In vitro analysis confirms talin binding is sufficient for activation and extension of membrane-embedded integrin.
Abstract
Increased affinity of integrins for the extracellular matrix (activation) regulates cell adhesion and migration, extracellular matrix assembly, and mechanotransduction. Major uncertainties concern the sufficiency of talin for activation, whether conformational change without clustering leads to activation, and whether mechanical force is required for molecular extension. Here, we reconstructed physiological integrin activation in vitro and used cellular, biochemical, biophysical, and ultrastructural analyses to show that talin binding is sufficient to activate integrin αIIbβ3. Furthermore, we synthesized nanodiscs, each bearing a single lipid-embedded integrin, and used them to show that talin activates unclustered integrins leading to molecular extension in the absence of force or other membrane proteins. Thus, we provide the first proof that talin binding is sufficient to activate and extend membrane-embedded integrin αIIbβ3, thereby resolving numerous controversies and enabling molecular analysis of reconstructed integrin signaling.
Introduction
Increased ligand binding affinity of integrins (“activation”) is central to cell migration, extracellular matrix assembly, immune response, and hemostasis (Hynes, 2002). There remain uncertainties about the final events that lead to activation, including whether talin binding to the integrin is sufficient for activation (Moser et al., 2009b), whether conformational changes lead to activation (Bazzoni and Hemler, 1998; Carman and Springer, 2003), the nature of such conformational changes (Takagi et al., 2002; Adair et al., 2005), and the role of mechanical force (Zhu et al., 2008). A critical barrier to answering these questions is the absence of systems that enable a complete recreation of the final steps in physiological integrin activation.
Integrins are noncovalent heterodimers of transmembrane α- and β-subunit, each with a single transmembrane and cytoplasmic domain (tail; Hynes, 2002); activation is initiated through interactions at the integrin tails (O’Toole et al., 1991, 1994). Both in vitro (Calderwood et al., 1999; Tadokoro et al., 2003) and in vivo (Nieswandt et al., 2007; Petrich et al., 2007a,b) studies reveal that the binding of the ∼50-kD talin head domain (THD) to the integrin-β tail is involved in integrin activation. Recent in vitro (Ma et al., 2008) and in vivo experiments (Montanez et al., 2008; Moser et al., 2008, 2009a) indicate that kindlins are important in integrin activation. Kindlins bind integrin-β tails (Kloeker et al., 2004) and this interaction is involved in activation (Shi et al., 2007; Ma et al., 2008; Moser et al., 2008), suggesting that talin requires kindlins to activate integrins (Moser et al., 2009b). The forgoing experiments used genetic modifications or expression of recombinant proteins in cells; such studies are subject to potential contributions of unknown cellular components and of complex effects of deletion or overexpression of proteins. Indeed, kindlin-3 deletion induces dramatic global changes in cytoskeletal composition (Krüger et al., 2008) and talin can regulate the biosynthesis of PIP2, a critical regulator of the cytoskeleton (Di Paolo et al., 2002). In consequence, a definitive test of the sufficiency of talin for integrin activation requires a means to analyze the activation of purified integrins.
Integrins exist in at least three functional states: inactive, active, and active and ligand-occupied (Frelinger et al., 1991), and long-range allosteric rearrangements underlie the transitions between these states (Du et al., 1993). The landmark structure of integrin αVβ3 ectodomain revealed a bent conformation (Xiong et al., 2001). An electron microscopy (EM) study proposed that activation required the bent integrin to assume an extended conformation (Takagi et al., 2002); however, another analysis revealed that the bent form could bind ligand and ascribed the earlier results to sampling bias (Adair et al., 2005). A recent structure of the integrin αIIbβ3 ectodomain was bent, suggesting that extension required tractional forces or collisions with other membrane proteins (Zhu et al., 2008). These studies used soluble integrins, lacking the transmembrane and cytoplasmic domains that regulate affinity state (Ginsberg et al., 2005). Studies on full-length integrins have been limited by a number of factors and have led to divergent results (Hantgan et al., 2001; Adair and Yeager, 2002; Iwasaki et al., 2005). Cryo-EM of lipid bilayer-embedded integrin αIIbβ3 (Ye et al., 2008) revealed an 11-nm height, consistent with the bent form; the height did not change in response to Mn++ activation, which alters the cation coordination in integrin A domains (Shimaoka et al., 2002). Furthermore, integrin clustering is linked to integrin activation (Li et al., 2003) and the relative contribution of clustering and conformational changes is hotly debated (Bazzoni and Hemler, 1998; Carman and Springer, 2003).
Here, we recreated the triggering event in physiological activation of integrin αIIbβ3 and performed cellular, biochemical, biophysical, and EM analyses. We find that THD binding to the integrin-β tail is sufficient for activation; however, efficient talin-induced activation requires embedding of the integrin in a phospholipid bilayer and a membrane-binding site on the talin. We used phospholipid nanodiscs to create lipid-embedded integrins in which simultaneous access to the extracellular and cytoplasmic domain is available and in which the unclustered state is enforced. Using these nanodiscs, we show that THD binding activates single monomeric integrins and that talin binding alone, in the absence of force or of other membrane proteins, is sufficient to induce the extended conformation. Thus, we provide the first proof that talin binding is sufficient to activate and extend membrane-embedded integrin αIIbβ3.
Results
THD activates purified integrin αIIbβ3
Expression of THD in cells activates integrins (Calderwood et al., 1999; Tadokoro et al., 2003); however, talin may not be sufficient because a kindlin is required for activation (Moser et al., 2009b). To ascertain whether THD is sufficient to activate integrin αIIbβ3, we reconstituted the highly purified integrin into phospholipid vesicles with or without THD (Fig. 1, A and B). The individual integrins inserted into the lipid bilayer were clearly visible, indicating successful integrin incorporation. The integrins were randomly oriented (35–50% were facing the outside) as judged by cryo-EM tomograms (Fig. 1, A and B). THD is too small to be visualized in the tomograms; however, we verified the coincorporation of THD and integrins by SDS-PAGE of the liposomes (Fig. 2 B) and quantification revealed an approximate 2:1 molar ratio of THD to integrin in the liposomes. No gross integrin clustering was observed either in the presence or absence of THD (Fig. 1, A and B).
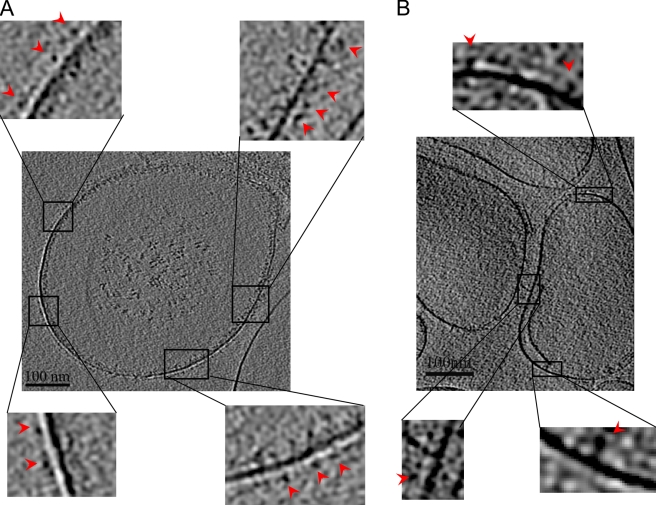
Figure 1.
Cryo-EM tomography reveals insertion of unclustered integrins into liposomes. (A) Central section of the cryo-EM tomogram of frozen hydrated liposomes with integrin αIIbβ3 and THD co-incorporated. (B) Central section of the cryo-EM tomogram of liposomes containing only integrin αIIbβ3. The individual integrins were clearly visible in both A and B with a head density connected by a narrow leg density to the lipid densities, indicating that the integrins were incorporated into the lipid bilayers. Insets show magnified views of a few regions in both A and B and indicate that the integrins are well resolved. No gross clustering was observed in the presence or absence of THD. The red arrows point to the individual integrin densities.
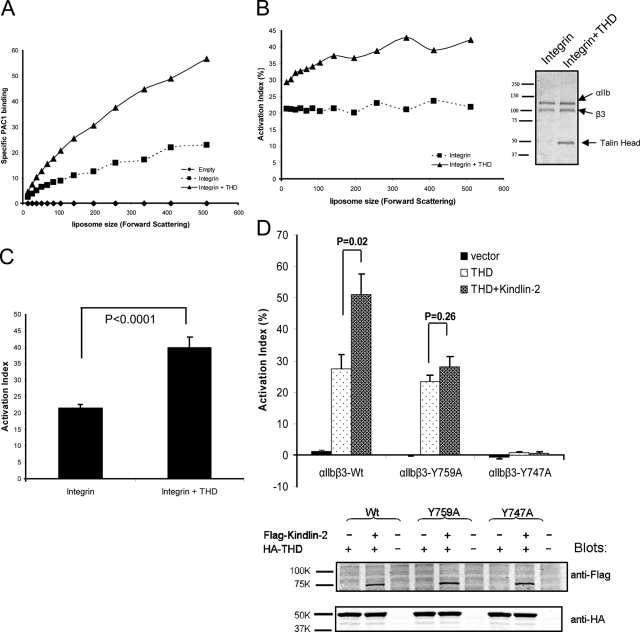
Figure 2.
Talin binding is sufficient to activate integrins. (A) Liposome size vs. PAC1 binding. Flow cytometric data were analyzed in 11 regions of forward scatter (FSC; a measure of particle size). THD increased PAC1 binding (▴) compared with integrin alone (■) or empty liposomes (♦). (B) Activation indices plotted against liposome size. The presence of THD (▴) increased activation relative to its absence (■). The inset depicts an SDS-PAGE of integrin liposomes in the presence and absence of THD. Data in A and B are from one of three independent experiments. (C) Activation indices of the liposomes at the mid-point of the forward scatter spectrum (mean FSC = 142) were averaged from three experiments. THD increased integrin activation. Unless otherwise indicated, error bars in the graphs shown here and below denote the standard error of three independent experiments. (D) THD activates an integrin mutant defective in kindlin binding. THD activated integrin αIIbβ3, and coexpression of kindlin-2 led to an additional increase in activation. In contrast, THD activated αIIbβ3(Y759A), a mutant defective in kindlin binding, whereas kindlin-2 did not augment the activation. αIIbβ3(Y747A), which is defective in talin binding, was not activated by THD nor by a combination of THD and kindlin 2. Activation index is calculated as in B. The bottom panel is a Western blot showing that the expression of THD and kindlin-2 are comparable in αIIbβ3, αIIbβ3(Y759A), or αIIbβ3(Y747A) cells.
Having confirmed the coincorporation of THD and integrin αIIbβ3 in liposomes, we next sought to test if the THD activated the purified integrins by measuring the binding of an activation-specific antibody, PAC1 (Shattil et al., 1985). Incorporation of THD into the liposomes increased PAC1 binding (Fig. 2 A) as judged by flow cytometry. Importantly, the increase in PAC1 binding was not due to altered αIIbβ3 content of the liposomes (Fig. 2, B and C), or increased liposome size or aggregation as judged by forward scatter (Fig. 2, B and C). Binding was specific, as it was blocked by a competitive antagonist, eptifibatide (Scarborough et al., 1993). We calculated an “activation index” (100 × (F − F0) / (Fmax − F0), where F = PAC1 binding, F0 = binding in the presence of 1 µM eptifibatide, and Fmax = binding in the presence of 1 mM MnCl2) and found that incorporation of THD resulted in a twofold increase in PAC1 binding (∼40% vs. ∼20% in the absence of THD). Furthermore, THD did not change the orientation of αIIbβ3 in the liposomes (Fig. 2, A and B). Thus, THD is sufficient to activate purified integrin αIIbβ3 in vitro.
