Abstract
Drug abuse has become a global health concern. Understanding how drug abuse modulates the immune system and how the immune system responds to pathogens associated with drug abuse, such hepatitis C virus (HCV) and human immunodeficiency virus (HIV-1), can be assessed by an integrated approach comparing proteomic analyses and quantitation of gene expression. Two-dimensional (2D) difference gel electrophoresis was used to determine the molecular mechanisms underlying the proteomic changes that alter normal biological processes when monocyte-derived mature dendritic cells were treated with cocaine or methamphetamine. Both drugs differentially regulated the expression of several functional classes of proteins including those that modulate apoptosis, protein folding, protein kinase activity, and metabolism and proteins that function as intracellular signal transduction molecules. Proteomic data were validated using a combination of quantitative, real-time PCR and Western blot analyses. These studies will help to identify the molecular mechanisms, including the expression of several functionally important classes of proteins that have emerged as potential mediators of pathogenesis. These proteins may predispose immunocompetent cells, including dendritic cells, to infection with viruses such as HCV and HIV-1, which are associated with drug abuse.
Keywords: Cocaine, Methamphetamine, Monocyte-derived mature dendritic cells, Difference gel electrophoresis (DIGE), High performance liquid chromatography-tandem mass spectrometry (HPLC-MS/MS)
INTRODUCTION
Cocaine and methamphetamine are widely abused, addictive drugs, both of which can be snorted, smoked or injected (NIDA Info Facts, 2005). The global annual prevalence of cocaine use and methamphetamine/amphetamine use is 0.37% and 0.6% respectively (World Drug Report (WDR), 2007). In the United States, in 2006, cocaine, heroin, marijuana and methamphetamine use were the leading causes of drug abuse-related visits to the emergency room (Drug abuse warning network (DAWN), 2006). In 2007, the number of new users of cocaine and methamphetamine among persons aged 12 years or older in the United States was 906,000 and 157,000, respectively (Substance Abuse and Mental Health Services Administration (SAMHSA), 2007).
Most newly acquired HCV and HIV-1 infections in the US, are associated with high-risk sexual activity and/or drug use (Centers for Disease Control (CDC), 2005). A large percentage of HIV-infected, injection drug users are also co-infected with HCV (CDC, 2005; Kim, 2008). Patients co-infected with HIV-1 and HCV have a greater mortality rate due to higher incidences of hepatocellular carcinoma (Parodi, 2007; Beretta, 2008). Additionally, co-infected patients may have a reduced response to highly active antiretroviral therapy (HAART) (Parodi 2007; Mathews, 2008; Thein, 2008).
Cocaine and methamphetamine have both been shown to enhance HIV-1 replication in macrophages and dendritic cells (Nair, 2005; Reynolds, 2007; Liang, 2008) while methamphetamine enhances HCV replication in hepatocytes (Ye, 2008). Cocaine has a direct effect on the function of lymphocytes, natural killer cells, neutrophils, and macrophage. Cocaine suppresses lymphocyte proliferation (Bayer, 1996; Bagasrsa, 1989), neutrophil activation (Haines, 1990; Mukunda, 2000) and the phagocytic activity of macrophages (Baldwin, 1997; Pellegrino, 2001) while increasing natural killer cell functions (Van Dyke, 1989; Poet, 1991). Methamphetamine modulates protein expression in monocyte derived dendritic cells (Nair, 2006; Mahajan, 2006; Reynolds, 2007) and inhibits endosomal antigen processing and MHC II restricted antigen presentation by bone marrow derived dendritic cells (Tallóczy, 2008).
Dendritic cells are antigen presenting cells that modulate innate and adaptive immune responses. Dendritic cells are present in the skin and the mucous membranes of the nose, lungs, stomach, and intestines; they are found in the blood in an immature state (Banchereau, 1998; Reis e Sousa, 2006; Steinman, 2007). Immature dendritic cells are recruited to and migrate through the bloodstream to sites of inflammation to confront invading pathogens. Immature dendritic cells capture antigen that subsequently induces maturation to mature, antigen-presenting, T cell-priming, dendritic cells. During their conversion from immature to mature cells, dendritic cells undergo phenotypic and functional alterations (e.g., redistribution of major histocompatibility complex (MHC) molecules to the cell surface; increase in the surface expression of costimulatory molecules, chemokine receptors and adhesion molecules; morphological changes; and secretion of chemokines, cytokines and proteases).
Mature dendritic cells activate T-cells and B-cells by presenting surface MHC-peptide complexes derived from the antigens captured during the immature stage (Banchereau, 1998; Reis e Sousa, 2006; Steinman, 2007; Mohamadzadeh, 2008). Mature dendritic cells are permissive for the growth of HIV-1 and HCV (Navas, 2002; Lo, 2003; Pachiadakis 2005; Piguet, 2007). Cell surface receptors and adhesion molecules on mature dendritic cells mediate physical contact between dendritic cells and T cells and may allow for more efficient viral transmission (Sanders, 2002; McDonald, 2003). HIV-1 transmission is increased by maturation of dendritic cells (Granelli-Piperno, 1998; Sanders, 2002; McDonald, 2003). Mature dendritic cells can also transfer HIV-1 to T cells even in the absence of productive dendritic cell infections (trans-infection) (Granelli-Piperno, 1998; Sanders, 2002; McDonald, 2003, Moris, 2004; Lore, 2005; Yu; 2008). In chronic HCV infections mature dendritic cells have impaired ability to stimulate T cells (Kanto 1999; Bain 2001; Kanto 2004). Dendritic cells not only direct the induction of adaptive immune responses but are also involved in the regulation of humoral-mediated immunity (Banchereau, 1998; Reis e Sousa, 2006; Steinman, 2007; Mohamadzadeh, 2008).
Considerable evidence exists linking drug abuse to susceptibility to and progression of various infections, particularly HIV-1 and HCV. In this paper, we test the hypothesis that cocaine and/or methamphetamine regulates the expression of various host proteins in monocyte derived, mature dendritic cells. These studies were undertaken to identify unrecognized biomarkers that might underlie the association of drug abuse and HIV-1 infections. Such marker proteins could subsequently become targets for prevention and therapy. We used 2 dimensional (2D) difference gel electrophoresis to demonstrate quantitative differences in the expression of proteins induced by cocaine or methamphetamine followed by tandem mass spectrometry to specifically identify proteins of special interest. Additionally, we validated our proteomic results using a combination of quantitative, real-time PCR and Western blot analysis.
METHODS
Human Subjects
Blood donors were apprised of this study and consents were obtained consistent with the policies of the appropriate local institutions and the National Institutes of Health. Peripheral blood samples from healthy, HIV-1 negative individuals were drawn into a syringe containing heparin (20 units/ml).
Isolation and Generation of Monocyte-Derived Mature Dendritic Cells
Dendritic cells were prepared as described (Dauer, 2003; Cao, 2000; Bender, 1996). Briefly, human peripheral blood mononuclear cells (PBMCs) were isolated by density gradient centrifugation on Ficoll-Paque (GE Healthcare, Piscataway, NJ). CD14+ monocytes were separated from PBMCs using plastic adherence; the purity of monocytes was >90%, verified by flow cytometry (data not shown). Adherent cells were cultured for 6 days in RPMI 1640 media supplemented with 10% fetal bovine serum (FBS) containing 100 U/ml recombinant human interleukin-4 (IL-4; R&D Systems, Minneapolis, MN) and 100 U/ml, recombinant human granulocyte-macrophage colony-stimulating factor (GMCSF; R&D Systems). After 6 days of culture, monocyte derived, mature dendritic cells were prepared by culturing cells for 5 days in media containing IL-4 and GMCSF as above. Maturation of monocyte derived, mature dendritic cells was verified by immunostaining with antibodies to CD83+ and the major histocompatibility complex (MHC-HLA-DR, DQ-DP) followed by flow cytometry (data not shown).
Drug Treatment
Monocyte-derived, mature dendritic cells were treated with cocaine (Sigma-Aldrich, St. Louis, Mo) at 1 µM for 48 hr or methamphetamine hydrochloride (Sigma-Aldrich) at 100 µM for 24 hr. Control cells were treated with media alone. The concentrations of cocaine and METH used were based on previous dose response studies that produced a maximum biological response without causing toxicity to the target cells and also were based on published in vitro studies (Nair, 2005, 2006; Reynolds, 2007; Lee, 2001; Tallocy, 2008). The concentrations of cocaine and METH are similar to levels found in the blood, urine or tissue samples of users that have been found to range from ≤ 1 µM to 600 µM (Edward, 1998; Takayasu, 1995; Kalasinsky, 2001; Schepers, 2003).
Two-Dimensional (2D) Difference Gel Electrophoresis (DIGE)
The Ettan DIGE technique (GE Healthcare) was used to detect differences in protein abundance between normal and experimental samples. The Ettan DIGE system uses three CyDye DIGE fluors (Cy2, Cy3, Cy5), each with a unique fluorescent wavelength, matched for mass and charge. CyDyes form a covalent bond with the free epsilon amino group on lysine residues from the sample proteins. CyDyes label approximately 2% of the lysine residues. This system allows for two experimental samples and an internal standard to be simultaneously separated on the same gel. The internal standard is comprised a pool of an equal amount of all the experimental samples. The use of an internal standard facilitates accurate inter-gel matching of spots, and allows for data normalization between gels to minimize gel to gel experimental variability (www.amershambiosciences.com).
Sample Preparation
After stimulation, cells were washed 2 times with 1X PBS (Invitrogen, Grand Island, NY). Total protein was extracted using standard cell lysis buffer [30 mM TrisCl; 8 M Urea; 4% (w/v) 3-[(3-cholamidopropyl) dimethylammonio]-1-propanesulfonate (CHAPS), adjusted to pH 8.5] for 10 min on ice. Cell lysate was centrifuged at 4°C for 10 min at 12000 g and the lysate was further purified by precipitation with chloroform/methanol precipitated as described (Wessel, 1984). Cell lysates were resuspended in standard cell lysis buffer. Final cell lysate protein concentrations were determined using Bio-Rad Coomassie Protein Reagent (Bio-Rad, Hercules, CA) and used for protein determination by DIGE analysis.
