An overabundance of UbcH10 disrupts mitotic checkpoint signaling as a result of a degradation of cyclin B, increasing spontaneous and carcinogen-induced tumor formation in transgenic mice.
Abstract
The anaphase-promoting complex/cyclosome (APC/C) E3 ubiquitin ligase functions with the E2 ubiquitin–conjugating enzyme UbcH10 in the orderly progression through mitosis by marking key mitotic regulators for destruction by the 26-S proteasome. UbcH10 is overexpressed in many human cancer types and is associated with tumor progression. However, whether UbcH10 overexpression causes tumor formation is unknown. To address this central question and to define the molecular and cellular consequences of UbcH10 overexpression, we generated a series of transgenic mice in which UbcH10 was overexpressed in graded fashion. In this study, we show that UbcH10 overexpression leads to precocious degradation of cyclin B by the APC/C, supernumerary centrioles, lagging chromosomes, and aneuploidy. Importantly, we find that UbcH10 transgenic mice are prone to carcinogen-induced lung tumors and a broad spectrum of spontaneous tumors. Our results identify UbcH10 as a prominent protooncogene that causes whole chromosome instability and tumor formation over a wide gradient of overexpression levels.
Introduction
The disruption of cell cycle control mechanisms is a recurrent theme in tumorigenesis. Uncontrolled progression through mitosis can result in the missegregation of whole chromosomes and production of progeny cells with an abnormal chromosome content, which is referred to as aneuploidy (King, 2008; Ricke et al., 2008). Because most tumors contain aneuploid cells, it has long been hypothesized that aneuploidy might be causally implicated in tumor development (Boveri, 1902, 1914). Studies of mutant mice that are prone to missegregate chromosomes have provided some support for this hypothesis (Michel et al., 2001; Babu et al., 2003; Dai et al., 2004; Jeganathan et al., 2007; Weaver et al., 2007; Li et al., 2009). However, these studies also underscore the highly complex nature of the relationship between aneuploidy and cancer. In particular, the effect of aneuploidy on tumor development seems to depend strongly on the chromosomal instability gene defect that causes the aneuploidy, the extent and the nature of the defect, the tissue or cell type, and the context of other cancer-causing gene mutations (Pellman, 2007; Ricke et al., 2008; Weaver and Cleveland, 2009). These initial findings point out that it is critically important to identify and characterize the components and networks that regulate chromosome segregation and to test whether their dysfunction can drive tumorigenesis.
To minimize chromosome missegregation, eukaryotic cells have evolved a multiprotein surveillance mechanism called the mitotic checkpoint or spindle assembly checkpoint. It delays anaphase onset until all chromosomes are properly attached to the mitotic spindle and aligned at the metaphase plate (Musacchio and Salmon, 2007). Shortly after mitosis onset, core mitotic checkpoint components such as Bub and Mad proteins accumulate at unattached kinetochores to create inhibitors of Cdc20, one of two activating subunits of the anaphase-promoting complex/cyclosome (APC/C; Peters, 2006; Yu, 2007; Kulukian et al., 2009). It is believed that a protein complex of Mad2, BubR1, and Bub3, referred to as the mitotic checkpoint complex (MCC), is the most potent kinetochore-derived inhibitor of Cdc20 (Sudakin et al., 2001; Herzog et al., 2009). Proper attachment of the last kinetochore to the mitotic spindle quenches the inhibitory signals and triggers the release of the MCC from Cdc20, thereby activating the APC/C and allowing it to catalyze the polyubiquitination and destruction of securin and cyclin B. The removal of these mitotic regulators then results in the activation of separase, a clan D protease of the caspase family which initiates anaphase by opening the cohesin ring structures that hold sister chromosomes together (Nasmyth and Haering, 2005).
Besides initiating anaphase, APC/C activity also guides the cell through other stages of mitosis, at each step triggering the destruction of specific mitotic regulators (Peters, 2006; Sullivan and Morgan, 2007). In prometaphase, for instance, APC/CCdc20 targets cyclin A for degradation, whereas in anaphase, Cdc20 itself becomes an APC/C substrate when the Cdc20-related coactivator Cdh1 binds to the APC/C, although recent evidence suggests that Cdc20 is already subjected to APC/C-mediated degradation at an earlier stage in mitosis (Nilsson et al., 2008). In late mitosis, APC/CCdh1 drives mitotic exit through degradation of several mitotic kinases, including Plk1 and the Aurora A and B kinases. At least two E2 ubiquitin–conjugating enzymes (E2s), Ubc5 and UbcH10, are thought to collaborate with the APC/C (Sullivan and Morgan, 2007; Summers et al., 2008). Ubc5 interacts with various other E3 ubiquitin ligases and is constitutively expressed throughout the cell cycle. In contrast, UbcH10 is believed to be APC/C specific and reaches peak expression in mitosis (Rape and Kirschner, 2004; Summers et al., 2008). Furthermore, UbcH10 has been proposed to operate as part of an E2 module in conjunction with Ube2S, in which UbcH10 acts to initiate ubiquitin chain formation and Ube2S serves in chain elongation (Garnett et al., 2009; Williamson et al., 2009).
UbcH10 is expressed at relatively high levels in many different types of human tumors, including prostate, lung, gastric, esophageal, bladder, breast, ovarian, and uterine carcinomas (Okamoto et al., 2003; Wagner et al., 2004; Pallante et al., 2005; Berlingieri et al., 2007a,b; Jiang et al., 2008). Amplification of the UbcH10 gene locus is frequently observed in gastric and esophageal carcinomas and has been proposed to underlie UbcH10 overexpression in these cancers (Wagner et al., 2004). Importantly, UbcH10 overexpression correlates with tumor grade and prognosis in several cancer types. In some cancer cell lines, knockdown of UbcH10 expression by RNA interference has been shown to inhibit cell proliferation (Wagner et al., 2004; Berlingieri et al., 2007a,b), identifying UbcH10 as a potential target for cancer therapy. However, because UbcH10 transcript levels are low or undetectable in quiescent and differentiated cells and relatively high in proliferating cells (Yamanaka et al., 2000; Okamoto et al., 2003), the high UbcH10 transcript levels seen in many human cancers might simply be a reflection of the relatively high mitotic index of neoplastic versus normal tissue. Thus, the key open question is whether elevated UbcH10 expression is a consequence or a cause of neoplastic growth. Besides this, the impact of UbcH10 overexpression on the chromosome segregation process is unclear. One study reports that overexpression of UbcH10 in HeLa cells compromises the mitotic checkpoint (Rape and Kirschner, 2004), with follow up work suggesting that UbcH10 might function to inactivate the mitotic checkpoint by releasing the MCC from APC/CCdc20 through polyubiquitination of Cdc20 (Reddy et al., 2007). However, an independent study found that the mitotic checkpoint is virtually unperturbed when UbcH10 is overexpressed, leading to the conclusion that UbcH10 is not crucial for checkpoint inactivation (Walker et al., 2008). In this study, we generated a series of transgenic mice in which UbcH10 was overexpressed in a graded manner to determine whether this E2 is causally involved in tumor development and to examine the molecular and cellular defects associated with UbcH10 overexpression.
Results
Generation of transgenic mice with a graded increase in UbcH10 expression
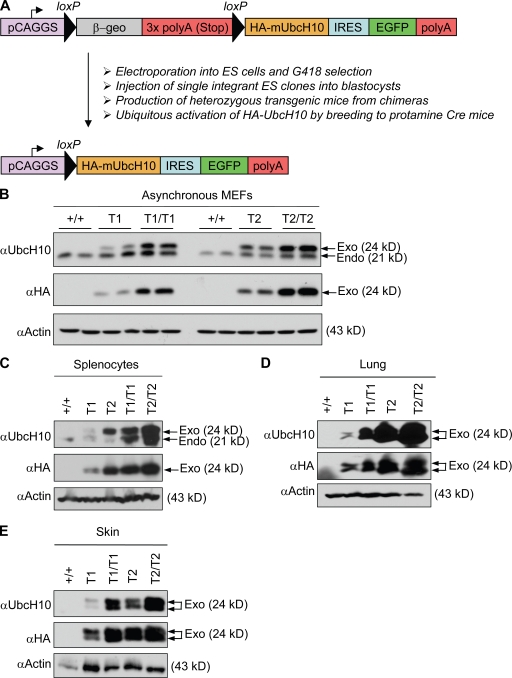
Because UbcH10 is overexpressed in a broad range of human cancer types, we sought to generate transgenic mice that overexpressed UbcH10 in a wide variety of tissues and organs. We used a transgenic vector containing the CAGGS promoter, which consists of the cytomegalovirus immediate enhancer and the chicken β-actin promoter (Novak et al., 2000). The CAGGS promoter drives a floxed β-geo-stop cassette consisting of a β-galactosidase and neomycin-resistance fusion gene and three polyadenylation sites (Fig. 1 A). Downstream of this cassette, we cloned the coding sequence for the murine UbcH10 protein, which we provided with a carboxy-terminal HA epitope tag sequence. The CAGGS promoter would express HA-UbcH10 only after Cre-mediated excision of the β-geo-stop cassette. EGFP was coexpressed from an internal ribosomal entry site to serve as a reporter for HA-UbcH10 expression.
Figure 1.
Generation of UbcH10 transgenic mice. (A) Overview of the generation of UbcH10 transgenic mice. Arrows indicate the direction of transcription. IRES, internal ribosomal entry site. (B) Western blot analysis of lysates from transgenic and control MEFs for endogenous (Endo) UbcH10 and exogenous (Exo) HA-UbcH10. Note that endogenous UbcH10 levels increase in transgenic MEFs. Actin was used as loading control. (C–E) Western blots of splenocyte (C), lung (D), and skin (E) extracts from mice of the indicated genotypes probed for UbcH10 and HA. Tissues were collected from 3-mo-old mice.
The vector was electroporated into mouse embryonic stem (ES) cells, and G418-resistant colonies were selected and clonally expanded. Part of each clone was infected with Cre-containing adenovirus to verify expression of EGFP and HA-UbcH10 by fluorescence microscopy and Western blotting, respectively. Positive clones were then further screened for single-copy transgene integration by Southern blotting to rule out that recombination between loxP sites from different integrations would cause chromosomal instability. Selected clones were injected into blastocysts to generate chimeric mice. Male chimeras from two independent ES clones, designated T1 and T2, produced transgenic offspring. Both strains were then crossed to protamine-Cre transgenic mice (Wagner et al., 1997) to excise the β-geo-stop cassette in the male germline. By breeding double transgenic males to wild-type females, we obtained offspring in which the CAGGS promoter was juxtaposed with the UbcH10 coding region in all cells. We refer to these transgenic strains as UbcH10T1 and UbcH10T2. By interbreeding UbcH10T1 animals, we generated wild-type, UbcH10T1, and UbcH10T1/T1 mice and mouse embryonic fibroblasts (MEFs). Likewise, wild-type, UbcH10T2, and UbcH10T2/T2 mice and MEFs were produced from UbcH10T2 intercrosses.
