COG subunits localize to the phagophore assembly site where they interact with autophagy proteins and are required for double-membrane Cvt vesicle and autophagosome formation.
Abstract
Macroautophagy is a catabolic pathway used for the turnover of long-lived proteins and organelles in eukaryotic cells. The morphological hallmark of this process is the formation of double-membrane autophagosomes that sequester cytoplasm. Autophagosome formation is the most complex part of macroautophagy, and it is a dynamic event that likely involves vesicle fusion to expand the initial sequestering membrane, the phagophore; however, essentially nothing is known about this process including the molecular components involved in vesicle tethering and fusion. In this study, we provide evidence that the subunits of the conserved oligomeric Golgi (COG) complex are required for double-membrane cytoplasm to vacuole targeting vesicle and autophagosome formation. COG subunits localized to the phagophore assembly site and interacted with Atg (autophagy related) proteins. In addition, mutations in the COG genes resulted in the mislocalization of Atg8 and Atg9, which are critical components involved in autophagosome formation.
Introduction
Macroautophagy (hereafter autophagy), an evolutionarily conserved catabolic pathway in eukaryotic cells, is involved in the degradation of long-lived proteins, organelles, and a large portion of the cytoplasm in response to internal and external stresses such as nutrient starvation (Levine and Klionsky, 2004; Reggiori and Klionsky, 2005). In addition, autophagy is involved in development and cellular remodeling and plays a cytoprotective role by removing protein aggregates and damaged or superfluous organelles, which may contribute to a role in life span extension (Klionsky and Emr, 2000; Ogier-Denis and Codogno, 2003; Shintani and Klionsky, 2004a; Yen and Klionsky, 2008). Defects in autophagy are associated with various diseases, such as cancer, gastrointestinal disorders, and neurodegeneration (Ogier-Denis and Codogno, 2003; Shintani and Klionsky, 2004a; Huang and Klionsky, 2007). The hallmark of autophagy is the sequestration of bulk cytoplasm into a double-membrane vesicle termed an autophagosome, which then fuses with the lysosome/vacuole, releasing the inner vesicle for degradation (Klionsky and Emr, 2000). Genetic screens in Saccharomyces cerevisiae and other fungi have identified >30 ATG (autophagy-related) genes that are involved in autophagy (Klionsky et al., 2003), and orthologues of many of the yeast ATG genes have been identified in higher eukaryotes.
Although autophagy is generally considered to be a nonselective process, several selective autophagic pathways for specific cargo delivery have been reported in both fungi and mammalian cells. One example in the yeast S. cerevisiae is a constitutive biosynthetic pathway that occurs under vegetative conditions, termed the cytoplasm to vacuole targeting (Cvt) pathway, in which certain vacuolar hydrolases such as precursor aminopeptidase I (Ape1 [prApe1]) are enwrapped in double-membrane Cvt vesicles, which fuse with the vacuole (Klionsky et al., 1992; Kim et al., 1997; Scott et al., 1997). The Cvt pathway shares similar morphological and mechanistic features with bulk autophagy and requires most of the same Atg proteins. Therefore, the study of the Cvt pathway and the analysis of the roles of the various Atg proteins have provided important information for understanding the molecular basis of autophagy; however, many questions remain to be addressed.
One of the unsolved mysteries with regard to autophagy is the origin of the membrane used to form the double-membrane sequestering vesicles and the mechanism of vesicle expansion (Reggiori and Klionsky, 2006). The autophagosome and Cvt vesicle are thought to form de novo, meaning they do not bud off intact from preexisting organelles as occurs with transport vesicles that function in the secretory pathway. Instead, the double-membrane vesicles appear to expand via the addition of membrane through vesicular fusion; this mode of formation is critical in allowing these sequestering vesicles to accommodate essentially any sized cargo. However, very little information is available regarding the presumed tethering and fusion components that are involved in this process.
In this paper, we screened the yeast deletion library and a set of temperature-sensitive mutants and found that mutants of the conserved oligomeric Golgi (COG) complex subunits were defective in the Cvt and autophagy pathways. In S. cerevisiae, the COG complex contains eight subunits (Cog1–8) and is composed of two lobes: lobe A (Cog2–4) and lobe B (Cog5–8) with Cog1 in the center (VanRheenen et al., 1998, 1999; Kim et al., 1999, 2001a; Whyte and Munro, 2001; Ram et al., 2002; Suvorova et al., 2002). Although only six of the yeast COG subunits show high sequence homology with their corresponding mammalian counterparts, the COG complex is structurally and functionally conserved (Ungar et al., 2002). The current evidence suggests that COG is mainly important for retrograde trafficking within the Golgi complex and possibly for ER to Golgi and endosome to Golgi transport as a tethering factor (VanRheenen et al., 1998, 1999; Kim et al., 1999; Whyte and Munro, 2001; Ram et al., 2002; Suvorova et al., 2002; Bruinsma et al., 2004; Oka et al., 2004; Zolov and Lupashin, 2005). Moreover, several pieces of evidence indicate that lobe A may be involved in different and more essential processes than lobe B: defects in any of the COG subunits in lobe A have more severe effects on cell growth than those in lobe B (Spelbrink and Nothwehr, 1999; Whyte and Munro, 2001; Bruinsma et al., 2004; Oka et al., 2004; Volchuk et al., 2004; Wu et al., 2004). In this study, we discovered that the lobe A but not the lobe B subunits are essential for autophagy. Additionally, the COG complex is involved in autophagosome formation and is required for correct sorting of various Atg membrane–associated proteins to the phagophore assembly site (PAS), the location of vesicle nucleation. COG subunits localize to the PAS and interact with Atg proteins. Our results show that the COG complex is involved in the formation of double-membrane sequestering vesicles during autophagy.
Results
The COG complex is involved in the Cvt pathway and autophagy
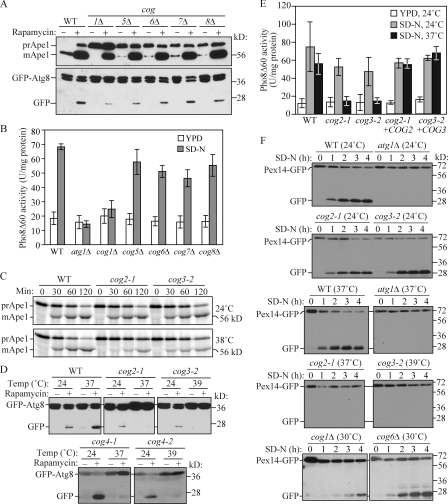
Previously, we screened the yeast deletion library to obtain mutants defective in the Cvt pathway (Nice et al., 2002; Wang et al., 2002; Reggiori et al., 2003; Strømhaug et al., 2004). Of the mutants we identified, the cog1Δ strain exhibited a relatively strong block in prApe1 maturation (Fig. 1 A). In rich medium in a wild-type strain, most of the Ape1 is present in the mature form as a result of delivery to the vacuole through the Cvt pathway. In the cog1Δ mutant, all of the Ape1 was present in the precursor form in rich medium conditions, indicating a block in the Cvt pathway.
Figure 1.
The Cvt, autophagy, and pexophagy pathways are defective in cog mutants. (A) The Cvt pathway is defective in COG deletion mutants. The wild-type (WT; BY4742), cog1Δ, cog5Δ, cog6Δ, cog7Δ, and cog8Δ cells were grown in rich medium, and rapamycin was added to the culture at a final concentration of 0.2 µg/ml. After 2 h, cell extracts were prepared and subjected to immunoblot analysis with anti-Ape1 or anti-YFP antisera. (B) Autophagic activity in the COG deletion mutants. The wild-type (YTS223), cog1Δ (YTS224), cog5Δ (YTS225), cog6Δ (YTS226), cog7Δ (YTS227), cog8Δ (YTS228), and atg1Δ (YTS243) cells expressing Pho8Δ60 were grown in YPD medium and incubated in SD-N for 3 h to induce autophagy. Cell extracts were prepared for measuring the Pho8Δ60-dependent alkaline phosphatase activity. Error bars indicate SD. (C) The Cvt pathway is defective in cog2-1 and cog3-2 mutants. The wild-type (BY4742), cog2-1, and cog3-2 cells were grown in rich medium at 24°C and shifted to SMD. After incubation at either 24 or 38°C for 30 min, cells were labeled with [35S]methionine/cysteine and subjected to a nonradioactive chase at the same temperature for 2 h. Ape1 was immunoprecipitated and subjected to SDS-PAGE. (D) GFP-Atg8 processing is defective in cog temperature-sensitive mutants. Wild-type (BY4742), cog2-1, cog3-2, cog4-1 (BCR29), and cog4-2 (BCR47) mutants were grown at permissive temperature until OD600 = 0.8. After incubation at either 24°C or nonpermissive temperatures (37 or 39°C) for 30 min, rapamycin was added for 2 h. Aliquots were collected, and protein extracts were subjected to immunoblotting analysis. (E) Autophagic activity in temperature-sensitive cog mutants. Wild-type (YTS223), cog2-1 (YTS230), cog3-2 (YTS231), and cog2-1 and cog3-2 strains harboring the pCuCOG2HA(416) and pCuCOG3HA(416) plasmids, respectively, expressing Pho8Δ60 were grown in YPD medium and incubated in SD-N at either 24 or 37°C for 3 h. Cell lysates were used to measure Pho8Δ60 activity. (F) COG complex subunits are required for pexophagy. The wild-type (IRA001), atg1Δ (IRA002), cog2-1 (WLY90), and cog3-2 (WLY91) cells expressing Pex14-GFP were grown under conditions to induce peroxisome proliferation as described in Materials and methods. After incubation at either 24°C or nonpermissive temperatures for 30 min, the cultures were shifted to SD-N. For cog1Δ (WLY175) and cog6Δ (WLY174) strains, the cells were grown at 30°C and after being induced for peroxisome proliferation were shifted to SD-N at the same temperature. Protein extracts were prepared at the indicated time points and resolved by SDS-PAGE.