Sample Labeling
All reagents used were from GE Healthcare. Briefly, 50 µg of cell lysate was labeled with 400 pmol of either Cy3 or Cy5 or Cy2 (Cy 2 was used to label the internal standard) on ice for 30 min and then quenched with a 50-fold molar excess of free lysine. Cy3, Cy5, Cy2 labeled samples and unlabeled protein (500–800 µg) were combined. An equal volume of 2X sample buffer [8 M Urea; 2% (v/v) Pharmalytes 3–10; 2% (w/v) dithiothreitol (DTT); 4% (w/v) CHAPS] was added and incubated on ice for 10 min. The total volume of sample was adjusted to 450 µl with rehydration buffer [4% (w/v) CHAPS; 8 M Urea; 1% (v/v) Pharmalytes 3–10 nonlinear (NL); 13 mM DTT].
DIGE
Samples were applied to immobilized pH gradient (IPG) strips (24cm, pH 3–10 NL), and absorbed by active rehydration at 30 V for 13 hr. Isoelectric focusing was carried out using an IPGphor IEF system with a three phase program; first phase at 500 V for 1 hr, second phase at 1000 V for 1 hr, and third phase (linear gradient) 8000 to 64,000 V for 2 hr (50 uA maximum per strip). Prior to separation in the second dimension, strips were equilibrated for 15 min in equilibration buffer I [50 mM Tris-HCl, 6 M Urea, 30% (v/v) glycerol, 2% (w/v) SDS, 0.5% (w/v) DTT]. The strips were again equilibrated for 15 min in equilibration buffer II [50 mM Tris-HCl, 6 M Urea, 30% (v/v) glycerol, 2% (w/v) SDS, 4.5% (w/v) iodoacetamide] and transferred onto 18 × 20 cm, 12.5% uniform polyacrylamide gels poured between low fluorescence glass plates. Gels were bonded to inner plates using Bind-Silane solution according to the manufacturer’s protocol. Strips were overlaid with 0.9% agarose in 1X running buffer containing bromophenol blue and were run for 16 hr (1.8 W per gel, overnight) at 15°C, in the Ettan DALT electrophoresis system.
Image Acquisition
Fluorescent images of each CyDye (Cy3, Cy5, Cy2) were acquired using a Typhoon 9410 variable mode imager. After scanning, gels were fixed in 30% (v/v) methanol, 7.5% (v/v) acetic acid for 3 hr and stained with SYPRO-Ruby dye (Molecular Probes, Eugene, OR) overnight at room temperature. Gels were de-stained in water and then scanned using the Typhoon 9410 scanner.
Image Analysis
Images were processed in ImageQuant v5.2 software, and imported into DeCyder differential in-gel analysis (DIA) software v5.0 from GE Healthcare for spot detection and normalization of spot intensities within each gel. Intergel matching was performed using the biological variation analysis (BVA) component of DeCyder for cross-gel statistical analysis. Standardized protein abundance was calculated by dividing each Cy3 or Cy5 spot volume by the corresponding Cy2 standard spot volume (internal control) within each gel, and the difference in standardized abundance between control and drug-treated mature dendritic cells were expressed as the average volume ratio (data represented as % change from control). Statistical analysis (Student’s t-test) was performed on spots matched across gels. Protein spots that showed statically significant differences between the control and drug-treated samples were picked using the Ettan Spot Picker (www.amershambiosciences.com; Lilley and Friedman, 2004).
High Performance Liquid Chromatography-Tandem Mass Spectrometry (HPLC-MS/MS)
Excised spots were sent for analysis to the Proteomic Analysis Laboratory at the University of Arizona. In gel digestion and HPLC-MS/MS were performed as described by Breci et al. (2005). Briefly, gel slices were destained (Gharahdaghi, 1999) and digested with trypsin (Cooper, 2003). The tryptic peptides were extracted with 5% formic acid/50% CH3CN. HPLC was performed using a microbore system (Surveyor, ThermoFinnigan, San Jose, CA). The HPLC column eluate was directed into a ThermoFinnigan LCQ Deca XP Plus ion trap mass spectrometer. Automated peak recognition, dynamic exclusion, and daughter ion scanning of the top two most intense ions were performed using Xcalibur software (Wilm, 1996). Spectra were scanned over the range 400–1400 mass units. MS/MS data were analyzed using SEQUEST, a computer program that allows the correlation of experimental data with theoretical spectra generated from known protein sequences.
A preliminary positive peptide identification for a doubly charged peptide was based upon a correlation factor (Xcorr) greater than 2.5, a delta cross-correlation factor (dCn) greater than 0.1 (indicating a significant difference between the best match reported and the next best match), a high preliminary scoring, and a minimum of one tryptic peptide terminus. For triply-charged peptides the correlation factor threshold was set at 3.5. All matched peptides were confirmed by visual examination of the spectra, and all spectra were searched against the latest version of the public, non-redundant protein database of the National Center for Biotechnology Information (NCBI) (Haynes, 1998; Andon, 2002).
Western Blots
Briefly, 40 µg of protein was separated by electrophoresis using 4–20% Tris-glycine Express gels (ISC Bioexpress, Kaysville, UT) and transferred to polyvinylidene fluoride (PVDF) membranes (Sigma-Aldrich). Membranes were blocked for 2 hr with 5% nonfat dry milk in Tris-buffered saline with Tween 20 [150 mM NaCl, 20 mM Tris, pH 7.5, 0.1% Tween 20] and incubated with primary antibodies overnight at 4°C with gentle rocking. Antibody concentrations used were based on the manufacturer’s specifications.
After incubation with primary antibodies, membranes were washed and incubated with an appropriate biotin-conjugated secondary antibody (Santa Cruz Biotech, Santa Cruz, CA). After secondary antibody incubations, the membranes were washed 3 times, for 10 min each, in 1X TBS with 0.5% Tween 20 and then incubated for another 30 min with a streptavidin-alkaline phosphatase conjugate (Invitrogen) followed by colorimetric detection using NBT/BCIP reagent (Roche, Indianapolis, IN). Densitometry analyses were done using a Syngene Image Analyzer with Gene Tools Analysis Software version 3.02.00 (Syngene, Frederick, MD). Data were normalized to protein expression levels of β-actin.
RNA Extraction and Real-Time, Quantitative PCR (Q-PCR)
Cytoplasmic RNA was extracted using TRIzol (Invitrogen). The final RNA pellet was dried and resuspended in diethyl pyrocarbonate (DEPC) water and the concentration of RNA was determined using a spectrophotometer at 260 nm. Any DNA contamination in the RNA preparation was removed by treating the RNA with DNAse (1 IU/µg of RNA, Promega, Madison WI) for 2 hr at 37°C, followed by proteinase K digestion at 37°C for 15 min and subsequent extraction with phenol/chloroform and NH4OAc/ETOH precipitation. The isolated RNA was stored at −70°C until used. DNA contamination of the RNA preparation was checked by including a control in which reverse transcriptase enzyme was not added to the PCR amplification procedure. Relative abundance of each mRNA species was assessed using the SYBR green master mix from Stratagene (La Jolla, CA) to perform Q-PCR. Differences in threshold cycle number were used to quantify the relative amount of PCR target contained within each tube. Relative mRNA species expression was quantitated and expressed as transcript accumulation index (TAI = 2−(ΔΔCT)), calculated using the comparative CT method. All data were controlled for quantity of RNA input by performing measurements on an endogenous reference gene, β-actin (Shively, 2003).
Statistics
For Western blot analyses and gene expression data, statistical significance was determined using Student’s t-test (SPSS Inc.).
RESULTS
Proteomic Analyses of Monocyte-Derived Mature Dendritic Cells
Cocaine Differentially Regulates the Expression of Several Proteins
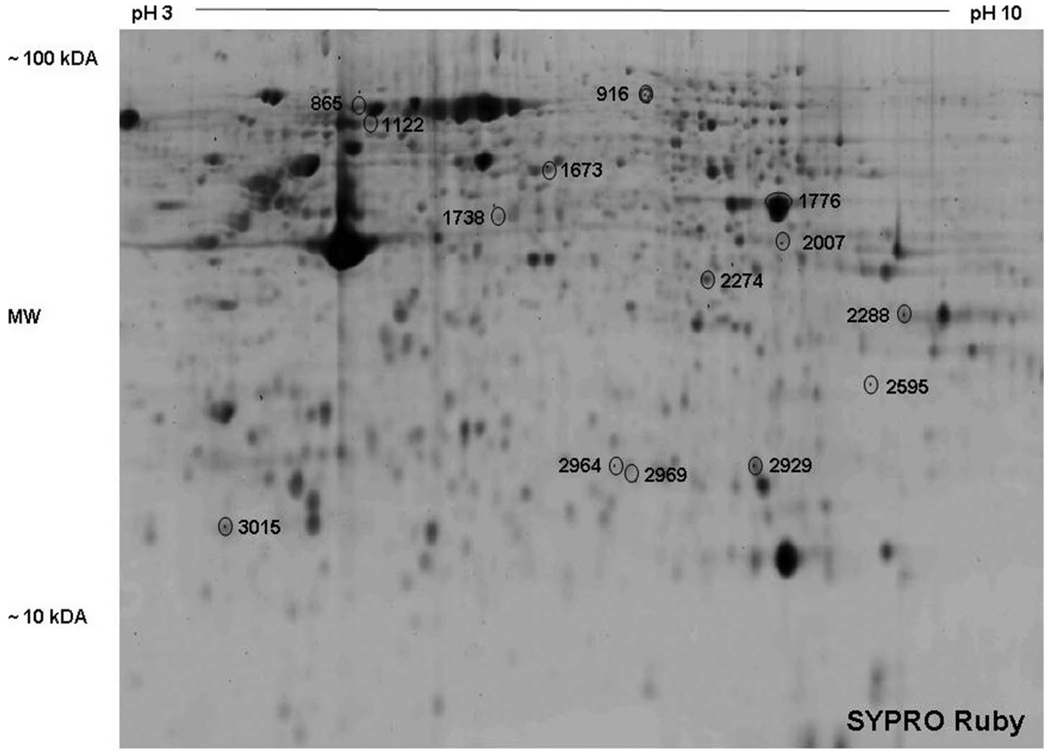
Figure 1 is a representative 2D gel image of SYPRO Ruby stained proteins. Comparing standardized protein abundance data generated from the Cy3, Cy5 and Cy2 images (DeCYDER software), several proteins spots were differentially regulated by cocaine. Protein spots were identified by HPLC-MS/MS. The protein spots that yielded positive identification are shown in Figure 1, labeled with arbitrary spot numbers. Respective protein identities are listed in Table 1.
