An 18-residue motif in the cytoplasmic tail of polycystic kidney disease gene product, fibrocystin, targets it to ciliary membranes through interactions with Rab8.
Abstract
Sensory functions of primary cilia rely on ciliary-localized membrane proteins, but little is known about how these receptors are targeted to the cilium. To further our understanding of this process, we dissected the ciliary targeting sequence (CTS) of fibrocystin, the human autosomal recessive polycystic kidney disease gene product. We show that the fibrocystin CTS is an 18-residue motif localized in the cytoplasmic tail. This motif is sufficient to target green fluorescent protein (GFP) to cilia of ciliated cells and targets GFP to lipid rafts if the cells are not ciliated. Rab8, but not several other Rabs implicated in ciliary assembly, binds to the CTS in a coimmunoprecipitation assay. Dominant-negative Rab8 interacts more strongly than wild-type or constitutively active Rab8, and coexpression of this dominant-negative mutant Rab8 blocks trafficking to the cilium. This suggests that the CTS functions by binding regulatory proteins like Rab8 to control trafficking through the endomembrane system and on to the cilium.
Introduction
The primary cilium is a ubiquitous eukaryotic organelle that plays vital roles in the development of mammals and in the etiology of diseases such as polycystic kidney disease and blindness. It is thought that primary cilia function as cellular antennae to monitor the extracellular environment and report this information back to the cell. This small organelle is composed of hundreds of proteins assembled onto a microtubule-based cytoskeleton that projects from the surface of the cell and is surrounded by an extension of the plasma membrane. Although contiguous with the plasma membrane, the ciliary membrane is unique, as cells have the ability to localize receptors and other membrane proteins specifically to this domain. This polarized distribution of proteins is required for the cilium to carry out its sensory function, but little is known about how the cell achieves this distribution.
To learn more about the mechanism of ciliary targeting of membrane proteins, we characterized the ciliary targeting sequence (CTS) in fibrocystin. Fibrocystin is the gene product of the human autosomal recessive polycystic kidney disease gene, PKHD1 (Onuchic et al., 2002; Ward et al., 2002). Patients with defects in this gene develop severe cystic kidney disease along with defects in the lung, pancreas, and liver. Fibrocystin is a large (>4,000 residues), single-pass transmembrane protein that is predicted to be entirely extracellular except for a short ∼190 residue C-terminal tail. Fibrocystin has been localized to cilia and centrosomes in mammalian cells (Ward et al., 2003, 2006; Menezes et al., 2004; Wang et al., 2004; Zhang et al., 2004), and a Chlamydomonas reinhardtii homologue was found in cilia (Pazour et al., 2005).
Results and discussion
The cytoplasmic tail of fibrocystin contains a ciliary targeting signal
To understand how fibrocystin is targeted to cilia, we characterized its CTS. To date, CTSs have been identified in a small number of proteins, but comparison of these does not reveal common motifs. However, all are found in intracellular domains (Pazour and Bloodgood, 2008). Thus, we reasoned that even though fibrocystin is large, it is mostly extracellular with only a short cytoplasmic tail, and this is the likely position of its CTS. To test this idea, we made two constructs fusing the C-terminal end of fibrocystin to reporter proteins (Fig. S1 A). In the first (JAF16), we fused the C-terminal 503 residues of fibrocystin to a fragment of CD8. This construct contains the extracellular domain of CD8 fused to fibrocystin just before its membrane-spanning domain and is predicted to have the same membrane topology as native fibrocystin. CD8 is a well-characterized membrane protein often used in chimerics to identify targeting domains (Xia et al., 2001). In the second construct (JAF99), we fused the last 193 residues of fibrocystin to the C-terminal end of GFP. This construct lacks most of the predicted membrane-spanning residues but contains the entire cytoplasmic tail. After transfection into cells, both constructs can localize to cilia (Fig. 1 A, a; and Fig. S1 B). In addition to cilia, JAF16 also is found in the endoplasmic reticulum. In nonciliated cells, JAF16 remains in the endoplasmic reticulum, whereas JAF99 is found throughout the cell in small punctate spots (Fig. 1 A, a′; and Fig. S1). These results indicate that a CTS is located within the C-terminal 193 residues of fibrocystin.
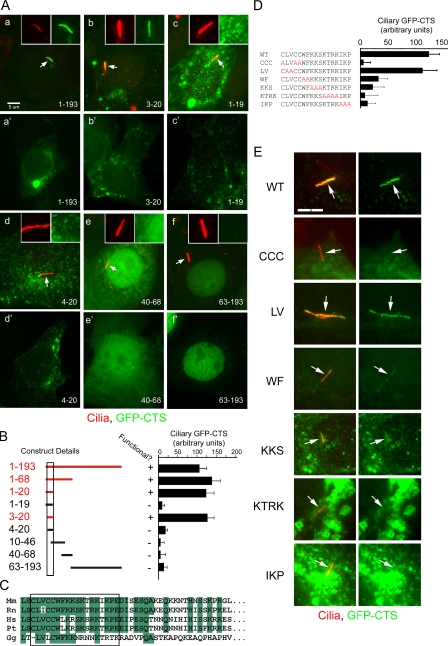
Figure 1.
