Abstract
The virulence of the opportunistic pathogen Pseudomonas aeruginosa involves the coordinate expression of many virulence factors, including type IV pili, which are required for colonization of host tissues and for twitching motility. Type IV pilus function is controlled in part by the Chp chemosensory system, which includes a histidine kinase, ChpA, and two CheY-like response regulators, PilG and PilH. How the Chp components interface with the type IV pilus motor proteins PilB, PilT, and PilU is unknown. We present genetic evidence confirming the role of ChpA, PilG, and PilB in the regulation of pilus extension and the role of PilH and PilT in regulating pilus retraction. Using informative double and triple mutants, we show that (i) ChpA, PilG, and PilB function upstream of PilH, PilT, and PilU; (ii) that PilH enhances PilT function; and (iii) that PilT and PilB retain some activity in the absence of signaling input from components of the Chp system. By site-directed mutagenesis, we demonstrate that the histidine kinase domain of ChpA and the phosphoacceptor sites of both PilG and PilH are required for type IV pilus function, suggesting that they form a phosphorelay system important in the regulation of pilus extension and retraction. Finally, we present evidence suggesting that pilA transcription is regulated by intracellular PilA levels. We show that PilA is a negative regulator of pilA transcription in P. aeruginosa and that the Chp system functionally regulates pilA transcription by controlling PilA import and export.
Pseudomonas aeruginosa is a ubiquitous environmental Gram-negative bacterium that is an opportunistic human pathogen and that has been shown to infect a diverse array of eukaryotes including amoebae, nematodes, insects, plants, and mice (9, 11, 28, 49). P. aeruginosa is the causative agent of both acute and chronic human infections, ranging from minor skin infections to persistent and often life-threatening disease in hospitalized or immunocompromised patients such as those suffering from AIDS, extensive burns, who are undergoing chemotherapy, or those who are recovering from major surgery (39). P. aeruginosa also chronically infects the cystic fibrosis lung and is the primary cause of respiratory morbidity and mortality in patients with this inherited disease (39).
P. aeruginosa virulence involves the coordinate expression of a wide range of secreted and cell-associated virulence factors (17). Key among these virulence factors are the type IV pili (Tfp). Tfp are polarly localized filamentous appendages synthesized by a variety of Gram-negative bacteria, including both pathogens and environmental species such as Myxococcus xanthus (40). Tfp constitute the major adhesin of P. aeruginosa and have been shown to play a role in adherence to epithelial cells in culture and in virulence in several animal models of infection (17). Tfp also function as receptors for certain bacteriophages (6) and allow for a type of flagellum-independent surface translocation termed twitching motility (TM). TM is propelled by the coordinated extension, tethering, and retraction of Tfp (41, 47) and has been shown to be involved in complex social behaviors, such as fruiting body development in M. xanthus (33, 34, 59) and biofilm formation in P. aeruginosa (3, 7, 35, 36, 43). P. aeruginosa biofilms have been implicated in chronic infections (10).
The biogenesis, assembly, and function of Tfp requires more than 40 genes (40). Regulation of Tfp function is complex and involves multiple signal transduction systems, including the two-component signaling systems (pilR/pilS and algR/fimS), the global carbon metabolism regulator crc, the virulence factor regulator vfr, and a putative chemosensory system encoded by the pilGHIJK-chpABC gene cluster and referred to as the Chp system. The proteins encoded by these genes appear to comprise a chemosensory signal transduction pathway similar to the Che system involved in the regulation of flagellar chemotaxis in Escherichia coli (2, 51). As is the case with a growing number of bacterial chemotaxis systems (33, 44), regulation of Tfp function by the Chp system appears to be considerably more complex than its E. coli counterpart.
The core signaling components of the Chp system include a putative histidine kinase, encoded by chpA (55) and two CheY-like response regulators, encoded by pilG and pilH (12, 13). Previous studies have suggested that chpA (55) and pilG (12) are involved in regulating pilus extension, whereas pilH (13) is involved in regulating pilus retraction. Similarly, the ATPases encoded by pilB and pilT (27) are postulated to be involved in mediating pilus extension and retraction, respectively. The role of the ATPase encoded by pilU is unclear (8). However, due to a high degree of similarity with pilT and in vitro ATPase activity, it has been hypothesized that pilU may play some role in regulating pilus retraction (8, 56).