THD activates an integrin mutant that is defective in kindlin binding
To see whether THD binding is sufficient to activate αIIbβ3 in vivo, we examined the effect of THD on integrins bearing β3 subunit mutations that selectively block talin or kindlin binding. The β3(Y747A) mutant impairs talin but not kindlin binding, whereas the β3(Y759A) mutant retains the ability to bind to talin but is defective in kindlin binding (Ma et al., 2008; Moser et al., 2008). We created CHO cells that stably express αIIbβ3, αIIbβ3(Y759A), or αIIbβ3(Y747A). Expression of THD activated αIIbβ3 but not αIIbβ3(Y747A) (Fig. 2 D). Consistent with our finding that THD is sufficient for integrin activation, THD activated αIIbβ3(Y759A) (Fig. 2 D). Kindlin-3 does not activate αIIbβ3 in CHO cells (Moser et al., 2008) or synergize with THD (unpublished data). We therefore examined the effect of the Y759A mutation on the capacity of kindlin-2 (Ma et al., 2008) to synergize with THD in CHO cells. THD and kindlin-2 synergized in activating αIIbβ3 but not αIIbβ3(Y759A) (Fig. 2 D), indicating that the mutant did indeed perturb the functional consequences of kindlin binding. Importantly, the expression levels of THD and kindlin-2 were comparable in αIIbβ3-, αIIbβ3(Y747A)-, and αIIbβ3(Y759A)-expressing cells (Fig. 2 D). Thus, both in vitro and in vivo, THD binding activates integrin αIIbβ3 independent of kindlin binding.
THD activates αIIbβ3 by binding the β3 tail
Talin activates αIIbβ3 by binding to the β3 tail (García-Alvarez et al., 2003; Wegener et al., 2007) in cells. We sought to ascertain whether THD activated purified αIIbβ3 by binding to the β3 tail. We first examined the capacity of added THD to activate αIIbβ3 in intact cells and found no effect on PAC1 binding at concentrations up to 25 µM. Second, we found that THD(W359A), a mutant with reduced affinity for the β3 tail (García-Alvarez et al., 2003), failed to activate the purified integrin (Fig. 3, A and B). The sharp melting point of THD(W359A) indicated that the protein was well folded, although the 1.7° lower melting point suggests it was slightly less stable than wild-type THD. (Fig. 3 F). In addition, THD(W359A) was incorporated into the liposomes (Fig. 3 A). Thus, THD required both access to the β3 tail and the capacity to bind to it in order to activate αIIbβ in vitro.
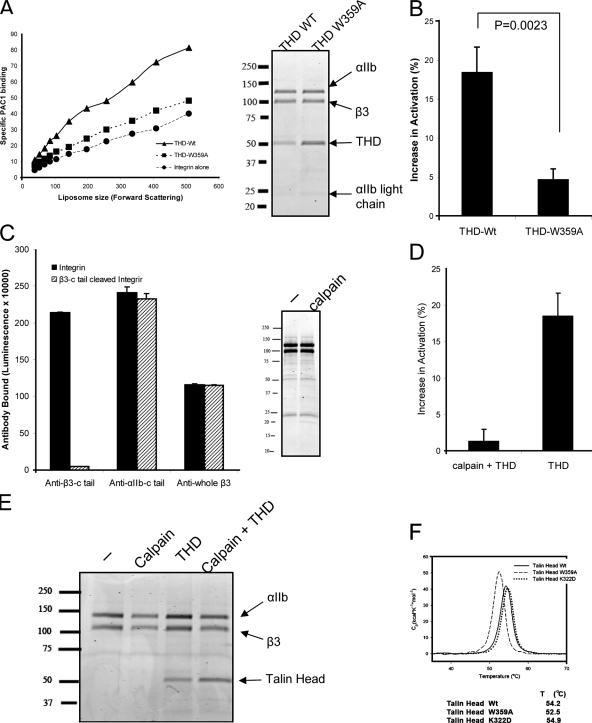
Figure 3.
THD activates purified αIIbβ3 by binding to the β3 tail. (A) THD induced an increase in PAC1 binding (▴), whereas THD-W359A (■), a mutant deficient in integrin tail binding, failed to do so. The inset on the right depicts SDS-PAGE showing that integrin and THD are comparable. Data are from one of three independent experiments. (B) Activation indices of the subset of liposomes at the mid-point of the forward scatter spectrum (mean FSC = 142) were averaged over three independent experiments. Increases in integrin activation (calculated as AIwith_THD − AIintegrin_alone, where AI stands for activation indices) are depicted. THD increased integrin activation compared with THD-W359A. (C) Requirement of the β3 tail for THD activation. Using the ELISA described in the Materials and methods section, calpain abolished anti–β3-c tail binding but had no effect on anti-β3 or anti–αIIb-c tail binding. SDS-PAGE revealed that calpain did not markedly shift mobility of β3 or αIIb. (D) THD fails to activate αIIbβ3 lacking the β3 C terminus. Increase in integrin activation was calculated as in B. (E) SDS-PAGE shows that integrin and THD incorporation are comparable for calpain-digested and intact integrins. (F) THD mutants do not disrupt protein folding. Differential scanning calorimetry of THD, THD-W359A, and THDF-K322D. All three proteins exhibited similar narrowly defined melting points, indicating that they were well folded.
As a further test of the mechanism of THD-induced activation, we assessed the requirement for the β3 tail. Mild calpain digestion selectively cleaves the β3 tail from purified αIIbβ3 (Du et al., 1995); calpain removed the β3 C terminus as judged by an anti-peptide antibody directed against this region (Du et al., 1995), but did not influence the binding of an antibody raised against full-length β3 (Fig. 3 C). Furthermore, this mild cleavage, which removes <5% of the mass of β3, produced no detectable change in the mobility of αIIb or β3 in SDS-PAGE (Fig. 3 C) and did not remove the C terminus of αIIb (Fig. 3 C). THD failed to activate the calpain-cleaved integrin (Fig. 3 D). Nevertheless, the levels of integrin and incorporated THD were similar in the liposomes formed with calpain-cleaved αIIbβ3 or intact αIIbβ3 (Fig. 3 E). Thus, activation required an intact β3 tail and THD access to and capacity to bind the β3 tail. We therefore conclude that in a purified system, THD is sufficient to activate αIIbβ3 by binding to the β3 tail.
The importance of lipids for THD activation
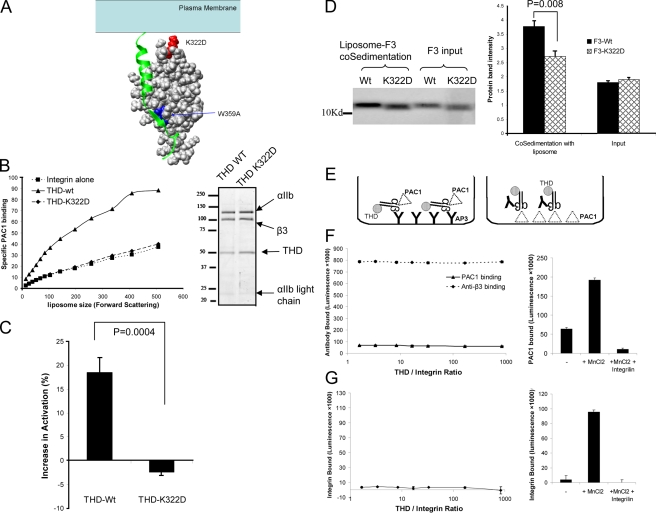
THD(K322D) fails to activate αIIbβ3 (Wegener et al., 2007) in cells; K322 is predicted to interact with phospholipid head groups rather than the β3 tail, an interaction proposed to orient the talin to favor activation (Fig. 4 A). To verify this contact with lipids, we tested the interaction of purified F3 or F3(K322D) with liposomes using a cosedimentation assay. The K322D mutation impaired F3 cosedimentation with lipid, thus providing direct evidence that the mutation impairs lipid binding (Fig. 4 D). Moreover, as predicted (Wegener et al., 2007), THD(K322D) binds to the β3 tail (Fig. S1). Purified recombinant THD(K322D) failed to activate αIIbβ3 in liposomes (Fig. 4, B and C) even though it was well folded (Fig. 3 F) and incorporated into the liposomes (Fig. 4 B). A previously proposed model (Wegener et al., 2007) suggested that the precise physical relationship of F3 relative to the integrin and the membrane controls activation. The present study shows that K322D mutation affects lipid binding and that it disrupts activation in a purified system even though the THD is well folded and binds the integrin.
Figure 4.
The importance of lipid embedding for THD activation. (A) A model of talin head–integrin-β3 tail interaction (Wegener et al., 2007). β3 tail is shown in ribbon diagram and the F3 domain of talin is shown in solid surface. The two mutations, W359A (blocks integrin binding) and K322D (predicted to reduce membrane binding), are highlighted in blue and red, respectively. (B) THD-K322D (♦) caused markedly reduced PAC1 binding relative to THD (▴). Data are from one of three independent experiments. The SDS-PAGE on the right confirms that the integrin and THD incorporation are comparable for THD-K322D and THD (both lanes were excised from the same gel image and were juxtaposed for clarity). (C) Activation indices of the subset of liposomes at the mid-point of the forward scatter spectrum were averaged over three independent experiments. Increases in integrin activation were calculated as in Fig. 3 B. THD but not THD-K322D induced increased integrin activation. (D) The K322D mutation reduces F3–membrane interaction. Co-sedimentation of F3 or F3-K322D with liposomes was assessed by SDS-PAGE. Densitometry was used to quantify these results, which are depicted in the bar graph to the right. Mass spectrometry revealed that both F3-WT and F3-K322D were within 0.1% of their predicted masses; hence, the increased mobility of F3-K322D is ascribed to its increased negative charge. (E) Schematic of assays for measuring activation of detergent-solubilized αIIbβ3. (F) THD failed to activate captured detergent-solubilized integrin. αIIbβ3 was mixed with various concentrations of THD and was captured on a plate coated with AP3, and activated αIIbβ3 was detected by PAC1 binding. Captured αIIbβ3 was detected by the binding of polyclonal anti-β3 antibody (anti-β3). PAC1 binding was low and did not change even with ∼1,000-fold molar excess of THD. The panel on the right shows that Mn++ activated the αIIbβ3 in the same experiment. (G) The plate was coated with PAC1 and αIIbβ3 mixed with various concentrations of THD was then added to the plate. The bound integrins were detected with polyclonal anti-β3 antibody. At a 1,000-fold molar excess, THD failed to induce binding of PAC1 to the αIIbβ3; in contrast, 1 mM Mn++ did so, and the binding was inhibited by a competitive antagonist, eptifibatide.
To ascertain if lipid-inserted integrins were required for THD-induced activation, we assessed the effect of THD on purified detergent-solubilized αIIbβ3. We first captured detergent-solubilized αIIbβ3 with an anti-β3 antibody and assayed its activation state by PAC1 binding (Fig. 4 E, left; Du et al., 1991). THD failed to increase PAC1 binding to detergent-solubilized αIIbβ3 at concentrations approaching a 1,000-fold molar excess to the integrin (Fig. 4 F). THD binds to αIIbβ3 with an affinity of Kd ∼400 nM (Goldmann, 2000), and we saw no effect of THD at concentrations almost 2 logs higher than this Kd. The assay was functioning because addition of 1 mM MnCl2 induced a marked increase in PAC1 binding that was blocked by eptifibatide (Fig. 4 F, right). We also developed an alternative assay format in which the detergent-solubilized αIIbβ3 was mixed with THD in solution and activation was measured by capture with immobilized PAC1 (Fig. 4 E, right). Again, THD failed to promote αIIbβ3 binding to PAC1, whereas Mn2+ resulted in a dramatic increase (Fig. 4 G). These results are in sharp contrast to those in the reconstituted liposomes wherein activation is observed at a 2:1 molar ratio of THD to αIIbβ3 (Fig. 2 B). Thus, efficient THD-induced αIIbβ3 integrin activation requires that the integrin be inserted into a lipid bilayer.