Cog1 is one of the components of the COG complex (Ungar et al., 2006). Therefore, we examined whether the other components of this complex were also involved in the Cvt pathway. Deletion of COG5, COG6, COG7, or COG8 caused an incomplete but substantial block in prApe1 maturation in rich medium (Fig. 1 A). Treatment of yeast cells with the Tor inhibitor rapamycin induces autophagy and causes an increase in prApe1 synthesis, and this cargo protein is now transported to the vacuole through autophagy. In wild-type cells treated with rapamycin, the majority of Ape1 is still detected as the mature form despite the large increase in synthesis. Rapamycin treatment of the COG deletion strains, particularly those of lobe B, largely rescued the defect in prApe1 transport to the vacuole, suggesting that the selective uptake of prApe1 via autophagy occurred in these strains (Fig. 1 A); the cog1Δ strain showed an ∼50% block in prApe1 maturation, suggesting a significant block in nonspecific autophagosome formation.
Because import of prApe1 is a selective process even under starvation conditions, it is not an adequate measure of nonspecific autophagy. Therefore, we measured nonspecific autophagy in the COG deletion cells by two established biochemical analyses, GPF-Atg8 processing and Pho8Δ60 activity assays. Atg8 is a ubiquitin-like protein, which remains associated with completed autophagosomes after phosphatidylethanolamine conjugation (Kirisako et al., 1999, 2000; Huang et al., 2000). After GFP-tagged Atg8 is delivered into the vacuole, Atg8 is degraded, whereas the GFP moiety remains relatively stable. Thus, autophagy progression can be monitored by free GFP accumulation (Shintani and Klionsky, 2004b; Cheong et al., 2005). As shown in Fig. 1 A, there was essentially no detectable free GFP in rich conditions in the wild-type strain (the level from the Cvt pathway is typically too low to detect), but there was a clear band after rapamycin treatment. All of the cog mutants displayed some level of GFP-Atg8 processing, with the cog1Δ mutant showing the strongest block, suggesting that nonspecific autophagy is severely impaired in this mutant, which is in agreement with the prApe1 processing phenotype. To quantitatively measure autophagy activity, we used Pho8Δ60, a mutant form of the vacuolar alkaline phosphatase in which the N-terminal 60 amino acids including the transmembrane domain are deleted. Pho8Δ60 localizes to the cytoplasm, and its activation by proteolytic removal of a C-terminal propeptide via a vacuolar protease depends on autophagy (Noda et al., 1995). To monitor Pho8Δ60-dependent alkaline phosphatase activity, cells were grown in a rich medium and incubated in a nitrogen-starvation medium for 3 h to induce autophagy. Wild-type cells showed a low level of activity in rich medium and a substantial increase after autophagy induction (Fig. 1 B). The negative control atg1Δ cells that are defective in autophagy showed a basal level of Pho8Δ60 activity even after autophagy was induced for 3 h. Autophagic activity in the cog5Δ, cog6Δ, cog7Δ, and cog8Δ cells was comparable with that in the wild-type cells, whereas in the cog1Δ cells, it was significantly decreased even after autophagy induction, which is similar to the atg1Δ cells.
The COG complex is proposed to be composed of two lobes: lobe A, containing the Cog2–4 subunits, and lobe B, containing the Cog5–8 subunits. Cog1 is the central subunit that connects the two lobes (Fotso et al., 2005; Ungar et al., 2006). In yeast, the lobe A components are essential for cell growth. Therefore, we used temperature-sensitive cog2-1 and cog3-2 mutants (Wuestehube et al., 1996) to assess their requirement in the Cvt pathway and autophagy. The Cvt pathway was measured by monitoring prApe1 maturation by a pulse-chase experiment. Cells were incubated at a nonpermissive temperature for 30 min before labeling and were subjected to a nonradioactive chase at the same temperature for 2 h. The prApe1 processing was significantly affected in the mutant strains at the nonpermissive temperature, although no obvious delay in processing was seen in these mutants compared with the wild type at a permissive temperature (Fig. 1 C). These results suggested a kinetic defect in the Cvt pathway.
Next, autophagic activities of the cog temperature-sensitive mutants were measured by both GFP-Atg8 processing and the Pho8Δ60 assay. Cells were transformed with plasmid-based GFP-Atg8 and grown in a rich medium at permissive temperature to early log phase. Cell cultures were split into two aliquots: one aliquot was incubated at a permissive temperature and the other at a nonpermissive temperature for 30 min. Rapamycin was added to induce autophagy for 2 h. Cell lysates were collected and subjected to Western blotting using anti-YFP antibody. In the wild-type strain, GFP-Atg8 processing occurred at both temperatures with a greater level of free GFP detected at the elevated temperature (Fig. 1 D). In contrast, in the cog2-1 and cog3-2 temperature-sensitive mutants, GFP-Atg8 processing was blocked at the nonpermissive temperature, suggesting that these mutants had autophagy defects. Transformation of these mutant strains with plasmids encoding the wild-type corresponding COG subunits rescued the GFP-Atg8 processing defects at the nonpermissive temperature (unpublished data). To extend our analysis to Cog4, we tested two temperature-sensitive mutants in the COG4 gene that were designed based on structural experiments of human Cog4 to disrupt binding of Cog4 to other proteins (Richardson et al., 2009). Similar to the results with cog2-1 and cog3-2, the cog4-1 and cog4-2 mutants displayed defects in GFP-Atg8 processing at the nonpermissive temperature (Fig. 1 D).
To make a quantitative measurement of autophagy in these mutants, we performed the Pho8Δ60 assay. Wild-type cells showed the expected increase in Pho8Δ60-dependent alkaline phosphatase activity after autophagy induction, but they displayed a slight decrease in activity at the elevated temperature (Fig. 1 E). When the cog mutant cells were incubated at a nonpermissive temperature, autophagic activity was severely compromised, whereas the level of activity was essentially normal at permissive temperature. As with the GFP-Atg8 processing assay, transformation with plasmids expressing the corresponding wild-type COG proteins rescued the defect seen in the Pho8Δ60 activity at the nonpermissive temperature. These data further suggest that the lobe A COG subunits are needed for nonspecific autophagy.
Considering the Cvt pathway defects in cog mutants, we decided to test whether COG subunits are required for the specific degradation of peroxisomes. To do this, we monitored the vacuolar delivery of the peroxisomal integral membrane protein Pex14 that was tagged with GFP at the C terminus (Albertini et al., 1997; Reggiori et al., 2005a). Similar to GFP-Atg8, delivery of Pex14-GFP–tagged peroxisomes into the vacuole would result in the release of the GFP moiety, allowing us to monitor pexophagy progression through the appearance of free GFP (Reggiori et al., 2005a). Pexophagy was induced as described in Materials and methods. Free GFP was detected in the wild-type cells after being transferred to SD-N medium, whereas no free GFP was seen in the atg1Δ strain (Fig. 1 F). In contrast to the wild-type cells, no free GFP was observed in the cog2-1 or cog3-2 mutant at nonpermissive temperature, even though processing was normal at the permissive temperature. We also tested Pex14-GFP degradation in COG1 and COG6 deletion strains. There was a strong kinetic delay in Pex14-GFP processing in the cog1Δ strain but normal pexophagy in cog6Δ. These results indicated that lobe A but not lobe B of the COG complex was essential for autophagy and pexophagy, although all of the components were required for an efficient Cvt pathway.
Atg9 cycling is affected in cog mutants
Atg9 is the only transmembrane Atg protein that is essential for double-membrane vesicle formation. Atg9 is also required for organization of the PAS and recruitment of certain Atg proteins to this specialized site (Suzuki et al., 2001). Atg9 localizes to and cycles between the PAS and peripheral sites (Reggiori et al., 2004a,b). Atg9 transport to the PAS is affected in mutants defective in ER to Golgi trafficking (Reggiori et al., 2003; Reggiori and Klionsky, 2006), and those mutants have previously been shown to block the Cvt pathway and autophagy (Ishihara et al., 2001; Hamasaki et al., 2003; Reggiori et al., 2004b; Reggiori and Klionsky, 2006). Because Golgi complex structure and function are defective in cog mutants (Whyte and Munro, 2001; Ram et al., 2002; Suvorova et al., 2002), it is possible that Golgi structure abnormality in cog mutants may result in an Atg9-sorting defect, thus affecting both the Cvt and autophagy pathways. Accordingly, we examined Atg9 localization in the cog2-1 strain.
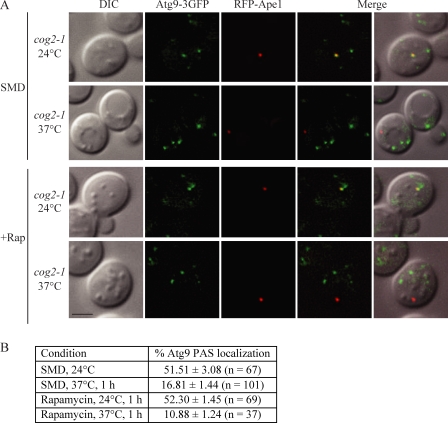
When the cog2-1 Atg9-3GFP strain carrying a plasmid expressing RFP-Ape1 was grown in rich medium or 1 h after autophagy was induced at permissive temperature, Atg9-3GFP was present in multiple puncta, one of which corresponds to the PAS as marked with RFP-Ape1 (Fig. 2 A). The frequencies of Atg9-3GFP PAS localization were 51.5 ± 3.08% and 52.3 ± 1.45% (Fig. 2 B). To test whether the COG complex is required for Atg9 transport to the PAS, we examined the Atg9-3GFP PAS localization frequency in cog2-1 at a nonpermissive temperature in both growing and autophagy-inducing conditions. When shifted to a nonpermissive temperature for 1 h, the Atg9-3GFP PAS localization frequency dropped to 16.81 ± 1.44%. Similarly, in the autophagy-induced condition, the PAS localization percentage dropped to 10.88 ± 1.24% (Fig. 2, A and B). These data suggest that the anterograde movement of Atg9 to the PAS was defective in the cog2-1 strain under both growing and autophagy-inducing conditions. Similar results were seen in cog1Δ and cog6Δ strains in vegetative conditions when we used an epistasis assay that relies on the atg1Δ phenotype; Atg9-3GFP localizes only to the PAS in the atg1Δ background unless a secondary mutation interferes with anterograde transport (Cheong et al., 2005). Atg9-3GFP was present in multiple puncta in atg1Δ cog1Δ or atg1Δ cog6Δ cells in synthetic minimal medium with dextrose (SMD), but single puncta were seen when the cells were shifted to SD-N (Fig. S1).
