mutants lacking genes known to be involved in spindle function. We focused on a subset of genes that appear to define a highly conserved mitotic spindle disassembly pathway, which is known to involve Ipl1p, the yeast aurora B kinase, as well as the cell cycle regulatory networks mitotic exit network (MEN) and fourteen early anaphase release (FEAR). We also dissected the function of the kinetochore protein Mcm21p, showing that sumoylation of Mcm21p regulates the enrichment of Ipl1p and other chromosomal passenger proteins to the spindle midzone to mediate spindle disassembly. Although we focused on spindle disassembly in a proof-of-principle study, our integrated HCS-SGA method can be applied to virtually any pathway, making it a powerful means for identifying specific cellular functions.
Introduction
A major challenge in post-genome biology is to exploit genome sequence information to produce reagents and technologies that decipher the molecular basis of gene function through an unbiased and systematic analysis. Atlhough functional genomic approaches have been applied productively with yeast, the integration of multiple datasets is typically required to accurately define gene function. Combining data from many large-scale studies remains problematic because individual screens may not be saturating or conducted under comparable experimental conditions. To facilitate integration of large-scale phenotypic and genetic datasets, we combined an automated form of yeast genetics, synthetic genetic array (SGA) analysis (Tong et al., 2001), with a high-content screening (HCS) system, which automates image acquisition and the quantification of specific morphological phenotypes.
We examined the morphological phenotypes of the growing mitotic spindle in both single gene deletion mutants and in selected double mutant arrays, sensitized for spindle defects. In addition, we also examined a subset of strains carrying conditional alleles of essential genes at both restrictive and permissive temperatures. For the implementation of the platform, each step, from sample processing to image acquisition and scoring of phenotypes, was automated and adapted for both live-cell and fixed-cell analysis. The cell biological phenotype of each yeast mutant was represented by a quantitative readout of cellular parameters, called a morphological profile. Using this information, we identified 182 mutants that influence spindle dynamics, 90 of which had defects apparent only in the double mutant backgrounds. Our results identify new genes involved in spindle disassembly and outline an intricate pathway involving the SUMO machinery required for efficient relocalization of the Ipl1p kinase to the spindle midzone. Our SGA-HCS approach offers a general and powerful method for quantifying the activity of specific pathways in the context of complex genetic backgrounds.
Results
Systematic identification of mutants with aberrant spindle morphology
SGA methodology enables marked genetic elements to be combined in a single haploid cell through standard yeast mating and meiotic recombination via an automated procedure (Boone et al., 2007). Here, our goal was to systematically survey the yeast deletion collection for defects in spindle morphogenesis. To do so, we applied SGA to introduce a GFP-tubulin (GFP-Tub1p) reporter into the arrayed collection of deletion mutants. To sharpen our focus on spindle function, we also constructed double mutant arrays harboring GFP-Tub1p as well as a deletion allele of BNI1, which encodes a formin protein that participates in spindle orientation by nucleating actin cables, or a deletion of BIM1, which encodes a protein that links microtubules to a myosin motor that walks along the actin cables (Pruyne et al., 2004). We chose to assay double mutants with bni1Δ and bim1Δ because genetic interactions involving BNI1 and BIM1 have been well characterized (Tong et al., 2001) and the mutants have subtle defects in spindle function that appear mechanistically distinct.
We used automated image acquisition and analysis to quantify cell shape with respect to spindle morphology and score aberrant spindle defects (Fig. 1 A and Fig. S1; MetaXpress version 1.63, see Materials and methods). In brief, we used background fluorescence and a low threshold for GFP intensity to identify the individual cells or objects in each image (whole cell segmentation). Next, we identified spindles in the same image by varying the GFP threshold (spindle segmentation). After this, a minimal set of features such as area and shape factor were used to train the imaging software so that it could efficiently classify an unseen segmented image into two categories such as budded and unbudded cells. Each budded cell or unbudded cell was earmarked as a region of interest, and corresponding morphometric features were logged separately using the built-in functions of MetaXpress. An image-processing tool was then used to identify the bud neck region. After object identification, quantitative measurements allowed us to extract numerous morphometric features (Fig. S1) to generate a unique profile for each mutant strain (Tables S3–S5; see Materials and methods for more details on image analysis and morphometric feature extraction; for review see Vizeacoumar et al., 2009).
Figure 1.
Automated image analysis and systematic identification of spindle-defective mutants using morphological profiles. (A) Illustration of image processing and segmentation approach. A representative fluorescent micrograph and corresponding segmented images from a high-content screen of cells expressing GFP-Tub1p are shown. The diagram on the right illustrates the attachment of specific labels to a region of interest (cell) to extract information for specific objects (the spindle) within the region. The numbers are automatically generated by the image analysis software. Bar, 5 µm. (B) Heat maps showing distribution of wild-type, bim1Δ, and bni1Δ cells for spindle orientation. 2D histograms of the distribution of ∼35,000 wild-type (left), and similar numbers of bim1Δ (middle) and bni1Δ (right) cells assessed for spindle orientation and budding index. The x axis represents the ratio of daughter to mother area (small to large budded cells) as an indication of cell cycle stage. The y axis represents the orientation of the spindle with respect to the orientation of the mother–bud axis of the cell. Colors on the heat map represent cell number as illustrated by the key to the right of the histograms. The yellow line is a contour line to indicate a boundary that contains the majority of the cells; with heat map representations, it is difficult to gauge the quantity of cells in lower density areas and some of the more subtle trends. To help visualize the overall trends in these heat maps and to make it easier to visually compare between the wild-type, bim1Δ, and bni1Δ plots, illustrative contour lines are plotted as yellow curves to indicate boundaries that roughly contain the majority of the cells below the curves. Given the binning of a 2D histogram, the curve is plotted so that the total number of cells in the bins above the curve is no more than 1,000, which is used so as to ignore the outliers with extreme spindle orientations. The curve is then smoothed by fitting the points using a linear/quadratic rational function. (C) Profiles of spindle orientation for rcr2Δ and dyn2Δ mutant cultures. The distribution of ∼60–100 budded cells from an rcr2Δ (control, left) or dyn2Δ (right) culture for spindle orientation relative to budding index is shown. Budded cells from the mutant culture are represented by blue “X’s” overlaid on an image of the 2D histogram from the wild-type culture (see B). Images corresponding to selected cells in each culture are shown. The increased representation of cells with misoriented spindles in the dyn2Δ mutant relative to the rcr2Δ culture is emphasized by the yellow contour line (see B). Axes are as in B. Bar, 5 µm. (D) Venn diagram summarizing the number of mutants with aberrant spindle morphology identified by high-content screens of single and double deletion collections.
To identify genes that play a role in spindle dynamics, we concentrated on three basic spindle-specific features derived from the GFP-Tub1p reporter: (1) the ratio of spindle to cell length, (2) the orientation of the spindle with respect to the cell axis, and (3) the distance between the center of the spindle and the bud neck. We assessed these features relative to cell cycle stage and searched for mutants that showed an altered distribution of cells for a given feature compared with wild type (see Materials and methods). For example, Fig. 1 B shows a 2D histogram of spindle orientation and budding index (cell cycle stage) for wild-type, bim1Δ, and bni1Δ cells. As expected, the bim1Δ culture showed a larger population of cells in the mid-upper region of the histogram (Fig. 1 B), representing cells that are in the medium-to-large budded stage with misoriented spindles. The distribution of bni1Δ cells was also deviant but more similar to that of wild-type cells due to a subtle spindle defect (Lee et al., 1999). Machine learning was used to classify the signal from the wild-type cells and used to rank the deviant mutants based on a significant p-value (see Materials and methods). Fig. 1 C shows the distribution of a collection of cells from two different mutant cultures: a control strain (rcr2Δ), which has normal spindle function, and dyn2Δ, a known spindle orientation mutant (Vallee et al., 2004). The budded cells examined from the mutant cultures are represented by a blue X and overlaid on the wild-type histogram. As expected, the rcr2Δ mutant fell into the wild-type category, with a p-value >0.1, whereas the dyn2Δ mutant was classified as having a spindle defect, with a p-value < 0.1.
Using a p-value cutoff of 0.1, we identified 1,962 mutants in our three screens as being defective in at least one of the three spindle-specific features (length, orientation, and distance to the bud neck). We applied a filter using 78 additional spindle-specific features, and by demanding a defect in at least 10 features, we honed our list to 745 mutants. Manual inspection confirmed obvious defects for 419/745 mutants (with a false negative rate of 44%). The known mutants that were missed using these criteria had p-values in the range of 0.1–0.5 and had significant p-values for some individual features.