Characterization of the CTS of fibrocystin. (A, a–f) Selected examples showing the distribution of subfragments of the cytoplasmic tail. Two different cells are shown for each construct. (a–f’) The first images (a–f) show a ciliated cell with the cilium marked with arrows, whereas the second images (a’–f’) show a nonciliated cell. Insets show the cilia (red) and GFP-CTS (green) channels alone. The amino acid fragments fused to GFP are listed at the bottom of each image and are shown graphically in B. (B) Graphical representation of the constructs and quantification of the ability of the constructs to function. The numbers on the left represent the amino acids included in the construct, and the box denotes the limits of the CTS. The graph shows the mean amount of GFP fluorescence per micrometer in cilia from 25 transfected cells. (C) Alignment of the CTS (box) and surrounding sequence of vertebrate fibrocystins (Mm, mouse; Rn, rat; Hs, human; Pt, chimp; Gg, chick). (D and E) Alanine-scanning mutagenesis of the CTS. (D) Sequence and quantification of the ability of the mutated CTSs to direct GFP to cilia. Quantification is described as in B. WT, wild type; CCC, LV, WF, KKS, KTRK, and IKP indicate which amino acids were mutated in each construct. Error bars indicate SEM. (E) Images illustrating the cellular distribution of constructs described in D. Arrows mark cilia. Bars, 5 µm.
To determine which part of the cytoplasmic tail is responsible for ciliary targeting, we constructed a series of GFP fusions containing smaller portions of the tail and quantitated their ability to localize to cilia (Fig. 1, A and B). This analysis indicated that 18 residues near the N-terminal end of the cytoplasmic tail were sufficient to target GFP to the cilium of ciliated cells or to punctate spots in nonciliated cells. The large size of fibrocystin prevented us from determining whether these residues are required for trafficking of native fibrocystin to cilia. Thus, it is currently unknown if this is the only CTS within the protein. If the GFP fusion construct did not contain the 18-residue CTS, the GFP was distributed throughout the cell or concentrated in the nucleus. We did not carefully demarcate the nuclear targeting sequence, but the fusion containing residues 63–193 was able to efficiently concentrate in the nucleus, whereas the 40–68 fusion could localize to the nucleus but was also found in the cytoplasm. Other work also mapped a nuclear targeting sequence to the region between residues 80 and 104 (Hiesberger et al., 2006).
Examination of the CTS in fibrocystins from other species indicates that it is highly conserved in mammals and moderately conserved in chicken (Fig. 1 C). The sequence is not conserved in the fibrocystin-related protein, fibrocystin-L from mammals, or other species. BLAST searches with the CTS did not identify any novel proteins containing similar sequences. The fibrocystin CTS does not contain a VxP motif that has been proposed to be a generic CTS (Deretic et al., 2005; Geng et al., 2006) nor does it contain an Ax[S/A]xQ motif identified in several G protein–coupled receptors (Berbari et al., 2008). To further our understanding of the CTS, we used alanine-scanning mutagenesis to mutate small blocks of residues (Fig. 1, D and E). Quantification of the effects of these mutations (Fig. 1 D) shows that most residues are important for function. The CCC and KTRK residues are most critical because mutating these to alanines almost completely blocks CTS function. At the other extreme, the LV mutation does little to the CTS function, whereas the other mutations reduce the ability to traffic to cilia but do not completely block function.
Although we did not detect any significant homology between the fibrocystin CTS and other nonfibrocystin sequences, we noted similarity between the CTS amino acid composition and a lipid raft targeting sequence in SNAP25 (Fig. 2 A; Salaün et al., 2005). This suggested that the punctuate spots to which the CTS localized might be lipid rafts. To test this, live CTS-GFP–expressing cells were labeled with fluorescent cholera toxin B (Fig. 2 B). Cholera toxin B binds GM1 gangliosides and is a marker for membrane domains enriched in these lipids. In nonciliated cells, there is strong colocalization between the CTS-GFP spots and the cholera toxin-binding sites (Fig. 2 B, a). The colocalization is also observed if the cholera toxin is cross-linked with an antibody and then fixed (Fig. 2 B, b). Cholera toxin B labels the cilium (Fig. 2 B, c), confirming a previous report that this organelle is enriched in GM1 gangliosides (Janich and Corbeil, 2007). SNAP25 has been reported to localize to cilia (Low et al., 1998), but the lipid raft targeting sequence of SNAP25 was not sufficient to target GFP to cilia (Fig. 2 B, d), indicating that a lipid raft targeting sequence alone is not sufficient for ciliary targeting.
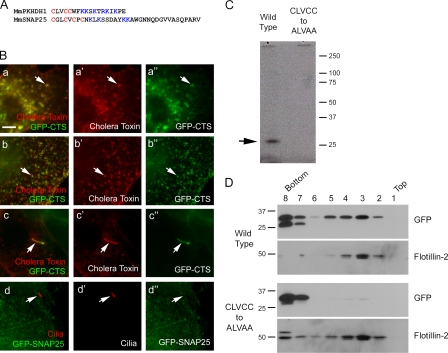
Figure 2.
The CTS is associated with lipid. (A) Comparison of the CTS from fibrocystin to the lipid raft targeting sequence of SNAP25 (Gonzalo et al., 1999). (B) Colocalization of GFP-CTS with lipid rafts. (a–a″) Live cells stained with Alexa Fluor 594–conjugated cholera toxin. In nonciliated cells, cholera toxin shows extensive colocalization with the GFP-CTS in the cell body (the arrows point at one example). (b–b″) Cholera toxin was cross-linked by antibody before fixation, which caused the toxin and GFP to cluster (arrows). (c–c″) In ciliated cells, GFP and cholera toxin colocalize in the cilium (arrows). (d–d″) The lipid raft targeting sequence of Snap25 does not target GFP to the cilium (arrows). Bar, 5 µm. (C) Tritiated palmitate is incorporated into the wild-type but not the cysteine-mutated CTS (arrow). (D) The CTS cysteines mediate interaction with membranes. Cells expressing either wild-type or cysteine-mutated CTS-GFP were lysed and fractionated by an OptiPrep gradient.