We first measured the level of exogenous UbcH10 expression in transgenic MEFs by Western blotting. Antibodies that detect either endogenous and exogenous UbcH10 or only exogenous UbcH10 were used in this analysis. Exogenous UbcH10 levels were highest in UbcH10T2/T2 MEFs and lowest in UbcH10T1 MEFs (Fig. 1 B). In turn, exogenous UbcH10 levels in UbcH10T1/T1 MEFs were higher than in UbcH10T2 MEFs. Furthermore, we noticed that endogenous UbcH10 levels were higher in transgenic MEF lines than in wild-type MEFs, suggesting that UbcH10 expression might be controlled by a positive autoregulatory feedback loop. As for MEFs, exogenous UbcH10 expression was highest in tissues and organs of UbcH10T2/T2 mice and lowest in those of UbcH10T1 mice (Fig. 1, C–E; and not depicted). Endogenous UbcH10 was undetectable in most wild-type and transgenic mouse tissues and organs. Thus, we obtained a series of transgenic mice and MEFs in which UbcH10 was overexpressed in graded fashion.
UbcH10 is overexpressed throughout the cell cycle
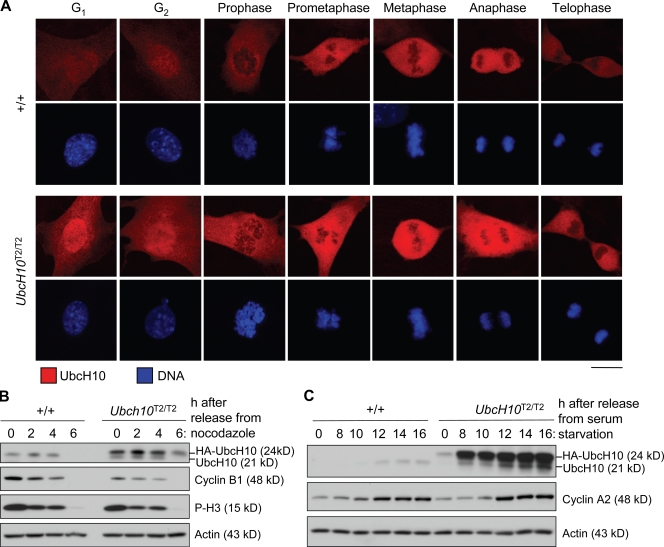
UbcH10 levels fluctuate during the mammalian cell cycle, with UbcH10 levels increasing before entry into mitosis and decreasing in late anaphase (Walker et al., 2008) or G1 (Rape and Kirschner, 2004). To examine whether the temporal expression pattern of UbcH10 was changed by UbcH10 overexpression, we immunostained wild-type and UbcH10T2/T2 MEFs with UbcH10 antibody (Fig. 2 A). Cells were costained for centrin 2 (not depicted) to allow for distinction between G1 and G2 phase. Wild-type MEFs showed low UbcH10 staining in G1 and G2. As expected, UbcH10 staining notably increased in mitosis and peaked between prometaphase and anaphase. It then dropped considerably, but substantial staining remained during telophase and early G1 phase. UbcH10T2/T2 MEFs showed a similar temporal expression pattern, but at each of the aforementioned stages, staining was noticeably higher than in wild-type MEFs (Fig. 2 A). Western blot analysis of extracts from nocodazole-arrested cells confirmed that UbcH10T2/T2 MEFs had much higher UbcH10 levels in midmitosis than wild-type MEFs (Fig. 2 B). After removal from nocodazole, UbcH10 levels declined in both UbcH10T2/T2 and wild-type MEFs simultaneously with cyclin B and phospho–histone H3 (P-H3) levels, although residual UbcH10 levels remained higher in UbcH10T2/T2 MEFs than in wild-type MEFs. To evaluate UbcH10 expression in interphase in greater detail, we arrested wild-type and UbcH10T2/T2 MEFs in G0 phase by serum starvation and then harvested cells at various time points after release in serum-containing medium. Both endogenous and transgenic UbcH10 levels were very low during quiescence (Fig. 2 C). After adding back serum, UbcH10 and cyclin A expression concomitantly increased in wild-type MEFs, indicating that endogenous UbcH10 levels normally rise as cells progress to S phase. In contrast, in UbcH10T2/T2 MEFs, UbcH10 overexpression resumed before S-phase entry and seemed to escalate further as cells entered S phase. Together, the aforementioned results demonstrate that transgenic MEFs overexpress UbcH10 throughout the cell cycle but that temporal fluctuations in protein levels remain. Proliferation assays suggested that UbcH10 overexpression had no impact on the duration of the cell cycle (Fig. S1).
Figure 2.
Transgenic UbcH10 protein is overexpressed throughout the cell cycle. (A) Immunostaining of wild-type and UbcH10T2/T2 MEFs with anti-UbcH10 antibody. DNA was visualized with Hoechst. (B) Western blot analysis of extracts from wild-type and UbcH10T2/T2 MEFs arrested in mitosis with nocodazole and released in fresh medium for the indicated times. Blots were probed for UbcH10 to monitor its degradation during mitotic exit. Cyclin B and P-H3 were markers for mitotic exit. (C) Analysis of UbcH10 levels in wild-type and UbcH10T2/T2 MEFs at various times after release from serum starvation. Cyclin A2 was a marker for S-phase entry. Bar, 10 µm.
UbcH10 overexpression causes chromosome lagging and aneuploidy
A key question is whether UbcH10 overexpression leads to inaccurate chromosome segregation and aneuploidy. To address this question, we performed chromosome counts on metaphase spreads of transgenic and wild-type MEFs. An abnormal number of chromosomes was found in 13% of wild-type MEFs (Table I). In contrast, aneuploidy was much more severe in transgenic MEFs, with UbcH10T1, UbcH10T2, UbcH10T1/T1, and UbcH10T2/T2 MEFs showing 28%, 29%, 31%, and 33% aneuploidy, respectively. The range of abnormal chromosome numbers was considerably broader in transgenic MEFs than in wild-type MEFs. UbcH10T1/T1, UbcH10T2, and UbcH10T2/T2 MEFs showed modest increases in incidence of premature sister chromatid separation (PMSCS), a defect which has been linked to precocious APC/C activity (Table I). To determine whether UbcH10 overexpression would also cause aneuploidy in vivo, we harvested splenocytes from 5-mo-old wild-type and transgenic mice and counted mitotic chromosomes. As expected, wild-type splenocytes had no aneuploidy (Table II). In contrast, UbcH10T1, UbcH10T2, UbcH10T1/T1, and UbcH10T2/T2 splenocytes had 4%, 6%, 13%, and 19% aneuploidy, respectively (Table II). Similar to MEFs, splenocytes showed small elevations of PMSCS at the highest levels of UbcH10 overexpression (Table II). Collectively, these experiments demonstrate that UbcH10 overexpression causes aneuploidy and indicate that there is a positive correlation between the level of UbcH10 overexpression and the degree of aneuploidy.
Table I.
Gradual overexpression of UbcH10 leads to progressive aneuploidy
| Mitotic MEF genotype | Aneuploid figures (SD) | Karyotypes with indicated chromosome number | Mitotic figures with PMSCS (SD) | |||||||||||
| 37 | 38 | 39 | 40 | 41 | 42 | 43 | 44 | 45 | 46 | 48 | 52 | |||
| % | % | |||||||||||||
| +/+ | 13 (1) | 1 | 4 | 6 | 130 | 6 | 3 | 2 (1) | ||||||
| UbcH10T1 | 28 (2) | 5 | 2 | 9 | 108 | 9 | 6 | 6 | 2 | 1 | 2 | 3 (0) | ||
| UbcH10T2 | 29 (2) | 2 | 4 | 10 | 107 | 6 | 9 | 3 | 4 | 1 | 1 | 1 | 2 | 6 (2) |
| UbcH10T1/T1 | 31 (2) | 3 | 4 | 9 | 103 | 16 | 5 | 7 | 3 | 5 (2) | ||||
| UbcH10T2/T2 | 33 (4) | 3 | 13 | 100 | 13 | 7 | 8 | 3 | 2 | 1 | 5 (2) | |||
Of each genotype, 50 spreads from three individual cell lines were counted. Aneuploidy and PMSCS were measured at passage 5. Empty cells indicate that there were no karyotypes with the indicated chromosome number.
Table II.
Gradual overexpression of UbcH10 in splenocytes leads to progressive aneuploidy
| Mouse genotype | Age | Aneuploid figures (SD) | Karyotypes with indicated chromosome number | Mitotic figures with PMSCS (SD) | |||||
| 37 | 38 | 39 | 40 | 41 | 42 | ||||
| mo | % | % | |||||||
| +/+ | 5 | 0 (0) | 150 | 0 (0) | |||||
| UbcH10T1 | 5 | 4 (0) | 1 | 144 | 4 | 1 | 0 (0) | ||
| UbcH10T2 | 5 | 6 (2) | 1 | 2 | 3 | 141 | 3 | 1 (1) | |
| UbcH10T1/T1 | 5 | 13 (1) | 2 | 4 | 130 | 12 | 2 | 4 (1) | |
| UbcH10T2/T2 | 5 | 19 (1) | 1 | 3 | 9 | 121 | 13 | 3 | 5 (1) |
Of each genotype, 50 spreads from three individual mice were counted. Empty cells indicate that there were no karyotypes with the indicated chromosome number.
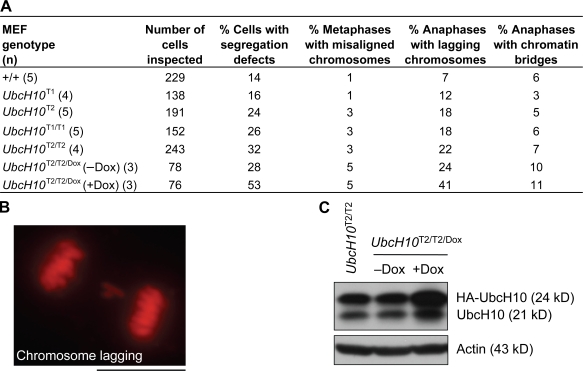
To determine the nature of the mitotic defects underlying the aneuploidy observed in cells with high UbcH10 levels, we followed the chromosome movements of transgenic MEFs through an unchallenged mitosis by time-lapse live microscopy. To visualize chromosomes, MEFs were transduced with lentivirus pTSIN–H2B–monomeric RFP (mRFP). We found that the percentage of cells with mitotic defects that can result in chromosome missegregation increased with increasing UbcH10 overexpression, with 32% of UbcH10T2/T2 MEFs showing errors compared with 14% of wild-type MEFs (Fig. 3 A). The main defect observed in all transgenic MEF lines was chromosome lagging (Fig. 3 B). Furthermore, chromosome misalignment was threefold higher in UbcH10T1/T1, UbcH10T2, and UbcH10T2/T2 MEFs than in wild-type MEFs but not elevated at the lowest level of UbcH10 expression (Fig. 3 A). To test whether further escalation of UbcH10 expression would exacerbate the observed mitotic defects, we transduced UbcH10T2/T2 MEFs with lentivirus containing a doxycycline (Dox)-inducible HA-tagged UbcH10 transgene (designated UbcH10Dox) and performed live cell imaging on induced and noninduced cells. Western blotting confirmed that Dox boosted UbcH10 overexpression of UbcH10T2/T2/Dox MEFs (Fig. 3 C). As expected, noninduced UbcH10T2/T2/Dox MEFs had similar mitotic error rates as UbcH10T2/T2 MEFs (Fig. 3 A). However, we observed a 25% increase in mitotic defects in Dox-treated UbcH10T2/T2/Dox MEFs, with chromosome lagging being the most prominently increased (Fig. 3 A).