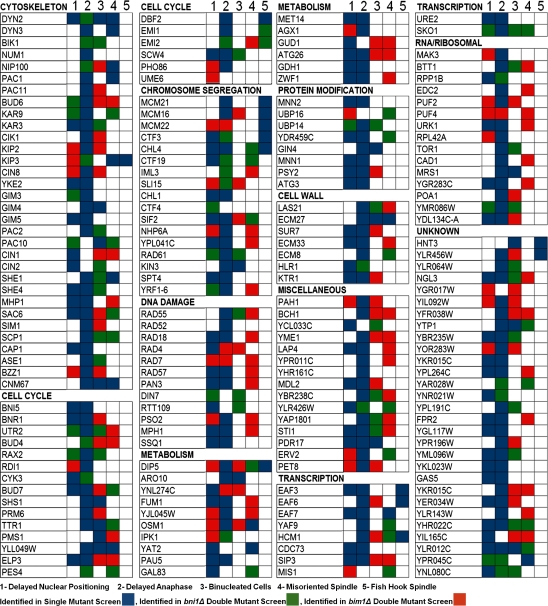
To hone our list for more detailed analysis, we selected mutants with a highly penetrant nuclear positioning or delayed anaphase phenotype, the two most common defects seen in our screens. We also selected any mutants that showed a relatively rare bi-nucleation, spindle misorientation, or fish hook spindle phenotypes (Fig. 2). In this way, we selected 182 mutants from manual inspection for more detailed analysis; 92 genes were identified from screening the single mutant deletion array. We scored double mutants that enhanced spindle phenotypes relative to either the bni1Δ or bim1Δ single mutants and identified an additional 90 mutants. As expected, most mutants identified in the single mutant screen were also identified in the double mutant screens, and the bni1Δ and bim1Δ double mutant screens overlapped extensively, by 122 genes (Fig. 1 D). Analysis of gene ontology annotations revealed a clear enrichment for genes encoding proteins implicated in spindle-related processes, including genes with roles in microtubule-based processes (P = 1.021 × 10−14), tubulin folding (P = 4.701 × 10−10), cytoskeleton organization and biogenesis (P = 5.702 × 10−11), mitotic anaphase (P = 9.394 × 10−7), and chromosome segregation (P = 7.699 × 10−7), among others (Robinson et al., 2002). We also observed enrichment for genes with annotated functions in metabolism, ribosomal biogenesis, and RNA processing, which is consistent with reported roles for nutrient signaling (Ras–cAMP pathway) and RNA-dependent protein complexes in regulating mitotic exit and mitotic spindle assembly, respectively (Morishita et al., 1995; Yoshida et al., 2003; Kittler et al., 2004; Blower et al., 2005).
Figure 2.
Categorization of top-ranked spindle mutants identified in high-content screens. The list of mutant strains identified computationally as spindle defective is shown. Mutants were manually assessed in a secondary assay and classified into five major categories: (1) delayed nuclear positioning; (2) delayed anaphase; (3) binucleated; (4) misoriented spindle; and (5) fish hook spindle. Blue, a phenotype seen only in the indicated single deletion mutant; green, a phenotype seen in the relevant bni1Δ double deletion strain; red, a phenotype for the corresponding bim1Δ double deletion mutant. Mutants are organized according to gene ontology “process” annotations.
To summarize the mutant phenotypes, we categorized each of the top-ranked mutants based on five major phenotypes by manual inspection (Fig. 2): (1) delayed nuclear positioning, in which the spindle fails to position close to the bud neck in medium/large budded cells; (2) delayed anaphase, in which the metaphase spindle persists in large budded cells; (3) bi-nucleated cells, where we observed two GFP dots, representing the spindle poles, within the mother of large budded cells; (4) misoriented spindle, in which the mitotic spindle is misaligned with the axis of the cell; and (5) fish hook spindle, in which the anaphase spindle is hyperelongated. Several genes with known roles in spindle dynamics were detected by our analysis, validating our approach (Fig. 2). Although our method allows rapid assessment of deletion collections and the detection of many new spindle mutants, false negatives may occur because of a cell-sampling issue, especially if the single or the double mutants show a severe fitness defect. For example, some known spindle mutants were just below the cutoff range as assessed by the p-values generated (Table S6).
Novel candidate genes required for spindle disassembly
We identified a relatively rare subset of mutants that exhibited fish hook spindles, including five members of the CTF19 complex (MCM21, MCM16, MCM22, CTF3, and CHL4), which is associated with the yeast kinetochore. This hyper-extended spindle phenotype resembles that of spindle disassembly mutants defective for IPL1, which encodes the Aurora B kinase (Buvelot et al., 2003), or KIP3, which encodes a kinesin that depolymerizes microtubules (Gupta et al., 2006). In addition, this hyperextended spindle phenotype is observed in several mitotic exit mutants as well (Toyn and Johnston, 1994; Jensen et al., 2004; Stoepel et al., 2005). We also identified three other genes that were not linked previously to spindle disassembly but whose deletion, either singly or in the context of a bni1Δ double mutant, resulted in a significant fish hook spindle phenotype (see Table S6). They were HNT3, which encodes a member of the HIT (histidine triad) superfamily of nucleotide-binding proteins, whose distant mammalian homologue, FHIT, binds microtubules (Chaudhuri et al., 1999); and EMI1 and EMI2, which were identified in our double deletion screen with BNI1 (Fig. 2) and have been implicated in control of the developmental program that activates the early meiotic-specific transcription factor, IME1 (Enyenihi and Saunders, 2003). We performed further analysis on these three genes, as well as MCM21, a nonessential component of the CTF19 complex, which had not been specifically linked to spindle disassembly.
We scored the fish hook spindle phenotype in a synchronized population of cells (Fig. 3 A) and by time-lapse imaging (Fig. S2 A, yellow arrows). Analysis of the kinetics of spindle elongation in mcm21Δ, hnt3Δ, emi1Δ, and emi2Δ mutant strains (Fig. S2, B–E and G) revealed a clear delay in anaphase spindle disassembly showing a fish hook spindle with varying degrees of defect. A small percentage of hnt3Δ bni1Δ double mutant cells also showed swapping of spindle poles between the mother and daughter cells after spindle disassembly (Fig. S2 A, red arrows), which is consistent with a loss of spindle pole polarity (Pereira et al., 2001) and consequent delay in cytokinesis (not depicted; Jensen et al., 2004). Time-lapse imaging also revealed cycles of partial elongation and collapse of the mitotic spindle without full elongation along the mother–bud axis in both mcm21Δ and hnt3Δ mutants (Fig. S2 A, blue arrows; and Fig. S2 F). Interestingly, we observed both the spindle instability and hyper-elongation phenotypes in the same cell (Fig. S2 A). We conclude that Mcm21p and Hnt3p may regulate both the stability of the growing anaphase spindle as well as spindle disassembly.
Figure 3.
hnt3Δ, mcm21Δ, emi1Δ, and emi2Δ mutants have spindle elongation phenotypes and components of the CPC become partially mislocalized in mcm21Δ mutant cells. (A) Quantitation of number of cells with fish hook spindles in single and double mutant cultures. A histogram illustrating the percentage of cells in the indicated mutant cultures with fish hook spindles is shown. Cells were synchronized with α factor for 90 min, released into fresh medium, and assessed for fish hook spindles after 60 min; images of at least 100 anaphase cells for each culture were captured and quantified. Error bars show the standard deviation from three independent experiments. (B, top) Ipl1p-GFP localization to the elongating spindle and spindle midzone is disrupted in mcm21Δ mutant cells. Mid-log phase cells were treated with nocodazole for 2.5 h at room temperature and released into fresh medium before imaging. DIC images, fluorescent micrographs, and merged images are shown of representative single cells of wild-type (left) or mcm21Δ (right) strains expressing Ipl1p-GFP and RFP-Tub1p. Yellow arrowheads indicate the diffused localization of Ipl1p-GFP (for the mcm21Δ cells) and the spindle midzone. Bar, 5 µm. (B, bottom) Sli15p-GFP is mislocalized, and Bir1p-GFP localization is unaffected in mcm21Δ mutant cells. Mid-log phase cells were treated with nocodazole for 2.5 h and released into fresh medium before imaging. DIC images, fluorescent micrographs, and merged images of a representative wild-type or mcm21Δ anaphase cell expressing Sli5p-GFP/Bir1p-GFP and RFP-Tub1p are shown. Yellow arrowheads indicate the localization of Sli15p-GFP. (C and D) Quantitation of the defect in Ipl1p-GFP localization at the kinetochore during metaphase and the spindle midzone during anaphase in wild-type and mcm21Δ mutant cells. (C) For kinetochore localization, cells with spindles between <2 and 2–4 µm were quantified, as there was an increased mislocalization of Ipl1p-GFP/Sli15p-GFP during early anaphase. (D) For midzone localization, cells with spindles 4 µm and longer were assessed. Cells with Ipl1p-GFP signal throughout the spindle were also considered as enriched for midzone in the quantitation. n > 60–100 cells in each spindle length category.