Cysteine residues near blocks of basic amino acids are often palmitoylated (Bijlmakers and Marsh, 2003). The fibrocystin CTS contains three cysteine residues followed by a block of basic residues (Fig. 2 A), and mutational analysis indicated that these cysteines are critical to CTS function (Fig. 1 D). To test whether the CTS cysteines are palmitoylated, we grew cells expressing either the wild-type or CCC-mutated GFP-CTS in radioactive palmitate, immunoprecipitated the GFP-CTS, and looked for the incorporation of isotope. The wild-type protein but not the CCC-mutated version incorporated radioactive palmitate (Fig. 2 C). This indicates that the CTS includes a palmitoylation motif, and because the mutation of the cysteines blocks function, this suggests that palmitoylation is important for targeting this protein to the cilia. Acylations like palmitoylation and myristoylation are common modifications of ciliary membrane proteins. The opsin photoreceptor contains two cysteine residues that are palmitoylated and needed for proper targeting to the cilium (Tam et al., 2000). A Trypanosoma ciliary calcium-binding protein contains a palmitoylated cysteine and a myristoylated glycine (Godsel and Engman, 1999) that are needed for targeting to the cilium.
It is likely that the palmitoylated cysteines of fibrocystin serve to link the GFP-CTS to lipid membranes, and this is responsible for the puncta that are observed in nonciliated cells. The fact that the GFP-CTS is evenly distributed in the cells when the cysteines are mutated (Fig. 1 E) supports this idea, but we tested this more directly by floatation analysis. Membranes from cells expressing either wild-type or the CCC-mutated GFP-CTS were loaded on the bottom of an OptiPrep gradient and centrifuged. Membranes floated up and carried along the lipid raft marker flotillin-2. The wild-type GFP-CTS was also carried up, whereas the CCC-mutated protein remained at the bottom where it was loaded (Fig. 2 D). This indicates that the palmitoylated cysteines link the GFP-CTS to the membrane. Thus, even though the protein no longer contains a transmembrane domain, it remains associated with the membrane.
To understand the cellular compartment to which the GFP-CTS localized, we labeled cells expressing the GFP-CTS with a variety of compartmental markers (Fig. S2). No significant colocalization was seen with Golgi, lysosome, and most endosome markers, but colocalization was seen with markers for the recycling endosome. This suggests that the palmitoylated GFP-CTS has affinity for the membranes of this compartment, but whether the native protein is trafficked through the recycling endosome remains to be determined.
Trafficking of the CTS is regulated by Rab8
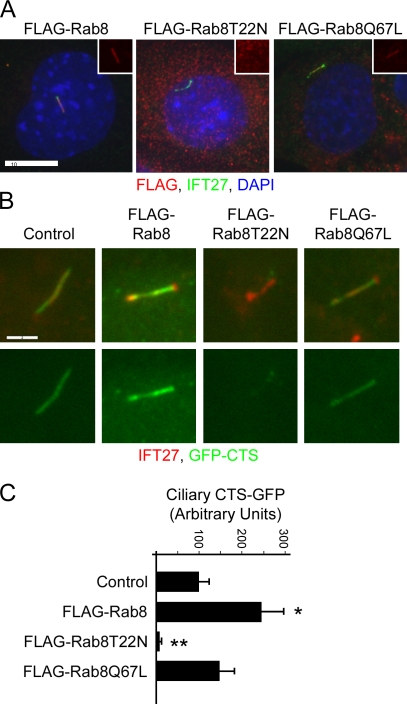
Work in frog retina indicates that Rab8 plays a key role in trafficking of opsin to the outer segment (Moritz et al., 2001), and recent work in mammalian cell culture has indicated that Rab8 and other Rab family proteins are critical for ciliary assembly (Nachury et al., 2007; Yoshimura et al., 2007). To test whether Rab8 plays a role in the trafficking of the fibrocystin CTS, we generated cell lines expressing Flag-tagged wild-type Rab8, dominant-negative Rab8T22N, and constitutively active Rab8Q67L (Fig. 3). These mutations are often used to perturb the GTP/GDP cycle of small G proteins. The T22N mutation is thought to keep the protein in the GDP-bound state, which binds guanine exchange factors to inhibit their activity on native substrates. The Q67L mutation reduces GTP hydrolysis, keeping the protein in the GTP-bound state (Feig, 1999). As previously reported (Nachury et al., 2007), wild-type Flag-Rab8 and Flag-Rab8Q67L localized to cilia, whereas Flag-Rab8T22N did not localize to the cilia that formed on these cells (Fig. 3 A). Cells expressing Flag-tagged wild-type Rab8 and Rab8Q67L ciliated fairly well, but cells expressing Rab8T22N did not ciliate nearly as well (wild type, 68 ± 12%; T22N, 2.7 ± 0.6%; Q67L, 46 ± 3.5%). The fibrocystin GFP-CTS did not traffic to the cilia that formed on the Flag-Rab8T22N cells (Fig. 3, B and C) but was trafficked to cilia on cells expressing Flag-Rab8 and Flag-Rab8Q67L. Interestingly, the amount of the GFP-CTS trafficked to cilia was higher in cells that expressed the wild-type Flag-Rab8 than in cells that were not transfected or in cells expressing either of the mutant forms. This suggests that Rab8 is a limiting factor in the amount of GFP-CTS that can be trafficked to cilia. The observation that the mutant forms either do not show the enhancement (Q67N) or reduce the amount of trafficking (T22N) suggests that Rab8 needs the normal GTP/GDP cycle to function properly.
Figure 3.
Effect of Rab8 on the trafficking of the fibrocystin CTS. (A) Flag-tagged Rab8 and Rab8Q67L are localized to cilia of IMCD3 cells, whereas Rab8T22N is not. Insets show the red (Flag) channels of the cilia. Bar, 10 µm. (B) The fibrocystin CTS targets GFP to cilia in control cells and cells expressing wild-type Flag-Rab8 and Flag-Rab8Q67L but not in cells expressing Flag-Rab8T22N. Bar, 2 µm. (C) Quantification of ciliary GFP-CTS fluorescence. Quantification was performed as described in Fig. 1. Error bars indicate SEM. Significance is shown as compared with control: *, P < 0.02; **, P < 0.001.