We used genetic approaches to test the hypothesis that phosphotransfer from ChpA to PilG and PilH is important for Tfp function and that PilG and PilH regulate pilus extension and retraction through PilB and PilT, respectively (Fig. 1). We demonstrate that ChpA, PilG, and PilB function upstream of PilH, PilT, and PilU and that PilH enhances PilT activity. We provide evidence that PilB and PilT retain some activity in the absence of upstream signaling input from ChpA and PilG, and PilH, respectively. We demonstrate that the histidine kinase domain of ChpA and the phosphoacceptor sites of both PilG and PilH are required for Tfp function. Finally, we present evidence suggesting that pilA transcription is regulated by intracellular PilA levels. We show that PilA is a negative regulator of pilA transcription in P. aeruginosa and that the Chp system functionally regulates pilA transcription by controlling PilA import and export.
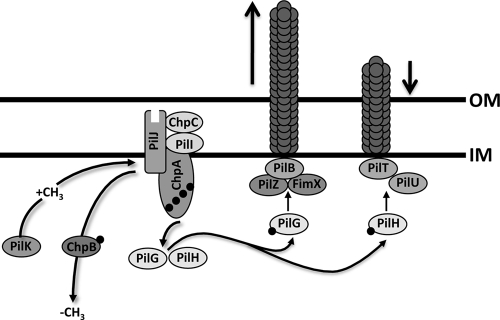
FIG. 1.
Model for the regulation of type IV pilus function by the Chp chemosensory system. The hybrid histidine kinase ChpA, likely associated with the inner membrane, is coupled to a methyl-accepting chemotaxis protein receptor, PilJ, by one of two CheW adaptor protein homologues, PilI and ChpC. Upon receipt of a yet-to-be-elucidated signal, PilJ undergoes a conformational change causing ChpA to autophosphorylate. Phosphate groups are transferred from ChpA to two CheY-like response regulator proteins, PilG and PilH. PilG-P interacts with a motor complex including PilZ, the diguanylate cyclase FimX, and ATPase PilB to mediate pilus extension. PilH-P interacts with ATPases PilT and/or PilU to mediate pilus retraction. Adaptation to the chemical signal is mediated through methylation of PilJ by the competing activities of the methyltransferase PilK and the methylesterase PilB.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
Escherichia coli strain DH5α (recA endA gyr96 hsdR17 thi-1 supE44 relA1 φ80dlacZΔM15) was used in all genetic manipulations and in the preparation of DNA sequencing templates. E. coli strains S17.1 (thi pro hsdR recA chr::RP4-2) and SM10 (thi thr leu tonA lacY supE recA::RP4-2-Tc::Mu Km λpir) were used as donor strains in bacterial conjugation. The P. aeruginosa strain used was PAO1 strain ATCC 15692 (American Type Culture Collection). Chemically competent E. coli cells (Invitrogen) were transformed according to the manufacturer's instructions. The bacterial strains and plasmids used in the present study are described in Table 1. E. coli and P. aeruginosa liquid cultures were maintained in Luria-Bertani (LB) broth, and solid medium was prepared by adding 0.8 to 1.5% agar (Bacto agar; Becton Dickinson). After mating with E. coli, P. aeruginosa strains were selected by growth on 1.5% Difco Pseudomonas isolation agar (Becton Dickinson). The following antibiotic concentrations were used for the selection of E. coli: tetracycline, 5 μg/ml; ampicillin, 100 μg/ml; gentamicin, 10 μg/ml; and kanamycin, 50 μg/ml. The following antibiotic concentrations were used for the selection of P. aeruginosa: tetracycline, 100 μg/ml; carbenicillin, 250 μg/ml; and gentamicin, 100 μg/ml.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype and relevant characteristicsa | Source or reference |
|---|---|---|
| Strains | ||
| P. aeruginosa | ||
| PAO1 | Wild type | |
| PAO1ΔchpA | In-frame deletion of chpA | 61 |