Figure 1.
Representative 2-D gel image of a lysate of monocyte derived, mature dendritic cell stained with SYPRO Ruby dye. Arbitrary spot numbers represent statistically significant, differentially expressed proteins between control and cocaine treatment. The identity of each protein is shown in Table 1. Four separate experiments gave similar results.
Table 1.
Mature dendritic cells were cultured with and without cocaine (1 µM) for 48 hr (n = 4 independent experiments).
| Spot # | Protein name | Gene accession number |
% change | p value | Theoretical mass |
Theoretical pI |
Biological function |
|---|---|---|---|---|---|---|---|
| 916 | programmed cell death 6 interacting protein (AIP1) |
gi|34866400| | 25 ↑ | 0.001 | 70545.76 | 7.45 | regulation of apoptosis actin binding protein |
| 1738 | cathepsin D preproprotein | gi|4503143| | 19 ↑ | 0.01 | 44552.22 | 6.10 | aspartic protease |
| 1766 | enolase | gi|4503571| | 16 ↑ | 0.05 | 47168.96 | 7.01 | glycolysis |
| 1766 | non-neural alpha enolase | gi|22096350| | 16 ↑ | 0.05 | 47200.91 | 5.84 | glycolysis |
| 1766 | nuclear factor NF-kappa-B p105 subunit |
gi|21542418| | 16 ↑ | 0.05 | 105356.00 | 5.20 | transcription factor |
| 2274 | D-2-hydroxy-acid dehydrogenase |
gi|11399466| | 28 ↑ | 0.01 | 27312.44 | 9.10 | oxidoreductase |
| 2274 | hypothetical protein DKFZp564C047.1 |
gi|11360180| | 28 ↑ | 0.01 | 58140.27 | 4.94 | unknown |
| 2274 | heterogeneous nuclear ribonucleoprotein H3 |
gi|14141157| | 28 ↑ | 0.01 | 36926.49 | 6.37 | RNA binding protein |
| 2929 | phosphoglycerate mutase 1 | gi|4505753| | 13 ↑ | 0.01 | 28803.93 | 6.67 | glycolysis/gluconeogenesis |
| 2969 | aldolase A (AA) | gi|229674| | 16 ↑ | 0.007 | 39288.83 | 8.39 | glycolysis |
| 2969 | aldolase C, fructose- bisphosphate |
gi|4885063| | 16 ↑ | 0.007 | 39455.87 | 6.41 | glycolysis |
| 3015 | tumor protein | gi|4507669| | 40 ↑ | 0.003 | 19595.34 | 4.84 | unknown |
| 865 | heat shock 90kDa protein (HSP90) |
gi|13129150| | 11 ↓ | 0.001 | 84659.71 | 4.94 | molecular chaperone |
| 1122 | heat shock 70 kDa (HSP70) |
gi|662802| | 16 ↓ | 0.004 | 71194.37 | 5.36 | molecular chaperone |
| 1122 | L-plastin | gi|4504965| | 16 ↓ | 0.004 | 70289.33 | 5.20 | actin-binding protein |
| 1673 | heterogeneous nuclear ribonucleoprotein H2 |
gi|9624998| | 11 ↓ | 0.005 | 49263.57 | 5.89 | ribonucleoprotein |
| 1673 | tryptophan–tRNA ligase | gi|135191| | 11 ↓ | 0.005 | 53165.42 | 5.83 | ligase activity |
| 2007 | protein phosphatase 1 regulatory inhibitor subunit 8(NIPP1) |
gi|13699256| | 22 ↓ | 0.002 | 38478.79 | 6.87 | RNA binding protein |
| 2007 | phosphoglycerate kinase 1 | gi|4505763| | 22 ↓ | 0.002 | 44614.69 | 8.30 | glycolysis |
| 2288 | glyceraldehyde-3- phosphate dehydrogenase |
gi|7669492| | 16 ↓ | 0.05 | 36053.21 | 8.57 | glycolysis/gluconeogenesis |
| 2595 | carbonyl reductase 1 | gi|4502599| | 27 ↓ | 0.004 | 30374.94 | 8.55 | KEGG pathway: |
| 2964 | similar to high mobility group box 1 |
gi|27479871| | 24 ↓ | 0.007 | 24665.60 | 5.92 | nonhistone chromosomal protein |
Listed are statistically significant, differentially expressed proteins (Student’s t-test) that were identified by HPLC-MS/MS (all proteins had an expect score >10). Theoretical mass and pI were calculated using: http://au.expasy.org/tools/pi_tool.html.
Proteins that had increased expression levels in cells treated with cocaine compared to control included: programmed cell death 6 interacting protein (AIP1), cathepsin D preproprotein, enolase, non-neural alpha enolase, nuclear factor (NF)-κβ p105 subunit, D-2-hydroxy-acid dehydrogenase, hypothetical protein DKFZp564C047.1, heterogeneous nuclear ribonucleoprotein H3, phosphoglycerate mutase 1, aldolase A (AA), aldolase C, fructose-bisphosphate and tumor protein. Proteins showing a decrease in expression were heat shock protein 90 (HSP90), heat shock protein 70 (HSP70), L-plastin, heterogeneous nuclear ribonucleoprotein H2, tryptophan-tRNA ligase, protein phosphatase 1 regulatory inhibitor (NIPP1), phosphoglycerate kinase 1, glyceraldehyde-3-phosphate dehydrogenase, carbonyl reductase 1 and a protein that is similar to high mobility group box 1.
Methamphetamine Differentially Regulates the Expression of Several Proteins
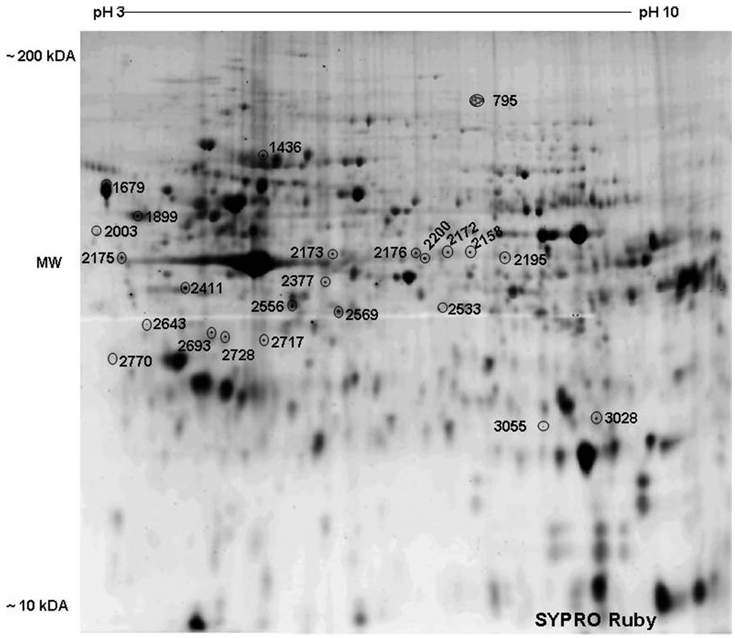
Figure 2 is a representative 2D gel image of SYPRO Ruby stained proteins. Several proteins spots were differentially expressed between methamphetamine treated and control cultures. Protein spots that were positively identified by HPLC-MS/MS are shown in Figure 2 labeled with arbitrary numbers. The identities of the respective proteins are listed in Table 2. Proteins showing increased levels of expression after treatment of monocyte derived mature dendritic cells with methamphetamine in comparison with control cultures include AIP1, heat shock protein 70kDa, protein 8 (HSPA8), L-plastin, laminin B1, calreticulin, nuclease sensitive element-binding protein 1 (YB-1), mitogen-activated protein kinase kinase 1 (MEK1), mitogen-activated protein kinase kinase 2 (MEK2), stress-induced-phosphoprotein 1, membrane-organizing extension spike protein (moesin), hydroxyacyl-coenzyme A dehydrogenase, and cathepsin S.
Figure 2.
Representative 2-D gel image of a lysate of monocyte derived mature dendritic cell stained with SYPRO Ruby dye. Arbitrary spot numbers represent statistically significant, differentially expressed proteins between control and methamphetamine treatment. The identity of each protein is shown in Table 2. Four separate experiments gave similar results test (n = 4 independent experiments).
Table 2.
Mature dendritic cells were cultured with and without methamphetamine (100 µM) for 24 hr (n = 4 independent experiments).