Talin activates unclustered integrins
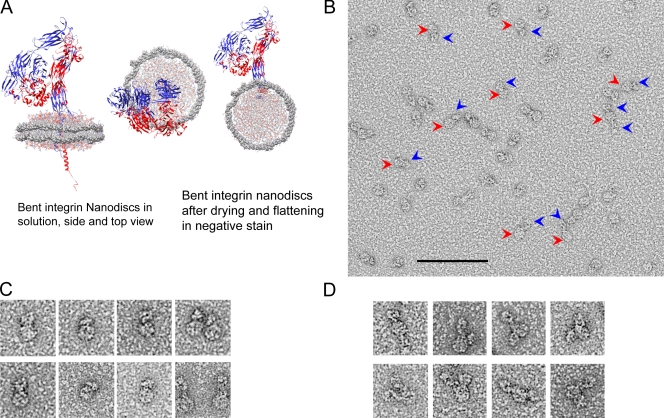
Having established THD is sufficient to activate membrane-embedded integrin αIIbβ3 by binding to the β3 tail, we examined the relative role of increased monomer affinity. As noted above, there was no gross integrin clustering when THD activated αIIbβ3 in liposomes (Fig. 1, A and B). To address this question definitively, we inserted single integrins into lipid while maintaining access to both extracellular and cytoplasmic domains by embedding αIIbβ3 into nanodiscs, derived from apolipoprotein A-1 (Denisov et al., 2004). The nanodiscs consist of lipid bilayers encircled by membrane scaffold proteins. The integrins are inserted in the lipid bilayer with both extracellular and cytoplasmic domain exposed and accessible (Fig. 5 A). We assembled and purified the integrin nanodiscs and characterized them by analytical ultracentrifugation (Table I, Fig. S2, A and B) and negative-stain electron microscopy (Fig. 5, B–D). The dominant species (77%; molecular weight 299 kD) was consistent with a single integrin (Mr ∼218 k) (Jennings and Phillips, 1982) inserted into a nanodisc (Mr ∼81 k). In negatively stained electron micrographs, we observed a round density for the empty nanodiscs, whereas in the integrin nanodiscs we observed additional densities (Fig. 5, A–D). Closer examination of the single-integrin nanodiscs revealed that they assembled as depicted (Fig. 5 A) with integrins inserted into the nanodiscs and confined to the segregated lipid bilayers of each individual nanodisc (Fig. 5 D).
Figure 5.
Embedding of integrin αIIbβ3 in phospholipid nanodiscs. (A) Model of integrin αIIbβ3 in a compact, bent conformation, inserted into a nanodisc. The membrane scaffold protein (MSP) is shown in solid surface view, lipid molecules are shown in colored wire, integrins are shown as ribbons. The proposed arrangement of the nanodiscs in solution and the likely arrangement of the integrin nanodiscs after drying and flattening in negative stain are shown. PDB coordinates for the integrin structures obtained from 3FCS (Zhu et al., 2008) (ectodomain), 2H7E (Wegener et al., 2007) (cytoplasmic domain), and 2K9J (Lau et al., 2009) (transmembrane domain). The nanodisc coordinates (Segrest et al., 1999) were obtained from Dr. S.C. Harvey’s website (http://rumour.biology.gatech.edu). (B) Negatively stained EM images of integrin nanodiscs. Several examples of integrin nanodiscs are highlighted with red arrows pointing to the nanodisc density and blue arrows pointing to the integrin densities. Bar, 100 nm. (C) Enlarged view of a representative gallery of single-integrin nanodiscs shown at higher magnification than in B. Examples of both compact bent-over and extended integrin conformations are included. Each panel in C and D is 50 × 50 nm. (D) Similar to C, except that occasional nanodiscs containing two integrins are shown.
Table I.
Characterization of integrin nanodiscs
| Samplea | MW | MW after subtract empty nanodiscs | Integrins per nanodiscs |
| Empty nanondiscs | 80,500 ± 582 D | 0 | 0 |
| Integrin nanodiscs, specie 1 | 299,000 ± 1,483 D | 218,500 | 1 |
| Integrin nanodiscs, specie 2 | 530,000 ± 5,451 D | 449,500 | 2 |
| Sampleb | Sedimentation coefficient (S*) | Areas under S* peak | Percentage of total |
| Nanodiscs, 1 integrin specie | 8.56E-13 | 2.86905 | 77.4 |
| Nanodiscs, 2 integrin specie | 1.47E-12 | 0.83728 | 22.6 |
Molecular weight measurement of empty nanodiscs, single-integrin nanodiscs and dual-integrin nanodiscs.
Sedimentation coefficient determination of single-integrin nanodiscs and dual-integrin nanodiscs, and their respective percentage in the total population.
Analytical ultracentrifugation revealed a minor (22.6%; Table I) specie of 530-kD mass, indicating an increment of 449 kD above the empty nanodisc. This increase suggests the presence of two integrins per nanodisc. Occasional EM images in which two additional densities were associated with the nanodisc confirmed this suggestion. These densities resembled those previously ascribed to integrins and were in two configurations. In one, the two integrins were on opposite sides of the nanodisc and therefore could not cluster. In others, both integrins faced the same way (Fig. 5 D) and were only ∼10 nm apart; hence, they were preclustered (Fig. 5 D). Thus, we have created integrin nanodiscs wherein the integrins are in three states of oligomerization: single integrins (the dominant species), and dual integrins in which the integrins are either preclustered or oriented in opposing directions.
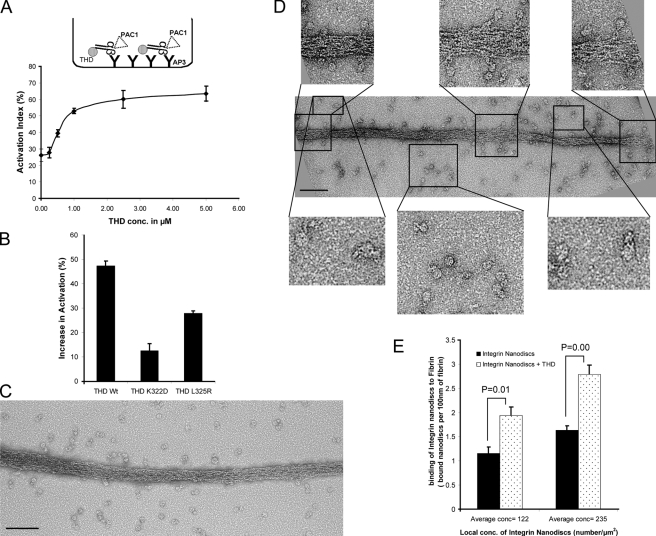
We captured the αIIbβ3 nanodiscs with an anti-β3 antibody and assessed their activation state by PAC1 binding (Fig. 6 A, top). We reasoned that because integrin nanodiscs are immobilized before addition of THD and ligands, neither THD nor ligand can induce further integrin clustering. THD induced a concentration-dependent increase in PAC1 binding (Fig. 6 A). To confirm that the increase in integrin activation is a result of physiologically relevant THD–β3 interaction, we compared the effect of THD, THD(L325R), a mutation that disrupts THD-β3 membrane proximal interaction site, and THD(K322D), a mutation that reduced THD–membrane interaction (Fig. 4 D) (Wegener et al., 2007). Both mutations reduced the capacity of THD to activate (Fig. 6 B). Thus, THD activates integrin αIIbβ3 in nanodiscs under conditions which disfavor clustering.
Figure 6.
THD activates integrins in nanodiscs. (A) THD increases the binding of PAC1 to captured integrin nanodiscs. The inset shows the configuration of the assay, which is described in the Materials and methods section. The activation indices were calculated as (L − L0) / (Lmax − L0) where L = luminescence, L0 = luminescence in presence of 1 µM eptifibatide, and Lmax = luminescence in the presence of anti-LIBS6 activating anti-β3 antibody. (B) Mutations of THD reduce its capacity to activate integrins in nanodiscs. The increase in activation is calculated as in Fig. 3 B. A mutation that reduces membrane binding (K322D) or one that blocks binding to the integrin-β3 tail (L325R) reduced capacity of THD to activate αIIbβ3 in nanodiscs. (C) EM images of negatively stained fibrin and integrin nanodiscs alone. Relatively few integrin nanodiscs are bound to the fibrin. Bar, 100 nm. (D) EM images of negatively stained fibrin and integrin nanodiscs in the presence of 5 µM THD showing an increase in the number of bound integrin nanodiscs when THD is present. The top insets show enlarged views of integrin nanodiscs bound to the fibrin. Single fibrin-bound integrin nanodiscs are clearly visible, indicating that the increase in binding is due to increased monomer affinity rather than clustering. The bottom insets show enlarged views of unbound integrin nanodiscs. Both compact and extended conformations are visible. Bar, 100 nm. (E) The local concentrations of nanodiscs were estimated from EM images by manual enumeration of nanodiscs/µm2. The number of bound integrin nanodiscs per 10-nm length of fibrin strand was plotted for each concentration. The presence of THD increased the number of bound integrin nanodiscs. Error bars = SE (n = 4) for each concentration.
As another test of the idea that THD-induced affinity changes do not require clustering, we directly imaged ligand binding to THD-activated integrins. We chose fibrin to enable visualization of an ordered integrin ligand (Fig. S2 C). When integrin nanodiscs were added to the fibrin, we observed few fibrin-bound nanodiscs (Fig. 6 C). In sharp contrast, a large number of unbound integrin nanodiscs were in the vicinity of the fibrin (Fig. 6 C). Addition of THD increased the number of integrin nanodiscs bound to the fibrin (Fig. 6 D), providing visual confirmation of the activation of the integrins. Moreover, this increase in binding was not due to an increase in the local concentration of nanodiscs, as the total densities of nanodiscs were comparable in the presence and absence of THD (Fig. 6, C and D). To quantify THD-induced activation, we measured the bound nanodiscs per nm length of fibrin at differing local nanodisc densities; there was a significant (P = 0.01) increase in the number of bound integrin nanodiscs in the presence of THD (Fig. 6 F). Discrete nanodiscs containing a single integrin (Fig. 6 D, top inserts) were bound to the fibrin. Thus, based on biochemical measurements under conditions that prevent clustering and on direct visualization of activated integrins, we conclude that the binding of THD induces an increase in the affinity of unclustered integrins.
