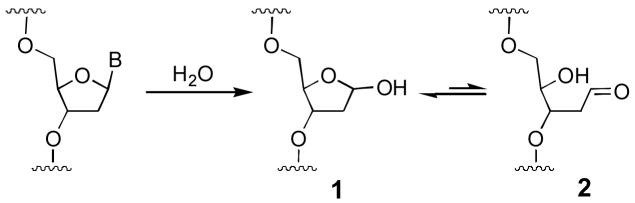
Abasic sites (1, Scheme 1), which are generated by hydrolysis of the glycosidic bonds connecting the heterocyclic nucleobases to the deoxyribose backbone, represent the most common type of damage suffered by genomic DNA.1 Spontaneous hydrolysis yields approximately 10,000 abasic sites per cell per day. In addition, exposure of DNA to radiation, mutagens, and anticancer drugs can lead to the generation of abasic sites.2 Unrepaired abasic sites are mutagenic or cytotoxic.1
Scheme 1.
Interestingly, an early report suggested that abasic sites can generate interstrand crosslinks in duplex DNA.3 Subsequent studies confirmed this finding but the structure of these lesions has remained unclear.4 This is a striking observation because interstrand DNA crosslinks have profound biological consequences.5 For example, a single unrepaired interstrand crosslink can be sufficient to kill a eukaryotic cell.6 The widespread occurrence of abasic sites in cellular DNA along with the potent biological activities associated with crosslinks make it important to characterize interstrand crosslinks derived from abasic sites in duplex DNA.
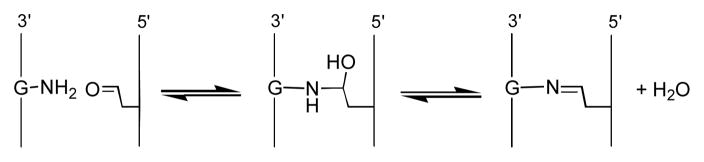
Abasic sites in duplex DNA consist of an equilibrating mixture of the ring-closed acetal (99%) and the ring-opened aldehyde (1%, Scheme 1).7 When considering the possible chemical structure(s) of abasic site-derived crosslinks, it is important to note that aldehydes can covalently modify DNA, with a preference for reactions involving the exocyclic N2-amino group of guanine residues.8 Furthermore, recent studies have shown that an aldehyde residue tethered in the minor groove of duplex DNA can generate an interstrand crosslink via carbinolamine/imine formation with the exocyclic N2-amino group of a guanine residue on the opposite strand of the double helix (Scheme 2).8c,9 In some cases, reduction of the initially-formed imine with NaCNBH3 can stabilize these crosslinks. This type of reductive amination is favored at pH ~ 5.10
Scheme 2.
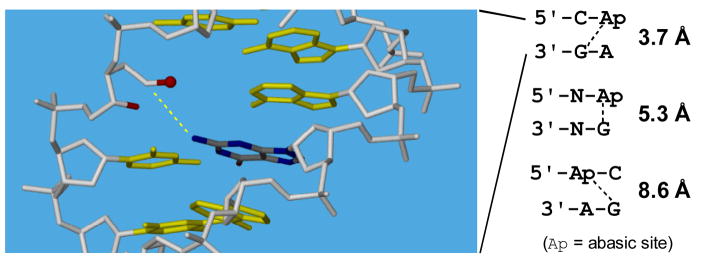
Based upon these literature precedents, we used molecular modeling to predict B-DNA sequences that might be preferred sites for interstrand crosslink formation involving reaction of the abasic aldehyde 2 with the exocyclic N2-amino group of an opposing guanine residue. The modeling results summarized in Figure 1 inspired us to investigate formation of interstrand DNA crosslinks in the context of duplexes A and B (Scheme 3) which were expected to place the reacting groups in close proximity.
Figure 1.
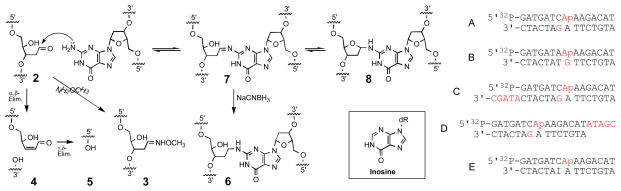
Scheme 3.
DNA duplexes containing abasic sites were prepared by treatment of 2′-deoxyuridine-containing duplexes with uracil deglycosylase.11 This enzyme hydrolytically removes uracil from the DNA backbone to generate the desired abasic site. To facilitate detection of interstrand DNA crosslinks, the abasic strand in these duplexes was 32P-labeled on the 5′-end. In the event, incubation of duplex A in MES buffer (pH 5.5) in the presence of NaCNBH3, followed by 20% denaturing polyacrylamide gel electrophoresis, reveals a substantial yield (3 ± 0.3%) of a slow-moving band in the region of the gel where interstrand crosslinks are expected5 to migrate (Figure 2). Consistent with the anticipated involvement of the abasic aldehyde residue in formation of the slow-moving band, addition of methoxyamine (20 mM) inhibits formation of this species. Methoxyamine “caps” abasic sites through conversion of the aldehyde group to the oxime derivative (3, Scheme 3).12 Importantly, when the reaction is carried out at pH 7 rather than 5.5, the slow-moving crosslink band is still observed in significant yield (~1%).