Figure 2.
Anterograde movement of Atg9 is defective in the cog2-1 strain. cog2-1 Atg9-3GFP (WLY227) cells expressing RFP-Ape1 were grown to OD600 = 0.8 at 24°C or shifted to 37°C for 1 h. For autophagy-inducing conditions, rapamycin was added for 1 h at either 24 or 37°C before imaging. DIC, differential interference contrast. Bar, 2.5 µm. (B) Quantification of Atg9 PAS localization. The colocalization percentages were quantified in cells containing both Atg9-3GFP and RFP-Ape1 signals from three independent repeat experiments.
COG is involved in double-membrane vesicle formation
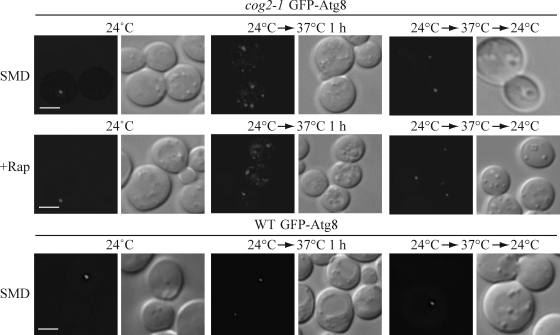
The inefficient transport of Atg9 to the PAS may be caused by indirect defects in the secretory pathway in cog mutants. Therefore, we next extended our study to test Atg8 PAS localization. Atg8 is synthesized in the cytosol, and its membrane association is through lipid conjugation (Kirisako et al., 1999, 2000; Huang et al., 2000); Atg8 does not transit through the secretory pathway. Because Atg8 forms a punctum at the putative Cvt vesicle and autophagosome assembly site and remains associated with the complete double-membrane vesicles, it serves as a marker for both Cvt vesicles and autophagosomes. Wild-type and two mutant strains, cog2-1 and cog3-2, were transformed with a GFP-Atg8 plasmid under the control of the CUP1 promoter and were incubated at 24°C and shifted to a nonpermissive temperature for 1 h. When the cells were grown at permissive temperature, the GFP-Atg8 localization in the cog2-1 and cog3-2 mutants was similar to the wild-type pattern, being diffuse in the cytosol and having one prominent PAS punctum (Fig. 3 and not depicted). After 1 h of shifting to the nonpermissive temperature, abnormal GFP-Atg8 fluorescent structures started to appear in the mutant strains. To examine the GFP-Atg8 localization during starvation in the cog2-1 and cog3-2 mutants, the same strains were grown at permissive temperature, either shifted to nonpermissive temperature or maintained at permissive temperature for 1 h, and autophagy was induced by adding rapamycin for 30 min. When grown at permissive temperature after autophagy induction, the localization pattern of the GFP-Atg8 chimera was one single punctum (Fig. 3 and not depicted). In contrast to cells incubated at the permissive temperature, during autophagy at the nonpermissive temperature, a mixture of phenotypes was observed: some cells showed abnormal GFP-Atg8 localization, whereas some had one single GFP-Atg8 punctum. After a shift back to the permissive temperature, GFP-Atg8 regained its normal localization within 30 min for both growing and autophagy-inducing conditions, implying that the aberrant GFP-Atg8 structures that formed during incubation at the nonpermissive temperature were not terminal structures. In contrast to the cog mutant strains, in wild-type cells, GFP-Atg8 remained localized to a single perivacuolar dot during autophagy-inducing conditions regardless of the temperature. The same localization patterns were also observed in wild-type and cog mutant strains when transformed with a plasmid expressing GFP-Atg8 driven by the endogenous ATG8 promoter (unpublished data), indicating that the localization phenotype was not a consequence of overexpressing Atg8. Finally, we tested the survival of the cog2-1 cells after a 1.5-h incubation at the nonpermissive temperature. The cells remained viable, ruling out the possibility that the GFP-Atg8 phenotype was a result of cell death.
Figure 3.
The cog mutants are defective in Atg8 localization to the PAS/phagophore. GFP-Atg8 localization is defective in cog2-1 and cog3-2 mutants. The wild-type (WT; BY4742) and cog2-1 strains carrying a plasmid expressing GFP-Atg8 (pCuGFP-AUT7(416)) were grown in SMD at 24°C to OD600 = 0.8 or shifted to nonpermissive temperature for 1 h before imaging. For starvation conditions, the cells were incubated at either 24°C or nonpermissive temperature for 60 min, rapamycin (final concentration 0.2 µg/ml) was added, and the culture was incubated for another 30 min before imaging. To reverse the temperature, cultures were shifted back to 24°C for 30 min in both growing and rapamycin-treated conditions. Essentially, the same results as shown for cog2-1 were seen with the cog3-2 mutant. DIC, differential interference contrast. Bars, 2.5 µm.
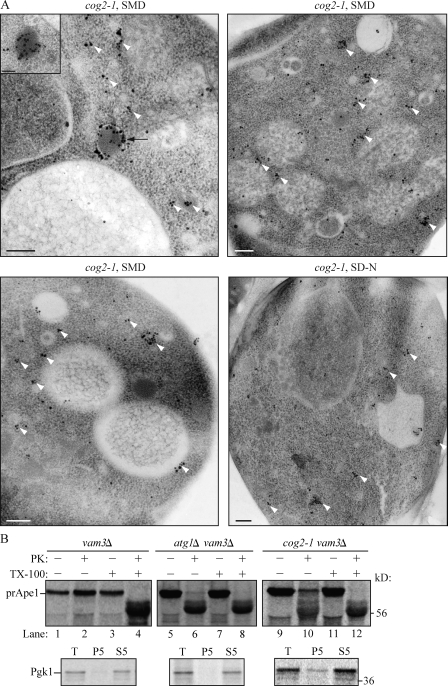
We extended this analysis by examining Atg8 localization through immunoelectron microscopy. In wild-type cells, Atg8 is detected primarily at the phagophore/PAS and on completed autophagosomes (Kirisako et al., 1999). In contrast, in the cog2-1 mutant at the nonpermissive temperature, Atg8 was largely dispersed throughout the cytosol in multiple clusters and dots (Fig. 4 A). To verify that the dispersed labeling was not the result of nonspecific binding by the antibody, we performed the same experiment in isogenic wild-type cells not expressing GFP-Atg8. In this strain, essentially no gold particles were observed (Fig. S2). Thus, lobe A cog mutants affect the normal localization of GFP-Atg8 during autophagy-inducing conditions.
Figure 4.
The cog mutants affect sequestering vesicle formation. (A) cog2-1 cells were grown in SMD and shifted to 38°C in SMD or SD-N for 1.5 h. The cells were analyzed by immunoelectron microscopy and labeled with anti-YFP antibody followed by immunogold as described in Materials and methods. The white arrowheads mark clusters of GFP-Atg8, and the black arrow marks a phagophore. Bars, 200 nm. The inset shows an image immunostained with affinity-purified antiserum to Ape1, demonstrating that the electron-dense structure is a Cvt complex. Bar, 100 nm. (B) Cog2 is involved in Cvt vesicle formation. Spheroplasts from the wild-type (vam3Δ; WLY118), cog2-1 (WLY117), and atg1Δ (WLY119) strains were incubated at 37°C for 20 min, pulse labeled with [35S]methionine/cysteine for 10 min, and subjected to a nonradioactive chase for 30 min. The prApe1-containing pellet fractions were obtained after osmotically lysing the spheroplasts and centrifugation at 5,000 g. The pellets were treated with proteinase K (PK) in the presence or absence of 0.2% Triton X-100 for 20 min on ice. The resulting samples were TCA precipitated and immunoprecipitated with Ape1 or Pgk1 antiserum and resolved by SDS-PAGE. T, total; P5 and S5, pellet and supernatant fractions, respectively, after 5,000 g centrifugation.
The analyses of Atg8 localization in cog mutants suggested that the COG complex may be involved in double-membrane vesicle formation. To further examine whether double-membrane vesicle formation is defective in cog mutants, we performed a prApe1 protease-sensitive assay. When prApe1 is enwrapped in a completed double-membrane vesicle, it is protected from exogenously added protease after lysis of spheroplasts in osmotic conditions that retain the integrity of the vacuole and other subcellular compartments including autophagosomes and Cvt vesicles. In contrast, when there are defects in vesicle formation and/or completion, the protease-sensitive propeptide domain of prApe1 is proteolytically cleaved, resulting in a molecular mass shift. We used the vam3Δ strain background in which the fusion of the autophagosome with the vacuole is blocked; in vam3Δ cells, prApe1 accumulates in cytosolic vesicles, and the prApe1 propeptide cannot be processed in the vacuolar lumen. Spheroplasts generated from vam3Δ, atg1Δ vam3Δ, and cog2-1 vam3Δ cells were incubated at nonpermissive temperature for 20 min, pulse labeled with [35S]methionine/cysteine for 10 min, and subjected to a nonradioactive chase for 30 min at nonpermissive temperature. The prApe1-containing low speed pellet fractions were prepared and subjected to proteinase K treatment with or without detergent as described in Materials and methods. In vam3Δ cells, prApe1 was protected from proteinase K and was only digested in the presence of detergent (Fig. 4 B, compare lane 2 with lane 4). In contrast, in the atg1Δ strain, which is defective in vesicle formation, prApe1 was sensitive to the proteinase K digestion independent of detergent (Fig. 4 B, lane 6). In the cog2-1 mutant, the prApe1 was partially sensitive to exogenously added proteinase K (Fig. 4 B, lane 10), indicating that the prApe1 was not completely enclosed within a completed autophagosome membrane. As a control, we examined the presence of a cytosolic marker protein, Pgk1, in the total cell lysate, low speed pellet, and supernatant fractions. Pgk1 was predominantly detected in the total and supernatant fractions (Fig. 4 B). The presence of a low level of Pgk1 in the cog2-1 pellet fraction indicated a low level of incomplete spheroplast lysis, which may account for some of the protease-insensitive prApe1 in this sample. Collectively, these results suggest that the COG complex is required for double-membrane vesicle biogenesis.