The CTS interacts with Rab8
Because Rab8 appeared to regulate the trafficking of the fibrocystin CTS, we sought to understand how this might be functioning and asked whether Rab8 or other Rabs could be physically connected to the CTS (Fig. 4). To do this, we coexpressed the cytoplasmic tail of fibrocystin with a series of Rab proteins (Rab6, Rab8, Rab11, Rab17, and Rab23) that have been implicated in trafficking of ciliary membrane proteins (Deretic and Papermaster, 1993; Moritz et al., 2001; Yoshimura et al., 2007). We also included IFT20, which is not a Rab protein, but is an intraflagellar transport subunit that we have implicated in trafficking proteins to the ciliary membrane (Follit et al., 2006, 2008). None of these proteins were completely colocalized with the GFP-CTS in nonciliated cells. However, Rab8 showed some colocalization, whereas Rab11 and Rab17 showed the most colocalization (Fig. S3). The colocalization with Rab11 and Rab17 is consistent with the results of the compartmental analysis (Fig. S2), as these two G proteins localize to the recycling endosome (Zerial and McBride, 2001). Rab8 was the only one of these Rabs to localize to cilia (Fig. 3 B and not depicted).
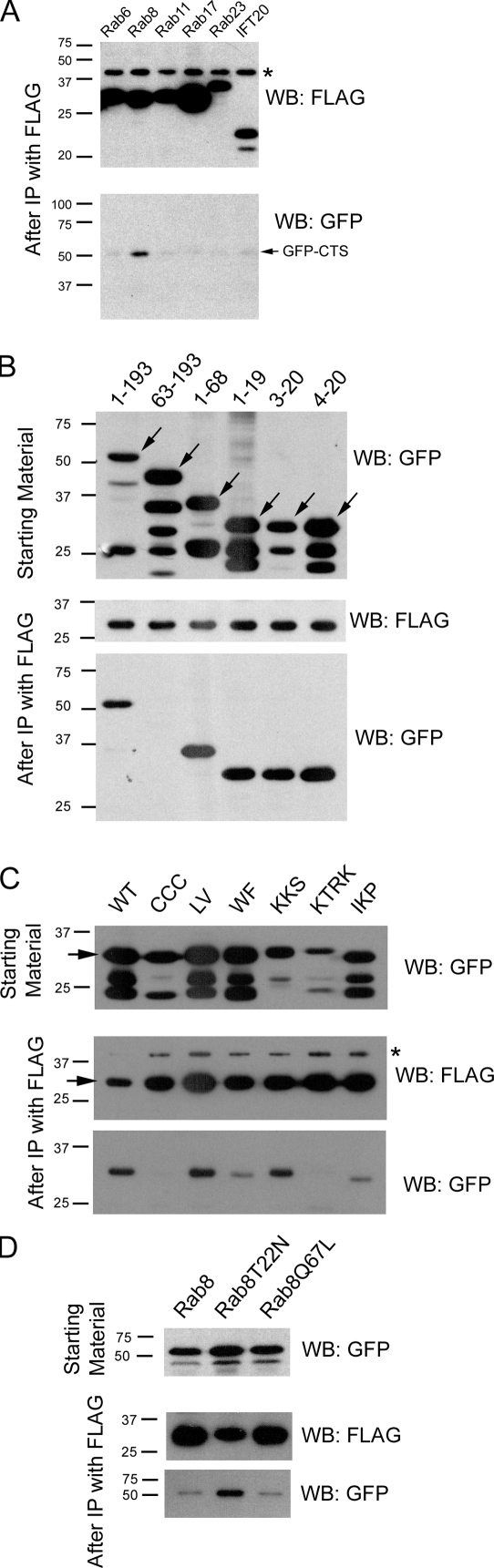
Figure 4.
Rab8 interacts with the fibrocystin CTS. (A) Flag-tagged Rab proteins were coexpressed with the GFP-CTS (1–193) and precipitated. The precipitates were analyzed by Western blotting with Flag and GFP antibodies. Positive interactions have a band in the GFP Western blot (WB). (B) The deletion constructs (Fig. 1 B) were tested for the ability to interact with Rab8. (C) The alanine scan mutations (Fig. 1 D) were examined for their ability to interact with Rab8. WT, wild type; CCC, LV, WF, KKS, KTRK, and IKP indicate which amino acids were mutated in each construct. (D) The ability of Rab8T22N and Rab8Q67L mutants to bind the GFP-CTS were compared with wild-type Rab8 in an analogous assay. Arrows mark the predicted size of the full-length proteins. IP, immunoprecipitation; *, nonspecific band precipitated by the Flag antibody.
The ability of these proteins to interact with the GFP-CTS was tested by a coimmunoprecipitation assay. In this assay, the GFP-CTS was coexpressed with each of the Flag-tagged Rabs in mouse kidney cells. The Rabs were precipitated via the Flag tag, and the precipitates were probed for the GFP-CTS. No interaction was seen between the GFP-CTS and Rab6, Rab11, Rab17, Rab23, or IFT20, but a significant amount of the GFP-CTS was precipitated by Rab8 (Fig. 4 A). This analysis was performed with the entire cytoplasmic tail, so we asked whether the Rab8-binding site overlapped the essential 18-residue CTS motif within the tail. To do this, we tested selected deletion constructs described in Fig. 1 B. The Rab8-binding site is located within the 18-residue minimal CTS, as fragments that contain this sequence are coprecipitated (Fig. 4 B, lanes 1, 3, and 5), whereas the C-terminal 130 residue (Fig. 4 B, lane 2), which does not target to cilia (Fig. 1, A and B), is not precipitated. The CTS is not simply a Rab8-binding site, as deletions of single residues from either end of the minimal CTS blocked the ability of the peptide to direct GFP to the cilium (Fig. 1, A and B) but did not block the binding to Rab8 (Fig. 4 B, lanes 4 and 6).