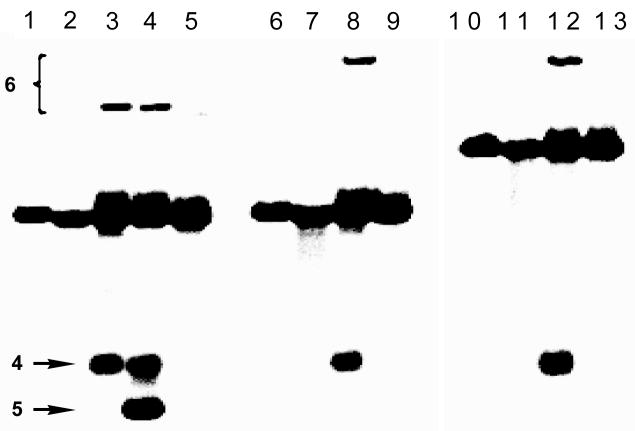
Figure 2.
Interstrand crosslinking of duplexes A, C, and D. In a typical crosslinking reaction, the abasic-site-containing DNA duplex was incubated in MES buffer (50 mM, pH 5.5) for 7 d in the presence of NaCNBH3 (270 mM) at 30 C. The sample was lyophilized, dissolved in formamide loading buffer, the DNA fragments resolved on a 20% denaturing polyacrylamide gel, and visualized by phosphorimaging. Lanes: 1, 6, and 10, uracil-containing precursors to duplexes A, C, and D, respectively (as size markers); lanes 2–5, duplex A; 7–9, duplex C; 11–13, duplex D. Lanes: 2, 7, 11, freshly prepared abasic duplex (without subsequent incubation); 3, 8, 12, standard crosslinking reaction; 4, standard crosslinking reaction+piperidine workup; 5, 9, 13, standard crosslinking reaction+methoxyamine (CH3ONH2). In lane 4, the non-piperidine-labile band co-migrating with 4 likely consists of the NaCNBH3-reduced, alcohol analog of 4.
Extension of the strand opposing the abasic oligonucleotide (duplex C) retards migration of the slow-moving band (lane 8, Figure 2), indicating that this band contains both strands of the starting duplex. Extension of the 3′-end of the abasic oligonucleotide (duplex D) similarly retards migration of the slow moving band, indicating that the slow-moving band contains the full length abasic oligo and not the truncated strands (4 or 5) stemming from elimination of the oligonucleotide fragment to the 3′-side of the abasic site (Scheme 3). Overall these results indicate that the slow-moving bands observed in Figure 2 are abasic-aldehyde-derived interstrand crosslinks.
Consistent with our expectation that interstrand crosslink formation would involve reaction of the abasic site with an opposing guanine residue, crosslinking is strongly dependent upon the location of this base. For example, a slow-moving crosslink band is not generated by duplex B, in which the guanine lies directly across from the abasic site (Figure 3). Evidence implicating the involvement of the guanine N2-amino group in the crosslinking reaction is provided by the observation that replacement of the key guanosine in duplex A with inosine (to yield duplex E) completely abrogates crosslink formation (Figure 3). This base substitution amounts to the targeted deletion of a single guanine N2-amino group from duplex A (for the structure of inosine, see inset Scheme 3). Finally, negative ion nanospray mass spectrometric analysis of the slow-moving band derived from the incubation of duplex A in the presence of NaCNBH3 reveals an m/z of 9018.2 ± 0.4 consistent with that expected (m/z 9018.0) for the crosslink structure 6, stemming from hydride reduction of the parent crosslink 7/8 (Scheme 3).
Figure 3.
Evidence for the role of the opposing guanine residue in the crosslinking reaction. Crosslinking reactions were conducted as described in the legend of Figure 2. Lanes 1, 5, 9, uracil-containing precursors to duplexes A, B, and E, respectively (as size markers). Lanes: 2–4, duplex A; 6–8, duplex B; 10–12, duplex E. Lanes: 2, 6, 10 freshly prepared abasic duplex (without subsequent incubation); 3, 7, 11, standard crosslinking conditions; 4, 8, 12, standard crosslinking conditions+CH3ONH2. The bands marked “4” likely consist of a mixture of 4 the NaCNBH3-reduced, alcohol analog of 4.
In summary, the results provide evidence that the most common type of cellular DNA damage, the abasic site, has the potential to generate one of the most biologically deleterious lesions, an interstrand DNA crosslink, via reaction of the abasic aldehyde group with the exocyclic N2-amino group of an opposing guanine residue in 5′-d(CAp) sequences. Further work is required to determine how these crosslinks contribute to the chemistry and biology of abasic sites in duplex DNA.
Supplementary Material
Acknowledgments
We are grateful to the National Institutes of Health for support of this work (CA 83925) and Dr. Beverly DaGue for mass spectrometry.