THD induces the extended integrin conformation in the absence of ligand or externally applied force
To examine the conformational effects of a physiological activator, talin, on a membrane-embedded integrin, we randomly extracted images of 1,283 individual integrin nanodiscs and 1,553 integrin nanodiscs in the presence of THD and subjected them to image analysis of alignment and classification. We set number of classes ≈ total images ÷ 50 to capture the variability of the integrin nanodiscs and to minimize the possibility of grouping different conformations in the same class. The class averages of integrin nanodiscs revealed two distinctive conformations (Fig. 7, A–C; Fig. S3). All of the class-averaged integrin nanodisc images in the absence of THD revealed a nanodisc density and a compact density that averaged a height of 11 ± 1 nm, thereby resembling the bent conformation (Takagi et al., 2002; Xiong et al., 2002) (Fig. 5 A, Fig. 7 A). Because overrepresented images tend to dominate the classes, leaving underrepresented images poorly classified (Shaikh et al., 2008) and because structural flexibility and heterogeneity can hinder the alignment and classification (Penczek et al., 2006; Herman and Kalinowski, 2008), manual selection of images of different structures are frequently warranted (Takagi et al., 2003; Nishida et al., 2006). Therefore, we also manually evaluated 1,283 images and observed that the vast majority (89.7%) were in the compact conformation, whereas 10.3% of integrin densities appeared to be extended as judged by their average height of 19 ± 1 nm. In sharp contrast, at least 40% of the fibrin-bound integrins were extended (Fig. 6 D, top insets), establishing that this conformer is associated with high affinity. When THD was added, 22% of integrins were now in the extended conformations in the class-averaged images (Fig. 7, C and D; Fig. S4); this result was confirmed by manual evaluation of 1,553 images in which there was a marked increase in unoccupied integrins in the extended conformation (25%). A globular protein the size of THD (∼5.1 nm; Winkler et al., 1997) couldn’t be resolved in these negative-stained images. Thus, THD induces the extended conformation; hence, this physiologically relevant activator can activate and extend integrins in the absence of other proteins, ligands, or imposed forces.
Figure 7.
THD induces the extended conformation in the absence of ligand or externally applied force. All panels are images of 50 × 50-nm fields. The number of images in each class is shown at the bottom left corner of each box. (A) Class averages of integrin nanodiscs in compact bent-over conformation. The class averages show a round-shaped nanodisc density and compact bent-over density for the integrins. All 18 classes are shown in Fig. S3. (B) Class averages of integrin nanodiscs in compact conformation in the presence of THD. The class averages show a round-shaped nanodisc density and compact density for the integrins. Although the integrin densities are compact, they appear slightly less compact than when THD is absent. The numbers of molecular images in each class are shown at the bottom left corner of each box. (C) Class averages of integrin nanodiscs in extended conformation in the presence of THD. All classes for the integrin nanodiscs with THD are shown in Fig. S4. (D) Total numbers of extended or compact integrin nanodiscs were obtained by summing the number of images in the classes showing extended or compact conformation. Only classes with clearly defined integrin density were included in the calculation of the percentages.
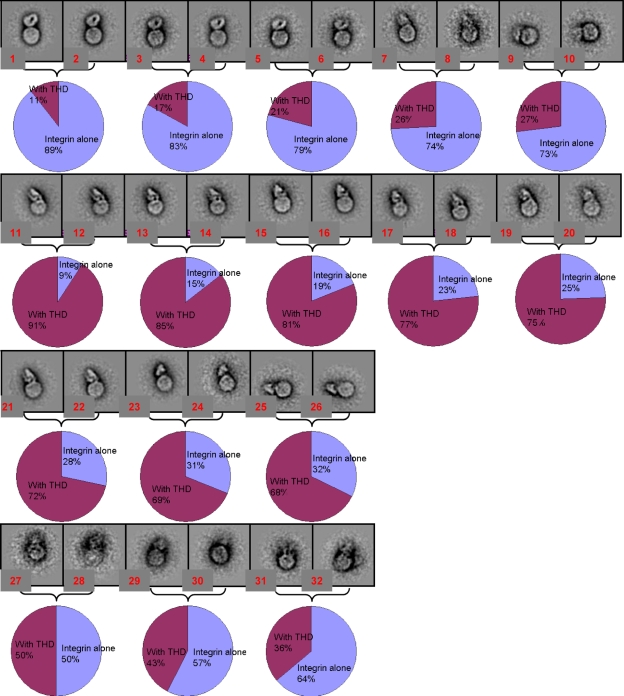
In the presence of THD, the compact integrin densities appeared slightly less compact than when THD is absent (Fig. 7, A and B). We verified this observation by pooling images of compact integrins in the presence or absence of THD, and subjecting this mixed pool to image analysis of alignment and classification. The slightly less compact forms were strongly associated with the presence of THD (Fig. 8). This result suggests that THD may also induce a more subtle difference in conformation.
Figure 8.
Compact integrin with and without THD roughly segregate into separate classes. Panels are images of 50 × 50-nm fields. Images of nanodiscs containing compact integrins from sample prepared in the presence or absence of THD were grouped together, and the pooled images were then classified and averaged iteratively. 20 classes were generated during the iterative process (parent classes) and at the final step, each class was split to two subclasses (daughter subclasses) to verify the internal consistency of the class averages. Each bracket contains two daughter subclasses from a parent class. Little or no difference between the daughter subclasses indicated robust alignment and classification by the iterative procedure. Within each class, the percentage of images from samples containing either integrin nanodiscs in the presence or absence of THD are depicted in a pie chart. Each resulting class average in which the integrin structure was clearly defined was greatly enriched in images from the integrin nanodiscs alone or from the THD-containing integrin nanodisc population. For example, the first row of images shows classes dominated by images from the integrin alone sample (up to 89%). In contrast, the second and third row are classes dominated by images from the sample containing added THD (up to 91%). In the classes in which the integrin structure is not clearly resolved, both samples were more evenly represented (fourth row). 8 classes of empty nanodiscs were omitted from this figure.
As noted above, the extended form was more abundant in the fibrin-bound integrins. We sought to generate class averages of THD-activated, fibrin-bound integrin nanodiscs; however, the alignment and class averaging procedure failed because the multiple sources of variability from fibrin, integrin, nanodiscs, and angle between fibrin and integrins rendered these molecular images too heterogeneous to converge. Nevertheless, the raw images (Fig. 6 D, top insets) revealed that, at a minimum, 40% of these integrins were in extended conformations (extended conformation defined as extending more than 15 nm, the mid-point height between bent-over and extended conformation).
Discussion
Integrin activation is central to metazoan development and function, yet there remain sharp divisions of opinion concerning the terminal steps in physiological activation, in large part because there were no means to study these events in purified systems. Here, we devised two in vitro methods to quantitatively study purified integrin activation by THD, a physiologically relevant activator. We report that THD binding to the integrin-β tail is sufficient for integrin activation in the absence of other proteins; however, efficient talin-induced activation requires a membrane-binding site on talin and membrane insertion of the integrin. Furthermore, THD binding can activate a kindlin binding–defective integrin mutant, further documenting the sufficiency of talin binding for activation. To precisely specify the concentrations of proteins accessible to the cytoplasmic side of a lipid-inserted integrin while measuring ligand binding to the extracellular domain, we embedded integrin αIIbβ3 in phospholipid nanodiscs. THD binding activated integrin αIIbβ3 in these nanodiscs, even when the unclustered state was both biochemically enforced and documented by EM. Quantitative analysis of EM images of these nanodiscs revealed that talin activation of unoccupied integrins shifted the conformational equilibrium toward extension in the absence of force or of other membrane proteins. Thus, we show that talin binding alone is sufficient to activate and extend membrane-embedded integrin αIIbβ3.
THD activation of integrin αIIbβ3 recapitulates physiological inside-out integrin signal transduction. In two different formats (liposomes, nanodiscs) and using three different measures (PAC1 binding by flow cytometry or ELISA and by visualizing fibrin binding), we found that THD increased the affinity of αIIbβ3 for both a model ligand (PAC1) and a physiological ligand (fibrin). Previous studies in cells (Calderwood et al., 1999; Tadokoro et al., 2003) and mice (Petrich et al., 2007a) show that THD binding to the β tail is required for activation. In the purified system, we found that cleavage of the β3 tail prevented activation as did THD mutations that disrupt either the membrane distal (W359A) or membrane proximal (L325R) β tail binding sites required for activation (García-Alvarez et al., 2003; Wegener et al., 2007). In the reconstituted liposomes, the calculated molar ratio of THD to αIIbβ3 was ∼2:1. In platelets, talin accounts for 3–8% of total proteins in platelet (Collier and Wang, 1982). Assuming 1.34 × 10−12 g of total protein per platelet (Karpatkin and Charmatz, 1970) and 80,000 copies of αIIbβ3 (Wagner et al., 1996), the calculated molar ratio of talin to αIIbβ3 is 1:1–3:1, similar to the ratio in the reconstituted liposomes. In the integrin nanodiscs, half-maximal αIIbβ3 activation was observed at a THD concentration of ∼700 nM, a value in remarkable agreement with the 400-nM Kd of talin for integrin αIIbβ3 measured by dynamic light scattering (Goldmann, 2000). Thus, based on the activation of αIIbβ3 by THD at realistic concentrations and the requirement for both the β3 tail and for two distinct β3 binding sites on THD, we conclude that we have recreated the triggering event in physiological integrin activation.
THD is sufficient to activate integrins in in vitro systems and THD can activate a kindlin binding–defective integrin mutant in cells. These results are in good agreement with the weak or absent effects of kindlin overexpression when it is not accompanied by THD overexpression (Kloeker et al., 2004; Shi et al., 2007; Ma et al., 2008). In sharp contrast, the genetic deletion of kindlins results in deficient integrin activation (Meves et al., 2009). Kindlin binds to the β tail at a site distinct from the talin-binding site and the β3(Y759A) mutation disrupts β3–kindlin interaction (Ma et al., 2008; Moser et al., 2008). Overexpression of the isolated integrin-β tails inhibit integrin activation by sequestering talin (Calderwood et al., 2004), and mutations in β1 (Zent et al., 2000) or β3 (Calderwood et al., 2004) that block talin binding reverse this process even though these β3 tails retain kindlin binding. Kindlins are important in the assembly of focal adhesions (Tu et al., 2003) and in the signaling events that follow integrin ligation (Montanez et al., 2008), and positive feedback from integrin ligation can help sustain activation (Shattil and Newman, 2004). Consequently, kindlins may sustain integrin activation by their ability to stabilize integrin-mediated cytoplasmic assemblies such as focal adhesions. Alternatively, as previously noted, kindlin deficiency may affect cytoskeletal composition, thereby affecting talin’s capacity to localize to and activate integrins. Filamin can compete with talin for binding to integrins, thereby blocking activation (Kiema et al., 2006). Migfilin, a protein recruited by kindlins (Tu et al., 2003), can displace integrins from filamin (Lad et al., 2008). These findings were used to advance the innovative hypothesis that kindlins recruit migfilin to integrins, thereby displacing filamin to favor talin–integrin interaction and activation (Ithychanda et al., 2009). Our data that talin is both necessary and sufficient for integrin activation are consistent with these hypotheses. These data are also consistent with location of the kindlin-binding site on the β3 tail, some distance from the membrane proximal site (Wegener et al., 2007) to which talin binds to perturbs the inner membrane clasp (Lau et al., 2009) that maintains the integrin in the low affinity state.
Talin activation of unoccupied integrins can lead to molecular extension of single integrins in the absence of force or of other membrane proteins. We found that THD activated integrin αIIbβ3 in immobilized nanodiscs and identified single-integrin nanodiscs bound to ligand, thereby establishing that the individual integrins were activated. Previous efforts to resolve this issue in cells made use of “monovalent” ligands; however, protein ligands may still contain multiple subsites. Furthermore, mutations that activate integrins may also lead to clustering in cells (Hughes et al., 1996); conversely, mutations designed to block activation by introducing disulfide bonds (Luo et al., 2004; Zhu et al., 2007) could also prevent transmembrane domain separation and resultant clustering (Li et al., 2003). Within cells, full-length talin, having multiple integrin-binding sites (Yan et al., 2001), may also cluster integrins as could the binding of multivalent ligands or the assembly of integrin-associated protein complexes. That said, our studies provide the first direct proof that THD, a physiologically relevant activator, activates demonstrably monomeric integrins.