We next examined the effect of the alanine-scanning mutations on the ability of the CTS to be coprecipitated by Rab8. Interestingly, there is a good correlation between the function of the CTS to traffic to the cilium (Fig. 1 D) and its ability to bind to Rab8 (Fig. 4 C). For example, the LV mutation does little to block function of the CTS, and this mutated protein binds Rab8. However, the CCC and KTRK mutations are most disruptive to the CTS and most significantly decrease binding to Rab8. Other mutations had intermediate effects on the targeting ability and have intermediate effects on the ability to bind Rab8. The KKS mutation is an exception, as this is fairly detrimental to CTS function but still binds to Rab8. This suggests that additional proteins may bind the CTS and require these residues for activity.
To begin to understand whether the interactions between the CTS and Rab8 may be regulated by the GTP/GDP state of Rab8, we compared the ability of the constitutively active and dominant-negative mutations of Rab8 to bind to the CTS (Fig. 4 D). Interestingly, the dominant-negative form bound more CTS than either the wild-type protein or the constitutively active form. This suggests that exchange of a GDP with a GTP would release Rab8 from the CTS. This behavior is different from classical Rab effectors, which bind more strongly when bound to GTP (Zerial and McBride, 2001) but is similar to what was previously observed in the interaction between Rab5 and the angiotensin receptor (Seachrist et al., 2002). In the case of the angiotensin receptor, it was proposed that the receptor anchors Rab5-GDP on the surface of the carrier vesicle so that once GTP is exchanged for GDP, efficient vesicle fusion could occur. Similarly, the fibrocystin CTS may bind Rab8-GDP to increase its local concentration to allow for efficient execution of the next step in transport when GTP exchange occurs. Based on work in frog photoreceptors, it is likely that the regulated step is the fusion of carrier vesicles at the base of the cilium, as expression of dominant-negative Rab8 mutants caused small vesicles to accumulate around the base of photoreceptors (Moritz et al., 2001). This process may be regulated by the BBSome, as the guanine exchange factor Rabin8 is associated with the BBSome, and this protein would be expected to exchange GDP with a GTP on Rab8 (Nachury et al., 2007).
Comparison of ciliary targeting to apical and basolateral targeting
Compared with trafficking to the ciliary membrane, much more is known about trafficking to apical and basal-lateral membranes (Rodriguez-Boulan et al., 2005). Basolateral targeting sequences consist of short motifs that interact with the adaptor protein complexes that bind to clathrin coats as part of the sorting mechanism. Apical targeting motifs are much more diverse and do not share significant sequence homology with each other but have been proposed to function by directing proteins to lipid rafts that are preferentially sorted to the apical membrane (van Meer and Simons, 1988). Ciliary targeting is similar to apical targeting in that no sequence similarity is seen between the known CTSs (Pazour and Bloodgood, 2008), and at least for fibrocystin and the Trypanosoma calcium-binding protein (Tyler et al., 2009), association with a lipid raft appears to be required for proper targeting. Thus, it appears that ciliary targeting is related to apical targeting but has additional components to direct the proteins into the ciliary membrane. It is interesting to note that on vertebrate epithelial cells, the ciliary membrane is a subdomain of the apical membrane, so perhaps it is logical that trafficking to these two domains share similar features.
Materials and methods
Plasmids
The cytoplasmic tail of mouse fibrocystin (GenBank/EMBL/DDBJ accession no. NM_153179.2) was amplified from mouse kidney cDNA using 5′-GGGAATTCGCTTTGACTGTGACATTTTCAGTCCTAG-3′ and 5′-GGGAATTCTTACTGGATGGTTTCTGGTGG-3′ and fused to the CD8 open reading frame to create JAF16. The CD8 open reading frame was amplified from pCMS-CD8-NR1C (provided by H. Xia, Stanford University, Palo Alto, CA; Xia et al., 2001). JAF16 contains the last 503 residues of fibrocystin fused in frame to the CD8 extracellular domain. The fusion junction reads LDFACDefALTVTF, with the first six residues being derived from CD8, the last six from MmPKHD1, and the two in the middle from the restriction enzyme site. Likewise, mouse kidney cDNA was amplified with 5′-GGGAATTCCTGAGCTGTCTCGTTTGCTG-3′ and 5′-GGGAATTCTTACTGGATGGTTTCTGGTGG-3′ and cloned into the EcoRI site of pEGFP-C2 (BD) to create pJAF99. This plasmid contains the last 193 amino acids of fibrocystin fused in frame to the C-terminal end of GFP. Deletion constructs were generated by similarly amplifying smaller fragments and cloning them into this vector. Point mutations were generated by inverse PCR. All plasmids were confirmed by DNA sequencing. Rab8, Rab8T22N, and Rab8Q67L were cloned into p3×FlagMyc-CMV26 (Sigma-Aldrich) for expression in mammalian cells.
Miscellaneous methods
All cell culture work used mouse kidney IMCD3 (Rauchman et al., 1993) cells cultured in 47.5% DME and 47.5% F12 supplemented with 5% FBS and penicillin/streptomycin (Cellgro) at 37°C in 5% CO2. For transfection, cells were electroporated using a Gene Pulser Xcell (200 V; 50-ms pulse; 4-mm cuvette; Bio-Rad Laboratories). Stable cell lines were selected with 400 µg/ml G418 (Sigma-Aldrich). Clonal lines were isolated by dilution cloning after drug selection.