Ipl1p-GFP association with the spindle midzone is delayed in mcm21Δ cells
As noted earlier, yeast strains carrying mutations in the gene encoding the essential Aurora B kinase, IPL1, exhibit fish hook spindles, which is consistent with a role for Ipl1p in anaphase spindle disassembly (Buvelot et al., 2003). The products of IPL1 (Aurora B), SLI15 (IN-CENP), and BIR1 (SURVIVIN) interact to form the chromosomal passenger complex (CPC), which performs key mitotic roles through dynamic relocalization in a dividing cell (Zeng et al., 1999; Sullivan et al., 2001; Buvelot et al., 2003; Pereira and Schiebel, 2003; Ruchaud et al., 2007). Given that Mcm21p functions at the yeast kinetochore, we wondered whether the fish hook spindle defect in mcm21Δ mutants might be associated with mislocalization of Ipl1p. As expected, in wild-type cells, Ipl1p-GFP was localized to discreet dots at the kinetochore during metaphase, along the spindle as it elongated, at the midzone of the spindle during late anaphase, and finally to the trailing end of the disassembling microtubules (Fig. 3 B). In mcm21Δ cells, during metaphase, Ipl1p-GFP association with the kinetochore was reduced and remained diffusely localized throughout the nucleus (Fig. 3, B and C). During late anaphase, although Ipl1p appeared properly relocated to the spindle, we observed a delay in its spindle midzone accumulation (Fig. 3, B and D). Similar results were obtained for Sli15p-GFP (Fig. 3 B, bottom left). In contrast, Bir1p-GFP localized to the anaphase spindle normally in mcm21Δ cells with a mild delay (Fig. 3 B, bottom right), which is consistent with previous work suggesting that CPC components may have distinct functions (Gassmann et al., 2004; Thomas and Kaplan, 2007). These findings suggest that the fish hook phenotype in mcm21Δ cells may be caused by the mislocalization of the CPC, particularly the failure to localize Ipl1p to the spindle midzone. Consistent with our results, Knockleby and Vogel (2009) recently reported that Sli15p association with the kinetochore was reduced in mutants lacking the COMA complex, which is composed of Mcm21p and three other members of the CTF19 complex.
Genetic interactions involving MCM21 and late mitotic regulators
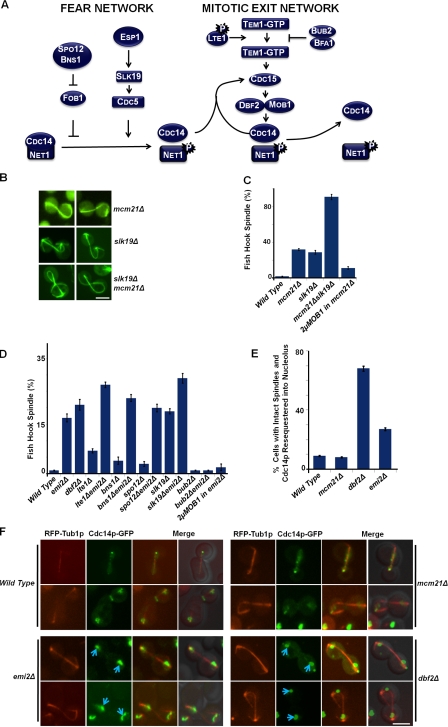
Sli15p dephosphorylation at the kinetochore is necessary for the relocalization of the CPC to the spindle midzone (Pereira and Schiebel, 2003). This reaction is mediated by the Cdc14p phosphatase, which is activated during early anaphase by the fourteen early anaphase release (FEAR) network proteins (Fig. 4 A). Although the release of Cdc14p from the nucleolus is initiated during early anaphase in a FEAR-dependent manner, maintenance of Cdc14p in the released state is controlled by the mitotic exit network (MEN) during the late stages of anaphase (Fig. 4 A; Buonomo et al., 2003; Pereira and Schiebel, 2003; Sullivan and Uhlmann, 2003; D’Amours and Amon, 2004). Ultimately, Cdc14p is resequestered back into the nucleolus concomitant with spindle disassembly after degradation of the Polo kinase Cdc5p, a key factor in releasing Cdc14p from its inhibitor in the nucleolus (Visintin et al., 2008).
Figure 4.
Genetic interactions involving the FEAR network, MEN, and kinetochore components. (A) Diagram of key components of the FEAR network and MEN. Cdc14p is released from the nucleolus during early anaphase by the FEAR network and maintained in a released state by the MEN proteins. (B) Exacerbation of the fish hook spindle phenotype of mcm21Δ mutant cells by deletion of a FEAR network activator. Representative fluorescence micrographs of GFP-Tub1p in mcm21Δ, slk19Δ, and slk19Δ mcm21Δ cells are shown. Bar, 5 µm. (C) Quantitation of the fish hook spindle phenotype for the mutants shown in B. The plot shows the percentage of anaphase cells for the indicated mutant cultures with a clear fish hook spindle phenotype. Quantitation of the defect in an mcm21Δ mutant overexpressing MOB1 is also shown. Error bars show the standard deviation from three independent experiments counting ∼100 cells for each strain. (D) Quantitation of the fish hook spindle phenotype for single and double mutants of EMI2 with FEAR and MEN components. The plot shows the percentage of anaphase cells for the indicated mutant cultures with a clear fish hook spindle phenotype. Error bars show the standard deviation from three independent experiments counting ∼100 cells for each strain. (E) Quantitation of the percentage of cells with intact spindles that show premature resequestration of Cdc14p-GFP into the nucleolus. Cells were synchronized using α factor, released into fresh medium, and monitored using live-cell imaging every 15 min for 180 min. RFP-Tub1p was used to monitor the spindle and identify the cell cycle stage. Error bars show the standard deviation from three independent experiments counting ∼80 cells for each strain in all the time points. (F) Release of Cdc14p-GFP in mcm21Δ, emi2Δ, and dbf2Δ mutant cells. Cells were synchronized using α factor, released into fresh medium, and monitored using live-cell imaging every 15 min for 180 min. RFP-Tub1p was used to monitor the spindle and identify the cell cycle stage. Representative wild-type, emi2Δ, mcm21Δ, and dbf2Δ mutant cells in late anaphase are shown. Blue arrows indicate the premature sequestration of Cdc14p to the nucleolus before spindle disassembly. Note that unlike emi2Δ and dbf2Δ cells, mcm21Δ cells maintained the released state of Cdc14p even when the spindle was hyperelongated. Bar, 5 µm.
To examine the functional relationship between Mcm21p and the FEAR network, we first tested for genetic interactions between MCM21 and SLK19, which encodes a signaling component of the FEAR network (Fig. 4 A; Stegmeier et al., 2002; Pereira and Schiebel, 2003). The mcm21Δ and slk19Δ mutations combined to cause a more extreme double mutant phenotype in comparison to the corresponding single mutants. The mcm21Δ slk19Δ double mutant had a significant fitness defect, and ∼90% of the anaphase cells exhibited severely coiled fish hook spindles (Fig. 4, B and C). This double mutant analysis highlights the critical roles of both the kinetochore and Cdc14p activation in spindle disassembly.
To generalize our conclusions, we examined spindle morphology in a panel of mutants defective for genes encoding other components of the CTF19 complex, the FEAR network, and the MEN network. All the mutants we examined exhibited fish hook spindles to varying degrees (Fig. S3 A and Fig. 4 D). As previously described (Buvelot et al., 2003), we also found that mutant alleles of IPL1 (ipl1-1 and ipl1-2) were associated with a fish hook spindle phenotype (Fig. S3 A). Our observations combined with previous findings (Pereira and Schiebel, 2003; Stegmeier and Amon, 2004; Stoepel et al., 2005) suggest a model in which MEN and FEAR pathway components activate Cdc14p, which regulates the CPC after its docking at the kinetochore in a CTF19 complex–dependent manner.
Emi2p participates in Cdc14p activation
Because the MEN and FEAR networks control spindle disassembly, we asked if the fish hook spindle mutants that we identified in our screens might participate in these pathways. Emi2p is a possible MEN network component because affinity purification/mass spectrometry suggested an interaction with two MEN network proteins, Dbf2p and Lte1p (Graumann et al., 2004). Emi2p encodes a candidate hexose kinase (Johnston, 1999), which suggests a potential connection between nutrient sensing and cell cycle control. To assay for a functional linkage between Emi2p and the MEN or FEAR pathway, we made emi2Δ double mutants with deletion alleles of genes encoding either MEN or FEAR components. Double mutants combining emi2Δ with deletion alleles of either MEN (lte1Δ, dbf2Δ) or FEAR (bns1Δ, spo12Δ, slk19Δ) components showed an exaggerated fish hook spindle (Fig. 4 D and Fig. S3 B). An emi2Δ dbf2Δ double mutant strain was inviable, providing genetic evidence that Emi2p may function in parallel with DBF2, perhaps in the FEAR pathway (unpublished data). These genetic data, and the possible physical interaction between Emi2p and MEN components, suggest involvement of Emi2p in both the MEN and FEAR networks.
To contrast these observations, we also examined genetic conditions that activate the MEN network. Bub2p and Bfa1p are inhibitors of MEN and the subsequent release of Cdc14p, as they form a GTPase-activating complex (GAP), or an inhibitor for the Tem1p GTPase (Fesquet et al., 1999; Pereira et al., 2000; Geymonat et al., 2002; Ro et al., 2002). In the emi2Δ bub2Δ double mutant, the anaphase spindles extended normally, and cells underwent mitosis like wild-type cells (Fig. 4 D and Fig. S3 B). Consistent with this finding, overexpression of MOB1, an activating MEN component, also rescued the fish hook spindle in emi2Δ cells (Fig. 4 D). Thus, defects in MEN and FEAR signaling accentuate the emi2Δ mutant phenotype, whereas activation of the MEN pathway appears to suppress it. Finally, we examined Cdc14p localization in emi2Δ cells to evaluate its role in Cdc14p release. Wild-type cells largely release Cdc14p from the nucleolus as the spindles start to elongate, and, concomitant with spindle disassembly, they resequester Cdc14p back into the nucleolus (Stegmeier et al., 2002; Visintin et al., 2008). In contrast, the phenotype of emi2Δ cells resembled that of dbf2Δ mutants; ∼21% of the cells showed Cdc14p-GFP resequestered back into the nucleolus even before spindle disassembly (Fig. 4, E and F, blue arrows). Thus, like mutants in the MEN and FEAR network genes, emi2Δ cells appear to have a defect in maintaining Cdc14p in an active released state, which suggests that Emi2p may modulate one of these pathways.