Figure 3.
Chromosome missegregation increases as UbcH10 levels rise. (A) Analysis of chromosome segregation defects in MEFs with increasing amounts of exogenous UbcH10. Cells scored as metaphases with misaligned chromosomes displayed congression failure at anaphase onset. UbcH10T2/T2/Dox (+Dox) MEFs were grown in medium containing 1 µg/ml Dox for 2 d before live cell imaging. (B) Image of a transgenic MEF with chromosome lagging. (C) Immunoblots of asynchronous UbcH10T2/T2/Dox MEFs cultured in the presence (+Dox) or absence (−Dox) of 1 µg/ml Dox for 2 d. Blots were probed for UbcH10 and actin. Bar, 10 µm.
UbcH10 overexpression promotes mitotic slippage
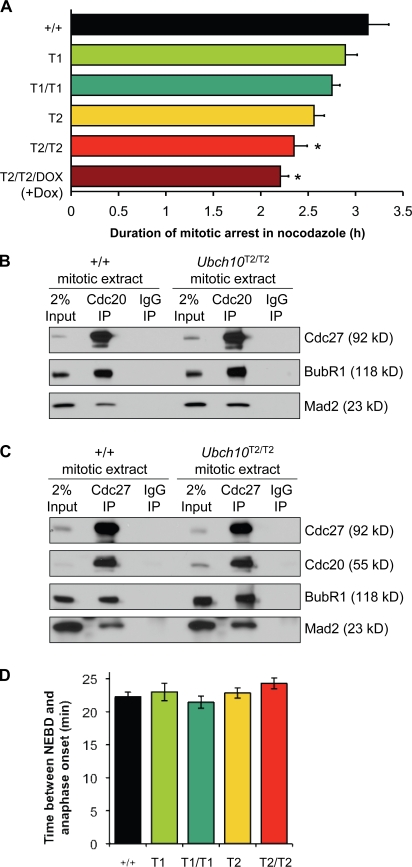
One possible explanation for the inaccurate chromosome segregation is that UbcH10 overexpression deregulates the mitotic checkpoint. However, published results regarding the effect of UbcH10 overexpression on mitotic checkpoint function are somewhat conflicting, with one study reporting that transient expression of UbcH10 in HeLa cells causes checkpoint inactivation (Reddy et al., 2007) and another reporting that it does not (Walker et al., 2008). To further examine the relationship between UbcH10 overexpression and mitotic checkpoint function, we performed a live cell imaging–based nocodazole challenge assay on mRFP-H2B–positive UbcH10 transgenic MEFs. In this assay, the mitotic checkpoint is activated, and the time between nuclear envelope breakdown (NEBD) and DNA decondensation is measured (Baker et al., 2006). Cells with an intact mitotic checkpoint typically arrest in prometaphase for a prolonged period of time but eventually exit mitosis and enter G1 phase without chromosome segregation. This process is referred to as mitotic slippage and requires the polyubiquitination and degradation of cyclin B (Brito and Rieder, 2006). Earlier studies have demonstrated that the rate of mitotic slippage is accelerated in cells with a defective mitotic checkpoint (Michel et al., 2001; Meraldi et al., 2004; Baker et al., 2006; Jeganathan et al., 2007; Perera et al., 2007). The time of arrest in nocodazole was reduced by 25% at the highest level of UbcH10 overexpression (UbcH10T2/T2) and by 7% at the lowest level of overexpression (UbcH10T1; Fig. 4 A). MEFs with intermediate levels of UbcH10 overexpression had reductions in the range of 12–18%. Furthermore, superinduction of UbcH10 in MEFs with the highest level of UbcH10 overexpression (UbcH10T2/T2/Dox MEFs [+Dox]) further reduced the time of arrest in mitosis from 25 to 29% (Fig. 4 A). Together, these data indicate that the rate of mitotic slippage increases with escalating levels of UbcH10 overexpression and imply that UbcH10 overexpression causes a mild mitotic checkpoint defect.
Figure 4.
UbcH10 overexpression enhances mitotic slippage. (A) Analysis of mitotic slippage rates of UbcH10 transgenic and wild-type MEFs by nocodazole challenge assay. At least three independent MEF lines per genotype were analyzed. *, P < 0.05 versus wild-type MEFs (Log-rank test). (B) UbcH10 overexpression does not seem to cause premature release of MCC proteins from Cdc20. MEFs were arrested in mitosis by nocodazole treatment and harvested by mitotic shake off. Lysates were prepared from equal amounts of wild-type and UbcH10T2/T2 cells and subjected to IP with antibodies against Cdc20 and analyzed by Western blotting with antibodies against Mad2, BubR1, and Cdc27. (C) Same as B, but IPs were performed with antibodies against Cdc27 instead of Cdc20. (D) Timing of mitosis is normal in UbcH10 transgenic MEFs. mRFP-H2B–expressing cells were followed through an unchallenged mitosis by live cell imaging, and the time between NEBD to anaphase onset was measured. At least three MEF lines per genotype were analyzed with a minimum of 43 total cells. (A and D) Error bars represent SEM.
It has recently been suggested that UbcH10 might function to inactivate the mitotic checkpoint by promoting the dissociation of Mad2 and BubR1 from Cdc20 or Cdc20 bound to APC/C (Reddy et al., 2007). To test whether UbcH10 overexpression causes the early release of Mad2 and BubR1 from Cdc20, we performed immunoprecipitations (IPs) for Cdc20 from nocodazole-arrested UbcH10T2/T2 and wild-type MEF extracts and analyzed bound proteins by immunoblotting using BubR1 and Mad2 antibodies. Cdc20 precipitated similar amounts of Mad2 and BubR1 from both extracts (Fig. 4 B). Furthermore, IPs for Cdc27 from mitotic UbcH10T2/T2 and wild-type extracts revealed that binding of Cdc20, Mad2, and BubR1 to APC/C was not overtly diminished by UbcH10 overexpression (Fig. 4 C). Thus, binding of inhibitory mitotic checkpoint proteins to Cdc20 or Cdc20 bound to APC/C seems largely unaffected by UbcH10 overexpression. Consistent with this, timing from NEBD to anaphase onset during an unchallenged mitosis, which is thought to depend on binding of BubR1 and Mad2 to Cdc20 (Meraldi et al., 2004; Malureanu et al., 2009), was unaltered in MEFs that overexpressed UbcH10 (Fig. 4 D). In addition, immunolocalization of core mitotic checkpoint proteins that accumulate at unattached kinetochores to propel MCC formation, including Bub1, BubR1, Mad1, Mad2, and CENP-E, was normal in UbcH10T2/T2 MEFs (Fig. S2 and not depicted).
UbcH10 overexpression reduces cyclin B stability
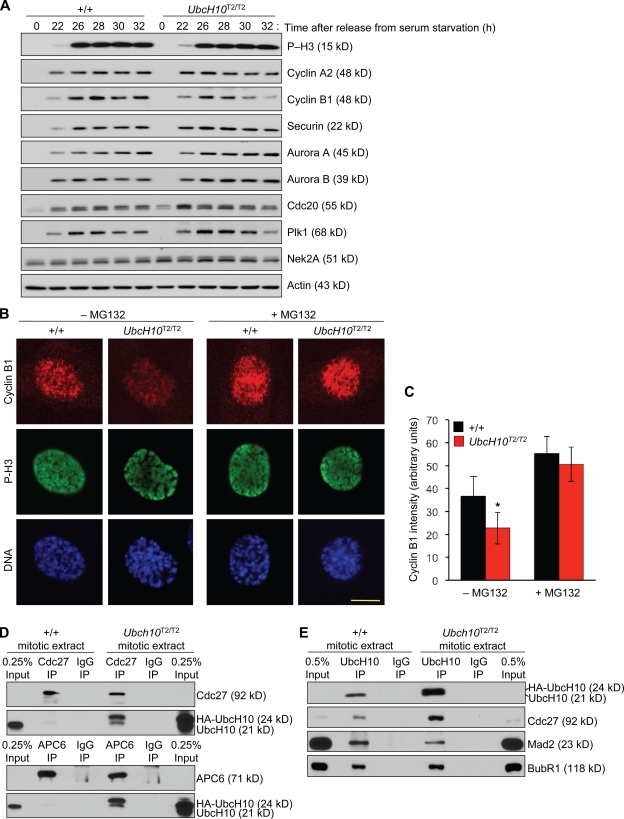
To better understand the molecular basis of the mitotic slippage and the chromosomal instability, we screened UbcH10T2/T2 MEFs for alterations in the levels of key regulators of chromosome segregation whose stability is determined by the APC/C. Western blot analysis revealed that cyclin B levels were consistently lower in mitotic UbcH10T2/T2 MEF extracts than in those of wild-type MEFs (Figs. 2 B and 5 A). Immunostainings of wild-type and UbcH10T2/T2 MEFs for cyclin B confirmed this reduction (Fig. 5, B and C). When UbcH10T2/T2 MEFs were treated for 1 h with the proteasome inhibitor MG132 before cell fixation, cyclin B levels increased to normal (Fig. 5, B and C), indicating that the observed decline of cyclin B in UbcH10T2/T2 MEFs was APC/C mediated. In contrast, cyclin A2, Nek2A, and securin, three APC/C substrates which are normally degraded in prometaphase or metaphase, were present at normal levels in mitotic UbcH10T2/T2 MEF extracts (Fig. 5 A and Fig. S3). This also holds true for Cdc20, Plk1, and Aurora A and B, four substrates which are normally targeted for degradation by APC/CCdh1 later in mitosis (Fig. 5 A; Sullivan and Morgan, 2007). Aurora B functions in the correction of merotelic kinetochore attachments (Andrews et al., 2004; Kline-Smith et al., 2004), and its premature degradation would have provided a plausible explanation for the high incidence of lagging chromosomes in UbcH10-overexpressing cells. We verified that Aurora B properly targeted to kinetochores of UbcH10T2/T2 MEFs in mitosis (Fig. S4 A). Furthermore, mitotic centromere-associated kinesin (MCAK), a microtubule-depolymerizing protein which is essential for resolving merotelic attachments and requires Aurora B activity for recruitment to kinetochores, was also properly targeted to kinetochores of UbcH10-overexpressing cells (Fig. S4 B), suggesting that key components of the mechanism that act to correct merotelic attachments are intact in cells that overexpress UbcH10.
Figure 5.