Percent ciliation was determined by counting cilia on cells stained with an IFT27 antibody after being grown for 48 h in low serum (0.25%). Results reported are the percentage of cilia ± standard deviation from three independent experiments in which >500 cells were counted.
Endocytosis assays were performed on IMCD3 cells electroporated with JAF99 48 h after electroporation. Cells were washed with KRH (125 mM NaCl, 5 mM KCl, 1.3 mM CaCl2, 1.2 mM MgSO4, 25 mM Hepes, pH 7.4, 2 mM sodium pyruvate, and 0.5% bovine serum albumin), incubated with Alexa Fluor 568–labeled EGF or transferrin for 5 min at 37°C, washed with KRH, and incubated at 37°C in KRH (Leonard et al., 2008). Coverslips were periodically removed and fixed during the chase period.
For visualization of lipid domains, live cells grown in glass-bottom dishes (MatTec) were incubated for 10 min at 37°C with Alexa Fluor 594–conjugated cholera toxin B (Invitrogen). Excess toxin was washed out, and the cells were imaged at 37°C in medium lacking Phenol red. Alternatively, cells were incubated for 10 min at 4°C with the cholera toxin followed by 15 additional min at 4°C with cholera toxin antibody before fixing and staining, following the manufacturer’s specifications (Vybrant Lipid Raft Labeling kit; Invitrogen).
Lipids were floated using the protocol of Macdonald and Pike (2005). In brief, cells were lysed by passing them through a needle, the postnuclear supernatant was mixed with OptiPrep (Sigma-Aldrich) to a final concentration of 25%, placed at the bottom of a centrifuge tube, and a gradient of 0–20% OptiPrep was layered on top. The gradient was centrifuged at 52,000 g for 90 min, and fractions were collected and analyzed by Western blotting.
To determine whether the CTS was palmitoylated, IMCD3 cells were electroporated with the wild-type (1–22) and equivalent cysteine-mutated GFP-CTS (1–22CCC) constructs. After 24 h, the medium was changed to one containing dialyzed serum (Invitrogen) supplemented with 0.25 mCi tritiated palmitate (PerkinElmer). After 16 h, cells were lysed, the GFP-CTS was precipitated with the JL8 GFP antibody (BD), and the eluates were separated by SDS-PAGE. The gel was fixed in 2:9:9 acetic acid/methanol/water for 1 h followed by 1 M sodium salicylate for 30 min, dried, and exposed to film.
Immunoprecipitations were performed using anti-Flag resin. To do this, cells expressing the tagged constructs were rinsed with cold PBS lysed in cell lytic M + 0.1% NP-40 (Sigma-Aldrich) and 0.1% CHAPSO (Bio-Rad Laboratories) with Complete Protease Inhibitor (Roche) at 4°C and clarified by centrifugation (18,000 g for 10 min). Clarified lysates were incubated with agarose beads coupled with Flag M2 antibody (Sigma-Aldrich) for 1 h. Flag beads were washed three times with wash buffer (50 mM Tris and 150 mM NaCl, pH 7.4) plus 1% NP-40. Bound Flag proteins were eluted with 200 µg/ml 3× Flag peptide (Sigma-Aldrich). Purified proteins were separated by SDS-PAGE and electrophoretically transferred to Immobilon-P (Millipore). After transfer, the membranes were incubated with antibodies to GFP (JL8; BD) and Flag (F1804; Sigma-Aldrich) followed by an HRP-conjugated anti–mouse IgG antibody (Thermo Fisher Scientific). The HRP conjugates were detected on film (BioMax XAR; Kodak) after LumiGLO (KPL) treatment.
Immunofluorescence microscopy
Cells were transfected by electroporation and seeded on coverslips. After 24 h, serum was reduced to 0.25% to promote ciliation, and the cells were grown for an additional 24–96 h before being fixed with paraformaldehyde and stained with primary antibodies as described previously (Follit et al., 2006). Primary antibodies were detected by Alexa Fluor 350–, 488–, and 594–labeled secondary antibodies (Invitrogen). Primary antibodies used included anti-acetylated tubulin (611β1; Sigma-Aldrich), anti-Flag (Sigma-Aldrich), anti-CD8 (Invitrogen), EEA1 (provided by S. Corvera, University of Massachusetts Medical School, Worcester, MA), giantin (provided by M. Fritzler, University of Calgary, Calgary, Alberta, Canada), golgin97 (CDF4; Invitrogen), flotillin-2 (BD), and mouse IFT20 and IFT27 (Follit et al., 2006).
Wide-field images were acquired by a camera (Orca ER; Hamamatsu Photonics) on a microscope (Axiovert 200M; Carl Zeiss, Inc.) equipped with a 100× Plan Apochromat 1.4 NA objective (Carl Zeiss, Inc.). If comparisons are to be made between images, the photos were taken with identical conditions and manipulated equally. For the quantification of GFP in the cilia, the length, area, and mean fluorescence intensity of the cilia were measured using the measurement tools of Openlab (PerkinElmer). Numbers reported are fluorescence intensity per micrometer of length.
Online supplemental material
Fig. S1 documents the initial pair of constructs used to identify the fibrocystin CTS. Fig. S2 examines the cellular compartment to which the GFP-CTS localizes in nonciliated cells. Fig. S3 documents the cellular distribution of the Flag-tagged proteins examined in Fig. 4. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200910096/DC1.
Acknowledgments
We thank Drs. N. Kennedy for assistance with the palmitoylation assay, D. Navaroli for assistance with the endocytosis assays, J. Jonassen for statistical analysis, and H. Xia, M. Fritzler, and S. Corvera for reagents.