Sumoylation of Mcm21p targets the CPC specifically to the spindle midzone
In addition to the nonessential gene set, we also screened a collection of temperature-sensitive mutants covering ∼100 essential genes for defects in spindle morphology (Table S7). By screening this collection, we identified a spindle disassembly defect (fish hook spindle) associated with a mutant allele of MMS21 (Fig. S3 A), which encodes one of the known E3 SUMO ligases (Zhao and Blobel, 2005). We were intrigued by this observation because recent work from our laboratory identified genetic interactions between MCM21 and genes encoding SUMO pathway components, which suggested a link between sumoylation and Mcm21p function (Makhnevych et al., 2009). Mcm21p is the only protein in the CTF19 complex that is known to be sumoylated (Panse et al., 2004; Wohlschlegel et al., 2004; Zhou et al., 2004; Denison et al., 2005; Hannich et al., 2005; Wykoff and O’Shea, 2005), raising the possibility that sumoylation may regulate CPC localization and that the sumoylation of Mcm21p may be regulated by the E3 ligase MMS21. Consistent with this idea, two observations suggested that factors in addition to Cdc14p influenced CPC localization in the mcm21Δ mutant. First, unlike emi2Δ cells, we found that the kinetics of Cdc14p localization were unaffected in mcm21Δ cells, yet Ipl1p-GFP failed to properly localize to the spindle (Fig. 4 F and Fig. 3 B). Second, hyperactivation of the Cdc14p signaling pathway through overexpression of a MEN component, MOB1 (Komarnitsky et al., 1998), resulted in only partial rescue of either the Ipl1p-GFP mislocalization phenotype or the fish hook spindle phenotype (Fig. 4 C and Fig. S3, C and D). These findings indicate that signaling from Cdc14p alone is not sufficient to target the CPC to the spindle midzone, which suggests that Mcm21p might play additional roles at the kinetochore.
We explored this idea by first assessing Mcm21p and its closely associated CTF19 complex proteins Okp1p, Ame1p, and Ctf19p (the COMA subcomplex), and confirmed that only Mcm21p was detectably sumoylated in our assay (Fig. 5 A). To dissect the role of Mcm21p sumoylation in spindle function, we first mutated the best matches to the consensus site for sumoylation in Mcm21p (Jeram et al., 2009), and observed no significant effect on Mcm21p sumoylation (unpublished data). Because >40% of the published yeast SUMO conjugation sites occur at nonconsensus lysine residues (Jeram et al., 2009), we next synthesized a sumoylation-deficient allele of MCM21, mcm21-32R. We made mutations that changed all 32 Mcm21p lysine residues to arginine (Mcm21p-32R), and we examined the functionality of the Mcm21p-32R mutant in three different ways. First, we found that the Mcm21p-32R mutant was expressed at normal levels but was not sumoylated (Fig. 5 A). Second, when expressed as a GFP chimera, Mcm21p-32R localized normally to the kinetochore (Fig. 5 B). Third, tetrad dissection revealed that expression of MCM21-32R rescued the synthetic lethality of a mcm21Δ mad1Δ double mutant strain (unpublished data).
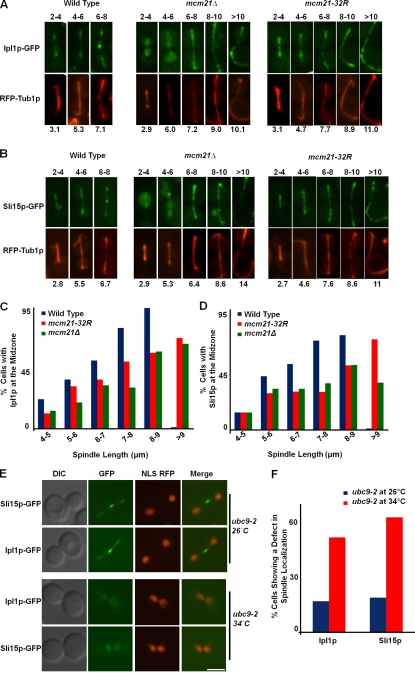
Figure 5.
Sumoylation of Mcm21p is not required for kinetochore localization of the CPC. (A) Sumoylation of CTF19 complex components. Immunoprecipitation/Western blotting was performed from protein extracts of wild-type or ubc9-2 strains expressing Mcm21p-TAP, Ame1p-TAP, Ctf19p-TAP, or Okp1p-TAP. A strain expressing a version of TAP-tagged Mcm21p lacking 32 lysine residues (mcm21-32R) was also analyzed for SUMO modification. The asterisk indicates sumoylated Mcm21p protein. The anti-SUMO antibody detects both sumoylated and unmodified proteins due to the associated TAP tag. (B) Subcellular localization of wild-type Mcm21p-GFP and Mcm21p-32R-GFP. DIC images, fluorescent micrographs, and merged images of representative wild-type or mcm21Δ anaphase cells expressing either wild-type Mcm21p-GFP or Mcm21p-32R-GFP and RFP-Tub1p are shown. Note the formation of a fish hook spindle in the mcm21-32R mutant background. Bar, 5 µm. (C and D) Ipl1p-GFP (C) and Sli15p-GFP (D) localization to the kinetochore in wild-type, mcm21-32R, and mcm21Δ mutant cells. Mid-log phase cells were treated with nocodazole for 2.5 h at room temperature and released into fresh medium before imaging. Fluorescent micrographs and merged images are shown of representative single cells of wild-type, mcm21Δ, or mcm21-32R strains expressing Ipl1p-GFP and RFP-Tub1p. Bar, 5 µm. (E and F) Quantitation of the defect in Ipl1p-GFP and Sli15p-GFP localization to the kinetochore during metaphase in wild-type, mcm21Δ, and mcm21-32R mutant cells. Cells with spindles between <2 and 2–4 µm were quantified for Ipl1p-GFP (E) or Sli15p-GFP localization to the kinetochore (F) as indicated. n > 200 cells in each spindle length category.
Unlike in the mcm21Δ deletion mutant, the kinetochore association of Ipl1p-GFP and Sli15p-GFP was not affected during metaphase in the mcm21-32R mutant, which suggests that Mcm21p-32R still retains the scaffolding function associated with wild-type Mcm21p at the kinetochore (Fig. 5, C–F). Nevertheless, like the mcm21Δ deletion mutant, the mcm21-32R mutant was defective for targeting Ipl1p-GFP and Sli15p-GFP to the spindle midzone, resulting in the formation of extended fish hook spindles (Fig. 6, A–D). Wild-type cells were able to target Ipl1p-GFP and Sli15p-GFP to the midzone, such that the spindle reached a maximum length of 6–8 µm before disassembly, whereas the mcm21Δ and the mcm21-32R mutant cells were defective in this localization and showed an extended fish hook spindle that often grew beyond 9 µm (Fig. 6, A–D). Invariably, every cell that formed a fish hook spindle did not localize Ipl1p-GFP to the midzone in both the mcm21Δ deletion mutant and the mcm21-32R mutant, which suggests that sumoylation of Mcm21p may signal the efficient relocation of the CPC to the spindle midzone.
Figure 6.
Sumoylation of Mcm21p regulates the localization of the CPC to the spindle midzone. (A and B) Ipl1p-GFP and Sli15p-GFP localization to the spindle midzone in wild-type, mcm21-32R, and mcm21Δ mutant cells. Mid-log phase cells were treated with nocodazole for 2.5 h at room temperature and released into fresh medium before imaging. Fluorescent micrographs of representative single cells of wild-type, mcm21Δ, or mcm21-32R strains expressing Ipl1p-GFP (A), Sli15p-GFP (B), and RFP-Tub1p are shown. Numbers at the top of each panel denote the spindle length (µm) range that each micrograph represents, and the numbers at the bottom represent the spindle length (µm) of the cell shown. (C and D) Quantitation of the defect in Ipl1p-GFP and Sli15p-GFP localization to the spindle midzone in wild-type, mcm21Δ mutant cells, and mcm21-32R mutant cells. Cells with spindles 4 µm and longer were used to quantify Ipl1p-GFP (C) or Sli15p-GFP (D) localization to the spindle midzone as indicated. n > 60–100 cells in each spindle length category. Cells with Ipl1p-GFP or Sli15p-GFP signal throughout the spindle were also counted as mid-zone enriched for the quantitation. (E) Localization of Ipl1p-GFP and Sli15p-GFP in ubc9-2 strains. Cells were grown at 34°C for 3 h and imaged immediately (34°C panels) or shifted to 26°C for 2 h before samples were imaged (26°C panels). DIC images, fluorescent micrographs, and merged images are shown of representative single cells. Bar, 5 µm. (F) Quantitation of the defect in spindle localization of Ipl1p-GFP and Sli15p-GFP for mutants represented in E. n > 100 cells.