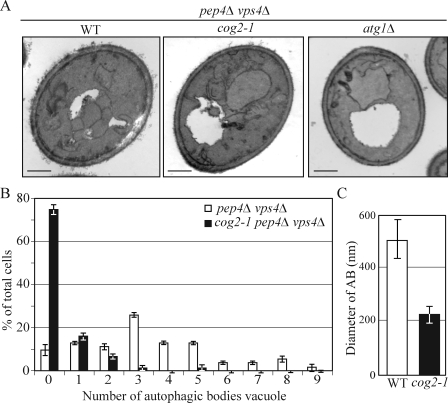
To determine the role of the COG complex in autophagosome biogenesis, we examined the ultrastructure of autophagic bodies that accumulated in the cog2-1 mutant strain vacuoles by electron microscopy. After the double-membrane autophagosomes fuse with the vacuole, the single-membrane inner vesicles, termed the autophagic bodies, are released into the vacuole lumen where their degradation is dependent on the activity of Pep4, the vacuolar proteinase A. In pep4Δ cells, the breakdown of autophagic bodies is blocked, allowing them to accumulate in the vacuole. To eliminate background vesicles targeted to the vacuole through the multivesicular body pathway, we also used a vps4Δ background (Reggiori et al., 2004b). Wild-type, atg1Δ, and cog2-1 cells additionally carrying pep4Δ vps4Δ mutations were grown in YPD medium at 24°C to OD600 = 0.8, shifted to SD-N at 37°C for 1.5 h, and prepared for electron microscopy as described in Materials and methods.
After 1.5 h starvation, wild-type (pep4Δ vps4Δ) cells contained about three autophagic bodies in 25% of the vacuoles, with a mean of 3.38 ± 2.23 per vacuole (n = 54; Fig. 5, A and B). As expected, no autophagic bodies accumulated in the control atg1Δ cells. The cog2-1 cells contained no autophagic bodies in 74.8% of the vacuoles, with a mean of 0.4 ± 0.84 per vacuole (n = 75; Fig. 5, A and B). To determine whether the cog2-1 cells accumulated normal-sized autophagic bodies, we quantified their diameters and compared them with the wild-type cells. The autophagic bodies accumulated in cog2-1 cells were smaller in diameter than those in the wild-type cells, being ∼202.21 ± 72.17 nm versus 506.27 ± 139 nm, respectively (Fig. 5 C). These results indicate that the COG complex is required for efficient autophagosome formation and may explain the observation that rapamycin treatment largely suppresses the prApe1 accumulation phenotype even though nonspecific autophagy still appears to be defective (Fig. 1); the reduced number of smaller autophagosomes was able to sequester prApe1, but the decreased volume resulted in Pho8Δ60 values that were essentially below the detectable range (Cheong et al., 2005).
Figure 5.
The COG complex is required for autophagosome formation. (A) The wild-type (WT; pep4Δ vps4Δ; FRY143), atg1Δ (JHY28), and cog2-1 (WLY221) strains were grown in YPD at 24°C to OD600 = 0.8, shifted to SD-N for 1.5 h at 37°C, and prepared for electron microscopy analysis as described in Materials and methods. Bars, 0.5 µm. (B) Quantification of autophagic body accumulation. The number of autophagic bodies in 54 or 75 cells containing vacuoles of similar size in wild-type and cog2-1 cells, respectively, was quantified. (C) Quantification of the diameter of autophagic bodies (AB). The diameter of autophagic bodies in wild-type (n = 45) and cog2-1 (n = 37) cells was measured and quantified. Error bars represent SD.
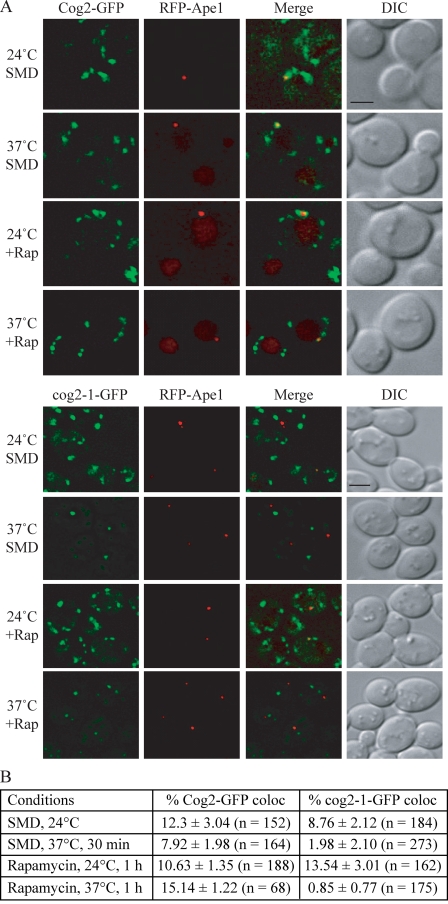
The COG complex localizes to the PAS and interacts with Atg proteins
The requirements for COG subunits in localization of Atg8 and Atg9 suggested that the COG complex may have a direct role in the Cvt pathway and autophagy. Therefore, we decided to examine whether the COG complex localized to the PAS. We note that the PAS is poorly defined and is thought to be the precursor to or organizing center for formation of the phagophore; we cannot distinguish between the PAS and the phagophore in these analyses. The chromosomally tagged Cog1-GFP, Cog2-GFP, and Cog6-GFP were distributed in several punctate structures, one of which either colocalized with or formed a cup-shaped structure around RFP-Ape1, the PAS/phagophore marker (Fig. 6 and Fig. S3). We further analyzed the PAS localization frequency of these COG components by quantifying the colocalization percentages from cells with both fluorescence signals. The PAS localization frequency of Cog1-GFP, Cog2-GFP, and Cog6-GFP were 11.1%, 12.3%, and 8%, respectively, in a wild-type strain (Fig. 6 B and not depicted). Next, we examined whether the PAS localization rate could be elevated by additional deletion of ATG1, which causes the accumulation of Atg proteins at the PAS. However, the PAS localization rates of Cog1-GFP and Cog6-GFP were not altered in the atg1Δ strain and were similar to that of the wild type, being 9.7% and 10.1%, respectively (Fig. S3 and not depicted).
Figure 6.
Cog2 localizes to the PAS. (A) Cog2-GFP (WLY190) and cog2-1–GFP (WLY188) strains expressing RFP-Ape1 were grown in SMD at 24°C or shifted to 37°C for 30 min before imaging. For autophagy-inducing conditions, cells were either grown at 24°C or shifted to 37°C for 30 min, and rapamycin was added for an additional 30 min. DIC, differential interference contrast. Bars, 2.5 µm. (B) Quantification of Cog2-GFP and cog2-1–GFP PAS localization. The colocalization percentages were quantified in cells containing both GFP and RFP-Ape1 signals from three independent repeat experiments.
To further verify the PAS localization of the COG proteins, we decided to compare the PAS localization rate between Cog2-GFP and the thermosensitive mutant cog2-1–GFP. cog2-1 bears a transversion mutation at nucleotide 624, thus encoding a truncated protein (VanRheenen et al., 1998). To monitor cog2-1–GFP localization, we chromosomally tagged GFP at the COG2 locus, resulting in a 194–amino acid truncated protein with a C-terminal GFP fusion. At the permissive temperature, both Cog2-GFP and cog2-1–GFP were localized to several punctate dots, one of which colocalized with RFP-Ape1 under both growing and rapamycin treatment conditions (Fig. 6). After shifting the cultures to nonpermissive temperature for 30 min, the cog2-1–GFP signal became one or two strong dots with several faint dots. The percentage of PAS localization of this protein dropped after the temperature shift (from 8.76 to 1.98%; Fig. 6 B). In contrast, the temperature shift did not dramatically change the PAS localization of wild-type Cog2-GFP. Similar results were seen when rapamycin was added to induce autophagy (Fig. 6). In this case, the PAS localization frequency of the mutant decreased from 13.54 to 0.85%. This result further verifies the PAS localization of the COG components. To determine whether the COG-GFP chimeras were localizing normally, we tested the Golgi localization of Cog1, Cog2, and Cog6. The majority of the Cog1-GFP, Cog2-GFP, and Cog6-GFP punctate dots colocalized with the Golgi marker Vrg4-RFP (Fig. S4), which does not colocalize with Ape1 (not depicted). At least one of the puncta did not localize to the Golgi complex, suggesting that a portion of the COG complex in the cell is not localized at this site.
Although the COG complex associates with the Golgi and COPI-containing vesicles (Whyte and Munro, 2001; Ungar et al., 2002; Shestakova et al., 2006; Vasile et al., 2006), it is still not clear which vesicles it tethers with regard to its role in autophagy. The PAS localization of the COG complex suggested that in addition to maintaining correct membrane flow through the secretory pathway that is important for autophagy, the COG complex may participate directly in double-membrane vesicle formation. To gain more insight into the function of the COG complex in the Cvt and autophagy pathways, we extended our study to test whether COG components interact with Atg proteins by yeast two-hybrid analysis.
Interactions between Atg24 and the Cog2 and Cog6 subunits as well as an Atg17–Cog2 interaction have been reported in high throughput screening studies (Uetz et al., 2000; Ito et al., 2001; Vollert and Uetz, 2004). In addition, we found that COG complex components showed weak interaction with Atg9 and stronger binding with Atg12, Atg17, Atg20, and Atg24 (Table I). To explore the physical interactions between COG and Atg proteins, we investigated the protein interactions under physiological conditions. First, we performed coimmunoprecipitation using endogenous COG subunits; however, we were unable to detect the endogenous COG proteins as a result of their low expression levels (unpublished data).
Table I.
Yeast two-hybrid interactions between lobe A COG components and Atg proteins
| Subunit | Atg5 | Atg7 | Atg8 | Atg9 | Atg11 | Atg12 | Atg13 | Atg16 | Atg17 | Atg18 | Atg20 | Atg24 |
| Cog1 | − | − | − | − | − | − | − | − | +++ | − | + | ++ |
| Cog2 | − | − | − | − | − | +++ | − | − | ++ | − | ++ | ++ |
| Cog3 | − | − | − | + | − | − | − | − | +++ | − | + | ++ |
| Cog4 | − | − | − | + | − | − | − | − | +++ | − | ++ | ++ |
The interaction strength is scored by the number of pluses: +, grows weakly on plates lacking histidine; ++, growth on plates lacking histidine but not on plates lacking adenine; +++, growth on plates lacking adenine; −, no growth. All the interactions were scored after growth for 3 d at 30°C.