This work was supported by grants from the National Institutes of Health (GM060992), the Worcester Foundation for Biomedical Research, and the Polycystic Kidney Foundation.
Footnotes
Abbreviations used in this paper:
- CTS
- ciliary targeting sequence
References
- Berbari N.F., Johnson A.D., Lewis J.S., Askwith C.C., Mykytyn K. 2008. Identification of ciliary localization sequences within the third intracellular loop of G protein-coupled receptors. Mol. Biol. Cell. 19:1540–1547 10.1091/mbc.E07-09-0942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bijlmakers M.J., Marsh M. 2003. The on-off story of protein palmitoylation. Trends Cell Biol. 13:32–42 10.1016/S0962-8924(02)00008-9 [DOI] [PubMed] [Google Scholar]
- Deretic D., Papermaster D.S. 1993. Rab6 is associated with a compartment that transports rhodopsin from the trans-Golgi to the site of rod outer segment disk formation in frog retinal photoreceptors. J. Cell Sci. 106:803–813 [DOI] [PubMed] [Google Scholar]
- Deretic D., Williams A.H., Ransom N., Morel V., Hargrave P.A., Arendt A. 2005. Rhodopsin C terminus, the site of mutations causing retinal disease, regulates trafficking by binding to ADP-ribosylation factor 4 (ARF4). Proc. Natl. Acad. Sci. USA. 102:3301–3306 10.1073/pnas.0500095102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feig L.A. 1999. Tools of the trade: use of dominant-inhibitory mutants of Ras-family GTPases. Nat. Cell Biol. 1:E25–E27 10.1038/10018 [DOI] [PubMed] [Google Scholar]
- Follit J.A., Tuft R.A., Fogarty K.E., Pazour G.J. 2006. The intraflagellar transport protein IFT20 is associated with the Golgi complex and is required for cilia assembly. Mol. Biol. Cell. 17:3781–3792 10.1091/mbc.E06-02-0133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Follit J.A., San Agustin J.T., Xu F., Jonassen J.A., Samtani R., Lo C.W., Pazour G.J. 2008. The Golgin GMAP210/TRIP11 anchors IFT20 to the Golgi complex. PLoS Genet. 4:e1000315 10.1371/journal.pgen.1000315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng L., Okuhara D., Yu Z., Tian X., Cai Y., Shibazaki S., Somlo S. 2006. Polycystin-2 traffics to cilia independently of polycystin-1 by using an N-terminal RVxP motif. J. Cell Sci. 119:1383–1395 10.1242/jcs.02818 [DOI] [PubMed] [Google Scholar]
- Godsel L.M., Engman D.M. 1999. Flagellar protein localization mediated by a calcium-myristoyl/palmitoyl switch mechanism. EMBO J. 18:2057–2065 10.1093/emboj/18.8.2057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalo S., Greentree W.K., Linder M.E. 1999. SNAP-25 is targeted to the plasma membrane through a novel membrane-binding domain. J. Biol. Chem. 274:21313–21318 10.1074/jbc.274.30.21313 [DOI] [PubMed] [Google Scholar]
- Hiesberger T., Gourley E., Erickson A., Koulen P., Ward C.J., Masyuk T.V., Larusso N.F., Harris P.C., Igarashi P. 2006. Proteolytic cleavage and nuclear translocation of fibrocystin is regulated by intracellular Ca2+ and activation of protein kinase C. J. Biol. Chem. 281:34357–34364 10.1074/jbc.M606740200 [DOI] [PubMed] [Google Scholar]
- Janich P., Corbeil D. 2007. GM1 and GM3 gangliosides highlight distinct lipid microdomains within the apical domain of epithelial cells. FEBS Lett. 581:1783–1787 10.1016/j.febslet.2007.03.065 [DOI] [PubMed] [Google Scholar]
- Leonard D., Hayakawa A., Lawe D., Lambright D., Bellve K.D., Standley C., Lifshitz L.M., Fogarty K.E., Corvera S. 2008. Sorting of EGF and transferrin at the plasma membrane and by cargo-specific signaling to EEA1-enriched endosomes. J. Cell Sci. 121:3445–3458 10.1242/jcs.031484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low S.H., Roche P.A., Anderson H.A., van Ijzendoorn S.C., Zhang M., Mostov K.E., Weimbs T. 1998. Targeting of SNAP-23 and SNAP-25 in polarized epithelial cells. J. Biol. Chem. 273:3422–3430 10.1074/jbc.273.6.3422 [DOI] [PubMed] [Google Scholar]
- Macdonald J.L., Pike L.J. 2005. A simplified method for the preparation of detergent-free lipid rafts. J. Lipid Res. 46:1061–1067 10.1194/jlr.D400041-JLR200 [DOI] [PubMed] [Google Scholar]
- Menezes L.F., Cai Y., Nagasawa Y., Silva A.M., Watkins M.L., Da Silva A.M., Somlo S., Guay-Woodford L.M., Germino G.G., Onuchic L.F. 2004. Polyductin, the PKHD1 gene product, comprises isoforms expressed in plasma membrane, primary cilium, and cytoplasm. Kidney Int. 66:1345–1355 10.1111/j.1523-1755.2004.00844.x [DOI] [PubMed] [Google Scholar]
- Moritz O.L., Tam B.M., Hurd L.L., Peränen J., Deretic D., Papermaster D.S. 2001. Mutant rab8 Impairs docking and fusion of rhodopsin-bearing post-Golgi membranes and causes cell death of transgenic Xenopus rods. Mol. Biol. Cell. 12:2341–2351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachury M.V., Loktev A.V., Zhang Q., Westlake C.J., Peränen J., Merdes A., Slusarski D.C., Scheller R.H., Bazan J.F., Sheffield V.C., Jackson P.K. 2007. A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell. 129:1201–1213 10.1016/j.cell.2007.03.053 [DOI] [PubMed] [Google Scholar]