To further assess the role of sumoylation in CPC localization, we examined a ubc9-2 temperature-sensitive mutant defective for the SUMO E2 ligase. Sumoylation is involved in many cellular processes (Melchior, 2000; Müller et al., 2001; Seeler and Dejean, 2003), and most ubc9-2 mutant cells arrest in metaphase and early anaphase (Fig. 6 E), precluding analysis of the fish hook spindle phenotype. Nonetheless, the large budded ubc9-2 cells clearly failed to efficiently localize both Ipl1p-GFP and Sli15p-GFP to the spindle at the restrictive temperature (Fig. 6, E and F). After release from the restrictive temperature, Ipl1p-GFP rapidly enriched along the spindles and specifically concentrated at the spindle midzone as expected (Fig. 6 E). Thus, sumoylation of Mcm21p appears to play a specific role in targeting the CPC to the spindle midzone.
Discussion
Integrating SGA analysis and HCS provides a general strategy for quantitative assessment of cell biological phenotypes on a genome-wide scale in sensitized genetic backgrounds. Automation and machine learning techniques are required to scale this analysis to multiple pathways, enabling objective, quantitative, and highly reproducible determination of detailed morphological phenotypes. Although the deletion collection has been assessed for morphological phenotypes through systematic analysis of fixed cells (Ohya et al., 2005), our study represents the first fully automated genome-scale mutant screen tracking a dynamic intracellular event. An unbiased quantitative readout allows for the identification of subtle but significant phenotypes, offering the potential to confirm and expand genetic networks based solely on fitness measurements. For example, with a BIM1 query mutation and combined SGA-HCS system, we scored 45 genes (∼50%) previously identified as BIM1 synthetic lethal–sick interactions (Tong et al., 2004), but we also identified 122 novel genes involved in spindle function. A limitation of our system is that true synthetic lethal double mutants fail to divide and cannot be examined by HCS (Fig. S4 A); however, this problem can be overcome if the double mutant is made conditional, as through the use of a temperature-sensitive allele of one member of the gene pair (Xu et al., 2007).
Morphological mutants, networks, and complexes
We compared our phenotypic profiling results to genetic and protein–protein interaction datasets, and found that mutant strains that had a high phenotypic deviation from wild type were more likely to be highly connected in the existing synthetic lethal genetic and protein–protein interaction networks (Fig. S4, B and C). Thus, an accurate assessment of phenotypic deviance from wild type may allow us to infer participation of proteins or genes in network hubs (Rutherford and Lindquist, 1998). The occurrence of a specific phenotypic change in response to a particular mutation was also correlated for proteins within the same complex (Fig. S4 D). For example, we observed that members of the CTF19 complex exhibit a similar fish hook spindle phenotype, as do components of the FEAR and MEN pathways (Fig. S3 A).
Spindle disassembly pathway
Our large-scale analysis of mutants displaying a fish hook spindle combined with previous observations elaborate a general model of spindle disassembly in yeast (Fig. 7; Pereira and Schiebel, 2003; Stegmeier and Amon, 2004; Stoepel et al., 2005). In particular, our study established a new role for sumoylation at the kinetochore associated with the CTF19 complex and modulation of spindle disassembly. We suggest that Mcm21p along with the members of the CTF19 complex may act as a scaffold for docking the CPC at the kinetochore. Our data suggest that signaling from Cdc14p alone is not sufficient and that sumoylation of Mcm21p is necessary for the movement of Ipl1p from the kinetochore to the spindle midzone. Alternatively, it might be that sumoylation could trigger release of Ipl1p from the kinetochore. This might facilitate Cdc14p-dependent dephosphorylation of Sli15p and the subsequent targeting of Ipl1p to the spindle midzone. This is evident from the observation that Ipl1p stably associates with the spindle in the mcm21-32R mutant, which suggests that the dynamic relocalization of Ipl1p is tightly regulated, presumably to avoid premature spindle disassembly.
Figure 7.
Model of mitotic exit and spindle disassembly. Major regulators of the CPC during mitosis are illustrated. For simplicity, MEN and FEAR pathway components are not shown. FEAR activates the partial release of Cdc14p during early anaphase, and MEN activates the complete release of Cdc14p during late anaphase. Cdc14p release from the nucleolus and sumoylation of Mcm21p at the kinetochore seem to be two important signals that regulate the dynamic relocalization of the CPC components to the spindle midzone from the kinetochore. We suggest that Mcm21p along with the members of the CTF19 complex may act as a scaffold for the CPC. Specifically, in cooperation with Cdc14p, sumoylation of Mcm21p may increase the efficiency of timed localization of the CPC to the spindle midzone for proper disassembly of the spindle.
We also discovered that Emi2p, a hexose kinase family member, is involved in the activation of Cdc14p. Hexose kinases participate in carbon sensing (Johnston, 1999), but several observations suggest an additional role for these enzymes in cytoskeletal regulation. For example, binding assays and electron microscopic studies revealed binding of bovine brain hexose kinase (HK1) to microtubules (Wágner et al., 2001). Also, hexose kinases may regulate the movement of kinesin motors on microtubules by quenching ATP molecules (Hess et al., 2001). Our observations, coupled with these results, suggest a possible role for Emi2p in linking nutrient sensing to mitotic exit. An emi2Δ dbf2Δ double deletion mutant is synthetic lethal, which suggests that Emi2p may play a role in the parallel FEAR pathway. However, the reported physical interaction of Emi2p with MEN components suggests that Emi2p may also be involved in the MEN pathway (Graumann et al., 2004). Genetic analyses of pathway components suggest that if we delete two nonessential components of a pathway containing at least one essential component, synthetic lethality is expected between the nonessential components (Bandyopadhyay et al., 2008). Accordingly, we suggest that Emi2p is a novel member of the MEN pathway.
Interestingly, Cdc14p is required for anaphase spindle stabilization (Khmelinskii et al., 2007; Khmelinskii and Schiebel, 2008), yet spindle disassembly defects are invariably found in all FEAR and MEN mutants (Toyn and Johnston, 1994; Jensen et al., 2004; Stoepel et al., 2005). Consistent with these observations, roles for Cdc14p in both meiotic and mitotic spindle disassembly have been described (Buvelot et al., 2003; Marston et al., 2003). Both Cdc14p and Ipl1p have several roles within the cell, and current literature suggests that there is a qualitative and quantitative difference in the Cdc14p released from the FEAR and MEN pathways (Stegmeier and Amon, 2004). Accordingly, Cdc14p released from the FEAR may stabilize the spindle, whereas Cdc14p released from the MEN may destabilize the spindle.
Although we used spindle morphology as a proof-of-principle, combining SGA analysis and HCS enables virtually any pathway that can be monitored with a fluorescent reporter to be assessed quantitatively within the context of numerous genetic and environmental perturbations. In theory, this general system should enable the yeast community to collect a wealth of quantitative HCS datasets under standardized formats. The potential for integration of hundreds of combined SGA-HCS analyses offers the possibility of a new global view of the cell.
Materials and methods
Automated image acquisition
A modification of the SGA method (Tong et al., 2001; Sopko et al., 2006) was used to introduce the GFP-Tub1p–expressing plasmid and the deletion allele of BNI1 into the deletion array. A strain harboring bni1Δ (Table S2) was transformed with a GFP-Tub1p plasmid and mated to the yeast deletion array (Giaever et al., 2002) by replica pinning. Diploids were selected and sporulated using standard SGA selections (Tong et al., 2001). MATa bni1Δ genexΔ haploids expressing GFP-Tub1p were isolated through successive pinning onto selective media and then into liquid selection medium for overnight growth. To optimize cell density for image analysis, we used liquid-handling robots (Biomek FX Laboratory automation work station; Beckman Coulter) to dispense sample volumes based on the optical density of each strain (SpectraMax Plus 384 Microplate Spectrophotometer; MDS Analytical Technologies). Plates were imaged in 96-well format using glass-bottomed plates (MMI Greiner M plates), and each plate contained control strains at 19 positions amounting to ∼1,064 controls per screen. A fluorescence microscopy system (ImageXpress 5000A; MDS Analytical Technologies) was used to acquire images. Images were acquired with 60× dry objective (Plan-fluor) with an NA of 0.85 and a working distance of 370 µm. Images were acquired at room temperature unless indicated using a 1,280 × 1,024 cooled charge-coupled device camera with 12-bit readout. Low fluorescent media (Sheff and Thorn, 2004) was used to resuspend the cells before imaging. To increase the throughput, we linked automated incubators (Cytomat; Thermo Fisher Scientific) to the Image Express system to allow automated loading of plates into the imager using a robotic arm (CRS Catalyst Express; Thermo Fisher Scientific) and integration software (Polara; Thermo Fisher Scientific).