| PAO1ΔchpA(comp) | PAO1ΔchpA complemented at the chpA locus | This study |
| PAO1ΔfliC | In-frame deletion of fliC | This study |
| PAO1ΔfliC(comp) | PAO1ΔfliC complemented at the fliC locus | This study |
| PAO1ΔpilA | In-frame deletion of pilA | This study |
| PAO1ΔpilA(comp) | PAO1ΔpilA complemented at the pilA locus | This study |
| PAO1ΔpilB | In-frame deletion of pilB | This study |
| PAO1ΔpilA(comp) | PAO1ΔpilB complemented at the pilB locus | This study |
| PAO1ΔpilG | In-frame deletion of pilG | This study |
| PAO1ΔpilG(comp) | PAO1ΔpilG complemented at the pilG locus | This study |
| PAO1ΔpilH | In-frame deletion of pilH | 3 |
| PAO1ΔpilH(comp) | PAO1ΔpilH complemented at the pilH locus | This study |
| PAO1ΔpilR | In-frame deletion of pilR | This study |
| PAO1ΔpilR(comp) | PAO1ΔpilR complemented at the pilR locus | This study |
| PAO1ΔpilTCTX-pilU | In-frame deletion of pilT; pilU and 1 kb upstream sequence at attB site | This study |
| PAO1pilTTrunc | pilT truncation mutant, deletes 429 bp of pilT including the start codon; stop codons in all three reading frames | This study |
| PAO1ΔpilT(comp) | PAO1ΔpilT complemented at the pilT locus | This study |
| PAO1ΔpilU | In-frame deletion of pilU | This study |
| PAO1ΔpilU(comp) | PAO1ΔpilU complemented at the pilU locus | This study |
| PAO1ΔchpAΔpilH | In-frame deletion of chpA and pilH | This study |
| PAO1ΔchpΔpilTCTX-pilU | In-frame deletion of chpA and pilT; pilU and 1 kb upstream sequence at attB site | This study |
| PAO1ΔchpAΔpilU | In-frame deletion of chpA and pilU | This study |
| PAO1ΔchpAΔpilTΔpilU | In-frame deletion of chpA, pilT, and pilU | This study |
| PAO1ΔpilBΔpilH | In-frame deletion of pilB and pilH | This study |
| PAO1ΔpilBΔpilTCTX-pilU | In-frame deletion of pilB and pilT; pilU and 1 kb upstream sequence at attB site | This study |
| PAO1ΔpilBΔpilU | In-frame deletion of pilB and pilU | This study |
| PAO1ΔpilBΔpilTΔpilU | In-frame deletion of pilB, pilT, and pilU | This study |
| PAO1ΔpilGΔpilH | In-frame deletion of pilG and pilH | This study |
| PAO1ΔpilGΔpilT-gent | PAO1ΔpilG with a Gmr-tagged insertion in pilT | This study |
| PAO1ΔpilGΔpilTCTX-pilU | In-frame deletion of pilG and pilT; pilU and 1 kb upstream sequence at attB site | This study |
| PAO1ΔpilGΔpilU-gent | PAO1ΔpilG with a Gmr-tagged insertion in pilU | This study |
| PAO1ΔpilGΔpilU | In-frame deletion of pilG and pilU | This study |
| PAO1ΔpilGΔpilTΔpilU | In-frame deletion of pilG, pilT, and pilU | This study |
| PAO1ΔpilHΔpilTCTX-pilU | In-frame deletion of pilH and pilT; pilU and 1 kb upstream sequence at attB site | This study |
| PAO1ΔpilTΔpilU | In-frame deletion of pilT and pilU | This study |
| PAO1ΔpilHΔpilU | In-frame deletion of pilH and pilU | This study |
| PAO1ΔpilHΔpilTΔpilU | In-frame deletion of pilH, pilT, and pilU | This study |
| PAO1chpA-FLAG | chpA with a double FLAG tag inserted into the NotI/KpnI sites of chpA at the chpA locus | This study |
| PAO1chpA(AAA)-FLAG | chpA-FLAG with ChpA D2091A, D2092A, and G2093A | This study |
| PAO1pilG-His | pilG with a C-terminal His6 tag at the pilG locus | This study |
| PAO1pilG(D58A)-His | pilG-His with PilG D58A | This study |
| PAO1pilH-His | pilH with a C-terminal His6 tag at the pilH locus | This study |
| PAO1pilH(D52A)-His | pilH-His with PilH D52A | This study |
| PAO1::CTX-PpilA-lacZ | PAO1; pilA promoter fused to lacZ at attB site | |