| Spot # | Protein name | Gene accession number |
% change | p value | Theoretical mass |
Theoretical pI |
Biological function |
|---|---|---|---|---|---|---|---|
| 795 | programmed cell death 6 interacting protein (AIP1) |
gi|31076784| | 61 ↑ | 0.001 | 44060.92 | 8.68 | signal transduction |
| 1436 | heat shock protein 70kDa, protein 8 (HSPA8) |
gi|5729877| | 21 ↑ | 0.028 | 70898.09 | 5.37 | molecular chaperone |
| 1436 | L-plastin | gi|4504965| | 21 ↑ | 0.028 | 70289.33 | 5.20 | calcium and actin binding |
| 1436 | laminin B1 | gi|5031877| | 21 ↑ | 0.028 | 66408.34 | 5.11 | protein binding |
| 1679 | calreticulin | gi|4757900| | 52 ↑ | 0.004 | 48141.56 | 4.29 | ER protein |
| 2175 | calreticulin | gi|4757900| | 67 ↑ | 0.003 | 48141.56 | 4.29 | ER protein |
| 2003 | nuclease sensitive element binding protein 1 (YB-1) |
gi|423015| | 70 ↑ | 0.023 | 34541.60 | 7.78 | DNA, RNA binding, Transcription factor |
| 2158 | mitogen-activated protein kinase kinase 2 (MEK1) |
gi|5579478| | 43 ↑ | 0.011 | 43439.01 | 6.18 | serine/threonine/ tyrosine kinase activity |
| 2158 | mitogen-activated protein kinase kinase 1 (MEK2) |
gi|13489054| | 43 ↑ | 0.011 | 44424.13 | 6.12 | serine/threonine/ tyrosine kinase activity |
| 2158 | stress induced phosphoprotein 1 |
gi|5803181| | 43 ↑ | 0.011 | 62639.26 | 6.40 | protein binding |
| 2176 | membrane-organizing extension spike protein (moesin) |
gi|4505257| | 36 ↑ | 0.008 | 67820.04 | 6.08 | cytoskeletal protein binding |
| 3028 | hydroxyacyl-Coenzyme A dehydrogenase |
gi|4758504| | 59 ↑ | 0.044 | 26923.08 | 7.65 | catalytic activity |
| 3055 | cathepsin S | gi|23200070| | 51 ↑ | 0.034 | 23947.91 | 7.66 | lysosomal cysteine proteinase |
| 1899 | lymphocyte-specific protein 1 |
gi|10880979| | 47 ↓ | 0.017 | 37191.59 | 4.69 | signal transduction |
| 2172 | pyruvate kinase | gi|4505839| | 39 ↓ | 0.026 | 57913.85 | 7.95 | kinase activity |
| 2200 | pyruvate kinase | gi|4505839| | 35 ↓ | 0.004 | 57913.85 | 7.95 | kinase activity |
| 2173 | heat shock 70kDa, protein 5 (HSPA5) |
gi|87528| | 32 ↓ | 0.037 | 72115.65 | 5.03 | molecular chaperone |
| 2195 | adenosine kinase | gi|7448823| | 23 ↓ | 0.036 | 37511.82 | 6.09 | phosphotransferase |
| 2377 | 3(2),5-bisphosphate nucleotidase |
gi|25282455| | 33 ↓ | 0.008 | 33174.03 | 5.58 | phosphatase activity |
| 2411 | heterogeneous nuclear ribonucleoprotein C (hRNPC) |
gi|4758544| | 26 ↓ | 0.003 | 31966.24 | 5.10 | RNA binding |
| 2411 | laminin receptor 1 | gi|9845502| | 58 ↓ | 0.003 | 32854.08 | 4.79 | receptor activity |
| 2411 | nucleophosmin (B23) | gi|112079| | 58 ↓ | 0.003 | 28385.22 | 4.54 | molecular chaperone |
| 2553 | 26S proteasome- associated pad1 homolog |
gi|34854852| | 26 ↓ | 0.042 | 41125.53 | 6.94 | unknown |
| 2556 | F-actin capping protein alpha-1 subunit |
gi|5453597| | 24 ↓ | 0.03 | 32922.77 | 5.45 | actin binding |
| 2569 | lactate dehydrogenase | gi|13786847| | 32 ↓ | 0.002 | 36507.30 | 5.72 | oxidoreductase activity |
| 2643 | gp96 | gi|6755863| | 73 ↓ | 0.041 | 92475.77 | 4.74 | molecular chaperone |
| 2693 | EF hand domain family member D2 |
gi|20149675| | 76 ↓ | 0.009 | 26697.28 | 5.15 | calcium ion binding |
| 2717 | alpha-SNAP | gi|4505329| | 54 ↓ | 0.009 | 33246.75 | 5.23 | intracellular transporter |
| 2728 | ribosomal protein P0 | gi|4506667| | 67 ↓ | 0.004 | 34273.51 | 5.72 | RNA and protein binding |
| 2770 | P32 A Doughnut-Shaped Acidic Mitochondrial Matrix Protein |
gi|4930073| | 86 ↓ | 0.021 | 23801.17 | 4.32 | Unknown |
Listed are statistically significant, differentially expressed proteins (Student’s t-test) that were identified using HPLC-MS/MS (all proteins had an expect score >10). Theoretical mass and pI were calculated using: http://au.expasy.org/tools/pi_tool.html.
Conversely, lymphocyte-specific protein 1, pyruvate kinase, heat shock 70kDa, protein 5 (HSPA5), adenosine kinase, 3(2),5-bisphosphate nucleotidase, heterogeneous nuclear ribonucleoprotein C (hRNPC), laminin receptor 1, nucleophosmin (B23), 26S proteasome-associated pad1 homolog, F-actin capping protein alpha-1 subunit, lactate dehydrogenase, gp96, EF hand domain family member D2, alpha-SNAP, ribosomal protein P0, and a P32 A Doughnut-Shaped Acidic Mitochondrial Matrix Protein showed decreased protein expression in cells treated with methamphetamine compared to the control culture.
For all 2D gels, several weak protein spots could not be identified due to insufficient amounts of peptides for identification and therefore are not shown in Figure 1 and Figure 2.
Western Blot and Gene Expression Analyses
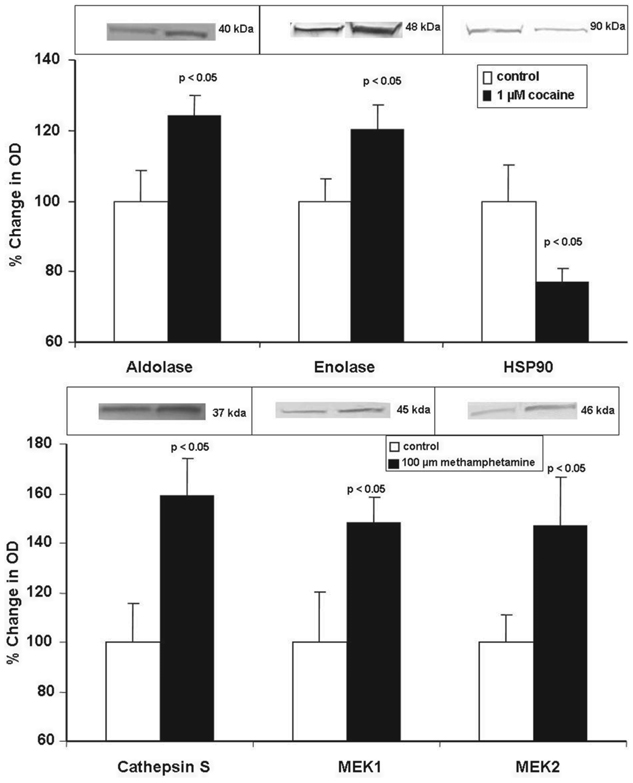
Modulation of protein and gene expression of select proteins from mature dendritic cells treated with cocaine or methamphetamine, respectively, were further examined by Western blots and Q-PCR. The proteins and genes that were selected for analysis showed statistically significant changes in expression on treatment with cocaine or methamphetamine. The specific proteins and genes that were validated were arbitrarily chosen based upon several logistical issues including the ready availability of primers and antibodies and a sufficient quantity of sample. Data shown in Figure 3 are representative Western blots and protein quantification by densitometry. Figure 3A demonstrates the effects of cocaine on aldolase, enolase and HSP90 protein expression.
Figure 3.
Western blot analyses of selected, differentially expressed proteins from mature dendritic cells treated with (A) cocaine or (B) methamphetamine. Data were normalized to protein expression levels of β-actin. The graphs show the mean ± SD of % change in OD as measured by densitometry of bands from Western blots from 4 independent experiments. Statistical significance was determined by Student’s t-test.
Cocaine induces a significant up-regulation in the protein expression of aldolase A (24%) and enolase (21%), while inducing a significant down-regulation in the expression of HSP90 (23%). Figure 3B shows that methamphetamine significantly up-regulates the protein expression of cathepsin S (59.2%), MEK1 (48.2%) and MEK2 (46.8%). Protein expression of β-actin was unchanged by treatment with cocaine or methamphetamine as compared to untreated cells (data not shown). These results confirm the effects of cocaine and methamphetamine on protein expression by monocyte derived, mature dendritic cells, as determined by DIGE and HPLC-MS/MS (Table 1 and Table 2).
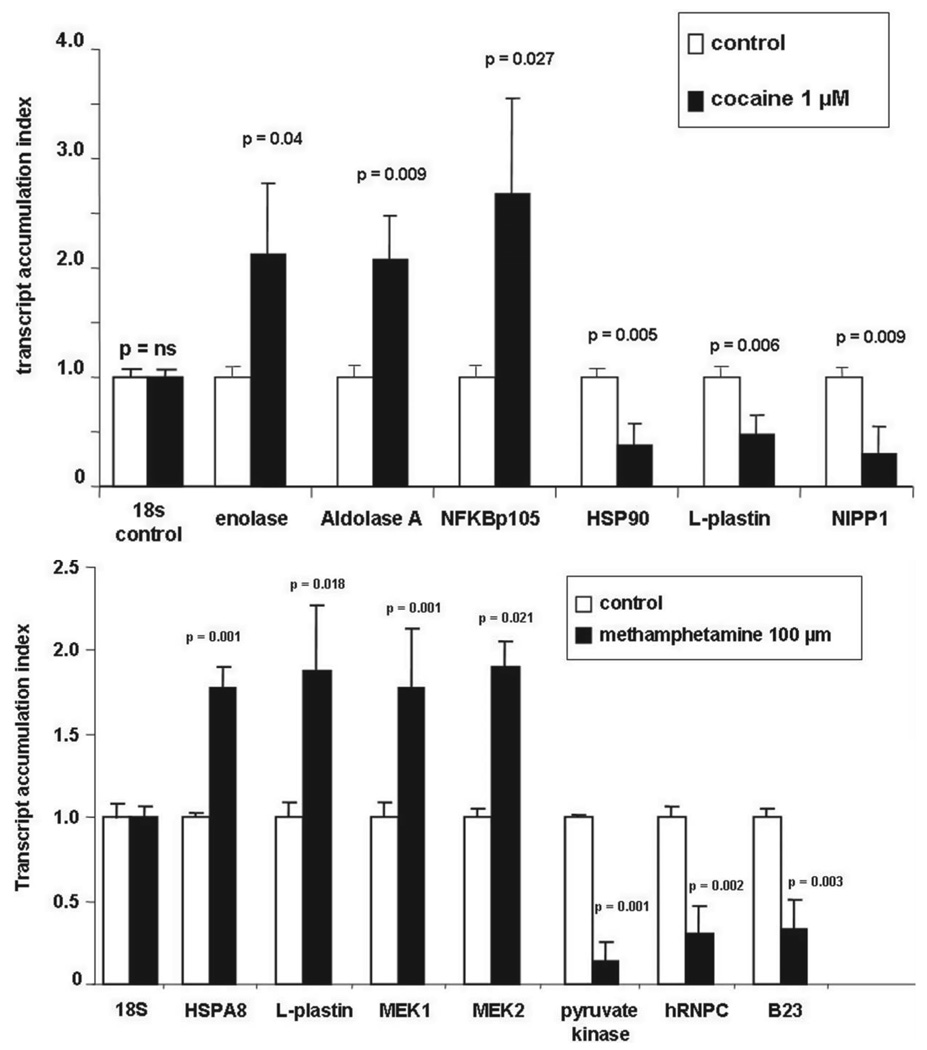
mRNA levels of arbitrarily chosen proteins were analyzed by Q-PCR using primers shown in Table 3. Data presented in Figure 4A show that cocaine treatment had no effect on 18s RNA expression used as control RNA. However, cocaine significantly up-regulated gene expression for enolase (TAI = 2.06 ± 0.74, p = 0.04), AA (TAI = 2.08 ± 0.39, p = 0.009), and NF-κβ p105 subunit (TAI = 2.69 ± 0.86, p = 0.027) compared to the respective untreated control culture. Similarly, cocaine significantly down-regulated the gene expression of HSP90 (TAI = 0.38 ± 0.19, p = 0.005), L-plastin (TAI = 0.48 ± 0.17, p = 0.006) and NIPP1 (TAI = 0. 29, p = 0.009) compared to the respective untreated control culture. Data presented in Figure 4B show that methamphetamine treatment had no effect on 18s control RNA gene expression. However, methamphetamine significantly up-regulated the gene expression of HSPA8 (TAI = 1.77 ± 0.025, p = 0.001), L-plastin (TAI = 1.87 ± 0.08, p = 0.018), MEK1 (TAI = 1.9 ± 0.16, p = 0.001) and MEK2 (TAI = 1.77 ± 0.36, p = 0.021) compared to respective controls. Methamphetamine significantly down-regulated the gene expression of pyruvate kinase (TAI = 0.14 ± 0.12, p = 0.001), hRNPC (TAI = 0.30 ± 0.16, p = 0.002) and B23 (TAI = 0.33 ± 0.06, p = 0.003), compared to respective controls.