THD shifts the conformational equilibrium of unoccupied integrin toward the extended form. Addition of THD increased the abundance of the extended conformation as judged by single-particle averaging analysis of negative-stained EM images. The effect of THD on extension is specific. There is much evidence that the extended conformation is an activated form (Takagi et al., 2002; Zhu et al., 2007). Here, we confirm this by showing that that more fibrin-bound integrins are in extended conformation. There is also a general agreement that the low affinity integrins are in a compact conformation (Xiong et al., 2001; Takagi et al., 2002). We found that in vitro THD activation requires the β3 tail and the integrin- and membrane-binding sites of talin, indicating that the effects of THD are specific. Furthermore, this effect of THD is not simply due to presence of recombinant protein because 85 µM recombinant membrane scaffold protein is present under all conditions; ∼100-fold molar excess relative to the 700-nM EC50 for THD. Extension was accompanied by both visual and biochemical evidence of activation, thus showing that a biological activator can induce extension in the absence of imposed external force. Pioneering modeling studies (Puklin-Faucher et al., 2006) suggested that such forces could contribute to activation; a prediction subsequently verified (Friedland et al., 2009); hence, cellular contractile forces may contribute to additional extension and force-induced stabilization of adhesion. We note that the conformation of the extended integrin observed in nanodiscs differs from soluble ectodomains (Takagi et al., 2002). The legs of the extended integrins are often crossed rather than separated (Fig. 7 C). Future structural assessments will be needed to ascertain whether these differences are seen as a consequence of an effect of the nanodisc on the orientation of the negative-stained integrin or due to the release of constraints imposed by the transmembrane and cytoplasmic domains in soluble integrin ectodomains (Takagi et al., 2002). Previous studies of Mn++ activated integrins (Xiong et al., 2002; Ye et al., 2008) reported no extension in Mn2+ activated integrins, and the present studies are consistent with a fraction of THD-activated integrins remaining in a bent, albeit subtly altered conformation. THD binds to the integrin cytoplasmic face to induce a long-range allosteric rearrangement, whereas Mn++ binds to the extracellular integrin A–like domain to induce activation (Shimaoka et al., 2002), and our studies now show that talin-induced activation can also lead to molecular extension. Finally, a previous structural study (Wegener et al., 2007) predicted that a talin membrane contact would be required for integrin activation, a prediction that was supported by the observation that joining a membrane-targeting sequence to the integrin-binding site of RIAM enables it to both induce talin membrane recruitment and integrin activation (Lee et al., 2009). We now establish that integrin embedding in a phospholipid bilayer is a prerequisite for talin-induced activation. Notably, the PH domain of kindlin-2 is required for its capacity to induce integrin activation, hinting that kindlins might also require membrane binding to influence integrins (Ma et al., 2008). Our establishment of this tractable system for analysis of the functions and structure of lipid-embedded integrins will enable a detailed examination of the role of membrane lipids in integrin function.
Integrin activation is of broad biological relevance and it represents a reversal of the direction of transmembrane signaling from the inside out (Hynes, 2002). Recent advances include high resolution crystal structures of integrin ectodomains (Xiong et al., 2001, 2002; Zhu et al., 2008), NMR structures of the heterodimeric transmembrane domains (Lau et al., 2009), and identification of critical cytoplasmic players in the final events of the activation process (Tadokoro et al., 2003; Han et al., 2006; Moser et al., 2009b) and the structures of some of them in complex with integrin tails (García-Alvarez et al., 2003; Wegener et al., 2007). The present study represents the next, critical step in our understanding of this process: the recreation of the last events in physiological inside-out integrin signaling. By resolving ongoing controversies about the sufficiency of talin for activation, the role of the membrane, and the extension of talin-activated unoccupied integrins, we set the stage for future studies to analyze these structural transitions, to examine the regulation of the process by other purified proteins or by changes in membrane lipid composition, and ultimately for the complete in vitro assembly of focal adhesions.
Materials and methods
Antibodies
The conformation-specific monoclonal antibody PAC1 (Shattil et al., 1985), monoclonal anti-β3 antibody AP3 (Newman et al., 1985), and rabbit polyclonal antibodies against whole-β3 (Ab8053), against αIIb-c tail (Ab8276), and against β3-c tail (Ab8275) have been described previously (Du et al., 1995).
Purification of inactive integrin αIIbβ3
Integrin αIIbβ3 was purified from outdated human platelets based on a modified protocol from Ye et al. (2008). In brief, outdated platelets were spun down first at 300 g to remove the red blood cells and leucocytes and subsequently at 1,800 g to pellet the platelets. The platelets were washed twice with Tris-buffered saline and then extracted overnight with lysis buffer: 20 mM Tris, pH 7.4, 150 mM NaCl, 1% Triton X-100, 5 mM PMSF, 0.5 mM CaCl2, 10 µM leupeptin, 10 µM protease inhibitor E64 (Sigma-Aldrich), and 2.76 µM calpeptin. The extracted integrin αIIbβ3 was first purified with a Con A column, and then passed through a heparin column to remove fibrinogen and thrombospondin 1. αIIbβ3 was further purified by gel filtration chromatography and the inactive fraction was isolated as the flow-through of an immobilized KYGRGDS affinity matrix as described by Kouns et al. (1992). The final purified integrins are in a buffer of 20 mM Tris, pH 7.4, 150 mM NaCl, 0.1% Triton X-100, 1 mM MgCl2, and 1 mM CaCl2. The pure inactive integrin was subjected to an integrin activity assay described below, performed to verify the inactive status of the integrins (Fig. S5). The abundance of contaminating fibrinogen was <5% and of kindlin-3 was <2% of that of the purified integrin based on the densitometry of the Coomassie blue staining of SDS-PAGE (Fig. S5).
Purification of recombinant talin head
The recombinant human talin head expression plasmid construct was obtained from Dr. Steve Lam (Patil et al., 1999). This construct, which contains amino acids 1–465 of human talin1, was truncated at residue 433 by introduction of a stop codon, and a his6 tag was added to the C terminus to facilitate purification. Mutations within the talin head were introduced with the QuikChange Mutagenesis kit (Stratagene). The wild-type and mutant proteins were expressed in Escherichia coli BL21-DE-pLys (which produced higher protein yields) and purified with His-binding beads according to the manufacturer’s instructions (EMD). The purified proteins were first dialyzed against 20 mM Tris, 150 mM NaCl, pH 7.4 (TBS buffer) with 2 mM EDTA to completely remove the residual Ni2+ on the his6 tag, which caused lipid precipitation problems, and then against EDTA-free TBS buffer.
Liposome reconstitution and purification
Integrin–talin head liposomes were made with a protocol modified from Ye et al. (2008). In brief, 450 nmol 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC), 450 nmol 1,2-dimyristoyl-sn-glycero-3-phospho-(1’-rac-glycerol) (DMPG), and 200 nmol cholesterol were solubilized in chloroform or a chloroform/methanol mixture, mixed thoroughly, and dried onto a glass tube under steady flow of nitrogen. The homogeneous lipid mixture was then solubilized with 20 mM Tris, 150 mM NaCl, pH 7.4, 0.1% Triton X-100, and 1 mM β-mercaptoethanol. 0.2 mg of αIIbβ3 with or without 0.5 mg of talin head construct in the same buffer was added to the lipid solution. The final volume of the entire mixture was 1 ml. To induce liposome formation, Triton X-100 in the solution was removed by several additions (in 3-h intervals) of 85 mg SM-2 Biobeads prewashed with methanol and water until the solution became visibly cloudy. The reconstituted liposomes were purified on a sucrose gradient to separate the free proteins from the reconstituted protein-liposome. The visible liposome band was extracted and dialyzed against three changes of 20 mM Tris and 150 mM NaCl, pH 7.4. Successful incorporations of talin head and integrins were verified by SDS-PAGE and Cryo-EM tomography.
Recombinant talin F3 expression and liposome cosedimentation assay
Talin F3 and F3(K322D) were expressed and purified as described previously (Wegener et al., 2007). In brief, GST-F3 or GST-F3(K322D) was expressed with pGEX-6P vector and purified with glutathione-conjugated agarose beads. F3 or F3(K322D) was then cleaved off from the GST using PreScission protease (GE Healthcare), and further purified with a Superdex 200 size exclusion column. The molecular weights of purified F3 or F3 mutant measured by matrix-assisted laser desorption mass spectrometry varied by less than 0.1% from calculated values. For the liposome cosedimentation assay, liposomes were prepared as described above except that no integrins were incorporated. 1 ml of 5 µg/ml purified F3 or F3 (K322D) was incubated with 100 µl of liposomes at room temperature for 1 h. The liposomes were centrifuged at 14,000 rpm for 30 min. The liposome pellets were washed and solubilized with 1× SDS-PAGE loading buffer and analyzed by SDS-PAGE. Coomassie-stained protein bands were scanned and quantified using an infrared fluorescence spectrometer (LI-COR Biosciences).
Liposome integrin activity assays
PAC1 binding was measured by fluorescence-activated cell sorting (FACS). For the analysis, 9 µl of purified liposomes were mixed with 6 µl of 25 µg/ml FITC-conjugated PAC1. After incubation for 30 min, the sample was diluted into 150 µl 20 mM Hepes and 150 mM NaCl, pH 7.4 (HBS buffer) and analyzed using FACSCalibur (BD). For each sample, separate FACS analysis was performed with addition of 1 mM MnCl2 as a control for full activation and MnCl2 plus 20 µM eptifibatide as a negative control. Because larger liposomes would have more integrin incorporated and thus exhibit higher PAC1 binding, we plotted PAC1 binding against liposome size using forward scattering (FSC) as a size measurement. This plot helped us compare the PAC1 binding of liposomes with the same size. The PAC1-FSC plots were further analyzed by dividing the liposomes into 11 subsets along the FSC axis. The mean FSC and mean fluorescent intensity (MFI) of PAC1 binding was calculated for each subset. The activation indexes were calculated as 100 × (F − F0) / (Fmax − F0), where F = PAC1 binding, F0 = binding in the presence of 20 µM eptifibatide, and Fmax = binding in the presence of 1 mM MnCl2. The PAC1 MFI or the activation index was then plotted against the FSC.
Assaying the effect of THD in CHO cells stably expressing αIIbβ3
Using a lentiviral cloning vector pRRLSIN.cPPT.PGK-IRES-GFP.WPRE. (Addgene, plasmid ID 12252), viruses containing αIIb or β3 gene were generated separately as described previously (Wiznerowicz and Trono, 2003). CHO cells stably expressing integrin αIIbβ3 were established by co-infection of CHO K1 cells with the αIIb and β3 viruses. The Y747A and Y759A mutations were introduced into the β3 gene with the QuikChange Mutagenesis kit. To assess the effects of THD on the activation of integrins, 3 µg THD expression construct and 0.1 µg D-tomato (a transfection marker) was cotransfected. After 24 h, the cells were trypsinized, stained with PAC1 for 30 min, washed, stained with APC-conjugated anti–mouse IgM, and then analyzed by FACS. Separate FACS was also performed in the presence of anti-LIBS6 as full activation control and in presence 20 µM eptifibatide as negative control. To assess the effect of kindlin-2 on those cells, 3 µg THD expression construct, 0.5 µg of kindlin-2 construct, and 0.1 µg D-tomato construct were cotransfected into the cells, and the cells were stained as described above and analyzed by FACS.