Automated image analysis
Automated image acquisition and analysis were performed with MetaXpress software version 1.63 (MDS Analytical Technologies). After images were shade-corrected and background-subtracted, objects were segmented and single cells were defined using background cell fluorescence in the GFP channel. We used a series of MetaXpress modules to segment whole cells as well as objects inside cells. The segmented images were then binarized to produce three files for each image: entire cells, spindle objects, and the bud neck regions (Fig. S1). Once cells were identified, dead cells were removed from further analysis by gating average grayscale, as they had high autofluorescence. Then a minimal set of features (dimension, shape factor, and elliptical form factor) were used to train the software to efficiently classify an unseen image into two categories such as budded and unbudded cells. Each budded or an unbudded cell was taken as a region of interest, and the morphometric features of each region of interest and relevant objects (spindle) were quantified individually for each cell. Water shedding was used to identify the bud neck region, and several morphometric readouts for the mother and daughter portions of the cells were also generated using the built-in options of MetaXpress.
We measured several morphometric characteristics of each cell and the spindle, applying appropriate statistical measurements. By dimensionality reduction (Kaufman and Rousseeuw, 1990), we chose 77 nonredundant “geometric features”, each of which contained four statistical parameters (mean, variance, minimum value, and maximum value). We also considered 13 features that calculate percentage values because of their biological relevance (for example, percentage of large budded cells with an anaphase spindle). Thus, the morphological profile of each strain includes 321 cellular measurements, 240 of which were attributed to cell morphology and 81 attributed to spindle morphology (Tables S3–S5; cataloged measurements are summarized in Table S1). Our measurements include a description of cell shape, a feature that can serve as a proxy for cell cycle position (unbudded, small budded, medium budded, and large budded cells), characteristics of spindle pole bodies, and the mitotic spindle. The cataloged measurements in Tables S3–S5 are a mean of all the values measured in each mutant.
Data analysis
To identify mutants whose morphological profiles differed from wild type, we used a mixture of Gaussian models to learn the probability density function of the control based on the four features (Bishop, 2006). The mother–daughter cell size ratio is used in the analysis as an indication of cell cycle stage (Byers and Goetsch, 1975; Kilmartin and Adams, 1984). Cross-validation revealed that 60 Gaussians was optimal for this dataset. Mutant strains were evaluated by computing the likelihood under this learned model. A low likelihood indicates that the cells of the mutant strain exhibit phenotypes noticeably different than that of the control. Because the number of identified and measured cells differs for each strain, the likelihoods must be normalized to compare the different strains as well as provide a measure of statistical significance. So for a strain with a sample size of n cells, a random sample of n cells from the mixture of Gaussians was drawn, and the likelihood of these randomly drawn cells was computed. This sampling procedure was conducted 2,000 times, and the likelihood of the mutant cells was compared with the likelihood of the random samples to provide a p-value for the mutant phenotype. The same procedure was applied to other measurements to find mutant strains for different phenotypes as well as for double knockout experiments.
To assess the correlation between gene connectivity and phenotype, we downloaded all available synthetic lethal interaction data from BioGrid (http://www.thebiogrid.org/) and protein–protein interaction data from the Database of Interacting Proteins (http://dip.doe-mbi.ucla.edu/). We identified the top 100 highly connected genes and the bottom 100 least-connected genes in each of these datasets. The mean phenotypic change and the 95% confidence interval of the mean are derived from 5,000 bootstrap resamplings. P-values were derived using a Mann-Whitney U test. Fig. S4 (B and C) shows the mean phenotypic alterations in these gene datasets. Mean phenotypic variation is defined as the mean number of altered morphometric measurements; for each gene, we counted the number of altered phenotypic features for both the single deletion and the double deletion. To determine whether or not the loss of proteins in the same complex tends to cause similar morphology profiles, we compared phenotypic correlation within complexes against random controls. A total of ∼274 literature-curated protein complexes and a similar set of randomly generated protein complexes were examined for the correlation of their morphological profile. Phenotypic correlation is defined as the mean pairwise correlation among complex subunits. We evaluated phenotypic correlation upon deletion of genes involved in the same protein complexes. In brief, we calculated pairwise Pearson’s correlation for genes involved in protein complexes, and mean pairwise correlation was then compared against a set of random controls in which complex memberships were randomly shuffled. Fig. S4 D shows the correlation between true complexes and random complexes.
Confocal microscopy and image quantitation
Images were captured using a spinning disc confocal system (WaveFX; Quorum) with an ultra-cooled 512 back-tinned EM charge-coupled device camera, or a microscrope (E-600FN; Nikon) with an OrcaII camera (Hamamatsu). Images were acquired with 63× HCX Plan-Apochromat oil objective with an NA of 1.4. Images were captured at room temperature after loading the cells on gelatin pads as described previously (Jones et al., 1993; Yeh et al., 1995). Stacks of 11 optical sections spaced 0.3 µm apart or 5 optical planes spaced 0.2 µm apart were captured every 1 min. GFP was excited using a 488-nm laser, and its emission was collected using a 505-nm long-pass filter. No further processing was needed, as raw images clearly showed the phenotype. The z-axis images were converted to a single composite image by using the brightest pixel at every position in each of the image planes. This maximum pixel projection technique produced a 2D representation of the GFP fusion proteins within the cell from the 3D dataset. A macro written with the MetaMorph scripting language was used to input the entire stack of background-subtracted images and output a new stack of only the projected images, and to measure the spindle length in every z-stack plane.
Detection of SUMO conjugates and Western blot analysis
The mcm21-32R mutant was made by gene synthesis (DNA2.0, Inc.). The mutated gene (1,190 bp) and its associated promoter (500 bp), with a C-terminal tandem affinity purification (TAP) tag (543 bp) and downstream sequences (200 bp), were cloned into the BamHI and HindIII sites of the pRS315 vector. To construct the mcm21-32R-GFP, the mutated gene was amplified with appropriate primers and integrated into the genome along with a GFP tag. wild-type or ubc9-2 strains expressing TAP-tagged versions of Mcm21p, Ctf19p, Ame1p, and Okp1p were grown in YPD to an OD600 of 0.4 at 25°C, then the temperature was shifted to 34°C for 3 h. Cells were collected by centrifugation and lysed by bead beating in 10% glycerol, 50 mM Hepes-KOH, pH 8.0, 0.1 M KCl, 2 mM EDTA, 0.1% NP-40, 2 mM DTT, 10 mM N-ethylmaleimide, and yeast protease inhibitor cocktail (Sigma-Aldrich). Samples were clarified by centrifugation (16,000 g for 5 min at 4°C), and TAP-tagged proteins were isolated using mouse IgG beads (Dynabeads; Invitrogen). Beads were washed extensively with wash buffer containing 0.25 M MgCl2, and bound proteins were eluted with Laemmli buffer. Specific rabbit polyclonal antibodies were used to detect Smt3p (Rockland Immunochemicals, Inc.). TAP-tagged proteins were detected with normal rabbit serum.
Online supplemental material
Fig. S1 shows steps involved in automated image analysis. Fig. S2 shows quantitation of spindle length defects in hnt3Δ, mcm21Δ, emi1Δ, and emi2Δ single and double mutant strains. Fig. S3 shows representative micrographs of fish hook spindle mutants. Fig. S4 shows correlation of the phenotypic changes in single mutant and bni1 or bim1 double mutant strains with other datasets. Table S1 lists the features measured. Table S2 shows strains and plasmids used in this study. Table S3 shows mean values of 90 features for single mutants. Table S4 shows mean values of 90 features for double deletion with bni1Δ. Table S5 shows mean values of 90 features for double deletion with bim1Δ. Table S6 shows p-values generated for single mutants and bni1Δ and bim1Δ double mutants on three spindle specific features. Table S7 lists essential genes analyzed for spindle defect by HCS. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200909013/DC1.
Acknowledgments
We thank Taras Makhenevych, Huiming Ding, Bilal Sheik, Grischa Chandy, Neal Gliksman, and Timothy Schwartz for technical help.
F.J. Vizeacoumar, C. Boone, and B.J. Andrews conceived and designed the experiments. F.J. Vizeacoumar and N. van Dyk performed the experiments. F.J. Vizeacoumar, F.S. Vizeacoumar, V. Cheung, and J. Li analyzed the data. Y. Sydorskyy generated Fig. 5 A. Z. Li, N. Case, F.S. Vizeacoumar, A. Datti, C. Nislow, B. Frey, B. Raught, Z. Zhang, and K. Bloom provided technical support/reagents/materials/analysis tools. F.J. Vizeacoumar, C. Boone, and B.J. Andrews wrote the paper.
This work was supported by a postdoctoral fellowship from the Best Foundation to F.J. Vizeacoumar and a grant from the Canadian Institutes of Health Research to B.J. Andrews and C. Boone (GSP-41567). Infrastructure for high content screening and SGA analysis in the Boone and Andrews laboratory was acquired with funds from the Canadian Foundation for Innovation and the Ontario Innovation Trust. Our SGA laboratories are supported by funds from Genome Canada through the Ontario Genomics Institute.