Cyclin B is precociously degraded in UbcH10-overexpressing cells. (A) Western blot analysis of wild-type and UbcH10T2/T2 MEFs for APC/C substrate levels. MEFs were synchronized in G0 by serum starvation and then released for the indicated durations in serum-containing medium. 100 ng/ml nocodazole was added 23 h after cells were released. Blots were probed with the indicated antibodies. We note that even though UbcH10T2/T2 MEFs show evidence of mitotic slippage, their P-H3 levels do not drop at the later time points, suggesting that P-H3 is not always a suitable marker for aberrant mitotic exit. (B) Immunostaining of wild-type and UbcH10T2/T2 prophases for cyclin B, P-H3, and DNA (Hoechst). P-H3 served as a marker for mitosis entry. (C) Cyclin B signals in prophase were quantified using ImageJ software (n = 10 cells per genotype). Error bars represent SD. *, P < 0.0001 versus wild type (unpaired t test). (D) Lysates prepared from equal amounts of mitotic wild-type and UbcH10T2/T2 cells obtained by mitotic shake off were subjected to IP with antibodies against Cdc27 or APC6 and analyzed by Western blotting with antibodies against the indicated proteins. (E) Lysates prepared as in D subjected to IP with antibodies against UbcH10 and analyzed by the indicated proteins. Results shown in D and E are representative for three independent experiments. Bar, 10 µm.
How then might APC/C become prematurely active against cyclin B in UbcH10T2/T2 MEFs if UbcH10 overexpression does not seem to interfere with the overall binding of MCC to APC/C? It has been proposed that even cells with a fully active mitotic checkpoint have a low rate of APC/C-mediated cyclin B degradation, implying that the MCC is unable to inhibit all of the APC/C molecules (Brito and Rieder, 2006). Binding of UbcH10 to APC/C is highly transient, and only a very small fraction of the APC/C is UbcH10 bound. UbcH10 overexpression is expected to shift the binding equilibrium of UbcH10 to APC/C to the active APC/C–UbcH10 complex, thereby perhaps increasing the activity of the uninhibited pool of APC/C molecules and destabilizing cyclin B. To examine the effect of UbcH10 overexpression on APC/C–UbcH10 complex formation, we performed IPs for APC/C components on mitotic extracts of UbcH10T2/T2 and wild-type MEFs and then used immunoblotting to determine the amount of coprecipitating UbcH10. Consistent with the transient nature of the UbcH10–APC/C interaction, Cdc27 and APC6 precipitated only a very small fraction of the cellular UbcH10 pool from wild-type extracts (Fig. 5 D). However, both APC/C components precipitated substantially more UbcH10 from UbcH10T2/T2 extracts. In the reverse experiment, UbcH10 precipitated much more Cdc27 from UbcH10T2/T2 extracts than from wild-type extracts (Fig. 5 E). These data indicate that APC/C–UbcH10 complex formation increases when UbcH10 is overexpressed, providing a plausible explanation for why cyclin B has reduced stability in UbcH10T2/T2 MEFs. Moreover, the amounts of Mad2 and BubR1 that precipitated with UbcH10 relative to Cdc27 were considerably lower in UbcH10T2/T2 extracts than in wild-type extracts (Fig. 5 E), which is consistent with the idea that UbcH10 overexpression increases the amount of active APC/C (Fig. S5).
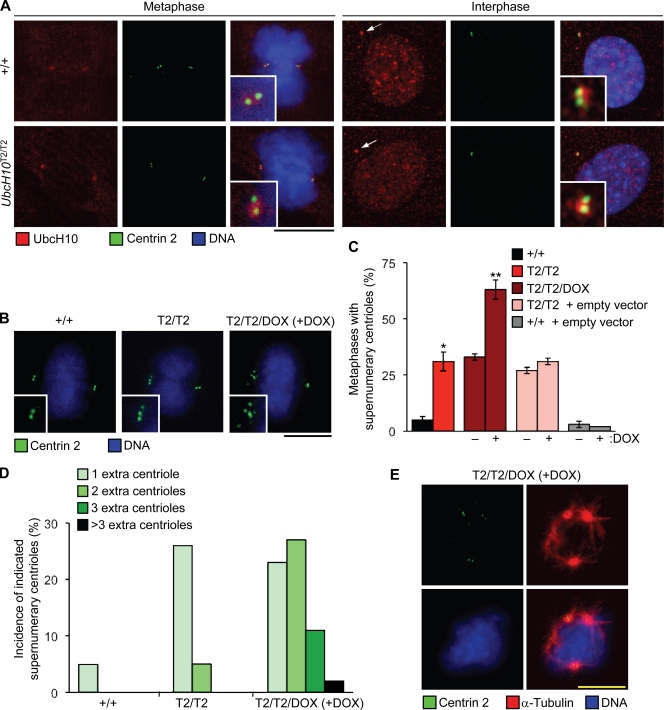
UbcH10-overexpressing cells have extra centrioles
Vihar E2-C, the Drosophila melanogaster homologue of UbcH10, localizes at centrosomes during mitosis (Máthé et al., 2004), which prompted us to investigate whether UbcH10 is also associated with centrosomes in mammalian cells. Using a mild fixation procedure that allows for the removal of soluble protein fractions before immunostaining, we observed UbcH10 localization at centrosomes of wild-type and UbcH10T2/T2 MEFs during mitosis and in interphase (Fig. 6 A). Staining intensities at centrosomes were similar for both genotypes. Although nearly all wild-type metaphases contained two centrioles per spindle pole, 31% of UbcH10T2/T2 metaphases had at least one spindle pole that consisted of three or more centrioles (Fig. 6, B and C). Superinduction of UbcH10 in these cells (UbcH10T2/T2/Dox MEFs [+Dox]) escalated this defect, with 63% of metaphases showing supernumerary centrioles. Moreover, superinduction considerably increased the number of extra centrioles per cell (Fig. 6 D). Extra centrosomes can increase the frequency of lagging chromosomes by promoting the formation of merotelic kinetochore–microtubule attachments (Ganem et al., 2009; Silkworth et al., 2009). Thus, it is possible that the observed increase in lagging chromosomes in UbcH10-overexpressing cells is caused by the numerical centriole abnormalities. Supernumerary centrioles (Fig. 6, B–D) and lagging chromosomes (Fig. 3 A) both correlated with UbcH10 overexpression, which supports this idea. It has been proposed that extra centrioles increase the incidence of merotelic kinetochore–microtubule attachments by inducing multipolar spindle intermediates that resolve into bipolar spindles through centrosome clustering (Ganem et al., 2009; Silkworth et al., 2009). To examine whether such intermediates are formed in UbcH10-overexpressing cells, we stained wild-type, UbcH10T2/T2, and superinduced UbcH10T2/T2 MEFs for centrioles, microtubules, and DNA. Although spindles of wild-type MEFs were exclusively bipolar, low percentages of multipolar spindles were indeed found in mitotic UbcH10T2/T2 and UbcH10T2/T2/Dox (+Dox) MEFs (1% and 7%, respectively; Fig. 6 E).
Figure 6.
UbcH10-overexpressing cells have supernumerary centrioles. (A) Wild-type and UbcH10T2/T2 MEFs during metaphase and interphase stained for UbcH10 and centrin 2. MEFs were fixed in 1% paraformaldehyde for 5 min and then permeabilized in 0.2% Triton X-100. Insets are an enlargement of the centrosome region, and arrows point to the centrosome. (B) Wild-type, UbcH10T2/T2, and superinduced UbcH10T2/T2 metaphases stained for centrin 2 and DNA. Spindle poles with extra centrioles are shown in the insets. (C) Percentage of wild-type, UbcH10T2/T2, and superinduced UbcH10T2/T2 metaphases with extra centrioles. Wild-type and UbcH10T2/T2 MEFs infected with lentivirus containing pTRIC empty vector served as controls for the impact of lentiviral infection on centrosome amplification. Error bars indicate SD. *, P = 0.0145 versus wild-type MEFs; and **, P = 0.0109 versus UbcH10T2/T2 MEFs (unpaired t test; n = 2 lines per genotype). (D) Incidence of wild-type, UbcH10T2/T2, and superinduced UbcH10T2/T2 metaphases with the indicated number of extra centrioles. (E) Tripolar UbcH10T2/T2/Dox (+Dox; superinduced UbcH10T2/T2) MEF stained for centrin 2, α-tubulin, and DNA. Bars, 10 µm.
UbcH10 overexpression causes tumor formation
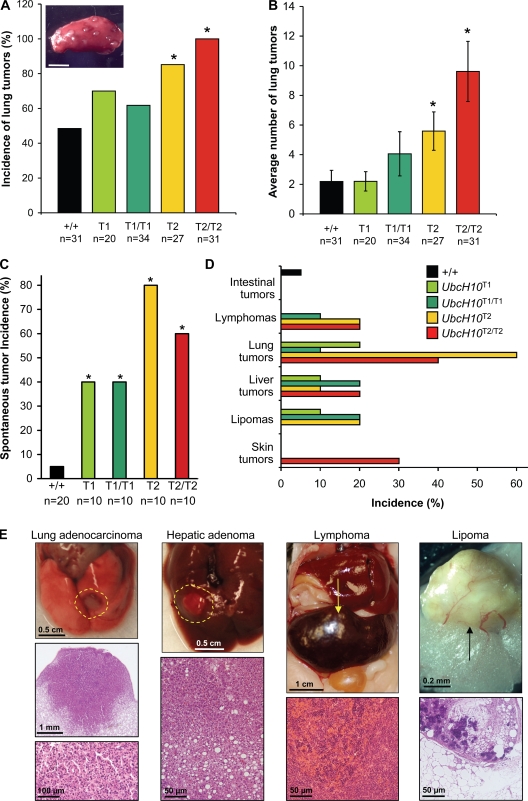
To address the central question as to whether UbcH10 overexpression can act to promote tumorigenesis, we first performed a tumor bioassay with 7,12-dimethylbenz(a)anthracene (DMBA), a carcinogen which, when applied to the skin at low dose, predisposes wild-type mice to lung tumors and skin tumors (Dawlaty et al., 2008). Pups from UbcH10T1 × UbcH10T1 and UbcH10T2 × UbcH10T2 intercrosses received a single application of 50 µl of 0.5% DMBA in acetone to the dorsal skin between postnatal day (P) 3 and P5. At 5 mo of age, animals were sacrificed and screened for lung and skin tumors. We observed at least one lung tumor in 50% of wild-type animals (Fig. 7 A). Lung tumor incidence was slightly elevated in both UbcH10T1 and UbcH10T1/T1 mice, although the differences were not statistically significant. However, as levels of UbcH10 overexpression progressively increased, the incidence of lung tumors concurrently inclined, with 84% of UbcH10T2 and 100% of UbcH10T2/T2 mice developing this tumor type (Fig. 7 A). The lung tumor burden also increased gradually along with increasing levels of UbcH10 overexpression, with UbcH10T2 and UbcH10T2/T2 mice developing, on average, nearly 6 and 10 tumors per mouse, respectively, compared with ∼2 in wild-type and UbcH10T1 mice (Fig. 7 B). Although UbcH10-overexpressing mice showed a trend toward increased DMBA-induced skin tumor incidence, there was no statistically significant difference versus wild-type mice (unpublished data).
Figure 7.