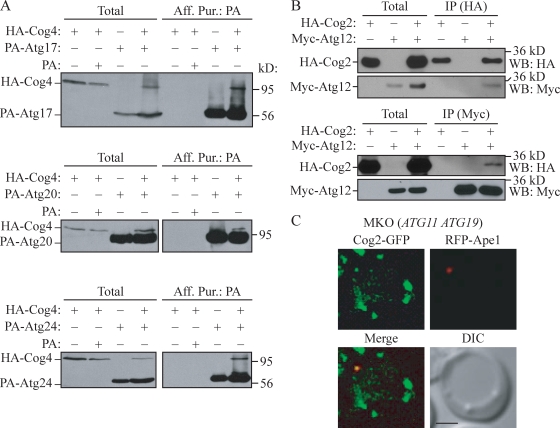
To overexpress COG subunits, we chromosomally replaced their endogenous promoters with the GAL1 promoter and an N-terminal HA tag and performed a series of protein A (PA) affinity purification experiments. Either a PA-tagged Atg protein or PA alone was coexpressed in combination with HA-COG subunits. Cells were grown in synthetic minimal medium with galactose (SMG) to induce COG subunit overexpression. Cell extracts were prepared, and PA-tagged Atg proteins and associated proteins were affinity isolated. The recovered immunocomplex was resolved by SDS-PAGE, and the presence of HA-COG subunits was analyzed by Western blotting using anti-HA antibody. HA-Cog1 bound to PA-Atg17 and PA-Atg20, whereas HA-Cog3 was coprecipitated with PA-Atg17 and PA-Atg24 (unpublished data). HA-Cog4 was able to bind PA-Atg17, PA-Atg20, and PA-Atg24 (Fig. 7 A). HA-COG subunits did not bind to PA alone, indicating that the interactions with HA-COG subunits were dependent on Atg17, Atg20, or Atg24 fused to PA. In addition, these interactions were absent when the affinity isolation was performed with combined cell lysates from two different strains each expressing an individual tagged protein; thus, the interactions we detected did not occur after lysis (unpublished data).
Figure 7.
COG subunits associate with Atg proteins. (A and B) HA-Cog4 (A; WLY208) and HA-Cog2 (B; WLY209) cells transformed with the indicated plasmids expressing tagged Atg proteins were grown in selective SMG medium to OD600 = 1.0. Cell lysates were prepared and subjected to affinity isolation or immunoprecipitation (IP) with either anti-HA or anti-Myc antibody as described in Materials and methods. Plasmids expressing PA (pRS424-CuProtA), PA-tagged Atg17 (pProtA-Apg17(424)), Atg20 (pProtA-Cvt20(424)), Atg24 (pProtA-Cvt13(424)), and Myc-tagged Atg12 (pMyc-Apg12(426)) were used as indicated. The eluted proteins were separated by SDS-PAGE and detected with monoclonal anti-HA antibody (A) or immunoblotting with anti-HA and anti-Myc antibodies (B). For each experiment, ∼1% of the total cell lysate or 10% of the total eluate was loaded. WB, Western blot. (C) Cog2-GFP colocalizes with RFP-Ape1 in the MKO (ATG11 ATG19) strain. The MKO (ATG11 ATG19; WLY205) cells expressing chromosomally tagged Cog2-GFP and a plasmid-based RFP-Ape1 were grown in selective SMD to OD600 = 0.8 and observed by fluorescence microscopy. DIC, differential interference contrast. Bar, 2.5 µm.
We also tested the interaction between Cog2 and Atg12. Lysates were prepared from yeast cells expressing HA-Cog2, Myc-Atg12, or both and incubated with PA-Sepharose beads and anti-HA or anti-Myc antibody. The resulting immunocomplex was subjected to Western blot analysis. In the cells expressing either HA-Cog2 or Myc-Atg12 alone, no Myc-Atg12 or HA-Cog2 was detected after immunoprecipitation, indicating that both proteins did not bind nonspecifically to the PA-Sepharose beads. In contrast, when both HA-Cog2 and Myc-Atg12 were coexpressed, Myc-Atg12 coimmunoprecipitated with HA-Cog2 and vice versa (Fig. 7 B). We were unable to detect an interaction between Cog2 and Atg12–Atg5 conjugates. This could be the result of either the absence of this interaction or the relatively low level of Atg12–Atg5 conjugates in our experimental conditions in which Atg12 was overexpressed. Atg9 weakly interacted with Cog3 and Cog4 in the yeast two-hybrid analysis; however, we could not verify these interactions by PA affinity isolation. Thus, the interaction between Atg9 and Cog3 or Cog4 might be transient or too weak to detect reproducibly.
The COG complex primarily localizes to the rims and tips of the Golgi membrane and their associated vesicles (Ungar et al., 2002; Vasile et al., 2006). However, none of the COG subunits have either a transmembrane domain or lipid-binding motif. Thus, it remains unclear whether COG subunits themselves or other factors mediate this membrane association. The interactions between COG and Atg proteins raise a possibility that PAS membrane association of the COG complex is through interaction with Atg proteins. To address this point, we used a multiple knockout (MKO) strain in which 24 ATG genes that are directly involved in autophagy and/or the Cvt pathway have been deleted. We used an MKO strain containing Atg19, the Ape1 cargo receptor, and reintroduced Atg11, an adaptor protein required for prApe1 cargo recruitment, to this strain, allowing the formation of the Cvt complex (Shintani et al., 2002; Yorimitsu and Klionsky, 2005; Suzuki et al., 2007; Cao et al., 2008). In this strain, chromosomally tagged Cog2-GFP and plasmid-based RFP-Ape1 were coexpressed, and the colocalization was examined by fluorescence microscopy. In the MKO (ATG11 ATG19) strain, Cog2-GFP colocalized with RFP-Ape1, the Cvt complex marker (Fig. 7 C). We further quantified the colocalization frequency of Cog2-GFP in the MKO (ATG11 ATG19) strain and found that the colocalization rate was similar to that of the wild type (MKO: 11.11%, n = 225; wild type: 12.5%, n = 152). Therefore, it is possible that the interactions between the COG and Atg subunits that we detected reflect functional interactions that are not involved in localization of the COG complex to the PAS/phagophore; unknown factors or the COG complex subunits themselves may direct the complex to this site.
Discussion
An autophagosome is formed through nucleation, membrane expansion, and completion. Because of the similarity between autophagy and the Cvt pathway, the Cvt vesicle is considered as a variant of an autophagosome. However, the Cvt vesicle (∼150 nm in diameter) is smaller than an autophagosome (300–900 nm in diameter) in size (Scott et al., 1996; Baba et al., 1997). Furthermore, during selective types of autophagy, the membrane may form in close apposition to the cargo, using receptors (e.g., Atg19) and adaptor proteins (e.g., Atg11) to link the two, whereas these types of proteins do not play an essential role in nonspecific autophagy. These observations suggest that the nature of Cvt vesicle and autophagosome formation may in part be different. Indeed, some components are involved in the formation of one but not the other, which may explain why lobe A subunits are required for both the Cvt and autophagy pathways, whereas the lobe B subunits of the COG complex are Cvt specific. This idea is supported by the normal Pho8Δ60-dependent alkaline phosphatase activity seen in lobe B but not lobe A mutants of the COG complex (Fig. 1, A, B, D, and E). Similarly, abnormal GFP-Atg8–positive structures were detected in cog2-1 and cog3-2 mutants during vegetative growth and after rapamycin treatment (Fig. 3), indicating that the lobe A subunits of COG may play a role in autophagosome formation.
The COG complex may be directly involved in autophagy
There may be two membrane fusion events during the process of double-membrane vesicle formation: the presumed condensation of transient vesicles with the growing phagophore at the PAS and the fusion of the completed double-membrane vesicles with the vacuole (Reggiori et al., 2004b). The last event is the best characterized and relies on components that are needed for other vesicle fusion events with the vacuole as well as homotypic vacuole fusion (Klionsky, 2005). In contrast, little is known about the role of SNAREs or tethering factors in the phagophore expansion step. Previous experiments were unable to detect any SNAREs that localize to the PAS (Reggiori et al., 2004b), possibly as a result of their presence at too low a level. Considering the tethering function of the COG complex in the secretory pathway and the localization of the COG complex at the PAS (Fig. 6), it is possible that the COG complex might participate in membrane tethering events, which are required for Cvt vesicle and autophagosome formation and completion. Although we have not directly shown a role for COG components in tethering, the dispersed localization of Atg8 in the cog2-1 mutant at the nonpermissive temperature (Fig. 3 and Fig. 4 A) suggests that it may function in this manner.
The interactions between the COG complex and Atg proteins suggest a direct involvement of the COG complex in autophagy and the Cvt pathway and also provide insight about the function of these Atg proteins. Atg17 showed strong interactions with many COG subunits by yeast two-hybrid analysis (Table I). Atg17 is an autophagy-specific protein and functions as a scaffold to recruit other Atg proteins to organize PAS formation (Cheong et al., 2005; Suzuki et al., 2007). Atg17 modulates the timing and magnitude of the autophagy response, such as the size of the sequestering vesicles, through interacting with and regulating Atg1 kinase activity (Kamada et al., 2000; Cheong et al., 2005). The interaction between Atg17 and COG subunits provides a link between the autophagy regulatory components and the phagophore expansion/vesicle fusion machinery and may provide a clue as to how the Atg1 kinase activity can be translated into the size of sequestering vesicles.