Footnotes
Abbreviations used in this paper:
- CPC
- chromosomal passenger complex
- FEAR
- fourteen early anaphase release
- HCS
- high-content screening
- MEN
- mitotic exit network
- SGA
- synthetic genetic array
- TAP
- tandem affinity purification
References
- Bandyopadhyay S., Kelley R., Krogan N.J., Ideker T. 2008. Functional maps of protein complexes from quantitative genetic interaction data. PLOS Comput. Biol. 4:e1000065 10.1371/journal.pcbi.1000065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop C. 2006. Pattern Recognition and Machine Learning. New York, Springer; 738 pp. [Google Scholar]
- Blower M.D., Nachury M., Heald R., Weis K. 2005. A Rae1-containing ribonucleoprotein complex is required for mitotic spindle assembly. Cell. 121:223–234 10.1016/j.cell.2005.02.016 [DOI] [PubMed] [Google Scholar]
- Boone C., Bussey H., Andrews B.J. 2007. Exploring genetic interactions and networks with yeast. Nat. Rev. Genet. 8:437–449 10.1038/nrg2085 [DOI] [PubMed] [Google Scholar]
- Buonomo S.B., Rabitsch K.P., Fuchs J., Gruber S., Sullivan M., Uhlmann F., Petronczki M., Tóth A., Nasmyth K. 2003. Division of the nucleolus and its release of CDC14 during anaphase of meiosis I depends on separase, SPO12, and SLK19. Dev. Cell. 4:727–739 10.1016/S1534-5807(03)00129-1 [DOI] [PubMed] [Google Scholar]
- Buvelot S., Tatsutani S.Y., Vermaak D., Biggins S. 2003. The budding yeast Ipl1/Aurora protein kinase regulates mitotic spindle disassembly. J. Cell Biol. 160:329–339 10.1083/jcb.200209018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers B., Goetsch L. 1975. Behavior of spindles and spindle plaques in the cell cycle and conjugation of Saccharomyces cerevisiae. J. Bacteriol. 124:511–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri A.R., Khan I.A., Prasad V., Robinson A.K., Ludueña R.F., Barnes L.D. 1999. The tumor suppressor protein Fhit. A novel interaction with tubulin. J. Biol. Chem. 274:24378–24382 10.1074/jbc.274.34.24378 [DOI] [PubMed] [Google Scholar]
- D’Amours D., Amon A. 2004. At the interface between signaling and executing anaphase—Cdc14 and the FEAR network. Genes Dev. 18:2581–2595 10.1101/gad.1247304 [DOI] [PubMed] [Google Scholar]
- Denison C., Rudner A.D., Gerber S.A., Bakalarski C.E., Moazed D., Gygi S.P. 2005. A proteomic strategy for gaining insights into protein sumoylation in yeast. Mol. Cell. Proteomics. 4:246–254 10.1074/mcp.M400154-MCP200 [DOI] [PubMed] [Google Scholar]
- Enyenihi A.H., Saunders W.S. 2003. Large-scale functional genomic analysis of sporulation and meiosis in Saccharomyces cerevisiae. Genetics. 163:47–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fesquet D., Fitzpatrick P.J., Johnson A.L., Kramer K.M., Toyn J.H., Johnston L.H. 1999. A Bub2p-dependent spindle checkpoint pathway regulates the Dbf2p kinase in budding yeast. EMBO J. 18:2424–2434 10.1093/emboj/18.9.2424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassmann R., Carvalho A., Henzing A.J., Ruchaud S., Hudson D.F., Honda R., Nigg E.A., Gerloff D.L., Earnshaw W.C. 2004. Borealin: a novel chromosomal passenger required for stability of the bipolar mitotic spindle. J. Cell Biol. 166:179–191 10.1083/jcb.200404001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geymonat M., Spanos A., Smith S.J., Wheatley E., Rittinger K., Johnston L.H., Sedgwick S.G. 2002. Control of mitotic exit in budding yeast. In vitro regulation of Tem1 GTPase by Bub2 and Bfa1. J. Biol. Chem. 277:28439–28445 10.1074/jbc.M202540200 [DOI] [PubMed] [Google Scholar]
- Giaever G., Chu A.M., Ni L., Connelly C., Riles L., Véronneau S., Dow S., Lucau-Danila A., Anderson K., André B., et al. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 418:387–391 10.1038/nature00935 [DOI] [PubMed] [Google Scholar]
- Graumann J., Dunipace L.A., Seol J.H., McDonald W.H., Yates J.R., III, Wold B.J., Deshaies R.J. 2004. Applicability of tandem affinity purification MudPIT to pathway proteomics in yeast. Mol. Cell. Proteomics. 3:226–237 10.1074/mcp.M300099-MCP200 [DOI] [PubMed] [Google Scholar]
- Gupta M.L., Jr., Carvalho P., Roof D.M., Pellman D. 2006. Plus end-specific depolymerase activity of Kip3, a kinesin-8 protein, explains its role in positioning the yeast mitotic spindle. Nat. Cell Biol. 8:913–923 10.1038/ncb1457 [DOI] [PubMed] [Google Scholar]
- Hannich J.T., Lewis A., Kroetz M.B., Li S.J., Heide H., Emili A., Hochstrasser M. 2005. Defining the SUMO-modified proteome by multiple approaches in Saccharomyces cerevisiae. J. Biol. Chem. 280:4102–4110 10.1074/jbc.M413209200 [DOI] [PubMed] [Google Scholar]
- Hess H., Clemmens J., Qin D., Howard J., Vogel V. 2001. Light-controlled molecular shuttles made from motor proteins carrying cargo on engineered surfaces. Nano Lett. 1:235–239 10.1021/nl015521e [DOI] [Google Scholar]
- Jensen S., Johnson A.L., Johnston L.H., Segal M. 2004. Temporal coupling of spindle disassembly and cytokinesis is disrupted by deletion of LTE1 in budding yeast. Cell Cycle. 3:817–822 [PubMed] [Google Scholar]
- Jeram S.M., Srikumar T., Pedrioli P.G., Raught B. 2009. Using mass spectrometry to identify ubiquitin and ubiquitin-like protein conjugation sites. Proteomics. 9:922–934 10.1002/pmic.200800666 [DOI] [PubMed] [Google Scholar]
- Johnston M. 1999. Feasting, fasting and fermenting. Glucose sensing in yeast and other cells. Trends Genet. 15:29–33 10.1016/S0168-9525(98)01637-0 [DOI] [PubMed] [Google Scholar]
- Jones H.D., Schliwa M., Drubin D.G. 1993. Video microscopy of organelle inheritance and motility in budding yeast. Cell Motil. Cytoskeleton. 25:129–142 10.1002/cm.970250203 [DOI] [PubMed] [Google Scholar]
- Kaufman L., Rousseeuw P.J. 1990. Finding groups in data: an introduction to cluster analysis. Wiley, New York: 342 pp. [Google Scholar]
- Khmelinskii A., Schiebel E. 2008. Assembling the spindle midzone in the right place at the right time. Cell Cycle. 7:283–286 [DOI] [PubMed] [Google Scholar]
- Khmelinskii A., Lawrence C., Roostalu J., Schiebel E. 2007. Cdc14-regulated midzone assembly controls anaphase B. J. Cell Biol. 177:981–993 10.1083/jcb.200702145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilmartin J.V., Adams A.E. 1984. Structural rearrangements of tubulin and actin during the cell cycle of the yeast Saccharomyces. J. Cell Biol. 98:922–933 10.1083/jcb.98.3.922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittler R., Putz G., Pelletier L., Poser I., Heninger A.K., Drechsel D., Fischer S., Konstantinova I., Habermann B., Grabner H., et al. 2004. An endoribonuclease-prepared siRNA screen in human cells identifies genes essential for cell division. Nature. 432:1036–1040 10.1038/nature03159 [DOI] [PubMed] [Google Scholar]
- Knockleby J., Vogel J. 2009. The COMA complex is required for Sli15/INCENP-mediated correction of defective kinetochore attachments. Cell Cycle. 8:2570–2577 [DOI] [PubMed] [Google Scholar]
- Komarnitsky S.I., Chiang Y.C., Luca F.C., Chen J., Toyn J.H., Winey M., Johnston L.H., Denis C.L. 1998. DBF2 protein kinase binds to and acts through the cell cycle-regulated MOB1 protein. Mol. Cell. Biol. 18:2100–2107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee L., Klee S.K., Evangelista M., Boone C., Pellman D. 1999. Control of mitotic spindle position by the Saccharomyces cerevisiae formin Bni1p. J. Cell Biol. 144:947–961 10.1083/jcb.144.5.947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makhnevych T., Sydorskyy Y., Xin X., Srikumar T., Vizeacoumar F.J., Jeram S.M., Li Z., Bahr S., Andrews B.J., Boone C., Raught B. 2009. Global map of SUMO function revealed by protein-protein interaction and genetic networks. Mol. Cell. 33:124–135 10.1016/j.molcel.2008.12.025 [DOI] [PubMed] [Google Scholar]
- Marston A.L., Lee B.H., Amon A. 2003. The Cdc14 phosphatase and the FEAR network control meiotic spindle disassembly and chromosome segregation. Dev. Cell. 4:711–726 10.1016/S1534-5807(03)00130-8 [DOI] [PubMed] [Google Scholar]
- Melchior F. 2000. SUMO—nonclassical ubiquitin. Annu. Rev. Cell Dev. Biol. 16:591–626 10.1146/annurev.cellbio.16.1.591 [DOI] [PubMed] [Google Scholar]