UbcH10 overexpression predisposes mice to DMBA-induced and spontaneous cancers. (A) Lung tumor incidence of DMBA-treated mice of the indicated genotypes. *, P < 0.05 versus wild-type mice (Fisher’s exact test). Note that UbcH10 expression levels in lung increase from +/+ < T1 < T1/T1 < T2 < T2/T2 (see Fig. 1 D). The inset shows gross appearance of DMBA-induced lung tumors. (B) Lung tumor burden of DMBA-treated mice of the indicated genotypes. Error bars indicate SEM. *, P < 0.05 versus wild-type mice (Mann-Whitney test). (C) Spontaneous tumor incidence of mice of the indicated genotypes. The average age of the wild-type, UbcH10T1, UbcH10T1/T1, UbcH10T2, and UbcH10T2/T2 mice used in the study was 15 mo, 16 mo, 12 mo, 16 mo, and 13 mo, respectively. *, P < 0.05 versus wild-type mice (Fisher’s exact test). (D) Tumor spectrum of mice of the indicated genotypes. (E) Gross images and histological analysis of selected spontaneous tumors from transgenic mice. Dashed lines encircle and arrows point to the tumor region. Bar, 3 mm.
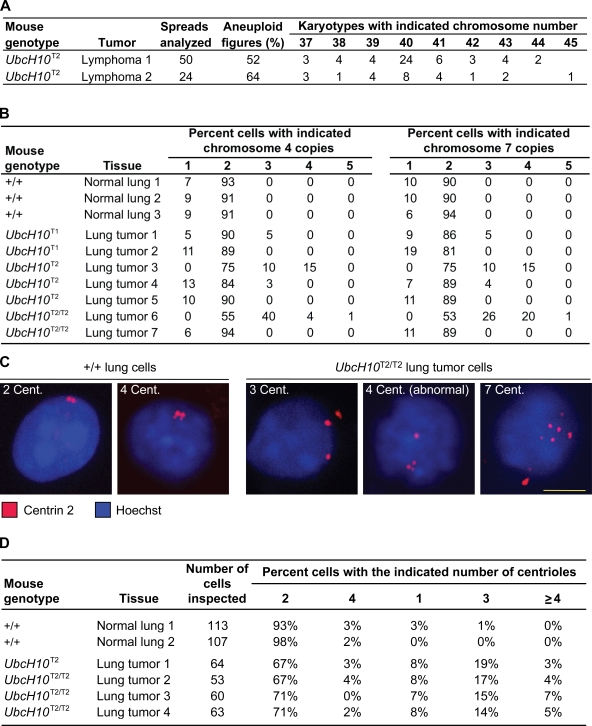
To determine whether UbcH10 overexpression predisposes mice to spontaneous tumors, cohorts of wild-type, UbcH10T1, UbcH10T1/T1, UbcH10T2, and UbcH10T2/T2 mice were aged to 12–16 mo and screened for the presence of tumors. Overt tumors were collected and subjected to histopathology. As shown in Fig. 7 C, all transgenic strains had marked and significant increases in tumor incidence compared with wild-type mice. The tumor spectrum of UbcH10 transgenic mice was broad and included lymphomas, lung adenomas, lipomas, and liver and skin tumors (Fig. 7, D and E). None of these tumor types were observed in mice of our wild-type cohort. Their overall tumor incidence (Fig. 7 C) did not correlate well with expected expression levels based on genotypes. Lung tumors were seen in all transgenic strains but were more frequent at the higher levels of UbcH10 overexpression (Fig. 7 D). Chromosome counts on metaphase spreads of two UbcH10T2/T2 lymphomas revealed that these tumors contained a high proportion of aneuploid cells (Fig. 8 A). Furthermore, interphase FISH on tumor sections using probes for chromosomes 4 and 7 provided evidence for aneuploidy in five out of seven lung tumors (Fig. 8 B) and two out of three liver tumors (not depicted). Immunostaining of lung tumor cells with an antibody against centrin 2 revealed evidence of substantial numerical centriole abnormalities in four out of four lung tumors analyzed (Fig. 8, C and D). Collectively, the aforementioned data demonstrate that UbcH10 overexpression is causally implicated in tumor formation and that it is associated with chromosome number instability. Furthermore, these data suggest that the range of overexpression levels at which UbcH10 drives tumorigenesis is rather wide.
Figure 8.
Evidence of numerical chromosome and centriole abnormalities in tumors from UbcH10 transgenic mice. (A) Chromosome counts on lymphoma cells from two UbcH10T2 transgenic mice. Empty cells in the table indicate that there were no karyotypes with the indicated chromosome number. (B) Interphase FISH for chromosomes 4 and 7 on 5-µm lung tumor sections of UbcH10 transgenic mice. Normal lung sections of wild-type mice were used as controls. The number of FISH signals was determined for 100 interphase cells per lung tumor/normal lung tissue sample. (C) Interphase cells from normal lung tissue of wild-type mice or lung tumors of UbcH10 transgenic mice stained for centrioles (cent.) and DNA. (D) Quantification of centriole numbers in normal lung cells and transgenic lung tumor cells. Bar, 10 µm.
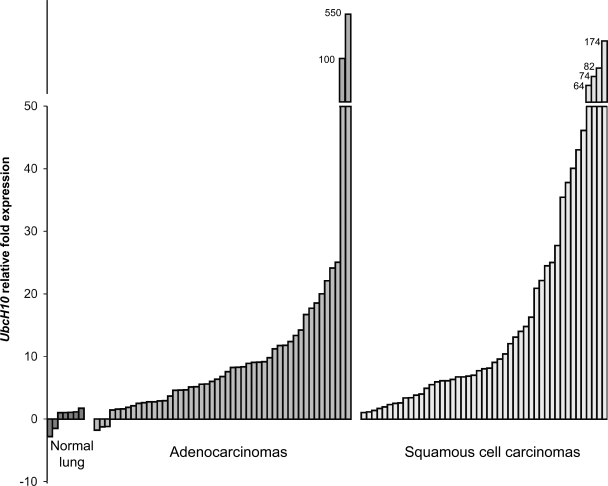
UbcH10 levels are high in human lung cancers
Using quantitative real-time PCR, we measured UbcH10 transcript levels in 49 lung adenocarcinomas, 47 human squamous cell carcinomas, and 7 normal lung samples. We found that mRNA levels were extremely high (>50-fold above normal) in two adenocarcinomas (4.1%) and four squamous cell carcinomas (8.5%; Fig. 9). High transcript levels (10–50-fold) were observed in 14 adenocarcinomas (28.6%) and in 16 squamous cell carcinomas (34%), whereas 15 adenocarcinomas (30.6%) and 14 squamous cell carcinomas (29.7%) showed moderate transcript levels (5–10-fold). The data suggest that human lung tumors have a high incidence of UbcH10 overexpression and that there is quite a wide range in the levels of UbcH10 overexpression in these tumors. However, because UbcH10 expression is induced in proliferating cells, it is important to consider that increases in UbcH10 transcript levels are at least partly caused by increased rates in mitosis of tumor versus normal tissue.
Figure 9.
UbcH10 is overexpressed in human primary lung cancers. UbcH10 expression in normal lung tissues, lung adenocarcinomas, and squamous cell lung carcinomas analyzed by quantitative RT-PCR. Data represent the mean of duplicate measurements. UbcH10 transcript levels were normalized to TBP. The relative fold UbcH10 expression was normalized to the average of normal tissue expression.
Discussion
UbcH10 is overexpressed in many human cancer types and is associated with tumor progression. In this study, we demonstrate that UbcH10 overexpression is causally implicated in tumor development. Direct evidence for this conclusion comes from the observation that UbcH10 overexpression in mice leads to a wide variety of spontaneous tumors, including lung adenomas and adenocarcinomas, hepatic adenomas and adenocarcinomas, lymphomas, skin tumors, and lipomas. These tumors were observed in most transgenic strains, indicating that a rather broad range of UbcH10 overexpression levels can initiate neoplastic transformation. Compared with most other animal models for aneuploidy, UbcH10 transgenic mice show a high incidence of tumor formation and a broad spectrum of spontaneous tumors (Ricke et al., 2008; Holland and Cleveland, 2009). Among the tissues that develop tumors, the lung seems to be particularly sensitive to the effects of UbcH10 overexpression, with both spontaneous and carcinogen-induced tumorigenesis being most profound at higher levels of UbcH10 overexpression. Given that UbcH10 transcript levels are commonly elevated in human lung adenocarcinomas and squamous cell carcinomas, it is tempting to speculate that at least a subset of these tumors expresses an oncogenic amount of UbcH10.
We find that UbcH10 overexpression causes whole chromosome instability. This, combined with data from recent studies of various mitotic checkpoint protein–defective mouse strains and human cells lines showing that aneuploidy can be causally linked to tumorigenesis (Ricke et al., 2008; Holland and Cleveland, 2009), suggests that the effect of UbcH10 overexpression on tumor formation results, at least in part, from chromosome missegregation and aneuploidy. Chromosome lagging, which is believed to be the primary source of aneuploidy in human cancers (Cimini, 2008; Thompson and Compton, 2008), is the main chromosome segregation error associated with UbcH10 overexpression. Chromosome lagging is caused by merotelic chromosome attachment (Cimini, 2008). Such attachments also occur at a low rate in normal cells but are efficiently corrected through a process that involves recruitment of Aurora B and MCAK to inner centromeric regions of duplicated chromosomes (Andrews et al., 2004; Lan et al., 2004). We find that both of these proteins properly target to the inner centromeric regions of UbcH10 transgenic MEFs, suggesting that two key components of the machinery that corrects merotelic attachments are functioning under conditions of supranormal UbcH10 levels. However, it cannot be excluded that other components are defective or that the efficiency with which attachment errors are corrected is reduced (Bakhoum et al., 2009a,b).
Interestingly, centrosome amplification has recently been identified as an underlying cause of merotely and chromosome lagging in cancer cells (Ganem et al., 2009; Silkworth et al., 2009). It has been proposed that extra centrosomes can lead to the formation of multipolar spindles, which, because of their abnormal spindle geometry, promote merotelic attachments. Centrosome clustering allows for bipolar cell division of these cells, but this frequently occurs while merotelic attachment errors persist, thereby yielding lagging chromosomes. This mechanism of chromosomal instability might apply to UbcH10 overexpression because supernumerary centrioles and multipolar spindles are features of cells with high UbcH10 levels. Furthermore, the incidence of both of these defects correlates well with the level of UbcH10 overexpression and the frequency of chromosome lagging. It should be noted that the incidence of multipolar spindles was considerably lower than that of centriole supernumerary, which may be because of the transient nature of multipolar spindle intermediates.