The significance of the interaction between Atg12 and Cog2 is not known. The finding that Atg9 interacts with COG subunits by yeast two hybrid is intriguing. Although we were unable to obtain consistent results from PA affinity isolation experiments, it is possible that the interaction between Atg9 and the COG complex is transient. Atg9 is an integral membrane protein that cycles between the PAS and peripheral sites and is directly involved in double-membrane vesicle formation (Noda et al., 2000; Reggiori et al., 2005b). During its cycling, it remains associated with lipid, which makes Atg9 a prime candidate for a carrier that brings membrane from sources to the PAS (Reggiori et al., 2005b). We hypothesize that the COG complex may function as a tethering factor, allowing Atg9-containing vesicles to fuse with the expanding phagophore at the PAS, which might explain the transient interaction between the tether and Atg9. In conclusion, our data suggest that the COG complex is required for the efficient fusion of transient vesicles with the phagophore at the PAS, which is required for Cvt vesicle and autophagosome formation and completion.
Materials and methods
Strains, media, and materials
The yeast strains used in this study are listed in Table S1. For gene disruption, the entire coding regions were replaced with the Kluyveromyces lactis URA3, LEU2, Saccharomyces kluyveri HIS3, Schizosaccharomyces pombe HIS5, Saccharomyces cerevisiae TRP1, HIS3, and the Escherichia coli kanr genes using PCR primers containing ∼50 bases identical to the flanking regions of the open reading frames. For PCR-based integrations of GFP at the 3′ end of PEX14, COG1, COG2, COG6, and cog2-1 and RFP tags at the 3′ end of VRG4, pFA6a-GFP-HIS3, pFA6a-GFP-KanMX, pFA6a-GFP-TRP1, pFA6a-mRFP-TRP1, pFA6a-mRFP-HIS5 S. pombe, and pFA6a-mRFP-KanMX were used as templates to generate strains expressing fusion proteins under the control of their native promoters (Longtine et al., 1998; Campbell and Choy, 2002; Gueldener et al., 2002). For PCR-based replacement of the GAL1 promoter with an N-terminal HA tag at the 5′ end of COG1, COG2, COG3, and COG4, pFA6a-KanMX6-PGAL1-3HA was used as a template (Longtine et al., 1998). PCR was used to verify the gene fusions. To generate an Atg9-3GFP fusion, the integrative plasmid pAtg9-3GFP(306) was linearized by StuI digestion and integrated into the URA3 gene locus.
Strains were grown in YPD (1% yeast extract, 2% peptone, and 2% glucose), SMD (2% glucose and 0.67% yeast nitrogen base without amino acids, supplemented with vitamins and appropriate amino acids), YPG (1% yeast extract, 2% peptone, and 2% galactose), or SMG (2% galactose and 0.67% yeast nitrogen base without amino acids, supplemented with vitamins and appropriate amino acids). Nitrogen starvation experiments were performed in synthetic medium lacking nitrogen (SD-N; 0.17% yeast nitrogen base without amino acids, ammonium sulfate, and vitamins, but containing 2% glucose).
Plasmids and constructions
The plasmids pGAD-Cog1, pGBDU-Cog1, pGAD-Cog3, and pGBDU-Cog3 were generated by PCR amplifying the full-length COG1 or COG3 gene and ligating into BamHI–PstI sites of the pGAD-C1 and pGBDU-C1 vectors. The pGAD-Cog2 and pGBDU-Cog2 plasmids were created by ligating the DNA fragment encoding full-length Cog2 with EcoRI and BglII sites into the pGAD-C1 and pGBDU-C1 plasmids. The pGAD-Cog4 and pGBDU-Cog4 plasmids were generated by amplifying the COG4 gene and cloning as BamHI–BglII fragments into pGAD-C1 and pGBDU-C1 vectors. The plasmids pCuCOG2-HA(416) and pCuCOG3-HA(416) were generated by PCR amplification of full-length COG2 and COG3 genes using primers containing a single HA sequence at the 3′ ends and ligated into a CUP1 promoter–driven pRS416 plasmid with EcoRI–XhoI and BamHI–XhoI, respectively. The plasmids pGFP-AUT7(416) (Kim et al., 2001b), pRFP-Ape1(305) (Shintani et al., 2002), pAtg9-3GFP(306) (Monastyrska et al., 2008), pSpo7-RFP(425) (Reggiori et al., 2005b), pProtA-CVT13(424) (Nice et al., 2002), pProtA-APG17(424) (Nice et al., 2002), pProtA-CVT20(424) (Nice et al., 2002), pRS424-CuProtA (Kim et al., 2002), and pMyc-APG12(426) (Mizushima et al., 1998) have been described previously. Plasmids expressing mutant forms of COG4, pBCR115 and pBCR137, were generated by PCR amplification of genomic COG4, including 400 bases of genomic DNA on either side of the gene, ligating the product into the pRS415 URA-CEN plasmid and introducing site-directed mutations. Residues shown to be both surface exposed (by x-ray crystallographic analysis of human Cog4) and well conserved across multiple species were selected for mutation by alanine truncation or charge introduction as appropriate (Richardson et al., 2009). To create pBCR115, a single QuikChange (Agilent Technologies) reaction was used to generate the double-substitution mutant N607A/T608A. To create pBCR137, six mutations were introduced using the QuikChange Multi kit (Agilent Technologies): N787A, R788A, G795D, C798D, R805A, and E806A. The corresponding residues in human Cog4 constitute a single large, conserved patch on the surface of the protein (Richardson et al., 2009). To generate an Atg11 integration plasmid, pATG11(414) was made by cloning the PCR fragment containing the ATG11 promoter, ORF, and terminator regions into the XmaI site of pRS414. A NotI–SalI fragment containing ATG11 was cut from pAtg11(414) and inserted into the corresponding sites of pRS307 to generate pATG11(307). pATG11(307) was linearized with PmlI and integrated into the LYS2 locus.
Ape1 pulse-chase radiolabeling
Cells were grown in SMD at 24°C to OD600 = 0.8. 20 OD600 U of cells were collected and converted to spheroplasts then incubated at 37°C for 20 min. Spheroplasts were harvested and resuspended in 200 µl of spheroplasting medium (1.2 M sorbitol, 2% glucose, and 0.67% yeast nitrogen base without amino acids, supplemented with vitamins and appropriate amino acids) without methionine and cysteine for 5 min at the same temperatures. Cells were labeled with Tran35S-Label ([35S]methionine/cysteine; MP Biomedicals) for 10 min (10 µCi/OD600). Chase was initiated by adding 4 ml of chase medium (SMD containing 0.2% yeast extract, 1.2 M sorbitol, and 2 mM cysteine and methionine). At each indicated time point, samples were collected, and the total cell lysates were prepared by adding ice-cold osmotic lysis buffer (20 mM Pipes, pH 6.8, 200 mM sorbitol, 5 mM MgCl2, and complete EDTA-free proteinase inhibitor cocktail) at a spheroplast density of 20 OD600/ml to lyse the cells. To obtain the prApe1-containing P5 fractions, cell lysates were subjected to a 5,000 g centrifugation for 5 min. The P5 fractions were resuspended in osmotic lysis buffer, split into four aliquots, and subjected to 100 µg/ml proteinase K and/or 0.2% Triton X-100 treatment on ice for 20 min. 10% TCA was added to precipitate the cell lysates and the precipitates were washed twice with 100% acetone and air dried. The pellets were resuspended in Tween immunoprecipitation buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.5% Tween 20, and 0.1 mM EDTA), PA-Sepharose and Ape1 antiserum were added, and the mixture was incubated overnight at 4°C. The PA-Sepharose–bound proteins were washed six times with Tween immunoprecipitation buffer, and the samples were eluted at 75°C for 10 min and resolved by SDS-PAGE.
Microscopy
For fluorescence microscopy, yeast cells expressing fusion proteins with fluorescence tags were grown to OD600 = 0.8 in YPD or SMD selective medium or starved in SD-N before imaging. To label the mitochondria, Mito Fluor red 589 (Invitrogen) was added to the growing culture with a final concentration of 1 µM for 30 min before imaging. Starved cells were stained with MitoFluor red for 1 h before imaging. For rapamycin treatment, 0.2 µg/ml rapamycin was added to the culture. Cells were visualized with a fluorescence microscope (Olympus) with DeltaVision (Spectris; Applied Precision) using a camera (CoolSNAP HQ; Photometrics) fitted with differential interference contrast optics. Images were taken using a 100× objective at the same temperature and in the same medium in which the cells were cultured. 15 z-section images were collected and were deconvolved using softWoRx software (Applied Precision). All fluorescence microscopy images show a single focal z section.
Electron microscopy
Transmission electron microscopy was performed as described previously (Kaiser and Schekman, 1990). For immunoelectron microscopy, cells were frozen in a freezing device (KF80; Leica), and the analysis was performed according to the procedures described previously (Baba, 2008). Ultrathin sections were stained with anti-YFP antibody or affinity purified anti-Ape1 antiserum. The anti-YFP was labeled with 0.8-nm ultrasmall gold particles (Aurion) that were detected by silver enhancement, whereas the anti-Ape1 was labeled with 10 nm colloidal gold–conjugated goat anti–rabbit IgG (British Biocell). Ultrathin sections were examined with an electron microscope (H-800; Hitachi High Technologies) at 125 kV. Images were prepared using a film scanner, and scale bars were added in Photoshop (Adobe).
PA affinity isolation and coimmunoprecipitation
50 ml of cells grown in SMG medium to OD600 = 1.0 was harvested and resuspended in 4 ml of lysis buffer (PBS, 200 mM sorbitol, 1 mM MgCl2, 0.2% Tween 20, 2 mM PMSF, and proteinase inhibitor cocktail [Roche]) and lysed by vortex after adding a 2-ml volume of acid-washed glass beads. IgG-Sepharose beads were added to the detergent extracts followed by incubation overnight at 4°C. The beads were washed eight times with 1 ml lysis buffer, and the proteins were resolved by SDS-PAGE. For the coimmunoprecipitation experiments, we used the same procedure as PA affinity isolation with only one exception: the cell lysates were incubated with PA-Sepharose beads (50% suspension) and 5 µl monoclonal anti-HA or anti-Myc antibody overnight at 4°C. The resulting immunocomplex was subjected to immunoblotting with monoclonal anti-HA or anti-Myc antibodies.
Additional assays
The GFP-Atg8 processing assay, Pex14-GFP processing to monitor pexophagy progression, and the alkaline phosphatase assay to measure Pho8Δ60 activity were performed as previously described (Noda et al., 1995; Tucker et al., 2003; Reggiori et al., 2005a).