Table 3.
Primer sequences for Real Time Q-PCR.
| Primer | Primer sequences |
|---|---|
| β-actin | 5’, 5’-TGACGGGGTCACCCACACTGTGCCCATCTA-3’ |
| 3’, 5-AGTCATAGTCCGCCTAGAAGCATTTGCGGT-3’ | |
| aldolase A (AA) | 5’, 5’-AAC ATG ACC CAC CTG TCC ATG CTA-3’ |
| 3’, 5’-TGG ATA TTG GTA GGG CAT GGT GCT-3’ | |
| B23 | 5’, 5’-GTT CAG GGC CAG TGC ATA TT-3’ |
| 3’, 5’-TTT CTT CAC TGG CGC TTT TT-3’ | |
| enolase | 5’, 5’-ATA AAG AAG GCC TGG AGC TGC TGA -3’ |
| 3’, 5’-TGC CCA GCT CCT CTT CAA TTC TGA-3’ | |
| hRNPC | 5’, 5’-AGAGGCTGAGGAAGGAGAGG-3’ |
| 3’, 5’-AGGAGCGTCAAAGGAAGTGA-3’ | |
| HSPA8 | 5’, 5’-GGA GGT GGC ACT TTT GAT GT-3’ |
| 3’, 5’-AGC AGT ACG GAG GCG TCT TA-3’ | |
| HSP90 | 5’, 5’-GAT TGG CCA GTT CGG TGT TGG TTT-3’ |
| 3’, 5’-GCT TGT TCC TGA TCG TTG GGC AAA-3’ | |
| L-plastin | 5’, 5’-TTA ACA GAT ACC CTG CCC TGC ACA-3’ |
| 3’, 5-TCA CTT CAA CCA GGT CTT CTG GCA-3’ | |
| MEK1 | 5’, 5’-GCT TGG GGC TAT TTG TGT GT-3’ |
| 3’, 5’-TCT CAC AAG GCT CCC TCC TA-3’ | |
| MEK2 | 5’,5’-CGT ACC TCC GAG AGA AGC AC-3’ |
| 3’, 5’-GGA GTT GGC CAT GGA GTC TA-3’ | |
| NFκB p105 subunit | 5’, 5’-AGG ATG AAG GAG TTG TGC CTG GAA-3’ |
| 3’, 5’-TGA GTT TGC GGA AGG ATG TCT CCA-3’ | |
| NIPP1 | 5’, 5’-TGC AGT GGT CCC AGT CAA GAA GAA-3’ |
| 3’, 5’-ATC CCA CCC TTC TCC ATG ACC AAA-3’ | |
| pyruvate kinase | 5’ 5′-ATC GTC CTC ACC AAG TCT GG-3’ |
| 3’, 5-GAA GAT GCC ACG GTA CAG GT-3’ |
Figure 4.
Q-PCR analyses of selected, differentially expressed genes from mature dendritic cells treated with (A) cocaine or (B) methamphetamine. Statistical significance was calculated by Student’s t-test (n = 4 independent experiments).
These results demonstrate concordance between both the gene and protein expression data from monocyte-derived mature dendritic cells treated with cocaine or methamphetamine, respectively.
DISCUSSION
Drug abuse has long been recognized as a significant co-morbidity associated with HIV-1 infections. In this study we attempted to gain insight into the molecular mechanisms underlying the effects of drug abuse on susceptibility to and progression of viral infections such HCV and HIV-1. Specifically we examined changes in the proteome of mature dendritic cells in response to treatment with the widely used, addictive drugs, cocaine and methamphetamine. Dendritic cells play an important role as a first line of defense of the immune system against pathogens. We tested the hypothesis that drugs of abuse regulate the expression of various host proteins in monocyte derived mature dendritic cells. We propose that these proteins may directly or indirectly play a significant role in increased susceptibility to various viral infections. The potential relevance of the differential expression of select regulated proteins is discussed below.
Effects of Cocaine on Mature Dendritic Cells
NFκB is a major, inducible transcription factors whose modulation triggers a cascade of intracellular signaling events. Activated NFκB stimulates the expression of genes involved in a wide variety of biological functions (Beinke, 2004; Celec, 2004). Previous studies have shown that cocaine activates NFκB in H9C2 cells (Hargrave, 2003), human brain endothelial cells (Lee, 2001) and in the nucleus accumbens in the mouse brain (Ang, 2001). In the present study we demonstrate that the p105 subunit of NFκB is up-regulated by cocaine in mature dendritic cells. HCV core protein promotes proliferation of human hepatoma cells via activation of NF-κB, suggesting a mechanism of hepatocarcinogenesis (Sato, 2006). U937 cells (immortalized cells of the monocyte-macrophage like lineage) infected with HIV-1 show increased levels of the p105 and p50 subunits of NF-κB, resulting in prolonged activation of NFκB, leading to viral persistence (McElhinny, 1995). Cocaine increases expression of NF-κB p105 in mature dendritic cells which may be associated with viral (e.g., HCV or HIV-1) persistence or an increase in the pathological effects.
Cocaine significantly down-regulated protein phosphatase 1 regulatory inhibitor subunit 8 also known as NIPP1, in monocyte derived mature dendritic cells. NIPP1 is a regulatory subunit of protein phosphatase (PP) and binds to the catalytic subunit of PP forming an inactive holoenzyme complex, inhibiting the dephosphorylation of a variety of substrates (Zolnierowicz, 2000). Previous studies have shown that overexpression of NIPP1 inhibits Tat-induced HIV-1 transcription in COS-7 cells (Ammosova, 2003, 2005). Since cocaine down-regulates NIPP1 and NIPP1 has been shown to inhibit Tat-induced HIV-1 transcription, it is possible that cocaine-induced down-regulation of NIPP1 may be a mechanism through which cocaine regulates HIV-1 transcription. HCV can upregulate PP expression (Duong, 2005). Cocaine and HCV may act synergistically through their actions on NIPP1 and PP, respectively, to interfere with the host’s cellular defenses against the virus.
Effects of Methamphetamine on Mature Dendritic Cells
Mitogen activated protein (MAP) kinase transduces extracellular signals into intracellular responses (Johnson, 2002; Nishimoto, 2006). HCV E2 envelope protein activates the MAP kinase pathway (Zhao, 2005, 2006, 2007). MAP kinase inhibitors block HIV-1 viral replication in T cells and in a monocyte cell line (Muthumani, 2004). HIV-1 viral proteins also regulate MAP kinase activity (Kumar, 1998; Robichaud, 2000; Rusnati, 2001; Kan, 2004). Suppression of virion-associated MAP kinases by specific inhibitors impaired HIV-1 infectivity (Jacque, 1998). Through the regulation of MAP kinases, methamphetamine may increase the susceptibility of mature dendritic cells to HCV or HIV-1 infection.
Nuclease sensitive element-binding protein 1 or Y box-binding protein 1 (YB-1) is a member of the Y-box DNA binding proteins located in the cytoplasm and nucleus of mammalian cells. Y-box proteins have multiple functions including the regulation of cell proliferation and gene transcription and translation (Wolffe, 1994; Kohno, 2003). YB-1 regulates the transcription of human T-cell lymphotropic virus type I (HTLV-I), adenoviruses, human polyomavirus, JC virus (JCV) and HIV-1 (Kashanchi, 1994; Holm, 2004; Safak, 2002). YB-1 binds to the C-terminal region of HIV-1 Tat protein which enhances activation of the HIV-1 LTR promoter (Ansari, 1999). Since YB-1 enhances viral transcription, our findings that methamphetamine increases the expression of YB-1 in mature dendritic cells also suggests that methamphetamine enhances viral replication by regulating the expression of transcription factors such as YB-1. Further studies are necessary to determine whether YB-1 regulates HCV transcription.
Common Proteins Whose Expression is Regulated by Cocaine and Methamphetamine
Cathepsins are lysosomal cysteine proteases released by dendritic cells and B cells. Cathepsins are located in the acid endosomal/lysosomal (low pH) compartment of dendritic cells and play a pivotal role in antigen processing (Riese, 2000). Previous studies have shown that cathepsins are associated with viral entry, dissemination and enhanced viral replication (El Messaoudi, 1999, 2000; Prin-Mathieu, 2001; Tscherene, 2006). Polymorphonuclear cells isolated from HIV-1 positive patients have significantly higher cathepsin D activity than cells from normal controls (Prin-Mathieu, 2001). Vaginal secretions contain cathepsin D and exposure of lymphocyte cultures to these secretions increased HIV-1 replication in these cells (El Messaoudi 1999, 2000). The current study demonstrates that cocaine and methamphetamine both enhance the expression of cathepsins in mature dendritic cells. An increase in expression of cathepsins may play a role in spreading HCV or HIV-1 infection to neighboring cells.