Footnotes
Supporting Information Available: Experimental procedures for all reactions and analyses. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Evans AR, Limp-Foster M, Kelley MR. Mutation Res. 2000;461:83–108. doi: 10.1016/s0921-8777(00)00046-x. [DOI] [PubMed] [Google Scholar]; Lhomme J, Constant JF, Demeunynck M. Biopolymers. 2000;52:65–83. doi: 10.1002/1097-0282(1999)52:2<65::AID-BIP1>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]; Loeb LA, Preston BD. Ann Rev Genet. 1986;20:201–230. doi: 10.1146/annurev.ge.20.120186.001221. [DOI] [PubMed] [Google Scholar]; Boiteux S, Guillet M. DNA Repair. 2004;3:1–12. doi: 10.1016/j.dnarep.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 2.For examples of abasic sites stemming from DNA alkylation: see: Gates KS, Nooner T, Dutta S. Chem Res Toxicol. 2004;17:839–856. doi: 10.1021/tx049965c.Nooner T, Dutta S, Gates KS. Chem Res Toxicol. 2004;17:942–949. doi: 10.1021/tx049964k.
- 3.Freese E, Cashel M. Biochim Biophys Acta. 1964;91:67–77. doi: 10.1016/0926-6550(64)90171-9. [DOI] [PubMed] [Google Scholar]
- 4.Burnotte J, Verly WG. Biochim Biophys Acta. 1972;262:449–452. doi: 10.1016/0005-2787(72)90488-1. [DOI] [PubMed] [Google Scholar]; Goffin C, Verly WG. FEBS Lett. 1983;161:140–144. doi: 10.1016/0014-5793(83)80747-9. [DOI] [PubMed] [Google Scholar]; Goffin C, Verly WG. Biochim Biophys Acta. 1984;783:1–5. doi: 10.1016/0167-4781(84)90071-x. [DOI] [PubMed] [Google Scholar]; Mittal A, Musarrat J. Med Sci Res. 1990;18:633–635. [Google Scholar]; Prakash AS, Gibson NW. Carcinogenesis. 1992;13:425–431. doi: 10.1093/carcin/13.3.425. [DOI] [PubMed] [Google Scholar]
- 5.Noll DM, Mason TM, Miller PS. Chem Rev. 2006;106:277–301. doi: 10.1021/cr040478b. [DOI] [PMC free article] [PubMed] [Google Scholar]; Schärer OD. ChemBioChem. 2005;6:27–32. doi: 10.1002/cbic.200400287. [DOI] [PubMed] [Google Scholar]; Rajski SR, Williams RM. Chem Rev. 1998;98:2723–2795. doi: 10.1021/cr9800199. [DOI] [PubMed] [Google Scholar]
- 6.Grossman KF, Ward AM, Matkovic ME, Folias AE, Moses RE. Mutation Res. 2001;487:73–83. doi: 10.1016/s0921-8777(01)00106-9. [DOI] [PubMed] [Google Scholar]; Reddy M, Vasquez KM. Radiation Res. 2005;164:345–356. doi: 10.1667/rr3419.1. [DOI] [PubMed] [Google Scholar]
- 7.Wilde JA, Bolton PH, Mazumdar A, Manoharan M, Gerlt JA. J Am Chem Soc. 1989;111:1894–1896. [Google Scholar]
- 8.(a) Riggins JN, Daniels JS, Rouzer CA, Marnett LJ. J Am Chem Soc. 2004;126:8237–8243. doi: 10.1021/ja040009r. [DOI] [PubMed] [Google Scholar]; (b) Wang M, McIntee EJ, Cheng G, Shi Y, Villalta PW, Hecht SS. Chem Res Toxicol. 2000;13:1149–1157. doi: 10.1021/tx000118t. [DOI] [PubMed] [Google Scholar]; (c) Chaw YFM, Crane LE, Lange P, Shapiro R. Biochemistry. 1980;19:5525–5531. doi: 10.1021/bi00565a010. [DOI] [PubMed] [Google Scholar]
- 9.Kozekov ID, Nechev LV, Moseley MS, Harris CM, Rizzo CJ, Stone MP, Harris TM. J Am Chem Soc. 2003;125:50–61. doi: 10.1021/ja020778f. [DOI] [PubMed] [Google Scholar]
- 10.For examples involving interstrand crosslinking of non-natural DNA via reductive amination, see: Manoharan M, Andrade LK, Cook PD. Org Lett. 1999;1:311–314. doi: 10.1021/ol9906209.Dohno C, Okamoto A, Saito I. J Am Chem Soc. 2005;127:16681–16684. doi: 10.1021/ja054618q.
- 11.Lindahl T, Ljunquist S, Siegert W, Nyberg B, Sperens B. J Biol Chem. 1977;252:3286–3294. [PubMed] [Google Scholar]
- 12.Talpaert-Borle M, Liuzzi M. Biochim Biophys Acta. 1983;740:410–16. doi: 10.1016/0167-4781(83)90089-1. [DOI] [PubMed] [Google Scholar]; Rosa S, Fortini P, Karran P, Bignami M, Dogliotti E. Nucleic Acids Res. 1991;19:5569–5574. doi: 10.1093/nar/19.20.5569. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.