Assay for calpain cleavage
To cleave β3 tail from the purified αIIbβ3, 25 µl of 1 mg/ml recombinant calpain-II (EMD) was added to 500 µl of 2 mg/ml purified αIIbβ3 in a buffer of TBS plus 0.1% Triton X-100, 1 mM CaCl2, and 1 mM MgCl2. The mixture was incubated at room temperature overnight. The next morning, the calpain was neutralized by adding protease inhibitor E-64 (Sigma-Aldrich) at a final concentration of 10 µM plus calpain inhibitor calpeptin at a final concentration of 5 µM. An ELISA assay was developed to measure the effectiveness of the cleavage. In brief, ELISA plates were coated with 5 µg/ml AP3 antibody overnight at 4°C, blocked with BSA for 1 h at 37°C, and incubated with 6 µg/ml of cleaved or uncleaved αIIbβ3 for 1 h at room temperature. After removing the protein solution and washing the ELISA plates, either Ab8053 against the whole protein, Ab8276 against the αIIb, or Ab8275 against the β3 tail were added to the captured αIIbβ3. After 1 h of incubation, the unbound antibodies were removed, the wells were washed again, and horseradish peroxidase (HRP)–conjugated goat anti–rabbit Ig antibodies were added for another hour of room temperature incubation. The amount of antibody binding was measured using enhanced chemiluminescence (ECL) reagent as peroxidase substrate (BD) and read on a Victor2 plate reader (PerkinElmer).
Integrin activity assay
For the integrin activity assay, ELISA plates were coated with 5 µg/ml AP3 antibody overnight at 4°C, blocked with BSA for 1 h at 37°C. After washing the plate, 6 µg/ml integrin with different amounts of talin head were added on to the plate. The plate was incubated for 2.5 h at room temperature. The wells were then washed and detected with either PAC1 antibody to measure the activity or anti-β3 Ab8053 to measure the capture. 1 mM MnCl2 and 1 mM MnCl2 plus 20 µM eptifibatide were used as positive and negative controls. After 1 h of incubation, the wells were again washed and HRP-conjugated anti–mouse IgM (for PAC1 wells) or HRP-conjugated anti–rabbit Igs (for Ab8053 wells) were added for 1 more hour of incubation. After the final wash, ECL reagent was added to the wells and the plate was read on a Victor2 plate reader (PerkinElmer). The procedure for the alternative activity assay configuration was similar to that described above, except that the plates were coated with 25 µg/ml of PAC1 antibody and that anti-β3 Ab8053 followed by HRP-conjugated anti–rabbit antibody was used to detect the amount of bound integrin.
Integrin nanodiscs assembly and purification
Integrin nanodiscs were assembled based on a protocol adapted from previous papers (Denisov et al., 2004; Nath et al., 2007). In brief, DMPC and DMPG were solubilized in chloroform or a chloroform/methanol mixture, mixed thoroughly, and dried onto a glass tube under steady flow of nitrogen. The homogeneous lipid mixture was then solubilized in 100 mM cholate in 10 mM Tris and 100 mM NaCl, pH 7.4, giving a lipid concentration of 50 mM. 72 µl of the lipid solution was then mixed with 200 µl of 200 µM membrane scaffold protein (MSP) in dH2O and 200 µl of 10 µM purified inactive integrin (described earlier). The final ratio of lipids/MSP/protein is 90:1:0.05 in a total volume of 472 µl. The integrin nanodiscs were assembled by removing the detergents with SM-2 Biobeads overnight at room temperature. The assembled integrin nanodiscs were then purified with a hi-load 16/60 Superdex 200 size exclusion column with 20 mM Tris, 150 mM NaCl, and 0.5 mM CaCl2, pH 7.4, as the column buffer. The integrin nanodiscs and empty nanodiscs were readily separated (Fig. S2) and the successful assembly was verified by SDS-PAGE analysis. Two MSP constructs, MSP1D1 and MSP1E3D1 (Denisov et al., 2004) expressed and purified from bacteria, were used to make integrin nanodiscs and similar patterns of integrin activation results were obtained with both constructs.
Analytical ultracentrifugation
Analytical ultracentrifugation was performed with a ProteomeLab (model XL-I) centrifuge according to the instructions from Beckman Coulter and established theories (Lebowitz et al., 2002). In brief, the integrin nanodiscs and empty nanodiscs were prepared and concentrated to 1 mg/ml using a Centricon concentrator with a molecular cut-off of 3 kD. For sedimentation equilibrium experiments, samples were analyzed in triplicate in three centrifuge chambers at different dilutions. The distributions of protein along the centrifugation force were determined by UV absorption at 280 nm. The measured protein distribution curves were then fitted to a theoretical model to determine the molecular weight. Buffer density calculated based on buffer composition and average protein partial specific volume was used as a parameter for the curve fitting. The molecular weight standard deviations were calculated from triplicates and the curve fitting residuals. The empty nanodiscs were analyzed at the centrifuge speed of 13,000 rpm. The integrin nanodiscs were first analyzed at a centrifuge speed of 5,000 rpm. We observed a high base absorption (the lowest 280-nm absorption at the start of the curve is 0.35 at 5,000 rpm; Fig. S2 B) and a persistent and high fitting residual, suggesting the presence of a second, lighter component in the solution. Therefore, we repeated the experiments to measure the lighter component at 10,000 rpm, at which the 530-kD component was calculated to be mostly at the bottom third of the centrifuge chamber and would not interfere with the measurement of the lighter component. For sedimentation velocity experiments, the samples were centrifuged at 20,000 rpm and the sedimentation coefficient curves were determined according to Beckman Coulter. The curves were then fitted to determine the sedimentation coefficient of each component in the solution and their ratios.
Integrin nanodisc activation assay
Integrin nanodisc activation assays were performed in a similar manner to that used to study the detergent-solubilized integrin as described above. In brief, ELISA plates were coated with 5 µg/ml AP3 antibody overnight at 4°C, blocked with BSA for 1 h at 37°C. After washing the plate, integrin nanodiscs were added to the plate. The plate was incubated for 3 h at room temperature. The integrin nanodiscs were then activated by various concentrations of THD as indicated, and activation was detected by binding of PAC1 antibody. PAC1 binding in the presence of anti-LIBS6 antibody was used as control for full activation and PAC1 binding in the presence of 20 µM eptifibatide was used as negative control. After 3 h of incubation, the wells were again washed and HRP-conjugated anti–mouse IgM (for PAC1 wells) was added for 1 more hour of incubation. After the final wash, ECL reagent was added to the wells and the luminescence of each well was read on a Victor2 plate reader. The activation indices were calculated as 100 × (L − L0) / (Lmax − L0), where L = luminescence intensity, L0 = luminescence in presence of 1 µM eptifibatide, and Lmax = luminescence in the presence of anti-LIBS6 antibody. Increase in activation was calculated as AIwith-THD − AIintegrin-alone, where AI stands for activation indices.
Integrin nanodiscs binding to fibrin and negatively stained EM images of integrin nanodiscs
2D fibrin was prepared by lipid monolayer technique as described previously (Taylor and Taylor, 1999; Taylor et al., 2007). In brief, wells in a Teflon block were filled with a fibrinogen solution. Thrombin was added to each well and a positively charged lipid solution was immediately applied to the solution surface to form the lipid monolayer. The samples were incubated at room temperature for 1 h while fibrin polymerized, and were then stored at 4°C. The fibrin was imaged to verify its 2D integrity and appropriate concentration. To test their binding to fibrin, the integrin nanodiscs were first incubated (with a THD concentration of either 0 or 5 µM) for 2 h then injected into the fibrin sample well where the final buffer concentration was 20 mM Tris, 50 mM NaCl, and 0.2 mM CalCl2, pH 7.6, and incubated for 24 h at 4°C. The resulting lipid monolayer sample was lifted on a reticulated carbon film and stained with 2% uranyl acetate. Images of the fibrin with integrin nanodiscs were taken on a transmission electron microscope (model 1200EX; JEOL) at a magnification of 65,000. The images were recorded on Kodak film and were digitized with a film scanner (Coolscan 9000; Nikon) to give a final pixel size of 3.9Å. The numbers of bound integrin nanodiscs and of unbound integrin nanodiscs were counted manually. The length of the fibrin was measured in ImageJ by tracing the fibrin fiber with segmented line.
Cryoelectron tomography
Specimens were prepared on glow-discharged, 200-mesh copper grids with reticulated carbon support film with a 2-µm hole size (Quantifoil, Inc.). The grids were plunge-frozen in liquid ethane and examined at a temperature of −180°C. The data were collected at a magnification of 24,000 on an electron microscope (model CM300-FEG; FEI) (integrin liposomes) and at a magnification of 31,000 on an electron microscope (Polara; FEI) (integrin + THD liposomes), each equipped with a 4 × 4-k CCD camera. The tomography tilt series was collected at an underfocus of 6–8 µm. The tilt range covered −65° to 65° with Saxton scheme angle increments with an initial angle of 2.0° and 1.5°, respectively (Saxton et al., 1984). The raw tilt images were processed and aligned with a marker-free alignment algorithm and the 3D reconstruction was calculated by weighted back projection (Winkler and Taylor, 2006).
Particle selection and 2D averaging
All images processing were performed with the EMAN1.9 package (Ludtke et al., 1999). Integrin nanodisc molecular images were identified in images. Images of well-resolved integrin nanodiscs not bound to fibrin were used for further image processing. In total, we obtained 1,283 images for integrin nanodiscs alone and 1,553 images for integrin nanodiscs in the presence of THD. The extracted molecular images were normalized, contrast transfer function (CTF) corrected by phase flipping, and low-pass filtered to 10Å resolution. The images were then subjected to an iterative classification, alignment, and averaging algorithm. No initial model or references were used and the data robustly converged to a final set of class averages. Minimum number of particles per class was set to be 20. Typical command used for 2D average and classification is: “refine2d.py–iter=41–ninitcls=10–finalsep=2–nbasis=64–minptcl=20”. The integrin nanodiscs activated by THD separated into class averages of either compact or extended conformations. However, due to the smaller number and the lower percentage of extended conformation in the sample of integrin nanodiscs alone, the algorithm failed to generate any classes attributable to the extended conformation despite repeated efforts with various parameter settings. The total numbers of extended or compact integrin nanodiscs were obtained by summing the number of images in the classes showing extended or compact conformation. The class averages showing only nanodisc density were treated as contaminating empty nanodiscs and their numbers were not included the activity calculation.
Online supplemental material
Fig. S1: THD(K322D) binds β3 tails. Fig. S2: Purification of integrin nanodiscs. Fig. S3: The entire 18 class averages of the integrin nanodiscs. Fig. S4: The entire 27 class averages of integrin nanodiscs in the presence of 5 µM THD. Fig. S5: Preparation and activity of inactive integrins. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200908045/DC1.
Acknowledgments
F. Ye and M.H. Ginsberg conceived the project, designed the experiments, and wrote the paper. F. Ye performed most of the experiments. B. Ratnikov performed preliminary studies of THD effect on purified detergent-soluble integrins. S.G. Sligar and M.A. McLean provided the membrane scaffold proteins for nanodisc construction and technical help. K.A. Taylor guided the EM experiments that were performed by F. Ye, G. Hu, and D. Taylor. A.A. Bobkov performed the DSC and analytical ultracentrifuge experiments. We thank Dr. Jun Liu for enabling use of the microscopy facility at The University of Texas Health Science Center at Houston.
This work was supported by National Institutes of Health (NIH) grants HL 078784, HL 31950, and AR 27214 to M.H. Ginsberg and by an American Heart Association post doctoral fellowship (09POST2180011) to F. Ye. The work was also supported by the NIH Cell Migration Consortium (NIH GM64346; to K.A. Taylor, S.G. Sligar, and M.H. Ginsberg).