Heat shock proteins (HSP), also known as stress proteins, belong to the family of molecular chaperones that can be induced upon cellular injury. HSP are classified by their molecular weight, comprising of six general families: HSP110, HSP90, HSP70, HSP60, small molecular weight HSP, and immunophilins (Prohaszka, 2004; Gullo, 2004). HSP have diverse functions including chaperone activity, regulation of redox state and regulation of protein turnover (Prohaszka, 2004). HSP90 plays an important role in HCV RNA replication (Okamoto, 2006). HSP70 and HSP27 expression are increased following HIV-1 infection (Wainberg, 1997) suggesting that these proteins are involved in innate immune responses to viruses. Findings from the current study demonstrate that cocaine and methamphetamine differentially regulate several types of HSP. This differential regulation may play a role in the ability of dendritic cells to initiate an anti-viral response, however the clinical significance of these data are yet to be determined.
Our studies provide a relationship of cocaine or methamphetamine in the immunomodulation of mature dendritic cells. The studies presented herein demonstrate that cocaine and methamphetamine can modulate the expression of various proteins within the proteome of mature dendritic cells. Future studies are necessary to determine the molecular mechanisms underlying the effects of these drugs mechanistically link these drugs of abuse to the immunomodulation of HCV and HIV-1. Identification of the various proteins whose expression is modulated by these drugs may yield new molecular targets for the prevention of infections such as HCV and HIV-1 that are associated with drug abuse.
ACKNOWLEDGMENT
Funding support K01 DA024577, Kaleida Health Foundation.
Footnotes
Full terms and conditions of use: http://www.informaworld.com/terms-and-conditions-of-access.pdf
This article may be used for research, teaching and private study purposes. Any substantial or systematic reproduction, re-distribution, re-selling, loan or sub-licensing, systematic supply or distribution in any form to anyone is expressly forbidden.
The publisher does not give any warranty express or implied or make any representation that the contents will be complete or accurate or up to date. The accuracy of any instructions, formulae and drug doses should be independently verified with primary sources. The publisher shall not be liable for any loss, actions, claims, proceedings, demand or costs or damages whatsoever or howsoever caused arising directly or indirectly in connection with or arising out of the use of this material.
Declaration of Interest: The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
REFERENCES
- Amersham Pharmaceuticals. Content/Proteomics+DIGE+Protocols http://www1.amershambiosciences.com/aptrix/upp00919.nsf/
- Ammosova T, Jerebtsova M, Beullens M, Voloshin Y, Ray PE, Kumar A, Bollen M, Nekhai S. Nuclear protein phosphatase-1 regulates HIV-1 transcription. J. Biol. Chem. 2003;278:32189–32194. doi: 10.1074/jbc.M300521200. [DOI] [PubMed] [Google Scholar]
- Ammosova T, Washington K, Debebe Z, Brady J, Nekhai S. Dephosphorylation of CDK9 by protein phosphatase 2A and protein phosphatase-1 in Tat-activated HIV-1 transcription. Retrovirology. 2005;2:47. doi: 10.1186/1742-4690-2-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andon NL, Hollingworth S, Koller A, Greenland AJ, Yates JR, Haynes PA. Proteomic characterization of wheat amyloplasts using identification of proteins by tandem mass spectrometry. Proteomics. 2002;2:1156–1168. doi: 10.1002/1615-9861(200209)2:9<1156::AID-PROT1156>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Ang E, Chen J, Zagouras P, Brady J, Holland J, Schaeffer E, Nestler EJ. Induction of nuclear factor-kappaB in nucleus accumbens by chronic cocaine administration. J. Neurochem. 2001;79:221–224. doi: 10.1046/j.1471-4159.2001.00563.x. [DOI] [PubMed] [Google Scholar]
- Ansari SA, Safak M, Gallia GL, Sawaya BE, Amini S, Khalili K. Interaction of YB-1 with human immunodeficiency virus type 1 Tat and TAR RNA modulates viral promoter activity. J. Gen. Virol. 1999;80:2629–2638. doi: 10.1099/0022-1317-80-10-2629. [DOI] [PubMed] [Google Scholar]
- Bagasra O, Forman L. Functional analysis of lymphocytes subpopulations in experimental cocaine abuse. I. Dose-dependent activation of lymphocyte subsets. Clin. Exp. Immunol. 1989;77:289–293. [PMC free article] [PubMed] [Google Scholar]
- Bain C, Fatmi A, Zoulim F, Zarski JP, Trépo C, Inchauspé G. Impaired allostimulatory function of dendritic cells in chronic hepatitis C infection. Gastroenterology. 2001;120:512–524. doi: 10.1053/gast.2001.21212. [DOI] [PubMed] [Google Scholar]
- Baldwin GC, Tashkin DP, Buckley DM, Park AN, Dubinett SM, Roth MD. Marijuana and cocaine impair alveolar macrophage function and cytokine production. Am. J. Respir. Crit. Care Med. 1997;156:1606–1613. doi: 10.1164/ajrccm.156.5.9704146. [DOI] [PubMed] [Google Scholar]
- Banchereau J, Steinman RM. DC and the control of immunity. Nature. 1998;392:245–252. doi: 10.1038/32588. [DOI] [PubMed] [Google Scholar]
- Bayer BM, Hernandez MC, Ding XZ. Tolerance and cross tolerance to the suppressive effects of cocaine and morphine on lymphocyte proliferation. Pharmacol. Biochem. Behav. 1996;53:227–234. doi: 10.1016/0091-3057(95)00188-3. [DOI] [PubMed] [Google Scholar]
- Beinke S, Ley SC. Functions of NF-kappaB1 and NF-kappaB2 in immune cell biology. Biochem. J. 2004;382:393–409. doi: 10.1042/BJ20040544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender A, Sapp M, Schuler G, Steinman RM, Bhardwaj N. Improved methods for the generation of dendritic cells from nonproliferating progenitors in human blood. J. Immunol. Meth. 1996;196:121–135. doi: 10.1016/0022-1759(96)00079-8. [DOI] [PubMed] [Google Scholar]
- Berretta M, Zanet E, Di Benedetto F, Simonelli C, Bearz A, Morra A, Bonanno S, Berretta S, Tirelli U. Unusual presentation of metastatic hepatocellular carcinoma in an HIV/HCV coinfected patient: case report and review of the literature. Tumori. 2008;94(4):589–591. doi: 10.1177/030089160809400424. [DOI] [PubMed] [Google Scholar]
- Breci L, Hattrup E, Keeler M, Letarte J, Johnson R, Haynes PA. Comprehensive proteomics in yeast using chromatographic fractionation, gas phase fractionation, protein gel electrophoresis, and isoelectric focusing. Proteomics. 2005;5(8):2018–2028. doi: 10.1002/pmic.200401103. [DOI] [PubMed] [Google Scholar]
- Celec P. Nuclear factor kappa B—molecular biomedicine: the next generation. Biomed. Pharmacother. 2004;58:365–371. doi: 10.1016/j.biopha.2003.12.015. [DOI] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention (CDC) Coinfection with HIV and Hepatitis C Virus. 2005 November [Google Scholar]
- Cooper B, Eckert D, Andon NL, Yates JR, Haynes PA. Investigative proteomics: identification of an unknown plant virus from infected plants using mass spectrometry. J. Am. Soc. Mass Spectrom. 2003;14:736–741. doi: 10.1016/S1044-0305(03)00125-9. [DOI] [PubMed] [Google Scholar]
- Dauer M, Obermaier B, Herten J, Haerle C, Pohl K, Rothenfusser S, Schnurr M, Endres S, Eigler A. Mature dendritic cells derived from human monocytes within 48 hours: a novel strategy for dendritic cell differentiation from blood precursors. J. Immunol. 2003;170:4069–4076. doi: 10.4049/jimmunol.170.8.4069. [DOI] [PubMed] [Google Scholar]
- Drug Abuse Warning Network (DAWN) http://dawninfo.samhsa.gov/files/ED2006/DAWN2k6ED.pdf.