- Morishita T., Mitsuzawa H., Nakafuku M., Nakamura S., Hattori S., Anraku Y. 1995. Requirement of Saccharomyces cerevisiae Ras for completion of mitosis. Science. 270:1213–1215 10.1126/science.270.5239.1213 [DOI] [PubMed] [Google Scholar]
- Müller S., Hoege C., Pyrowolakis G., Jentsch S. 2001. SUMO, ubiquitin’s mysterious cousin. Nat. Rev. Mol. Cell Biol. 2:202–210 10.1038/35056591 [DOI] [PubMed] [Google Scholar]
- Ohya Y., Sese J., Yukawa M., Sano F., Nakatani Y., Saito T.L., Saka A., Fukuda T., Ishihara S., Oka S., et al. 2005. High-dimensional and large-scale phenotyping of yeast mutants. Proc. Natl. Acad. Sci. USA. 102:19015–19020 10.1073/pnas.0509436102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panse V.G., Hardeland U., Werner T., Kuster B., Hurt E. 2004. A proteome-wide approach identifies sumoylated substrate proteins in yeast. J. Biol. Chem. 279:41346–41351 10.1074/jbc.M407950200 [DOI] [PubMed] [Google Scholar]
- Pereira G., Schiebel E. 2003. Separase regulates INCENP-Aurora B anaphase spindle function through Cdc14. Science. 302:2120–2124 10.1126/science.1091936 [DOI] [PubMed] [Google Scholar]
- Pereira G., Höfken T., Grindlay J., Manson C., Schiebel E. 2000. The Bub2p spindle checkpoint links nuclear migration with mitotic exit. Mol. Cell. 6:1–10 10.1016/S1097-2765(00)00002-2 [DOI] [PubMed] [Google Scholar]
- Pereira G., Tanaka T.U., Nasmyth K., Schiebel E. 2001. Modes of spindle pole body inheritance and segregation of the Bfa1p-Bub2p checkpoint protein complex. EMBO J. 20:6359–6370 10.1093/emboj/20.22.6359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruyne D., Legesse-Miller A., Gao L., Dong Y., Bretscher A. 2004. Mechanisms of polarized growth and organelle segregation in yeast. Annu. Rev. Cell Dev. Biol. 20:559–591 10.1146/annurev.cellbio.20.010403.103108 [DOI] [PubMed] [Google Scholar]
- Ro H.S., Song S., Lee K.S. 2002. Bfa1 can regulate Tem1 function independently of Bub2 in the mitotic exit network of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 99:5436–5441 10.1073/pnas.062059999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M.D., Grigull J., Mohammad N., Hughes T.R. 2002. FunSpec: a web-based cluster interpreter for yeast. BMC Bioinformatics. 3:35 10.1186/1471-2105-3-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruchaud S., Carmena M., Earnshaw W.C. 2007. The chromosomal passenger complex: one for all and all for one. Cell. 131:230–231 10.1016/j.cell.2007.10.002 [DOI] [PubMed] [Google Scholar]
- Rutherford S.L., Lindquist S. 1998. Hsp90 as a capacitor for morphological evolution. Nature. 396:336–342 10.1038/24550 [DOI] [PubMed] [Google Scholar]
- Seeler J.S., Dejean A. 2003. Nuclear and unclear functions of SUMO. Nat. Rev. Mol. Cell Biol. 4:690–699 10.1038/nrm1200 [DOI] [PubMed] [Google Scholar]
- Sheff M.A., Thorn K.S. 2004. Optimized cassettes for fluorescent protein tagging in Saccharomyces cerevisiae. Yeast. 21:661–670 10.1002/yea.1130 [DOI] [PubMed] [Google Scholar]
- Sopko R., Huang D., Preston N., Chua G., Papp B., Kafadar K., Snyder M., Oliver S.G., Cyert M., Hughes T.R., et al. 2006. Mapping pathways and phenotypes by systematic gene overexpression. Mol. Cell. 21:319–330 10.1016/j.molcel.2005.12.011 [DOI] [PubMed] [Google Scholar]
- Stegmeier F., Amon A. 2004. Closing mitosis: the functions of the Cdc14 phosphatase and its regulation. Annu. Rev. Genet. 38:203–232 10.1146/annurev.genet.38.072902.093051 [DOI] [PubMed] [Google Scholar]
- Stegmeier F., Visintin R., Amon A. 2002. Separase, polo kinase, the kinetochore protein Slk19, and Spo12 function in a network that controls Cdc14 localization during early anaphase. Cell. 108:207–220 10.1016/S0092-8674(02)00618-9 [DOI] [PubMed] [Google Scholar]
- Stoepel J., Ottey M.A., Kurischko C., Hieter P., Luca F.C. 2005. The mitotic exit network Mob1p-Dbf2p kinase complex localizes to the nucleus and regulates passenger protein localization. Mol. Biol. Cell. 16:5465–5479 10.1091/mbc.E05-04-0337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan M., Uhlmann F. 2003. A non-proteolytic function of separase links the onset of anaphase to mitotic exit. Nat. Cell Biol. 5:249–254 10.1038/ncb940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan M., Lehane C., Uhlmann F. 2001. Orchestrating anaphase and mitotic exit: separase cleavage and localization of Slk19. Nat. Cell Biol. 3:771–777 10.1038/ncb0901-771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas S., Kaplan K.B. 2007. A Bir1p Sli15p kinetochore passenger complex regulates septin organization during anaphase. Mol. Biol. Cell. 18:3820–3834 10.1091/mbc.E07-03-0201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong A.H., Evangelista M., Parsons A.B., Xu H., Bader G.D., Pagé N., Robinson M., Raghibizadeh S., Hogue C.W., Bussey H., et al. 2001. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 294:2364–2368 10.1126/science.1065810 [DOI] [PubMed] [Google Scholar]
- Tong A.H., Lesage G., Bader G.D., Ding H., Xu H., Xin X., Young J., Berriz G.F., Brost R.L., Chang M., et al. 2004. Global mapping of the yeast genetic interaction network. Science. 303:808–813 10.1126/science.1091317 [DOI] [PubMed] [Google Scholar]
- Toyn J.H., Johnston L.H. 1994. The Dbf2 and Dbf20 protein kinases of budding yeast are activated after the metaphase to anaphase cell cycle transition. EMBO J. 13:1103–1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallee R.B., Williams J.C., Varma D., Barnhart L.E. 2004. Dynein: An ancient motor protein involved in multiple modes of transport. J. Neurobiol. 58:189–200 10.1002/neu.10314 [DOI] [PubMed] [Google Scholar]
- Visintin C., Tomson B.N., Rahal R., Paulson J., Cohen M., Taunton J., Amon A., Visintin R. 2008. APC/C-Cdh1-mediated degradation of the Polo kinase Cdc5 promotes the return of Cdc14 into the nucleolus. Genes Dev. 22:79–90 10.1101/gad.1601308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizeacoumar F.J., Chong Y., Boone C., Andrews B.J. 2009. A picture is worth a thousand words: genomics to phenomics in the yeast Saccharomyces cerevisiae. FEBS Lett. 583:1656–1661 10.1016/j.febslet.2009.03.068 [DOI] [PubMed] [Google Scholar]
- Wágner G., Kovács J., Löw P., Orosz F., Ovádi J. 2001. Tubulin and microtubule are potential targets for brain hexokinase binding. FEBS Lett. 509:81–84 10.1016/S0014-5793(01)03136-2 [DOI] [PubMed] [Google Scholar]
- Wohlschlegel J.A., Johnson E.S., Reed S.I., Yates J.R., III 2004. Global analysis of protein sumoylation in Saccharomyces cerevisiae. J. Biol. Chem. 279:45662–45668 10.1074/jbc.M409203200 [DOI] [PubMed] [Google Scholar]
- Wykoff D.D., O’Shea E.K. 2005. Identification of sumoylated proteins by systematic immunoprecipitation of the budding yeast proteome. Mol. Cell. Proteomics. 4:73–83 [DOI] [PubMed] [Google Scholar]
- Xu H., Boone C., Brown G.W. 2007. Genetic dissection of parallel sister-chromatid cohesion pathways. Genetics. 176:1417–1429 10.1534/genetics.107.072876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh E., Skibbens R.V., Cheng J.W., Salmon E.D., Bloom K. 1995. Spindle dynamics and cell cycle regulation of dynein in the budding yeast, Saccharomyces cerevisiae. J. Cell Biol. 130:687–700 10.1083/jcb.130.3.687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S., Ichihashi R., Toh-e A. 2003. Ras recruits mitotic exit regulator Lte1 to the bud cortex in budding yeast. J. Cell Biol. 161:889–897 10.1083/jcb.200301128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X., Kahana J.A., Silver P.A., Morphew M.K., McIntosh J.R., Fitch I.T., Carbon J., Saunders W.S. 1999. Slk19p is a centromere protein that functions to stabilize mitotic spindles. J. Cell Biol. 146:415–425 10.1083/jcb.146.2.415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Blobel G. 2005. A SUMO ligase is part of a nuclear multiprotein complex that affects DNA repair and chromosomal organization. Proc. Natl. Acad. Sci. USA. 102:4777–4782 10.1073/pnas.0500537102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W., Ryan J.J., Zhou H. 2004. Global analyses of sumoylated proteins in Saccharomyces cerevisiae. Induction of protein sumoylation by cellular stresses. J. Biol. Chem. 279:32262–32268 10.1074/jbc.M404173200 [DOI] [PMC free article] [PubMed] [Google Scholar]