Online supplemental material
Fig. S1 shows the localization of Atg9-3GFP in wild-type, atg1Δ, cog1Δ, cog6Δ, and double-mutant strains analyzed by fluorescence microscopy. Fig. S2 shows an immunoelectron microscopy analysis of wild-type cells as a control for the specificity of the anti-YFP antibody. Fig. S3 is a fluorescence microscopy analysis of Cog1-GFP and Cog6-GFP in wild-type and atg1Δ strains relative to the PAS marker RFP-Ape1. Fig. S4 examines the colocalization of Cog1-GFP, Cog2-GFP, and Cog6-GFP with the Golgi marker Vrg4-RFP by fluorescence microscopy. Table S1 lists the yeast strains used in this study. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200904075/DC1.
Acknowledgments
This work was supported by a National Institutes of Health Public Health Service grant (GM53396) to D.J. Klionsky.
Footnotes
Abbreviations used in this paper:
- Ape1
- aminopeptidase I
- COG
- conserved oligomeric Golgi
- Cvt
- cytoplasm to vacuole targeting
- MKO
- multiple knockout
- PA
- protein A
- PAS
- phagophore assembly site
- prApe1
- precursor Ape1
- SMD
- synthetic minimal medium with dextrose
- SMG
- synthetic minimal medium with galactose
References
- Albertini M., Rehling P., Erdmann R., Girzalsky W., Kiel J.A., Veenhuis M., Kunau W.H. 1997. Pex14p, a peroxisomal membrane protein binding both receptors of the two PTS-dependent import pathways. Cell. 89:83–92 10.1016/S0092-8674(00)80185-3 [DOI] [PubMed] [Google Scholar]
- Baba M. 2008. Electron microscopy in yeast. Methods Enzymol. 451:133–149 10.1016/S0076-6879(08)03210-2 [DOI] [PubMed] [Google Scholar]
- Baba M., Osumi M., Scott S.V., Klionsky D.J., Ohsumi Y. 1997. Two distinct pathways for targeting proteins from the cytoplasm to the vacuole/lysosome. J. Cell Biol. 139:1687–1695 10.1083/jcb.139.7.1687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruinsma P., Spelbrink R.G., Nothwehr S.F. 2004. Retrograde transport of the mannosyltransferase Och1p to the early Golgi requires a component of the COG transport complex. J. Biol. Chem. 279:39814–39823 10.1074/jbc.M405500200 [DOI] [PubMed] [Google Scholar]
- Campbell T.N., Choy F.Y. 2002. Expression of two green fluorescent protein variants in citrate-buffered media in Pichia pastoris. Anal. Biochem. 311:193–195 10.1016/S0003-2697(02)00409-8 [DOI] [PubMed] [Google Scholar]
- Cao Y., Cheong H., Song H., Klionsky D.J. 2008. In vivo reconstitution of autophagy in Saccharomyces cerevisiae. J. Cell Biol. 182:703–713 10.1083/jcb.200801035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong H., Yorimitsu T., Reggiori F., Legakis J.E., Wang C.-W., Klionsky D.J. 2005. Atg17 regulates the magnitude of the autophagic response. Mol. Biol. Cell. 16:3438–3453 10.1091/mbc.E04-10-0894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fotso P., Koryakina Y., Pavliv O., Tsiomenko A.B., Lupashin V.V. 2005. Cog1p plays a central role in the organization of the yeast conserved oligomeric Golgi complex. J. Biol. Chem. 280:27613–27623 10.1074/jbc.M504597200 [DOI] [PubMed] [Google Scholar]
- Gueldener U., Heinisch J., Koehler G.J., Voss D., Hegemann J.H. 2002. A second set of loxP marker cassettes for Cre-mediated multiple gene knockouts in budding yeast. Nucleic Acids Res. 30:e23 10.1093/nar/30.6.e23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamasaki M., Noda T., Ohsumi Y. 2003. The early secretory pathway contributes to autophagy in yeast. Cell Struct. Funct. 28:49–54 10.1247/csf.28.49 [DOI] [PubMed] [Google Scholar]
- Huang J., Klionsky D.J. 2007. Autophagy and human disease. Cell Cycle. 6:1837–1849 [DOI] [PubMed] [Google Scholar]
- Huang W.-P., Scott S.V., Kim J., Klionsky D.J. 2000. The itinerary of a vesicle component, Aut7p/Cvt5p, terminates in the yeast vacuole via the autophagy/Cvt pathways. J. Biol. Chem. 275:5845–5851 10.1074/jbc.275.8.5845 [DOI] [PubMed] [Google Scholar]
- Ishihara N., Hamasaki M., Yokota S., Suzuki K., Kamada Y., Kihara A., Yoshimori T., Noda T., Ohsumi Y. 2001. Autophagosome requires specific early Sec proteins for its formation and NSF/SNARE for vacuolar fusion. Mol. Biol. Cell. 12:3690–3702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T., Chiba T., Ozawa R., Yoshida M., Hattori M., Sakaki Y. 2001. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. USA. 98:4569–4574 10.1073/pnas.061034498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser C.A., Schekman R. 1990. Distinct sets of SEC genes govern transport vesicle formation and fusion early in the secretory pathway. Cell. 61:723–733 10.1016/0092-8674(90)90483-U [DOI] [PubMed] [Google Scholar]
- Kamada Y., Funakoshi T., Shintani T., Nagano K., Ohsumi M., Ohsumi Y. 2000. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J. Cell Biol. 150:1507–1513 10.1083/jcb.150.6.1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Scott S.V., Oda M.N., Klionsky D.J. 1997. Transport of a large oligomeric protein by the cytoplasm to vacuole protein targeting pathway. J. Cell Biol. 137:609–618 10.1083/jcb.137.3.609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.W., Sacher M., Scarpa A., Quinn A.M., Ferro-Novick S. 1999. High-copy suppressor analysis reveals a physical interaction between Sec34p and Sec35p, a protein implicated in vesicle docking. Mol. Biol. Cell. 10:3317–3329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.W., Massey T., Sacher M., Pypaert M., Ferro-Novick S. 2001a. Sgf1p, a new component of the Sec34p/Sec35p complex. Traffic. 2:820–830 10.1034/j.1600-0854.2001.21111.x [DOI] [PubMed] [Google Scholar]
- Kim J., Huang W.-P., Klionsky D.J. 2001b. Membrane recruitment of Aut7p in the autophagy and cytoplasm to vacuole targeting pathways requires Aut1p, Aut2p, and the autophagy conjugation complex. J. Cell Biol. 152:51–64 10.1083/jcb.152.1.51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Huang W.-P., Stromhaug P.E., Klionsky D.J. 2002. Convergence of multiple autophagy and cytoplasm to vacuole targeting components to a perivacuolar membrane compartment prior to de novo vesicle formation. J. Biol. Chem. 277:763–773 10.1074/jbc.M109134200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirisako T., Baba M., Ishihara N., Miyazawa K., Ohsumi M., Yoshimori T., Noda T., Ohsumi Y. 1999. Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J. Cell Biol. 147:435–446 10.1083/jcb.147.2.435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirisako T., Ichimura Y., Okada H., Kabeya Y., Mizushima N., Yoshimori T., Ohsumi M., Takao T., Noda T., Ohsumi Y. 2000. The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J. Cell Biol. 151:263–276 10.1083/jcb.151.2.263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky D.J. 2005. The molecular machinery of autophagy: unanswered questions. J. Cell Sci. 118:7–18 10.1242/jcs.01620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky D.J., Emr S.D. 2000. Autophagy as a regulated pathway of cellular degradation. Science. 290:1717–1721 10.1126/science.290.5497.1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky D.J., Cueva R., Yaver D.S. 1992. Aminopeptidase I of Saccharomyces cerevisiae is localized to the vacuole independent of the secretory pathway. J. Cell Biol. 119:287–299 10.1083/jcb.119.2.287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky D.J., Cregg J.M., Dunn W.A., Jr., Emr S.D., Sakai Y., Sandoval I.V., Sibirny A., Subramani S., Thumm M., Veenhuis M., Ohsumi Y. 2003. A unified nomenclature for yeast autophagy-related genes. Dev. Cell. 5:539–545 10.1016/S1534-5807(03)00296-X [DOI] [PubMed] [Google Scholar]
- Levine B., Klionsky D.J. 2004. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev. Cell. 6:463–477 10.1016/S1534-5807(04)00099-1 [DOI] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie A., III, Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., Pringle J.R. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 14:953–961 [DOI] [PubMed] [Google Scholar]
- Mizushima N., Noda T., Yoshimori T., Tanaka Y., Ishii T., George M.D., Klionsky D.J., Ohsumi M., Ohsumi Y. 1998. A protein conjugation system essential for autophagy. Nature. 395:395–398 10.1038/26506 [DOI] [PubMed] [Google Scholar]
- Monastyrska I., He C., Geng J., Hoppe A.D., Li Z., Klionsky D.J. 2008. Arp2 links autophagic machinery with the actin cytoskeleton. Mol. Biol. Cell. 19:1962–1975 10.1091/mbc.E07-09-0892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nice D.C., Sato T.K., Stromhaug P.E., Emr S.D., Klionsky D.J. 2002. Cooperative binding of the cytoplasm to vacuole targeting pathway proteins, Cvt13 and Cvt20, to phosphatidylinositol 3-phosphate at the pre-autophagosomal structure is required for selective autophagy. J. Biol. Chem. 277:30198–30207 10.1074/jbc.M204736200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda T., Matsuura A., Wada Y., Ohsumi Y. 1995. Novel system for monitoring autophagy in the yeast Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 210:126–132 10.1006/bbrc.1995.1636 [DOI] [PubMed] [Google Scholar]
- Noda T., Kim J., Huang W.-P., Baba M., Tokunaga C., Ohsumi Y., Klionsky D.J. 2000. Apg9p/Cvt7p is an integral membrane protein required for transport vesicle formation in the Cvt and autophagy pathways. J. Cell Biol. 148:465–480 10.1083/jcb.148.3.465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogier-Denis E., Codogno P. 2003. Autophagy: a barrier or an adaptive response to cancer. Biochim. Biophys. Acta. 1603:113–128 [DOI] [PubMed] [Google Scholar]