- Duong FH, Christen V, Berke JM, Penna SH, Moradpour D, Heim MH. Upregulation of protein phosphatase 2Ac by hepatitis C virus modulates NS3 helicase activity through inhibition of protein arginine methyltransferase 1. J. Virol. 2005;79(24):15342–15350. doi: 10.1128/JVI.79.24.15342-15350.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards DJ, Bowles SK. Protein binding of cocaine in human serum. Pharm. Res. 1988;5:440–442. doi: 10.1023/a:1015992502509. [DOI] [PubMed] [Google Scholar]
- El Messaoudi K, Thiry L, Van Tieghem N, Liesnard C, Englert Y, Moguilevsky N, Bollen A. HIV-1 infectivity and host range modification by cathepsin D present in human vaginal secretions. AIDS. 1999;13:333–339. doi: 10.1097/00002030-199902250-00005. [DOI] [PubMed] [Google Scholar]
- El Messaoudi K, Thiry VF, Liesnard C, Van Tieghem N, Bollen A, Moguilevsky N. A human milk factor susceptible to cathepsin D inhibitors enhances human immunodeficiency virus type 1 infectivity and allows virus entry into a mammary epithelial cell line. J. Virol. 2000;74:1004–1007. doi: 10.1128/jvi.74.2.1004-1007.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garderet L, Cao H, Salamero J, Vergé V, Tisserand E, Scholl S, Gorin NC, Lopez M. In vitro production of dendritic cells from human blood monocytes for therapeutic use. J. Hematother. Stem Cell Res. 2001;10(4):553–567. doi: 10.1089/15258160152509163. [DOI] [PubMed] [Google Scholar]
- Gharahdaghi F, Weinberg CR, Meagher DA, Imai BS, Mische SM. Mass spectrometric identification of proteins from silver-stained polyacrylamide gel: a method for the removal of silver ions to enhance sensitivity. Electrophoresis. 1999;20:601–605. doi: 10.1002/(SICI)1522-2683(19990301)20:3<601::AID-ELPS601>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Granelli-Piperno A, Delgado E, Finkel V, Paxton W, Steinman RM. Immature dendritic cells selectively replicate macrophagetropic (M-tropic) human immunodeficiency virus type 1, while mature cells efficiently transmit both M- and T-tropic virus to T cells. J. Virol. 1998;72:2733–2737. doi: 10.1128/jvi.72.4.2733-2737.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gullo CA, Teoh G. Heat shock proteins: to present or not, that is the question. Immunol. Lett. 2004;94:1–10. doi: 10.1016/j.imlet.2004.04.002. [DOI] [PubMed] [Google Scholar]
- Haines KA, Reibman J, Callegari PE, Abramson SB, Philips MR, Weissmann G. Cocaine and its derivatives blunt neutrophil functions without influencing phosphorylation of a 47-kilodalton component of the reduced nicotinamide-adenine dinucleotide phosphate oxidase. J. Immunol. 1990;144(12):4757–4764. [PubMed] [Google Scholar]
- Hargrave BY, Tiangco DA, Lattanzio FA, Beebe SJ. Cocaine, not morphine, causes the generation of reactive oxygen species and activation of NF-kappaB in transiently cotransfected heart cells. Cardiovasc. Toxicol. 2003;3:141–151. doi: 10.1385/ct:3:2:141. [DOI] [PubMed] [Google Scholar]
- Haynes PA, Gygi SP, Figeys D, Aebersold R. Proteome analysis: biological assay or data archive? Electrophoresis. 1998;19:1862–1871. doi: 10.1002/elps.1150191104. [DOI] [PubMed] [Google Scholar]
- Holm PS, Bergmann S, Jurchott K, Lage H, Brand K, Ladhoff A, Mantwill K, Curiel DT, Dobbelstein M, Dietel M, Gansbacher B, Royer HD. YB-1 relocates to the nucleus in adenovirus-infected cells and facilitates viral replication by inducing E2 gene expression through the E2 late promoter. J. Biol. Chem. 2002;277(12):10427–10434. doi: 10.1074/jbc.M106955200. [DOI] [PubMed] [Google Scholar]
- Jacque JM, Mann A, Enslen H, Sharova N, Brichacek B, Davis RJ, Stevenson M. Modulation of HIV-1 infectivity by MAPK, a virion-associated kinase. EMBO J. 1998;17:2607–2618. doi: 10.1093/emboj/17.9.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson GL, Lapadat R. Mitogen-activated protein kinase pathways mediated by ERK, JNK, and p38 protein kinases. Science. 2002;298:1911–1912. doi: 10.1126/science.1072682. [DOI] [PubMed] [Google Scholar]
- Kalasinsky KS, Bosy TZ, Schmunk GA, Reiber G, Anthony RM, Furukawa Y, Guttman M, Kish SJ. Regional distribution of methamphetamine in autopsied brain of chronic human methamphetamine users. Forensic Sci. Int. 2001;116:163–169. doi: 10.1016/s0379-0738(00)00368-6. [DOI] [PubMed] [Google Scholar]
- Kan H, Xie Z, Finkel MS. p38 MAP kinase-mediated negative inotropic effect of HIV gp120 on cardiac myocytes. Am. J. Physiol. Cell Physiol. 2004;286:C1–C7. doi: 10.1152/ajpcell.00059.2003. [DOI] [PubMed] [Google Scholar]
- Kanto T, Hayashi N, Takehara T, Tatsumi T, Kuzushita N, Ito A, Sasaki Y, Kasahara A, Hori M. Impaired allostimulatory capacity of peripheral blood dendritic cells recovered from hepatitis C virus-infected individuals. J. Immunol. 1999;162:5584–5591. [PubMed] [Google Scholar]
- Kanto T, Inoue M, Miyatake H, Sato A, Sakakibara M, Yakushijin T, Oki C, Itose I, Hiramatsu N, Takehara T, Kasahara A, Hayashi N. Reduced numbers and impaired ability of myeloid and plasmacytoid dendritic cells to polarize T helper cells in chronic hepatitis C virus infection. J. Infect. Dis. 2004;190:1919–1926. doi: 10.1086/425425. [DOI] [PubMed] [Google Scholar]
- Kashanchi F, Duvall JF, Dittmer J, Mireskandari A, Reid RL, Gitlin SD, Brady JN. Involvement of transcription factor YB-1 in human T-cell lymphotropic virus type I basal gene expression. J. Virol. 1994;68:561–565. doi: 10.1128/jvi.68.1.561-565.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Psevdos G, Suh J, Sharp VL. Co-infection of hepatitis B and hepatitis C virus in human immunodeficiency virus-infected patients in New York City, United States. World J. Gastroenterol. 2008;14(43):6689–6693. doi: 10.3748/wjg.14.6689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohno K, Izumi H, Uchiumi T, Ashizuka M, Kuwano M. The pleiotropic functions of the Y-box-binding protein, YB-1. Bioessays. 2003;25:691–698. doi: 10.1002/bies.10300. [DOI] [PubMed] [Google Scholar]
- Kumar A, Manna SK, Dhawan S, Aggarwal BB. HIV-Tat protein activates c-Jun N-terminal kinase and activator protein-1. J. Immunol. 1998;161:776–781. [PubMed] [Google Scholar]
- Lee YW, Hennig B, Yao J, Toborek M. Methamphetamine induces AP-1 and NF-kappaB binding and transactivation in human brain endothelial cells. J. Neurosci. Res. 2001;66:583–591. doi: 10.1002/jnr.1248. [DOI] [PubMed] [Google Scholar]
- Liang H, Wang X, Chen H, Song L, Ye L, Wang SH, Wang YJ, Zhou L, Ho WZ. Methamphetamine enhances HIV infection of macrophages. Am. J. Pathol. 2008;172(6):1617–1624. doi: 10.2353/ajpath.2008.070971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley KS, Friedman DB. All about DIGE: quantification technology for differential-display 2D-gel proteomics. Expert Rev. Proteomics. 2004;1:401–409. doi: 10.1586/14789450.1.4.401. [DOI] [PubMed] [Google Scholar]
- Lo P. HIV hijacks dendritic cells. Nat. Med. 2003;9(6):650. doi: 10.1038/nm0603-650. [DOI] [PubMed] [Google Scholar]
- Loré K, Smed-Sörensen A, Vasudevan J, Mascola JR, Koup RA. Myeloid and plasmacytoid dendritic cells transfer HIV-1 preferentially to antigen-specific CD4+ T cells. J. Exp. Med. 2005;201:2023–2033. doi: 10.1084/jem.20042413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan SD, Hu Z, Reynolds JL, Aalinkeel R, Schwartz SA, Nair MP. Methamphetamine modulates gene expression patterns in monocyte derived mature dendritic cells: implications for HIV-1 pathogenesis. Mol. Diagn. Ther. 2006;10(4):257–269. doi: 10.1007/BF03256465. [DOI] [PubMed] [Google Scholar]
- Matthews GV, Dore GJ. HIV and hepatitis C coinfection. J. Gastroenterol. Hepatol. 2008;23(7 Pt 1):1000–1008. doi: 10.1111/j.1440-1746.2008.05489.x. [DOI] [PubMed] [Google Scholar]
- McDonald D, Wu L, Bohks SM, Kewal Ramani VM, Unutmaz D, Hope TJ. Recruitment of HIV and its receptors to dendritic cell-T cell junctions. Science. 2003;300:1295–1297. doi: 10.1126/science.1084238. [DOI] [PubMed] [Google Scholar]
- McElhinny JA, MacMorran WS, Bren GD, Ten RM, Israel A, Paya CV. Regulation of I kappa B alpha and p105 in monocytes and macrophages persistently infected with human immunodeficiency virus. J. Virol. 1995;69:1500–1509. doi: 10.1128/jvi.69.3.1500-1509.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamadzadeh M, Duong T, Hoover T, Klaenhammer TR. Targeting mucosal dendritic cells with microbial antigens from probiotic lactic acid bacteria. Expert Rev. Vaccines. 2008;7(2):163–174. doi: 10.1586/14760584.7.2.163. [DOI] [PubMed] [Google Scholar]
- Moris A, Nobile C, Buseyne F, Porrot F, Abastado JP, Schwartz O. DCSIGN promotes exogenous MHC-I-restricted HIV-1 antigen presentation. Blood. 2004;103:2648–2654. doi: 10.1182/blood-2003-07-2532. [DOI] [PubMed] [Google Scholar]
- Mukunda BN, Callahan JM, Hobbs MS, West BC. Cocaine inhibits human neutrophil phagocytosis and phagolysosomal acidification in vitro. Immunopharmacol. Immunotoxicol. 2000;22(2):373–386. doi: 10.3109/08923970009016426. [DOI] [PubMed] [Google Scholar]
- Muthumani K, Wadsworth SA, Dayes NS, Hwang DS, Choo AY, Abeysinghe HR, Siekierka JJ, Weiner DB. Suppression of HIV-1 viral replication and cellular pathogenesis by a novel p38/JNK kinase inhibitor. AIDS. 2004;18:739–748. doi: 10.1097/00002030-200403260-00004. [DOI] [PubMed] [Google Scholar]
- Nair MP, Mahajan S, Sykes D, Bapardekar MV, Reynolds JL. Methamphetamine modulates DC-SIGN expression by mature dendritic cells. J. Neuroimmune Pharmacol. 2006;1(3):296–304. doi: 10.1007/s11481-006-9027-1. [DOI] [PubMed] [Google Scholar]
- Nair MPN, Mahajan SD, Schwartz SA, Reynolds J, Whitney R, Berstein Z, Chawda RP, Sykes D, Hewett R, Hsiao CB. Cocaine modulates dendritic cell-specific C type intercellular adhesion molecule-3-grabbing nonintegrin expression by dendritic cells in HIV-1 patients. J. Immunol. 2005;174:6617–6626. doi: 10.4049/jimmunol.174.11.6617. [DOI] [PubMed] [Google Scholar]
- Navas MC, Fuchs A, Schvoerer E, Bohbot A, Aubertin AM, Stoll-Keller F. Dendritic cell susceptibility to hepatitis C virus genotype 1 infection. J. Med. Virol. 2002;67(2):152–161. doi: 10.1002/jmv.2204. [DOI] [PubMed] [Google Scholar]
- [accessed May 2005];NIDA (National Institute of Drug Abuse) INFO FACTS, Methamphetamine. from http://www.drugabuse.gov/Infofacts/Infofaxindex.html.