Failed cytokinesis and acentrosomal spindles can both cause numerical centriole anomalies (Nigg, 2002), but no such defects were observed in UbcH10-overexpressing cells. This implies that abnormal duplication of centrosomes, commonly referred to as centrosome amplification (Fukasawa, 2007), is the most plausible mechanism by which extra centrioles arise in transgenic cells. How UbcH10 overexpression might act to perturb the centrosome duplication process remains to be determined. A wide variety of proteins can cause centrosome number instability when defective, including the APC/C substrates cyclin B, Nek2A, Aurora A, and Plk1 (Fukasawa, 2007). Of these, cyclin B is prematurely degraded by the APC/C in UbcH10-overexpressing cells during early mitosis. Thus, one potential scenario is that UbcH10 overexpression affects numeral centrosome integrity by altering the timing of APC/C-mediated degradation of cyclin B and possibly other substrates. The centrosome duplication cycle spans the entire cell cycle, including mitosis, the stage in which sister centriole disengagement (separation) takes place (Salisbury, 2008). Disengagement is essential for proper centriole duplication during interphase and depends on separase and Plk1 activity (Tsou et al., 2009). Although Plk1 is a direct target of APC/C activity, our data suggest that its degradation is unperturbed by supranormal UbcH10 levels. In contrast, separase activity is partly controlled by Cdk1–cyclin B phosphorylation (Peters, 2006) and might be impaired in UbcH10-overexpressing cells because of reduced cyclin B stability. Alternatively, the formation of extra centrioles might originate from an interphase defect. Normally, UbcH10 levels sharply decline upon exit from mitosis. However, in transgenic cells, UbcH10 expression remains relatively high throughout the cell cycle, which might lead to the destabilization of key regulators of centrosome duplication in interphase.
Given that UbcH10 functions as an E2 for APC/C, it is reasonable to assume that UbcH10 overexpression leads to uncontrolled APC/C activity. The observation that UbcH10-overexpressing cells have reduced cyclin B levels in early mitosis supports this assumption. Furthermore, mitotic slippage, a mitotic exit process which requires APC/CCdc20-mediated degradation of cyclin B (Andreassen and Margolis, 1994; Brito and Rieder, 2006), is accelerated when UbcH10 is overexpressed. The idea that UbcH10 overexpression would cause premature cyclin B degradation by challenging the mitotic checkpoint seemed plausible given that UbcH10 has been suggested to activate APC/CCdc20 by triggering the release of BubR1 and Mad2 (Reddy et al., 2007). However, we found no evidence for overt changes in the binding of Mad2 and BubR1 to Cdc20 or APC/CCdc20 in response to UbcH10 overexpression. There was also no evidence for mislocalization of mitotic checkpoint proteins to unattached kinetochores in early mitosis, suggesting that kinetochore-derived checkpoint signaling was intact. So, how then might high UbcH10 levels act to destabilize cyclin B in the presence of an intact mitotic checkpoint? We propose that normally only a small fraction of APC/C is bound to UbcH10. However, UbcH10 overexpression appears to alter this equilibrium, thereby increasing the total pool of active APC/C. On the surface, this result appears to challenge our finding that UbcH10 overexpression has no impact on the binding of BubR1 and Mad2 to APC/C. However, because the UbcH10-bound fraction is small relative to the total APC/C pool even in transgenic cells, any changes in BubR1/Mad2 binding within this fraction would be difficult to detect by an IP–Western blot approach (Fig. S5).
Interestingly, of the comprehensive set of known APC/C substrates whose levels we analyzed under conditions of high levels of UbcH10, only cyclin B showed reduced stability. Although this certainly implies that UbcH10 overexpression does not cause widespread substrate destabilization, the possibility of premature degradation of other regulators of chromosome segregation by APC/C cannot be ruled out at this time. That activated APC/C can only act against particular substrates at certain stages of mitosis is well known (Peters, 2006; Sullivan and Morgan, 2007). The molecular basis for this selectivity is currently unclear and is a subject of intense investigation in the field.
Materials and methods
Generation of UbcH10 transgenic mice and DMBA treatment
UbcH10 transgenic mice were generated via a previously described procedure developed by the laboratory of C. Lobe (Novak et al., 2000; Liu and Lobe, 2007). PCR-amplified mouse UbcH10 cDNA with a carboxy-terminal HA tag was cloned into a unique XhoI site of the ZEG expression vector and sequence verified. ES cells were electroporated with ScaI-linearized ZEG HA-UbcH10 plasmid, and G418-resistant colonies were selected as previously described (van Deursen, 2003). ES colonies were isolated and grown in a 96-well plate. At confluence, each colony was split into four 96-well plates. Plate 1 was frozen at −80°C, and plate 2 was stained for β-galactosidase. Plate 3 was infected with Cre-expressing adenovirus to activate HA-UbcH10 and EGFP expression by excision of the floxed β-geo-stop cassette. EGFP-positive clones were identified by fluorescence microscopy and verified for expression of HA-UbcH10 by Western blotting using 3F10 antibody. Plate 4 was used to extract genomic DNA from clones that were triple positive for β-galactosidase, EGFP, and HA-UbcH10. This DNA was subjected to Southern blot analysis to select clones with a single-copy ZEG HA-UbcH10 integration. UbcH10 cDNA was used as a probe in this analysis. Single integrants were karyotyped and injected into C57BL/6 blastocysts to generate chimeric animals. Chimeric males from two ES clones were bred to C57BL/6 females, and offspring were screened for the presence of the transgene by PCR. Transgene-positive males were bred to protamine-Cre transgenic mice (purchased from The Jackson Laboratory) to excise the β-geo-stop cassette in the male germline. Protamine-Cre/HA-UbcH10 double mutant mice were bred to C57BL/6 females, and offspring were screened for transgene activation using a fluorescence stereomicroscope (MZ16F; Leica). All transgenic mice were maintained on a mixed 129SV/E × C57BL/6 genetic background. DMBA tumor bioassays were performed as previously described (Babu et al., 2003). Spontaneous tumors were collected and processed for standard histopathology. Tumor sections were evaluated with the assistance of board-certified Mayo Clinic pathologists. All mice were housed in a pathogen-free barrier environment. Mouse protocols were reviewed and approved by the institutional animal care and use committee.
Generation and culture of MEFs
UbcH10 transgenic MEFs were generated and cultured as previously described (Baker et al., 2004). MEFs were frozen at P2 or P3 and used for experimentation between P4 and P6. At least three independently generated MEF lines per genotype were used. Mitotic MEFs were prepared by culturing asynchronous cells for 5 h in medium containing 100 ng/ml nocodazole (Sigma-Aldrich) and harvesting cells by shake off. To synchronize MEFs in G0, confluent cultures were washed three times with PBS and then cultured in DME containing 0.1% FBS for 14 h. Quiescent MEFs were trypsinized and reseeded in DME with 20% FBS to allow their reentry into the cell cycle.
Western blotting, co-IP, and immunofluorescence
Western blot analyses, co-IPs, and indirect immunofluorescence were performed as previously described (Kasper et al., 1999). Standard fixations for immunostainings were with 3% paraformaldehyde for 12 min at RT. For kinetochore and centriole localization experiments, cells were fixed with 1% paraformaldehyde for 5 min at RT. For MCAK staining, cells were fixed with 4% paraformaldehyde for 10 min at RT followed by incubation with methanol for 10 min at −20°C. For microtubule–centriole double stainings, cells were permeabilized in PHEM buffer (25 mM Hepes, 10 mM EGTA, 60 mM Pipes, and 2 mM MgCl2, pH 6.9) containing 0.5% Triton X-100 for 5 min at RT followed by incubation with ice-cold 100% methanol for 10 min at −20°C. Centriole quantitation on single-cell suspensions of normal lung tissue and lung tumors were as follows. Normal lung tissue of wild-type mice was collected and rinsed with 20 ml PBS. About 0.5 g of lung tissue was cut into ∼20–30 pieces and transferred into a gentleMACS C tube containing 1 ml PBS/0.5% bovine serum albumin/2 mM EDTA (PEB). Individual lung tumors were collected using a dissection microscope. Each tumor was cut into small pieces and transferred into a gentleMACS C tube containing PEB. All samples were minced in a gentleMACS Dissociator using program m_impTumor_03 (Miltenyi Biotech; the program was run twice). 25 µl of 7 mg/ml Liberase blendzyme 3 solution (Roche) was added, and samples were incubated for 30 min at 37°C. Samples were run once more in the gentleMACS Dissociator and centrifuged at 1,000 rpm for 15 s. Cell suspensions were collected and cleared from undigested tissue using a 70-µm cell strainer. Cells were pelleted (1,000 rpm for 5 min) and resuspended in 1 ml DME/10% FCS. After two washes in 1 ml DME/10% FCS fixative, cells were resuspended in 250 µl DME/10% FCS and attached to chambered microscope slides coated with 1% polyethylene amine microscope slides. Fixation and visualization of centrioles were performed as described above for kinetochore and centriole localization experiments.
A laser-scanning microscope (LSM 510 v3.2SP2; Carl Zeiss, Inc.) with Axiovert 100M (Carl Zeiss, Inc.) with a c-Apochromat 100× oil immersion objective was used to analyze immunostained cells and capture representative images. For quantification of cyclin B levels, 10 prophases were analyzed per MEF line. The mean fluorescence intensity was determined after background subtraction of images transformed to 8-bit grayscale using ImageJ software (National Institutes of Health). Antibodies used were as follows: rabbit anti-UbcH10 (Boston Biochem), mouse anti–cyclin B1 (Santa Cruz Biotechnology, Inc.), rabbit anti–cyclin A2 (Santa Cruz Biotechnology, Inc.), rabbit anti-Cdc20 (Santa Cruz Biotechnology, Inc.), mouse anti-Mad2 (BD), mouse anti–Aurora B (BD), mouse anti–β-actin (Sigma-Aldrich), mouse anti–α-tubulin (Sigma-Aldrich), mouse anti-HA (3F10; Roche), rabbit anti-Nek2 (Abgent), rabbit anti–human BubR1 (Baker et al., 2004), human anticentromere antibody (Antibodies, Inc.), rabbit anti–P-H3Ser10 (Millipore), mouse anti-Plk1 (Santa Cruz Biotechnology, Inc.), mouse antisecurin (Abcam), mouse anti-Cdc27 (BD), goat anti-APC6 (Santa Cruz Biotechnology, Inc.), rabbit anti–Aurora A (Cell Signaling Technology), rabbit anti–centrin 2 (Santa Cruz Biotechnology, Inc.), mouse anti–centrin 2 (provided by J. Salisbury, Mayo Clinic College of Medicine, Rochester, MN; Hart et al., 1999), sheep anti–hamster MCAK (provided by L. Wordeman, University of Washington School of Medicine, Seattle, WA; Andrews et al., 2004), rabbit anti–mouse Bub1 (Jeganathan et al., 2007), and rabbit anti–CENP-E (provided by D. Cleveland, Ludwig Institute for Cancer Research, La Jolla, CA).
Inducible UbcH10 expression
For inducible expression of UbcH10, the HA-UbcH10 fragment from ZEG HA-UbcH10 was cloned into the pTRIC (for tightly regulatable inducible cDNA) lentiviral expression vector. pTRIC was generated by replacing the AgeI–MluI fragment of pTRIPZ (Thermo Fisher Scientific) containing the turboRFP tag and the 5′mir30/3′mir30 sequences by an AgeI–HpaI–XhoI–EcoR1–MluI polylinker sequence. UbcH10 expression was induced by culturing the cells for at least 48 h in the presence of 1 µg/ml Dox.