- Oka T., Ungar D., Hughson F.M., Krieger M. 2004. The COG and COPI complexes interact to control the abundance of GEARs, a subset of Golgi integral membrane proteins. Mol. Biol. Cell. 15:2423–2435 10.1091/mbc.E03-09-0699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ram R.J., Li B., Kaiser C.A. 2002. Identification of Sec36p, Sec37p, and Sec38p: components of yeast complex that contains Sec34p and Sec35p. Mol. Biol. Cell. 13:1484–1500 10.1091/mbc.01-10-0495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F., Klionsky D.J. 2005. Autophagosomes: biogenesis from scratch? Curr. Opin. Cell Biol. 17:415–422 10.1016/j.ceb.2005.06.007 [DOI] [PubMed] [Google Scholar]
- Reggiori F., Klionsky D.J. 2006. Atg9 sorting from mitochondria is impaired in early secretion and VFT-complex mutants in Saccharomyces cerevisiae. J. Cell Sci. 119:2903–2911 10.1242/jcs.03047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F., Wang C.-W., Stromhaug P.E., Shintani T., Klionsky D.J. 2003. Vps51 is part of the yeast Vps fifty-three tethering complex essential for retrograde traffic from the early endosome and Cvt vesicle completion. J. Biol. Chem. 278:5009–5020 10.1074/jbc.M210436200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F., Tucker K.A., Stromhaug P.E., Klionsky D.J. 2004a. The Atg1-Atg13 complex regulates Atg9 and Atg23 retrieval transport from the pre-autophagosomal structure. Dev. Cell. 6:79–90 10.1016/S1534-5807(03)00402-7 [DOI] [PubMed] [Google Scholar]
- Reggiori F., Wang C.-W., Nair U., Shintani T., Abeliovich H., Klionsky D.J. 2004b. Early stages of the secretory pathway, but not endosomes, are required for Cvt vesicle and autophagosome assembly in Saccharomyces cerevisiae. Mol. Biol. Cell. 15:2189–2204 10.1091/mbc.E03-07-0479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F., Monastyrska I., Shintani T., Klionsky D.J. 2005a. The actin cytoskeleton is required for selective types of autophagy, but not nonspecific autophagy, in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell. 16:5843–5856 10.1091/mbc.E05-07-0629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F., Shintani T., Nair U., Klionsky D.J. 2005b. Atg9 cycles between mitochondria and the pre-autophagosomal structure in yeasts. Autophagy. 1:101–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson B.C., Smith R.D., Ungar D., Nakamura A., Jeffrey P.D., Lupashin V.V., Hughson F.M. 2009. Structural basis for a human glycosylation disorder caused by mutation of the COG4 gene. Proc. Natl. Acad. Sci. USA. 106:13329–13334 10.1073/pnas.0901966106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott S.V., Hefner-Gravink A., Morano K.A., Noda T., Ohsumi Y., Klionsky D.J. 1996. Cytoplasm-to-vacuole targeting and autophagy employ the same machinery to deliver proteins to the yeast vacuole. Proc. Natl. Acad. Sci. USA. 93:12304–12308 10.1073/pnas.93.22.12304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott S.V., Baba M., Ohsumi Y., Klionsky D.J. 1997. Aminopeptidase I is targeted to the vacuole by a nonclassical vesicular mechanism. J. Cell Biol. 138:37–44 10.1083/jcb.138.1.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shestakova A., Zolov S., Lupashin V. 2006. COG complex-mediated recycling of Golgi glycosyltransferases is essential for normal protein glycosylation. Traffic. 7:191–204 10.1111/j.1600-0854.2005.00376.x [DOI] [PubMed] [Google Scholar]
- Shintani T., Klionsky D.J. 2004a. Autophagy in health and disease: a double-edged sword. Science. 306:990–995 10.1126/science.1099993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani T., Klionsky D.J. 2004b. Cargo proteins facilitate the formation of transport vesicles in the cytoplasm to vacuole targeting pathway. J. Biol. Chem. 279:29889–29894 10.1074/jbc.M404399200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani T., Huang W.-P., Stromhaug P.E., Klionsky D.J. 2002. Mechanism of cargo selection in the cytoplasm to vacuole targeting pathway. Dev. Cell. 3:825–837 10.1016/S1534-5807(02)00373-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spelbrink R.G., Nothwehr S.F. 1999. The yeast GRD20 gene is required for protein sorting in the trans-Golgi network/endosomal system and for polarization of the actin cytoskeleton. Mol. Biol. Cell. 10:4263–4281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strømhaug P.E., Reggiori F., Guan J., Wang C.-W., Klionsky D.J. 2004. Atg21 is a phosphoinositide binding protein required for efficient lipidation and localization of Atg8 during uptake of aminopeptidase I by selective autophagy. Mol. Biol. Cell. 15:3553–3566 10.1091/mbc.E04-02-0147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suvorova E.S., Duden R., Lupashin V.V. 2002. The Sec34/Sec35p complex, a Ypt1p effector required for retrograde intra-Golgi trafficking, interacts with Golgi SNAREs and COPI vesicle coat proteins. J. Cell Biol. 157:631–643 10.1083/jcb.200111081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K., Kirisako T., Kamada Y., Mizushima N., Noda T., Ohsumi Y. 2001. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 20:5971–5981 10.1093/emboj/20.21.5971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K., Kubota Y., Sekito T., Ohsumi Y. 2007. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells. 12:209–218 10.1111/j.1365-2443.2007.01050.x [DOI] [PubMed] [Google Scholar]
- Tucker K.A., Reggiori F., Dunn W.A., Jr., Klionsky D.J. 2003. Atg23 is essential for the cytoplasm to vacuole targeting pathway and efficient autophagy but not pexophagy. J. Biol. Chem. 278:48445–48452 10.1074/jbc.M309238200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetz P., Giot L., Cagney G., Mansfield T.A., Judson R.S., Knight J.R., Lockshon D., Narayan V., Srinivasan M., Pochart P., et al. 2000. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 403:623–627 10.1038/35001009 [DOI] [PubMed] [Google Scholar]
- Ungar D., Oka T., Brittle E.E., Vasile E., Lupashin V.V., Chatterton J.E., Heuser J.E., Krieger M., Waters M.G. 2002. Characterization of a mammalian Golgi-localized protein complex, COG, that is required for normal Golgi morphology and function. J. Cell Biol. 157:405–415 10.1083/jcb.200202016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungar D., Oka T., Krieger M., Hughson F.M. 2006. Retrograde transport on the COG railway. Trends Cell Biol. 16:113–120 10.1016/j.tcb.2005.12.004 [DOI] [PubMed] [Google Scholar]
- VanRheenen S.M., Cao X., Lupashin V.V., Barlowe C., Waters M.G. 1998. Sec35p, a novel peripheral membrane protein, is required for ER to Golgi vesicle docking. J. Cell Biol. 141:1107–1119 10.1083/jcb.141.5.1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanRheenen S.M., Cao X., Sapperstein S.K., Chiang E.C., Lupashin V.V., Barlowe C., Waters M.G. 1999. Sec34p, a protein required for vesicle tethering to the yeast Golgi apparatus, is in a complex with Sec35p. J. Cell Biol. 147:729–742 10.1083/jcb.147.4.729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasile E., Oka T., Ericsson M., Nakamura N., Krieger M. 2006. IntraGolgi distribution of the conserved oligomeric Golgi (COG) complex. Exp. Cell Res. 312:3132–3141 10.1016/j.yexcr.2006.06.005 [DOI] [PubMed] [Google Scholar]
- Volchuk A., Ravazzola M., Perrelet A., Eng W.S., Di Liberto M., Varlamov O., Fukasawa M., Engel T., Söllner T.H., Rothman J.E., Orci L. 2004. Countercurrent distribution of two distinct SNARE complexes mediating transport within the Golgi stack. Mol. Biol. Cell. 15:1506–1518 10.1091/mbc.E03-08-0625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollert C.S., Uetz P. 2004. The phox homology (PX) domain protein interaction network in yeast. Mol. Cell. Proteomics. 3:1053–1064 10.1074/mcp.M400081-MCP200 [DOI] [PubMed] [Google Scholar]
- Wang C.-W., Stromhaug P.E., Shima J., Klionsky D.J. 2002. The Ccz1-Mon1 protein complex is required for the late step of multiple vacuole delivery pathways. J. Biol. Chem. 277:47917–47927 10.1074/jbc.M208191200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte J.R., Munro S. 2001. The Sec34/35 Golgi transport complex is related to the exocyst, defining a family of complexes involved in multiple steps of membrane traffic. Dev. Cell. 1:527–537 10.1016/S1534-5807(01)00063-6 [DOI] [PubMed] [Google Scholar]
- Wu X., Steet R.A., Bohorov O., Bakker J., Newell J., Krieger M., Spaapen L., Kornfeld S., Freeze H.H. 2004. Mutation of the COG complex subunit gene COG7 causes a lethal congenital disorder. Nat. Med. 10:518–523 10.1038/nm1041 [DOI] [PubMed] [Google Scholar]
- Wuestehube L.J., Duden R., Eun A., Hamamoto S., Korn P., Ram R., Schekman R. 1996. New mutants of Saccharomyces cerevisiae affected in the transport of proteins from the endoplasmic reticulum to the Golgi complex. Genetics. 142:393–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen W.-L., Klionsky D.J. 2008. How to live long and prosper: autophagy, mitochondria, and aging. Physiology (Bethesda). 23:248–262 [DOI] [PubMed] [Google Scholar]
- Yorimitsu T., Klionsky D.J. 2005. Atg11 links cargo to the vesicle-forming machinery in the cytoplasm to vacuole targeting pathway. Mol. Biol. Cell. 16:1593–1605 10.1091/mbc.E04-11-1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolov S.N., Lupashin V.V. 2005. Cog3p depletion blocks vesicle-mediated Golgi retrograde trafficking in HeLa cells. J. Cell Biol. 168:747–759 10.1083/jcb.200412003 [DOI] [PMC free article] [PubMed] [Google Scholar]