- Nishimoto S, Nishida E. MAPK signaling: ERK5 versus ERK1/2. EMBO Rep. 2006;7:782–786. doi: 10.1038/sj.embor.7400755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto T, Nishimura Y, Ichimura T, Suzuki K, Miyamura T, Suzuki T, Moriishi K, Matsuura Y. Hepatitis C virus RNA replication is regulated by FKBP8 and Hsp90. EMBO J. 2006;25(20):5015–5025. doi: 10.1038/sj.emboj.7601367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pachiadakis I, Pollara G, Chain BM, Naoumov NV. Is hepatitis C virus infection of dendritic cells a mechanism facilitating viral persistence? Lancet Infect. Dis. 2005;5(5):296–304. doi: 10.1016/S1473-3099(05)70114-6. [DOI] [PubMed] [Google Scholar]
- Parodi C, Belmonte L, Baré P, de Bracco MM, Ruibal-Ares B. Impact of human immune deficiency virus infection on hepatitis C virus infection and replication. Curr. HIV Res. 2007;5(1):55–67. doi: 10.2174/157016207779316341. [DOI] [PubMed] [Google Scholar]
- Pellegrino TC, Dunn KL, Bayer BM. Mechanisms of cocaine-induced decreases in immune cell function. Int. Immunopharmacol. 2001;1:665–675. doi: 10.1016/s1567-5769(00)00051-5. [DOI] [PubMed] [Google Scholar]
- Piguet V, Steinman RM. The interaction of HIV with dendritic cells: outcomes and pathways. Trends Immunol. 2007;228(11):503–510. doi: 10.1016/j.it.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poet TS, Pillai R, Wood S, Watson RR. Stimulation of natural killer cell activity by murine retroviral infection and cocaine. Toxicol. Lett. 1991;59(1–3):147–152. doi: 10.1016/0378-4274(91)90066-f. [DOI] [PubMed] [Google Scholar]
- Prin-Mathieu C, Baty V, Faure G, Schumacher H, Kolopp-Sarda MN, May T, Canton P, Bene MC. Assessment by flow cytometry of peripheral blood leukocyte enzymatic activities in HIV patients. J. Immunol. Meth. 2001;252:139–146. doi: 10.1016/s0022-1759(01)00348-9. [DOI] [PubMed] [Google Scholar]
- Prohaszka Z, Fust G. Immunological aspects of heat-shock proteins-the optimum stress of life. Mol. Immunol. 2004;41:29–44. doi: 10.1016/j.molimm.2004.02.001. [DOI] [PubMed] [Google Scholar]
- Reis e Sousa C. Dendritic cells in a mature age. Nat Rev. Immunol. 2006;6:476–483. doi: 10.1038/nri1845. [DOI] [PubMed] [Google Scholar]
- Reynolds JL, Mahajan SD, Sykes DE, Schwartz SA, Nair MP. Proteomic analyses of methamphetamine (METH)-induced differential protein expression by immature dendritic cells (IDC) Biochim. Biophys. Acta. 2007;1774(4):433–442. doi: 10.1016/j.bbapap.2007.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riese RJ, Chapman HA. Cathepsins and compartmentalization in antigen presentation. Curr. Opin. Immunol. 2000;12:107–113. doi: 10.1016/s0952-7915(99)00058-8. [DOI] [PubMed] [Google Scholar]
- Robichaud GA, Poulin L. HIV type 1 nef gene inhibits tumor necrosis factor alpha-induced apoptosis and promotes cell proliferation through the action of MAPK and JNK in human glial cells. AIDS Res. Hum. Retrovir. 2000;16:1959–1965. doi: 10.1089/088922200750054684. [DOI] [PubMed] [Google Scholar]
- Rusnati M, Urbinati C, Musulin B, Ribatti D, Albini A, Noonan D, Marchisone C, Waltenberger J, Presta M. Activation of endothelial cell mitogen activated protein kinase ERK(1/2) by extracellular HIV-1 Tat protein. Endothelium. 2001;8(1):65–74. doi: 10.3109/10623320109063158. [DOI] [PubMed] [Google Scholar]
- Safak M, Sadowska B, Barrucco R, Khalili K. Functional interaction between JC virus late regulatory agnoprotein and cellular Y-box binding transcription factor, YB-1. J. Virol. 2002;76:3828–3838. doi: 10.1128/JVI.76.8.3828-3838.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders RW, de Jong EC, Baldwin CE, Schuitemaker JH, Kapsenberg ML, Berkhout B. Differential transmission of human immunodeficiency virus type 1 by distinct subsets of effector dendritic cells. J. Virol. 2002;76:7812–7821. doi: 10.1128/JVI.76.15.7812-7821.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y, Kato J, Takimoto R, Takada K, Kawano Y, Miyanishi K, Kobune M, Sato Y, Takayama T, Matunaga T, Niitsu Y. Hepatitis C virus core protein promotes proliferation of human hepatoma cells through enhancement of transforming growth factor alpha expression via activation of nuclear factor-kappaB. Gut. 2006;55(12):1801–1808. doi: 10.1136/gut.2005.070417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepers RJF, Oyler JM, Joseph RE, Cone EJ, Moolchan ET, Huestis MA. Methamphetamine and amphetamine pharmacokinetics in oral fluid and plasma after controlled oral methamphetamine administration to human volunteers. Clin. Chem. 2003;49:121–132. doi: 10.1373/49.1.121. [DOI] [PubMed] [Google Scholar]
- Schramm W, Craig PA, Smith RH, Berger GE. Cocaine and benzoylecgonine in saliva, serum, and urine. Clin. Chem. 1993;39:481–487. [PubMed] [Google Scholar]
- Shively L, Chang L, LeBon JM, Liu Q, Riggs AD, Singer-Sam J. Real-time PCR assay for quantitative mismatch detection. Biotechniques. 2003;34:498–502. doi: 10.2144/03343st01. [DOI] [PubMed] [Google Scholar]
- Steinman RM, Banchereau J. Taking dendritic cells into medicine. Nature. 2007;449(7161):419–426. doi: 10.1038/nature06175. [DOI] [PubMed] [Google Scholar]
- Substance Abuse and Mental Health Services Administration SAMSHA) 2007 September [Google Scholar]
- Takayasu T, Ohshima T, Nishigami J, Kondo T, Nagaon T. Screening and determination of methamphetamine and amphetamine in the blood, urine and stomach contents in emergency medical care and autopsy cases. J. Clin. Forens. Med. 1995;2:25–33. doi: 10.1016/1353-1131(95)90036-5. [DOI] [PubMed] [Google Scholar]
- Tallóczy Z, Martinez J, Joset D, Ray Y, Gácser A, Toussi S, Mizushima N, Nosanchuk J, Goldstein H, Loike J, Sulzer D, Santambrogio L. Methamphetamine inhibits antigen processing, presentation, and phagocytosis. PLoS Pathog. 2008;4(2):e28. doi: 10.1371/journal.ppat.0040028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thein HH, Yi Q, Dore GJ, Krahn MD. Natural history of hepatitis C virus infection in HIV-infected individuals and the impact of HIV in the era of highly active antiretroviral therapy: a meta-analysis. AIDS. 2008;22(15):1979–1991. doi: 10.1097/QAD.0b013e32830e6d51. [DOI] [PubMed] [Google Scholar]
- Tscherne DM, Jones CT, Evans MJ, Lindenbach BD, McKeating JA, Rice CM. Time- and temperature-dependent activation of hepatitis C virus for low-pH-triggered entry. J. Virol. 2006;80(4):1734–1741. doi: 10.1128/JVI.80.4.1734-1741.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UNODC. http://www.unodc.org/unodc/en/data-and-analysis/WDR-2007.html. [Google Scholar]
- Van Dyke C, Stesin A, Jones R, Chuntharapai A, Seaman W. Cocaine increases natural killer cell activity. J. Clin. Invest. 1986;77(4):1387–1390. doi: 10.1172/JCI112445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wainberg Z, Oliveira M, Lerner S, Tao Y, Brenner BG. Modulation of stress protein (hsp27 and hsp70) expression in CD4+ lymphocytic cells following acute infection with human immunodeficiency virus type-1. Virology. 1997;233:364–373. doi: 10.1006/viro.1997.8618. [DOI] [PubMed] [Google Scholar]
- Wessel D, Flugge UI. Method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 1984;138:141–143. doi: 10.1016/0003-2697(84)90782-6. [DOI] [PubMed] [Google Scholar]
- Wilm M, Shevchenko A, Houthaeve T, Breit S, Schweigerer L, Fotsis T, Mann M. Femtomole sequencing of proteins from polyacrylamide gels by nanoelectrospray mass spectrometry. Nature. 1996;379:466–469. doi: 10.1038/379466a0. [DOI] [PubMed] [Google Scholar]
- Wolffe AP. Structural and functional properties of the evolutionarily ancient Y-box family of nucleic acid binding proteins. Bioessays. 1994;6:245–251. doi: 10.1002/bies.950160407. [DOI] [PubMed] [Google Scholar]
- Ye L, Peng JS, Wang X, Wang YJ, Luo GX, Ho WZ. Methamphetamine enhances Hepatitis C virus replication in human hepatocytes. J. Viral. Hepat. 2008;15(4):261–270. doi: 10.1111/j.1365-2893.2007.00940.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu HJ, Reuter MA, McDonald D. HIV traffics through a specialized, surface-accessible intracellular compartment during trans-infection of T cells by mature dendritic cells. PLoS Pathog. 2008;4:100–134. doi: 10.1371/journal.ppat.1000134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao LJ, Wang L, Ren H, Cao J, Li L, Ke JS, Qi ZT. Hepatitis C virus E2 protein promotes human hepatoma cell proliferation through the MAPK/ERK signaling pathway via cellular receptors. Exp. Cell Res. 2005;305(1):23–32. doi: 10.1016/j.yexcr.2004.12.024. [DOI] [PubMed] [Google Scholar]
- Zhao LJ, Zhang XL, Zhao P, Cao J, Cao MM, Zhu SY, Liu HQ, Qi ZT. Up-regulation of ERK and p38 MAPK signaling pathways by hepatitis C virus E2 envelope protein in human T lymphoma cell line. J. Leukoc. Biol. 2006;80(2):424–432. doi: 10.1189/jlb.0106014. [DOI] [PubMed] [Google Scholar]
- Zhao LJ, Zhao P, Chen QL, Ren H, Pan W, Qi ZT. Mitogen-activated protein kinase signalling pathways triggered by the hepatitis C virus envelope protein E2: implications for the prevention of infection. Cell Prolif. 2007;40(4):508–521. doi: 10.1111/j.1365-2184.2007.00453.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolnierowicz S. Type 2A protein phosphatase, the complex regulator of numerous signaling pathways. Biochem. Pharmacol. 2000;60:1225–1235. doi: 10.1016/s0006-2952(00)00424-x. [DOI] [PubMed] [Google Scholar]