Karyotyping and FISH analysis
Chromosome counts were performed on metaphase spreads of P5 MEFs or splenocytes of 5-mo-old mice as previously described (Babu et al., 2003). Interphase FISH analysis was performed in the Mayo Clinic Cytogenetics Core Facility as described in detail by Bayani and Squire (2004). Tissue sections were 5 µm. Probes for chromosomes 4 and 7 were generated from the following chromosome-specific BAC clones cocktails: RP23-358H13, RP23-364K8, and RP23-208A12 (chromosome 4); and RP23-373E17, RP23-162M11, and RP24-176A23 (chromosome 7).
Live cell imaging
Nocodazole challenge assays were performed as follows. MEFs were first transduced with a lentivirus encoding an mRFP-tagged H2B to allow visualization of chromosomes by fluorescence microscopy. Cells were then seeded onto 35-mm glass-bottomed culture dishes (MatTek Corporation) and cultured in DME/10% FBS. Approximately 24 h later, experiments were performed using an AxioObserver Z1 system (Carl Zeiss, Inc.) with CO2 Module S, TempModule S, Heating Unit XL S, Plan-Apochromat 63× NA 1.4 oil DICIII objective, a camera (AxioCam MRm; Carl Zeiss, Inc.), and AxioVision 10.6 software (Carl Zeiss, Inc.). The imaging medium was DME/10% FBS. The temperature of the imaging medium was kept at 37°C. The exposure times in nocodazole challenge experiments were identical among experiments. Nocodazole was added to a final concentration of 100 ng/ml. Cells undergoing NEBD were marked and monitored at 30-min intervals to determine when they decondensed their chromosomes. The duration of arrest in mitosis, which is defined as the interval between NEBD (onset of mitosis) and chromatin decondensation (exit from mitosis without cytokinesis), was then calculated and plotted. The time at which 50% of the cells had exited mitosis was used for comparison (see also Baker et al., 2006). For mitotic timing experiments, the time interval between NEBD and anaphase onset was measured as H2B-mRFP–positive cells progressed through an unchallenged mitosis. For chromosome missegregation analysis, H2B-mRFP–positive cells progressing through an unchallenged mitosis were followed at interframe intervals of 3 min. Monitoring of securin stability by live cell imaging was performed as described previously (Malureanu et al., 2009), with the exception that pECFP-securin was used instead of pYFP-securin (transgenic MEFs already express EGFP). The pECFP-N1-securin expression vector that was used was produced by ligating the BamHI and XhoI securin cDNA fragment of pYFP-N1-securin (from J. Pines, Gurdon Institute, Cambridge, England, UK) into the BamHI and XhoI sites of pECFP-N1. H2B-mRFP–expressing UbcH10+/+ and UbcH10T2/T2 MEFs were nucleofected with 2 µg pECFP-N1-securin expression plasmid. For image processing, we used AxioVision 10.6 software and PowerPoint (Microsoft) for Mac.
Quantitative real-time PCR
Total RNA was extracted from cryosections of normal human lung tissue (n = 7), human adenocarcinomas (n = 49), and human lung squamous cell carcinomas (n = 47) using the RNeasy RNA Isolation kit (QIAGEN) according to the manufacturer’s protocol (specimens provided by V. Shidhar, Mayo Clinic College of Medicine). Complementary DNA was produced using random hexamers and SuperScript III reverse transcription (Invitrogen) according to the manufacturer’s instructions. Transcript levels were determined using the ABI PRISM Sequence Detection System 7900 (Applied Biosystems). Oligonucleotides were obtained from Applied Biosystems. Measurements were performed in duplicate. Amplification curves and gene expression were normalized to the housekeeping gene TBP. TaqMan primer IDs were Hs 00738962-m1 for UbcH10 and 4333769 for TBP. The relative fold expression was normalized to the average of normal tissue expression.
Online supplemental material
Fig. S1 shows that growth rates are normal in UbcH10-overexpressing MEFs. Fig. S2 shows that key mitotic checkpoint proteins properly accumulate at kinetochores during early mitosis in UbcH10-overexpressing cells. Fig. S3 shows that timing of securin degradation is normal when UbcH10 is overexpressed. Fig. S4 shows that Aurora B and MCAK properly target to inner centromeric regions in UbcH10-overexpressing cells. Fig. S5 shows a model for increased APC/C activity in UbcH10 transgenic cells. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200906147/DC1.
Acknowledgments
For technical assistance, we thank D. Norris, W. Zhou, and M. Li, and for helpful discussions, we thank Drs. R. Ricke, D. Baker, R. Bram, P. Galardy, and J. Salisbury. We also thank Drs. F. Jin, T. Wu, and A. Folpe for help with tumor histopathology, Dr. V. Shidhar for providing the human lung cancer specimens, and Drs. J. Salisbury and L. Wordeman for antibodies.
The National Cancer Institute (grants CA96985 and CA91956) and the Ellison Medical Foundation supported this work.
Footnotes
Abbreviations used in this paper:
- APC/C
- anaphase-promoting complex/cyclosome
- DMBA
- 7,12-dimethylbenz(a)anthracene
- Dox
- doxycycline
- ES
- embryonic stem
- IP
- immunoprecipitation
- MCAK
- mitotic centromere-associated kinesin
- MCC
- mitotic checkpoint complex
- MEF
- mouse embryonic fibroblast
- mRFP
- monomeric RFP
- NEBD
- nuclear envelope breakdown
- P-H3
- phospho–histone H3
- PMSCS
- premature sister chromatid separation
References
- Andreassen P.R., Margolis R.L. 1994. Microtubule dependency of p34cdc2 inactivation and mitotic exit in mammalian cells. J. Cell Biol. 127:789–802 10.1083/jcb.127.3.789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews P.D., Ovechkina Y., Morrice N., Wagenbach M., Duncan K., Wordeman L., Swedlow J.R. 2004. Aurora B regulates MCAK at the mitotic centromere. Dev. Cell. 6:253–268 10.1016/S1534-5807(04)00025-5 [DOI] [PubMed] [Google Scholar]
- Babu J.R., Jeganathan K.B., Baker D.J., Wu X., Kang-Decker N., van Deursen J.M. 2003. Rae1 is an essential mitotic checkpoint regulator that cooperates with Bub3 to prevent chromosome missegregation. J. Cell Biol. 160:341–353 10.1083/jcb.200211048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker D.J., Jeganathan K.B., Cameron J.D., Thompson M., Juneja S., Kopecka A., Kumar R., Jenkins R.B., de Groen P.C., Roche P., van Deursen J.M. 2004. BubR1 insufficiency causes early onset of aging-associated phenotypes and infertility in mice. Nat. Genet. 36:744–749 10.1038/ng1382 [DOI] [PubMed] [Google Scholar]
- Baker D.J., Jeganathan K.B., Malureanu L., Perez-Terzic C., Terzic A., van Deursen J.M. 2006. Early aging-associated phenotypes in Bub3/Rae1 haploinsufficient mice. J. Cell Biol. 172:529–540 10.1083/jcb.200507081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakhoum S.F., Genovese G., Compton D.A. 2009a. Deviant kinetochore microtubule dynamics underlie chromosomal instability. Curr. Biol. 19:1937–1942 10.1016/j.cub.2009.09.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakhoum S.F., Thompson S.L., Manning A.L., Compton D.A. 2009b. Genome stability is ensured by temporal control of kinetochore-microtubule dynamics. Nat. Cell Biol. 11:27–35 10.1038/ncb1809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayani J., Squire J.A. 2004. Fluorescence in situ Hybridization (FISH). Curr. Protoc. Cell Biol. Chapter 22:Unit 22.4 10.1002/0471143030.cb2204s23 [DOI] [PubMed] [Google Scholar]
- Berlingieri M.T., Pallante P., Guida M., Nappi C., Masciullo V., Scambia G., Ferraro A., Leone V., Sboner A., Barbareschi M., et al. 2007a. UbcH10 expression may be a useful tool in the prognosis of ovarian carcinomas. Oncogene. 26:2136–2140 10.1038/sj.onc.1210010 [DOI] [PubMed] [Google Scholar]
- Berlingieri M.T., Pallante P., Sboner A., Barbareschi M., Bianco M., Ferraro A., Mansueto G., Borbone E., Guerriero E., Troncone G., Fusco A. 2007b. UbcH10 is overexpressed in malignant breast carcinomas. Eur. J. Cancer. 43:2729–2735 [DOI] [PubMed] [Google Scholar]
- Boveri T. 1902. Uber mehrpolige Mitosen als Mittel zur Analyse der Zelkerns. Vehr d phys Med Ges zu Wurzburg Neu Folge. 35:67–90 [Google Scholar]
- Boveri T. 1914. Zur Frage der Entstehung Maligner Tumoren. Gustav Fisher, Jena, Germany: 64 pp [Google Scholar]
- Brito D.A., Rieder C.L. 2006. Mitotic checkpoint slippage in humans occurs via cyclin B destruction in the presence of an active checkpoint. Curr. Biol. 16:1194–1200 10.1016/j.cub.2006.04.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimini D. 2008. Merotelic kinetochore orientation, aneuploidy, and cancer. Biochim. Biophys. Acta. 1786:32–40 [DOI] [PubMed] [Google Scholar]
- Dai W., Wang Q., Liu T., Swamy M., Fang Y., Xie S., Mahmood R., Yang Y.M., Xu M., Rao C.V. 2004. Slippage of mitotic arrest and enhanced tumor development in mice with BubR1 haploinsufficiency. Cancer Res. 64:440–445 10.1158/0008-5472.CAN-03-3119 [DOI] [PubMed] [Google Scholar]
- Dawlaty M.M., Malureanu L., Jeganathan K.B., Kao E., Sustmann C., Tahk S., Shuai K., Grosschedl R., van Deursen J.M. 2008. Resolution of sister centromeres requires RanBP2-mediated SUMOylation of topoisomerase IIalpha. Cell. 133:103–115 10.1016/j.cell.2008.01.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukasawa K. 2007. Oncogenes and tumour suppressors take on centrosomes. Nat. Rev. Cancer. 7:911–924 10.1038/nrc2249 [DOI] [PubMed] [Google Scholar]
- Ganem N.J., Godinho S.A., Pellman D. 2009. A mechanism linking extra centrosomes to chromosomal instability. Nature. 460:278–282 10.1038/nature08136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnett M.J., Mansfeld J., Godwin C., Matsusaka T., Wu J., Russell P., Pines J., Venkitaraman A.R. 2009. UBE2S elongates ubiquitin chains on APC/C substrates to promote mitotic exit. Nat. Cell Biol. 11:1363–1369 10.1038/ncb1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart P.E., Glantz J.N., Orth J.D., Poynter G.M., Salisbury J.L. 1999. Testis-specific murine centrin, Cetn1: genomic characterization and evidence for retroposition of a gene encoding a centrosome protein. Genomics. 60:111–120 10.1006/geno.1999.5880 [DOI] [PubMed] [Google Scholar]