Footnotes
Abbreviations used in this paper:
- THD
- talin head domain
References
- Adair B.D., Yeager M. 2002. Three-dimensional model of the human platelet integrin alpha IIbbeta 3 based on electron cryomicroscopy and x-ray crystallography. Proc. Natl. Acad. Sci. USA. 99:14059–14064 10.1073/pnas.212498199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adair B.D., Xiong J.P., Maddock C., Goodman S.L., Arnaout M.A., Yeager M. 2005. Three-dimensional EM structure of the ectodomain of integrin alphaVbeta3 in a complex with fibronectin. J. Cell Biol. 168:1109–1118 10.1083/jcb.200410068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzoni G., Hemler M.E. 1998. Are changes in integrin affinity and conformation overemphasized? Trends Biochem. Sci. 23:30–34 10.1016/S0968-0004(97)01141-9 [DOI] [PubMed] [Google Scholar]
- Calderwood D.A., Zent R., Grant R., Rees D.J., Hynes R.O., Ginsberg M.H. 1999. The Talin head domain binds to integrin beta subunit cytoplasmic tails and regulates integrin activation. J. Biol. Chem. 274:28071–28074 10.1074/jbc.274.40.28071 [DOI] [PubMed] [Google Scholar]
- Calderwood D.A., Tai V., Di Paolo G., De Camilli P., Ginsberg M.H. 2004. Competition for talin results in trans-dominant inhibition of integrin activation. J. Biol. Chem. 279:28889–28895 10.1074/jbc.M402161200 [DOI] [PubMed] [Google Scholar]
- Carman C.V., Springer T.A. 2003. Integrin avidity regulation: are changes in affinity and conformation underemphasized? Curr. Opin. Cell Biol. 15:547–556 10.1016/j.ceb.2003.08.003 [DOI] [PubMed] [Google Scholar]
- Collier N.C., Wang K. 1982. Purification and properties of human platelet P235. A high molecular weight protein substrate of endogenous calcium-activated protease(s). J. Biol. Chem. 257:6937–6943 [PubMed] [Google Scholar]
- Denisov I.G., Grinkova Y.V., Lazarides A.A., Sligar S.G. 2004. Directed self-assembly of monodisperse phospholipid bilayer Nanodiscs with controlled size. J. Am. Chem. Soc. 126:3477–3487 10.1021/ja0393574 [DOI] [PubMed] [Google Scholar]
- Di Paolo G., Pellegrini L., Letinic K., Cestra G., Zoncu R., Voronov S., Chang S., Guo J., Wenk M.R., De Camilli P. 2002. Recruitment and regulation of phosphatidylinositol phosphate kinase type 1 gamma by the FERM domain of talin. Nature. 420:85–89 10.1038/nature01147 [DOI] [PubMed] [Google Scholar]
- Du X.P., Plow E.F., Frelinger A.L., III, O’Toole T.E., Loftus J.C., Ginsberg M.H. 1991. Ligands “activate” integrin alpha IIb beta 3 (platelet GPIIb-IIIa). Cell. 65:409–416 10.1016/0092-8674(91)90458-B [DOI] [PubMed] [Google Scholar]
- Du X., Gu M., Weisel J.W., Nagaswami C., Bennett J.S., Bowditch R., Ginsberg M.H. 1993. Long range propagation of conformational changes in integrin alpha IIb beta 3. J. Biol. Chem. 268:23087–23092 [PubMed] [Google Scholar]
- Du X., Saido T.C., Tsubuki S., Indig F.E., Williams M.J., Ginsberg M.H. 1995. Calpain cleavage of the cytoplasmic domain of the integrin beta 3 subunit. J. Biol. Chem. 270:26146–26151 10.1074/jbc.270.46.27766 [DOI] [PubMed] [Google Scholar]
- Frelinger A.L., III, Du X.P., Plow E.F., Ginsberg M.H. 1991. Monoclonal antibodies to ligand-occupied conformers of integrin alpha IIb beta 3 (glycoprotein IIb-IIIa) alter receptor affinity, specificity, and function. J. Biol. Chem. 266:17106–17111 [PubMed] [Google Scholar]
- Friedland J.C., Lee M.H., Boettiger D. 2009. Mechanically activated integrin switch controls alpha5beta1 function. Science. 323:642–644 10.1126/science.1168441 [DOI] [PubMed] [Google Scholar]
- García-Alvarez B., de Pereda J.M., Calderwood D.A., Ulmer T.S., Critchley D., Campbell I.D., Ginsberg M.H., Liddington R.C. 2003. Structural determinants of integrin recognition by talin. Mol. Cell. 11:49–58 10.1016/S1097-2765(02)00823-7 [DOI] [PubMed] [Google Scholar]
- Ginsberg M.H., Partridge A., Shattil S.J. 2005. Integrin regulation. Curr. Opin. Cell Biol. 17:509–516 10.1016/j.ceb.2005.08.010 [DOI] [PubMed] [Google Scholar]
- Goldmann W.H. 2000. Kinetic determination of focal adhesion protein formation. Biochem. Biophys. Res. Commun. 271:553–557 10.1006/bbrc.2000.2653 [DOI] [PubMed] [Google Scholar]
- Han J., Lim C.J., Watanabe N., Soriani A., Ratnikov B., Calderwood D.A., Puzon-McLaughlin W., Lafuente E.M., Boussiotis V.A., Shattil S.J., Ginsberg M.H. 2006. Reconstructing and deconstructing agonist-induced activation of integrin alphaIIbbeta3. Curr. Biol. 16:1796–1806 10.1016/j.cub.2006.08.035 [DOI] [PubMed] [Google Scholar]
- Hantgan R.R., Rocco M., Nagaswami C., Weisel J.W. 2001. Binding of a fibrinogen mimetic stabilizes integrin alphaIIbbeta3’s open conformation. Protein Sci. 10:1614–1626 10.1110/ps.3001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman G.T., Kalinowski M. 2008. Classification of heterogeneous electron microscopic projections into homogeneous subsets. Ultramicroscopy. 108:327–338 10.1016/j.ultramic.2007.05.005 [DOI] [PubMed] [Google Scholar]
- Hughes P.E., Diaz-Gonzalez F., Leong L., Wu C., McDonald J.A., Shattil S.J., Ginsberg M.H. 1996. Breaking the integrin hinge. A defined structural constraint regulates integrin signaling. J. Biol. Chem. 271:6571–6574 10.1074/jbc.271.12.6571 [DOI] [PubMed] [Google Scholar]
- Hynes R.O. 2002. Integrins: bidirectional, allosteric signaling machines. Cell. 110:673–687 10.1016/S0092-8674(02)00971-6 [DOI] [PubMed] [Google Scholar]
- Ithychanda S.S., Das M., Ma Y.Q., Ding K., Wang X., Gupta S., Wu C., Plow E.F., Qin J. 2009. Migfilin, a molecular switch in regulation of integrin activation. J. Biol. Chem. 284:4713–4722 10.1074/jbc.M807719200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki K., Mitsuoka K., Fujiyoshi Y., Fujisawa Y., Kikuchi M., Sekiguchi K., Yamada T. 2005. Electron tomography reveals diverse conformations of integrin alphaIIbbeta3 in the active state. J. Struct. Biol. 150:259–267 10.1016/j.jsb.2005.03.005 [DOI] [PubMed] [Google Scholar]
- Jennings L.K., Phillips D.R. 1982. Purification of glycoproteins IIb and III from human platelet plasma membranes and characterization of a calcium-dependent glycoprotein IIb-III complex. J. Biol. Chem. 257:10458–10466 [PubMed] [Google Scholar]
- Karpatkin S., Charmatz A. 1970. Hetrogeneity of human platelets. 3. Glycogen metabolism in platelets of different sizes. Br. J. Haematol. 19:135–143 10.1111/j.1365-2141.1970.tb01612.x [DOI] [PubMed] [Google Scholar]
- Kiema T., Lad Y., Jiang P., Oxley C.L., Baldassarre M., Wegener K.L., Campbell I.D., Ylänne J., Calderwood D.A. 2006. The molecular basis of filamin binding to integrins and competition with talin. Mol. Cell. 21:337–347 10.1016/j.molcel.2006.01.011 [DOI] [PubMed] [Google Scholar]
- Kloeker S., Major M.B., Calderwood D.A., Ginsberg M.H., Jones D.A., Beckerle M.C. 2004. The Kindler syndrome protein is regulated by transforming growth factor-beta and involved in integrin-mediated adhesion. J. Biol. Chem. 279:6824–6833 10.1074/jbc.M307978200 [DOI] [PubMed] [Google Scholar]
- Kouns W.C., Kirchhofer D., Hadváry P., Edenhofer A., Weller T., Pfenninger G., Baumgartner H.R., Jennings L.K., Steiner B. 1992. Reversible conformational changes induced in glycoprotein IIb-IIIa by a potent and selective peptidomimetic inhibitor. Blood. 80:2539–2547 [PubMed] [Google Scholar]
- Krüger M., Moser M., Ussar S., Thievessen I., Luber C.A., Forner F., Schmidt S., Zanivan S., Fässler R., Mann M. 2008. SILAC mouse for quantitative proteomics uncovers kindlin-3 as an essential factor for red blood cell function. Cell. 134:353–364 10.1016/j.cell.2008.05.033 [DOI] [PubMed] [Google Scholar]
- Lad Y., Jiang P., Ruskamo S., Harburger D.S., Ylänne J., Campbell I.D., Calderwood D.A. 2008. Structural basis of the migfilin-filamin interaction and competition with integrin beta tails. J. Biol. Chem. 283:35154–35163 10.1074/jbc.M802592200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau T.L., Kim C., Ginsberg M.H., Ulmer T.S. 2009. The structure of the integrin alphaIIbbeta3 transmembrane complex explains integrin transmembrane signalling. EMBO J. 28:1351–1361 10.1038/emboj.2009.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebowitz J., Lewis M.S., Schuck P. 2002. Modern analytical ultracentrifugation in protein science: a tutorial review. Protein Sci. 11:2067–2079 10.1110/ps.0207702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.S., Lim C.J., Puzon-McLaughlin W., Shattil S.J., Ginsberg M.H. 2009. RIAM activates integrins by linking talin to ras GTPase membrane-targeting sequences. J. Biol. Chem. 284:5119–5127 10.1074/jbc.M807117200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Mitra N., Gratkowski H., Vilaire G., Litvinov R., Nagasami C., Weisel J.W., Lear J.D., DeGrado W.F., Bennett J.S. 2003. Activation of integrin alphaIIbbeta3 by modulation of transmembrane helix associations. Science. 300:795–798 10.1126/science.1079441 [DOI] [PubMed] [Google Scholar]
- Ludtke S.J., Baldwin P.R., Chiu W. 1999. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 128:82–97 10.1006/jsbi.1999.4174 [DOI] [PubMed] [Google Scholar]
- Luo B.H., Springer T.A., Takagi J. 2004. A specific interface between integrin transmembrane helices and affinity for ligand. PLoS Biol. 2:e153 10.1371/journal.pbio.0020153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y.Q., Qin J., Wu C., Plow E.F. 2008. Kindlin-2 (Mig-2): a co-activator of beta3 integrins. J. Cell Biol. 181:439–446 10.1083/jcb.200710196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meves A., Stremmel C., Gottschalk K., Fässler R. 2009. The Kindlin protein family: new members to the club of focal adhesion proteins. Trends Cell Biol. 19:504–513 10.1016/j.tcb.2009.07.006 [DOI] [PubMed] [Google Scholar]
- Montanez E., Ussar S., Schifferer M., Bösl M., Zent R., Moser M., Fässler R. 2008. Kindlin-2 controls bidirectional signaling of integrins. Genes Dev. 22:1325–1330 10.1101/gad.469408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser M., Nieswandt B., Ussar S., Pozgajova M., Fässler R. 2008. Kindlin-3 is essential for integrin activation and platelet aggregation. Nat. Med. 14:325–330 10.1038/nm1722 [DOI] [PubMed] [Google Scholar]