- Onuchic L.F., Furu L., Nagasawa Y., Hou X., Eggermann T., Ren Z., Bergmann C., Senderek J., Esquivel E., Zeltner R., et al. 2002. PKHD1, the polycystic kidney and hepatic disease 1 gene, encodes a novel large protein containing multiple immunoglobulin-like plexin-transcription-factor domains and parallel beta-helix 1 repeats. Am. J. Hum. Genet. 70:1305–1317 10.1086/340448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour G.J., Bloodgood R.A. 2008. Targeting proteins to the ciliary membrane. Curr. Top. Dev. Biol. 85:115–149 10.1016/S0070-2153(08)00805-3 [DOI] [PubMed] [Google Scholar]
- Pazour G.J., Agrin N., Leszyk J., Witman G.B. 2005. Proteomic analysis of a eukaryotic cilium. J. Cell Biol. 170:103–113 10.1083/jcb.200504008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauchman M.I., Nigam S.K., Delpire E., Gullans S.R. 1993. An osmotically tolerant inner medullary collecting duct cell line from an SV40 transgenic mouse. Am. J. Physiol. 265:F416–F424 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Boulan E., Kreitzer G., Müsch A. 2005. Organization of vesicular trafficking in epithelia. Nat. Rev. Mol. Cell Biol. 6:233–247 10.1038/nrm1593 [DOI] [PubMed] [Google Scholar]
- Salaün C., Gould G.W., Chamberlain L.H. 2005. The SNARE proteins SNAP-25 and SNAP-23 display different affinities for lipid rafts in PC12 cells. Regulation by distinct cysteine-rich domains. J. Biol. Chem. 280:1236–1240 10.1074/jbc.M410674200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seachrist J.L., Laporte S.A., Dale L.B., Babwah A.V., Caron M.G., Anborgh P.H., Ferguson S.S. 2002. Rab5 association with the angiotensin II type 1A receptor promotes Rab5 GTP binding and vesicular fusion. J. Biol. Chem. 277:679–685 10.1074/jbc.M109022200 [DOI] [PubMed] [Google Scholar]
- Tam B.M., Moritz O.L., Hurd L.B., Papermaster D.S. 2000. Identification of an outer segment targeting signal in the COOH terminus of rhodopsin using transgenic Xenopus laevis. J. Cell Biol. 151:1369–1380 10.1083/jcb.151.7.1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler K.M., Fridberg A., Toriello K.M., Olson C.L., Cieslak J.A., Hazlett T.L., Engman D.M. 2009. Flagellar membrane localization via association with lipid rafts. J. Cell Sci. 122:859–866 10.1242/jcs.037721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Meer G., Simons K. 1988. Lipid polarity and sorting in epithelial cells. J. Cell. Biochem. 36:51–58 10.1002/jcb.240360106 [DOI] [PubMed] [Google Scholar]
- Wang S., Luo Y., Wilson P.D., Witman G.B., Zhou J. 2004. The autosomal recessive polycystic kidney disease protein is localized to primary cilia, with concentration in the basal body area. J. Am. Soc. Nephrol. 15:592–602 10.1097/01.ASN.0000113793.12558.1D [DOI] [PubMed] [Google Scholar]
- Ward C.J., Hogan M.C., Rossetti S., Walker D., Sneddon T., Wang X., Kubly V., Cunningham J.M., Bacallao R., Ishibashi M., et al. 2002. The gene mutated in autosomal recessive polycystic kidney disease encodes a large, receptor-like protein. Nat. Genet. 30:259–269 10.1038/ng833 [DOI] [PubMed] [Google Scholar]
- Ward C.J., Yuan D., Masyuk T.V., Wang X., Punyashthiti R., Whelan S., Bacallao R., Torra R., LaRusso N.F., Torres V.E., Harris P.C. 2003. Cellular and subcellular localization of the ARPKD protein; fibrocystin is expressed on primary cilia. Hum. Mol. Genet. 12:2703–2710 10.1093/hmg/ddg274 [DOI] [PubMed] [Google Scholar]
- Wu Y., Dai X.Q., Li Q., Chen C.X., Mai W., Hussain Z., Long W., Montalbetti N., Li G., Glynne R., et al. 2006. Kinesin-2 mediates physical and functional interactions between polycystin-2 and fibrocystin. Hum. Mol. Genet. 15:3280–3292 10.1093/hmg/ddl404 [DOI] [PubMed] [Google Scholar]
- Xia H., Hornby Z.D., Malenka R.C. 2001. An ER retention signal explains differences in surface expression of NMDA and AMPA receptor subunits. Neuropharmacology. 41:714–723 10.1016/S0028-3908(01)00103-4 [DOI] [PubMed] [Google Scholar]
- Yoshimura S., Egerer J., Fuchs E., Haas A.K., Barr F.A. 2007. Functional dissection of Rab GTPases involved in primary cilium formation. J. Cell Biol. 178:363–369 10.1083/jcb.200703047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerial M., McBride H. 2001. Rab proteins as membrane organizers. Nat. Rev. Mol. Cell Biol. 2:107–117 10.1038/35052055 [DOI] [PubMed] [Google Scholar]
- Zhang M.Z., Mai W., Li C., Cho S.Y., Hao C., Moeckel G., Zhao R., Kim I., Wang J., Xiong H., et al. 2004. PKHD1 protein encoded by the gene for autosomal recessive polycystic kidney disease associates with basal bodies and primary cilia in renal epithelial cells. Proc. Natl. Acad. Sci. USA. 101:2311–2316 10.1073/pnas.0400073101 [DOI] [PMC free article] [PubMed] [Google Scholar]