- Herzog F., Primorac I., Dube P., Lenart P., Sander B., Mechtler K., Stark H., Peters J.M. 2009. Structure of the anaphase-promoting complex/cyclosome interacting with a mitotic checkpoint complex. Science. 323:1477–1481 10.1126/science.1163300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland A.J., Cleveland D.W. 2009. Boveri revisited: chromosomal instability, aneuploidy and tumorigenesis. Nat. Rev. Mol. Cell Biol. 10:478–487 10.1038/nrm2718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeganathan K., Malureanu L., Baker D.J., Abraham S.C., van Deursen J.M. 2007. Bub1 mediates cell death in response to chromosome missegregation and acts to suppress spontaneous tumorigenesis. J. Cell Biol. 179:255–267 10.1083/jcb.200706015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L., Huang C.G., Lu Y.C., Luo C., Hu G.H., Liu H.M., Chen J.X., Han H.X. 2008. Expression of ubiquitin-conjugating enzyme E2C/UbcH10 in astrocytic tumors. Brain Res. 1201:161–166 10.1016/j.brainres.2008.01.037 [DOI] [PubMed] [Google Scholar]
- Kasper L.H., Brindle P.K., Schnabel C.A., Pritchard C.E., Cleary M.L., van Deursen J.M. 1999. CREB binding protein interacts with nucleoporin-specific FG repeats that activate transcription and mediate NUP98-HOXA9 oncogenicity. Mol. Cell. Biol. 19:764–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King R.W. 2008. When 2+2=5: the origins and fates of aneuploid and tetraploid cells. Biochim. Biophys. Acta. 1786:4–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kline-Smith S.L., Khodjakov A., Hergert P., Walczak C.E. 2004. Depletion of centromeric MCAK leads to chromosome congression and segregation defects due to improper kinetochore attachments. Mol. Biol. Cell. 15:1146–1159 10.1091/mbc.E03-08-0581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulukian A., Han J.S., Cleveland D.W. 2009. Unattached kinetochores catalyze production of an anaphase inhibitor that requires a Mad2 template to prime Cdc20 for BubR1 binding. Dev. Cell. 16:105–117 10.1016/j.devcel.2008.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan W., Zhang X., Kline-Smith S.L., Rosasco S.E., Barrett-Wilt G.A., Shabanowitz J., Hunt D.F., Walczak C.E., Stukenberg P.T. 2004. Aurora B phosphorylates centromeric MCAK and regulates its localization and microtubule depolymerization activity. Curr. Biol. 14:273–286 [DOI] [PubMed] [Google Scholar]
- Li M., Fang X., Wei Z., York J.P., Zhang P. 2009. Loss of spindle assembly checkpoint–mediated inhibition of Cdc20 promotes tumorigenesis in mice. J. Cell Biol. 185:983–994 10.1083/jcb.200904020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Lobe C.G. 2007. Cre-conditional expression of constitutively active Notch1 in transgenic mice. Genesis. 45:259–265 10.1002/dvg.20282 [DOI] [PubMed] [Google Scholar]
- Malureanu L.A., Jeganathan K.B., Hamada M., Wasilewski L., Davenport J., van Deursen J.M. 2009. BubR1 N terminus acts as a soluble inhibitor of cyclin B degradation by APC/C(Cdc20) in interphase. Dev. Cell. 16:118–131 10.1016/j.devcel.2008.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Máthé E., Kraft C., Giet R., Deák P., Peters J.M., Glover D.M. 2004. The E2-C vihar is required for the correct spatiotemporal proteolysis of cyclin B and itself undergoes cyclical degradation. Curr. Biol. 14:1723–1733 10.1016/j.cub.2004.09.023 [DOI] [PubMed] [Google Scholar]
- Meraldi P., Draviam V.M., Sorger P.K. 2004. Timing and checkpoints in the regulation of mitotic progression. Dev. Cell. 7:45–60 10.1016/j.devcel.2004.06.006 [DOI] [PubMed] [Google Scholar]
- Michel L.S., Liberal V., Chatterjee A., Kirchwegger R., Pasche B., Gerald W., Dobles M., Sorger P.K., Murty V.V., Benezra R. 2001. MAD2 haploinsufficiency causes premature anaphase and chromosome instability in mammalian cells. Nature. 409:355–359 10.1038/35053094 [DOI] [PubMed] [Google Scholar]
- Musacchio A., Salmon E.D. 2007. The spindle-assembly checkpoint in space and time. Nat. Rev. Mol. Cell Biol. 8:379–393 10.1038/nrm2163 [DOI] [PubMed] [Google Scholar]
- Nasmyth K., Haering C.H. 2005. The structure and function of SMC and kleisin complexes. Annu. Rev. Biochem. 74:595–648 10.1146/annurev.biochem.74.082803.133219 [DOI] [PubMed] [Google Scholar]
- Nigg E.A. 2002. Centrosome aberrations: cause or consequence of cancer progression? Nat. Rev. Cancer. 2:815–825 10.1038/nrc924 [DOI] [PubMed] [Google Scholar]
- Nilsson J., Yekezare M., Minshull J., Pines J. 2008. The APC/C maintains the spindle assembly checkpoint by targeting Cdc20 for destruction. Nat. Cell Biol. 10:1411–1420 10.1038/ncb1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak A., Guo C., Yang W., Nagy A., Lobe C.G. 2000. Z/EG, a double reporter mouse line that expresses enhanced green fluorescent protein upon Cre-mediated excision. Genesis. 28:147–155 [DOI] [PubMed] [Google Scholar]
- Okamoto Y., Ozaki T., Miyazaki K., Aoyama M., Miyazaki M., Nakagawara A. 2003. UbcH10 is the cancer-related E2 ubiquitin-conjugating enzyme. Cancer Res. 63:4167–4173 [PubMed] [Google Scholar]
- Pallante P., Berlingieri M.T., Troncone G., Kruhoffer M., Orntoft T.F., Viglietto G., Caleo A., Migliaccio I., Decaussin-Petrucci M., Santoro M., et al. 2005. UbcH10 overexpression may represent a marker of anaplastic thyroid carcinomas. Br. J. Cancer. 93:464–471 10.1038/sj.bjc.6602721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellman D. 2007. Cell biology: aneuploidy and cancer. Nature. 446:38–39 10.1038/446038a [DOI] [PubMed] [Google Scholar]
- Perera D., Tilston V., Hopwood J.A., Barchi M., Boot-Handford R.P., Taylor S.S. 2007. Bub1 maintains centromeric cohesion by activation of the spindle checkpoint. Dev. Cell. 13:566–579 10.1016/j.devcel.2007.08.008 [DOI] [PubMed] [Google Scholar]
- Peters J.M. 2006. The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat. Rev. Mol. Cell Biol. 7:644–656 10.1038/nrm1988 [DOI] [PubMed] [Google Scholar]
- Rape M., Kirschner M.W. 2004. Autonomous regulation of the anaphase-promoting complex couples mitosis to S-phase entry. Nature. 432:588–595 10.1038/nature03023 [DOI] [PubMed] [Google Scholar]
- Reddy S.K., Rape M., Margansky W.A., Kirschner M.W. 2007. Ubiquitination by the anaphase-promoting complex drives spindle checkpoint inactivation. Nature. 446:921–925 10.1038/nature05734 [DOI] [PubMed] [Google Scholar]
- Ricke R.M., van Ree J.H., van Deursen J.M. 2008. Whole chromosome instability and cancer: a complex relationship. Trends Genet. 24:457–466 10.1016/j.tig.2008.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salisbury J.L. 2008. Breaking the ties that bind centriole numbers. Nat. Cell Biol. 10:255–257 10.1038/ncb0308-255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silkworth W.T., Nardi I.K., Scholl L.M., Cimini D. 2009. Multipolar spindle pole coalescence is a major source of kinetochore mis-attachment and chromosome mis-segregation in cancer cells. PLoS One. 4:e6564 10.1371/journal.pone.0006564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudakin V., Chan G.K., Yen T.J. 2001. Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20, and MAD2. J. Cell Biol. 154:925–936 10.1083/jcb.200102093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan M., Morgan D.O. 2007. Finishing mitosis, one step at a time. Nat. Rev. Mol. Cell Biol. 8:894–903 10.1038/nrm2276 [DOI] [PubMed] [Google Scholar]
- Summers M.K., Pan B., Mukhyala K., Jackson P.K. 2008. The unique N terminus of the UbcH10 E2 enzyme controls the threshold for APC activation and enhances checkpoint regulation of the APC. Mol. Cell. 31:544–556 10.1016/j.molcel.2008.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson S.L., Compton D.A. 2008. Examining the link between chromosomal instability and aneuploidy in human cells. J. Cell Biol. 180:665–672 10.1083/jcb.200712029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsou M.F., Wang W.J., George K.A., Uryu K., Stearns T., Jallepalli P.V. 2009. Polo kinase and separase regulate the mitotic licensing of centriole duplication in human cells. Dev. Cell. 17:344–354 10.1016/j.devcel.2009.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Deursen J. 2003. Gene targeting in mouse embryonic stem cells. Methods Mol. Biol. 209:145–158 [DOI] [PubMed] [Google Scholar]
- Wagner K.U., Wall R.J., St-Onge L., Gruss P., Wynshaw-Boris A., Garrett L., Li M., Furth P.A., Hennighausen L. 1997. Cre-mediated gene deletion in the mammary gland. Nucleic Acids Res. 25:4323–4330 10.1093/nar/25.21.4323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner K.W., Sapinoso L.M., El-Rifai W., Frierson H.F., Butz N., Mestan J., Hofmann F., Deveraux Q.L., Hampton G.M. 2004. Overexpression, genomic amplification and therapeutic potential of inhibiting the UbcH10 ubiquitin conjugase in human carcinomas of diverse anatomic origin. Oncogene. 23:6621–6629 10.1038/sj.onc.1207861 [DOI] [PubMed] [Google Scholar]
- Walker A., Acquaviva C., Matsusaka T., Koop L., Pines J. 2008. UbcH10 has a rate-limiting role in G1 phase but might not act in the spindle checkpoint or as part of an autonomous oscillator. J. Cell Sci. 121:2319–2326 10.1242/jcs.031591 [DOI] [PubMed] [Google Scholar]
- Weaver B.A., Cleveland D.W. 2009. The role of aneuploidy in promoting and suppressing tumors. J. Cell Biol. 185:935–937 10.1083/jcb.200905098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver B.A., Silk A.D., Montagna C., Verdier-Pinard P., Cleveland D.W. 2007. Aneuploidy acts both oncogenically and as a tumor suppressor. Cancer Cell. 11:25–36 10.1016/j.ccr.2006.12.003 [DOI] [PubMed] [Google Scholar]
- Williamson A., Wickliffe K.E., Mellone B.G., Song L., Karpen G.H., Rape M. 2009. Identification of a physiological E2 module for the human anaphase-promoting complex. Proc. Natl. Acad. Sci. USA. 106:18213–18218 10.1073/pnas.0907887106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka A., Hatakeyama S., Kominami K., Kitagawa M., Matsumoto M., Nakayama K. 2000. Cell cycle-dependent expression of mammalian E2-C regulated by the anaphase-promoting complex/cyclosome. Mol. Biol. Cell. 11:2821–2831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H. 2007. Cdc20: a WD40 activator for a cell cycle degradation machine. Mol. Cell. 27:3–16 10.1016/j.molcel.2007.06.009 [DOI] [PubMed] [Google Scholar]