- Moser M., Bauer M., Schmid S., Ruppert R., Schmidt S., Sixt M., Wang H.V., Sperandio M., Fässler R. 2009a. Kindlin-3 is required for beta2 integrin-mediated leukocyte adhesion to endothelial cells. Nat. Med. 15:300–305 10.1038/nm.1921 [DOI] [PubMed] [Google Scholar]
- Moser M., Legate K.R., Zent R., Fässler R. 2009b. The tail of integrins, talin, and kindlins. Science. 324:895–899 10.1126/science.1163865 [DOI] [PubMed] [Google Scholar]
- Nath A., Atkins W.M., Sligar S.G. 2007. Applications of phospholipid bilayer nanodiscs in the study of membranes and membrane proteins. Biochemistry. 46:2059–2069 10.1021/bi602371n [DOI] [PubMed] [Google Scholar]
- Newman P.J., Allen R.W., Kahn R.A., Kunicki T.J. 1985. Quantitation of membrane glycoprotein IIIa on intact human platelets using the monoclonal antibody, AP-3. Blood. 65:227–232 [PubMed] [Google Scholar]
- Nieswandt B., Moser M., Pleines I., Varga-Szabo D., Monkley S., Critchley D., Fässler R. 2007. Loss of talin1 in platelets abrogates integrin activation, platelet aggregation, and thrombus formation in vitro and in vivo. J. Exp. Med. 204:3113–3118 10.1084/jem.20071827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida N., Xie C., Shimaoka M., Cheng Y., Walz T., Springer T.A. 2006. Activation of leukocyte beta2 integrins by conversion from bent to extended conformations. Immunity. 25:583–594 10.1016/j.immuni.2006.07.016 [DOI] [PubMed] [Google Scholar]
- O’Toole T.E., Mandelman D., Forsyth J., Shattil S.J., Plow E.F., Ginsberg M.H. 1991. Modulation of the affinity of integrin alpha IIb beta 3 (GPIIb-IIIa) by the cytoplasmic domain of alpha IIb. Science. 254:845–847 10.1126/science.1948065 [DOI] [PubMed] [Google Scholar]
- O’Toole T.E., Katagiri Y., Faull R.J., Peter K., Tamura R., Quaranta V., Loftus J.C., Shattil S.J., Ginsberg M.H. 1994. Integrin cytoplasmic domains mediate inside-out signal transduction. J. Cell Biol. 124:1047–1059 10.1083/jcb.124.6.1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil S., Jedsadayanmata A., Wencel-Drake J.D., Wang W., Knezevic I., Lam S.C. 1999. Identification of a talin-binding site in the integrin beta(3) subunit distinct from the NPLY regulatory motif of post-ligand binding functions. The talin n-terminal head domain interacts with the membrane-proximal region of the beta(3) cytoplasmic tail. J. Biol. Chem. 274:28575–28583 10.1074/jbc.274.40.28575 [DOI] [PubMed] [Google Scholar]
- Penczek P.A., Frank J., Spahn C.M. 2006. A method of focused classification, based on the bootstrap 3D variance analysis, and its application to EF-G-dependent translocation. J. Struct. Biol. 154:184–194 10.1016/j.jsb.2005.12.013 [DOI] [PubMed] [Google Scholar]
- Petrich B.G., Fogelstrand P., Partridge A.W., Yousefi N., Ablooglu A.J., Shattil S.J., Ginsberg M.H. 2007a. The antithrombotic potential of selective blockade of talin-dependent integrin alpha IIb beta 3 (platelet GPIIb-IIIa) activation. J. Clin. Invest. 117:2250–2259 10.1172/JCI31024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrich B.G., Marchese P., Ruggeri Z.M., Spiess S., Weichert R.A., Ye F., Tiedt R., Skoda R.C., Monkley S.J., Critchley D.R., Ginsberg M.H. 2007b. Talin is required for integrin-mediated platelet function in hemostasis and thrombosis. J. Exp. Med. 204:3103–3111 10.1084/jem.20071800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puklin-Faucher E., Gao M., Schulten K., Vogel V. 2006. How the headpiece hinge angle is opened: New insights into the dynamics of integrin activation. J. Cell Biol. 175:349–360 10.1083/jcb.200602071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton W.O., Baumeister W., Hahn M. 1984. Three-dimensional reconstruction of imperfect two-dimensional crystals. Ultramicroscopy. 13:57–70 10.1016/0304-3991(84)90057-3 [DOI] [PubMed] [Google Scholar]
- Scarborough R.M., Naughton M.A., Teng W., Rose J.W., Phillips D.R., Nannizzi L., Arfsten A., Campbell A.M., Charo I.F. 1993. Design of potent and specific integrin antagonists. Peptide antagonists with high specificity for glycoprotein IIb-IIIa. J. Biol. Chem. 268:1066–1073 [PubMed] [Google Scholar]
- Segrest J.P., Jones M.K., Klon A.E., Sheldahl C.J., Hellinger M., De Loof H., Harvey S.C. 1999. A detailed molecular belt model for apolipoprotein A-I in discoidal high density lipoprotein. J. Biol. Chem. 274:31755–31758 10.1074/jbc.274.45.31755 [DOI] [PubMed] [Google Scholar]
- Shaikh T.R., Trujillo R., LeBarron J.S., Baxter W.T., Frank J. 2008. Particle-verification for single-particle, reference-based reconstruction using multivariate data analysis and classification. J. Struct. Biol. 164:41–48 10.1016/j.jsb.2008.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shattil S.J., Newman P.J. 2004. Integrins: dynamic scaffolds for adhesion and signaling in platelets. Blood. 104:1606–1615 10.1182/blood-2004-04-1257 [DOI] [PubMed] [Google Scholar]
- Shattil S.J., Hoxie J.A., Cunningham M., Brass L.F. 1985. Changes in the platelet membrane glycoprotein IIb.IIIa complex during platelet activation. J. Biol. Chem. 260:11107–11114 [PubMed] [Google Scholar]
- Shi X., Ma Y.Q., Tu Y., Chen K., Wu S., Fukuda K., Qin J., Plow E.F., Wu C. 2007. The MIG-2/integrin interaction strengthens cell-matrix adhesion and modulates cell motility. J. Biol. Chem. 282:20455–20466 10.1074/jbc.M611680200 [DOI] [PubMed] [Google Scholar]
- Shimaoka M., Takagi J., Springer T.A. 2002. Conformational regulation of integrin structure and function. Annu. Rev. Biophys. Biomol. Struct. 31:485–516 10.1146/annurev.biophys.31.101101.140922 [DOI] [PubMed] [Google Scholar]
- Tadokoro S., Shattil S.J., Eto K., Tai V., Liddington R.C., de Pereda J.M., Ginsberg M.H., Calderwood D.A. 2003. Talin binding to integrin beta tails: a final common step in integrin activation. Science. 302:103–106 10.1126/science.1086652 [DOI] [PubMed] [Google Scholar]
- Takagi J., Petre B.M., Walz T., Springer T.A. 2002. Global conformational rearrangements in integrin extracellular domains in outside-in and inside-out signaling. Cell. 110:599–11 10.1016/S0092-8674(02)00935-2 [DOI] [PubMed] [Google Scholar]
- Takagi J., Strokovich K., Springer T.A., Walz T. 2003. Structure of integrin alpha5beta1 in complex with fibronectin. EMBO J. 22:4607–4615 10.1093/emboj/cdg445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor K.A., Taylor D.W. 1999. Structural studies of cytoskeletal protein arrays formed on lipid monolayers. J. Struct. Biol. 128:75–81 10.1006/jsbi.1999.4167 [DOI] [PubMed] [Google Scholar]
- Taylor D.W., Kelly D.F., Cheng A., Taylor K.A. 2007. On the freezing and identification of lipid monolayer 2-D arrays for cryoelectron microscopy. J. Struct. Biol. 160:305–312 10.1016/j.jsb.2007.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu Y., Wu S., Shi X., Chen K., Wu C. 2003. Migfilin and Mig-2 link focal adhesions to filamin and the actin cytoskeleton and function in cell shape modulation. Cell. 113:37–47 10.1016/S0092-8674(03)00163-6 [DOI] [PubMed] [Google Scholar]
- Wagner C.L., Mascelli M.A., Neblock D.S., Weisman H.F., Coller B.S., Jordan R.E. 1996. Analysis of GPIIb/IIIa receptor number by quantification of 7E3 binding to human platelets. Blood. 88:907–914 [PubMed] [Google Scholar]
- Wegener K.L., Partridge A.W., Han J., Pickford A.R., Liddington R.C., Ginsberg M.H., Campbell I.D. 2007. Structural basis of integrin activation by talin. Cell. 128:171–182 10.1016/j.cell.2006.10.048 [DOI] [PubMed] [Google Scholar]
- Winkler H., Taylor K.A. 2006. Accurate marker-free alignment with simultaneous geometry determination and reconstruction of tilt series in electron tomography. Ultramicroscopy. 106:240–254 10.1016/j.ultramic.2005.07.007 [DOI] [PubMed] [Google Scholar]
- Winkler J., Lünsdorf H., Jockusch B.M. 1997. Energy-filtered electron microscopy reveals that talin is a highly flexible protein composed of a series of globular domains. Eur. J. Biochem. 243:430–436 10.1111/j.1432-1033.1997.0430a.x [DOI] [PubMed] [Google Scholar]
- Wiznerowicz M., Trono D. 2003. Conditional suppression of cellular genes: lentivirus vector-mediated drug-inducible RNA interference. J. Virol. 77:8957–8961 10.1128/JVI.77.16.8957-8951.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong J.P., Stehle T., Diefenbach B., Zhang R., Dunker R., Scott D.L., Joachimiak A., Goodman S.L., Arnaout M.A. 2001. Crystal structure of the extracellular segment of integrin alpha Vbeta3. Science. 294:339–345 10.1126/science.1064535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong J.P., Stehle T., Zhang R., Joachimiak A., Frech M., Goodman S.L., Arnaout M.A. 2002. Crystal structure of the extracellular segment of integrin alpha Vbeta3 in complex with an Arg-Gly-Asp ligand. Science. 296:151–155 10.1126/science.1069040 [DOI] [PubMed] [Google Scholar]
- Yan B., Calderwood D.A., Yaspan B., Ginsberg M.H. 2001. Calpain cleavage promotes talin binding to the beta 3 integrin cytoplasmic domain. J. Biol. Chem. 276:28164–28170 10.1074/jbc.M104161200 [DOI] [PubMed] [Google Scholar]
- Ye F., Liu J., Winkler H., Taylor K.A. 2008. Integrin alpha IIb beta 3 in a membrane environment remains the same height after Mn2+ activation when observed by cryoelectron tomography. J. Mol. Biol. 378:976–986 10.1016/j.jmb.2008.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zent R., Fenczik C.A., Calderwood D.A., Liu S., Dellos M., Ginsberg M.H. 2000. Class- and splice variant-specific association of CD98 with integrin beta cytoplasmic domains. J. Biol. Chem. 275:5059–5064 10.1074/jbc.275.7.5059 [DOI] [PubMed] [Google Scholar]
- Zhu J., Boylan B., Luo B.H., Newman P.J., Springer T.A. 2007. Tests of the extension and deadbolt models of integrin activation. J. Biol. Chem. 282:11914–11920 10.1074/jbc.M700249200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Luo B.H., Xiao T., Zhang C., Nishida N., Springer T.A. 2008. Structure of a complete integrin ectodomain in a physiologic resting state and activation and deactivation by applied forces. Mol. Cell. 32:849–861 10.1016/j.molcel.2008.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]