| PAO1ΔchpA::CTX-PpilA-lacZ | ΔchpA; pilA promoter fused to lacZ at attB site | This study |
| PAO1ΔchpA(comp)::CTX-PpilA-lacZ | ΔchpA(comp); pilA promoter fused to lacZ at attB site | This study |
| PAO1ΔfliC::CTX-PpilA-lacZ | ΔfliC; pilA promoter fused to lacZ at attB site | This study |
| PAO1ΔfliC(comp)::CTX-PpilA-lacZ | ΔfliC(comp); pilA promoter fused to lacZ at attB site | This study |
| PAO1ΔpilA::CTX-PpilA-lacZ | ΔpilA; pilA promoter fused to lacZ at attB site | This study |
| PAO1ΔpilA(comp)::CTX-PpilA-lacZ | ΔpilA(comp); pilA promoter fused to lacZ at attB site | This study |
| PAO1ΔpilB::CTX-PpilA-lacZ | ΔpilB; pilA promoter fused to lacZ at attB site | This study |
| PAO1ΔpilA(comp)::CTX-PpilA-lacZ | ΔpilB(comp); pilA promoter fused to lacZ at attB site | This study |
| PAO1ΔpilG::CTX-PpilA-lacZ | ΔpilG; pilA promoter fused to lacZ at attB site | This study |
| PAO1ΔpilG(comp)::CTX-PpilA-lacZ | ΔpilG(comp); pilA promoter fused to lacZ at attB site | This study |
| PAO1ΔpilH::CTX-PpilA-lacZ | ΔpilH; pilA promoter fused to lacZ at attB site | This study |
| PAO1ΔpilH(comp)::CTX-PpilA-lacZ | ΔpilH(comp); pilA promoter fused to lacZ at attB site | This study |
| PAO1ΔpilR::CTX-PpilA-lacZ | ΔpilR; pilA promoter fused to lacZ at attB site | This study |
| PAO1ΔpilR(comp)::CTX-PpilA-lacZ | ΔpilR(comp); pilA promoter fused to lacZ at attB site | This study |
| PAO1pilTTrunc::CTX-PpilA-lacZ | pilT-Trunc; pilA promoter fused to lacZ at attB site | This study |
| PAO1ΔpilT(comp)::CTX-PpilA-lacZ | ΔpilT(comp); pilA promoter fused to lacZ at attB site | This study |
| PAO1ΔpilU::CTX-PpilA-lacZ | ΔpilU; pilA promoter fused to lacZ at attB site | This study |
| PAO1ΔpilU(comp)::CTX-PpilA-lacZ | ΔpilU(comp); pilA promoter fused to lacZ at attB site | This study |
| E. coli | ||
| DH5α | hsdR rec lacZYA φ80 lacZΔM15 | Invitrogen |
| S17.1λpir | Thi pro hsdR recA RP4-2(Tc::Mu)(Km::Tn7) | Stratagene |
| SM10 | thi thr leu tonA lacY supE recA::RP4-2-Tc::Mu Km λpir | 15 |
| Plasmids | ||
| pOK12 | E. coli cloning vector; Kmr | 54 |
| pUCPSK | P. aeruginosa-E. coli shuttle vector; Apr | 56 |
| pGEM-T | E. coli cloning vector; Apr | Promega |
| pEX100T | Allelic replacement suicide plasmid; Apr (Cbr) | 49 |
| pX1918GT | Source of Gmr cassette; Apr | 49 |
| mCTX-lacZ | Contains promoterless lacZ for constructing transcriptional fusions at the attB site; Tcr | 4 |
| mCTX2 | Construct for introduction of exogenous DNA fragments at the attB site | 24 |
| pFLP2 | Source of Flp recombinase; Apr | 23 |
| pAL64 | pUCPSK carrying chpA with an internal double FLAG tag inserted between the native NotI/NruI sites | 39 |
| pEN34 | Allelic replacement suicide plasmid; Tcr | 61 |
| pJEN34PAO1KO3 | pJEN34 carrying ΔpilA on SpeI fragment | This study |
| pJB4 | pOK12 carrying ΔchpA on EcoRI/XbaI fragment | 61 |
| pJB43 | pGEM-T containing 1-kb region 5′ of pilG | This study |
| pJB44 | pGEM-T containing 1-kb region 3′ of pilG | This study |
| pJB48 | pGEM-T containing 1-kb region 3′ of pilH | 3 |
| pJB60 | XhoI/HindIII fragment from pJB43 and HindIII/XbaI from pJB44 concatamerized and cloned into XhoI/XbaI sites of pOK12 | This study |
| pJB62 | pOK12 carrying ΔpilH on KpnI/XbaI fragment | 3 |
| pJB87 | pJEN34 carrying ΔpilG on SpeI fragment | This study |
| pJB96 | XhoI/HindIII fragment from pJB43 and HindIII/XbaI from pJB48 concatamerized and cloned into XhoI/XbaI sites of pOK12 | This study |
| pJB97 | pJEN34 carrying ΔpilGΔpilH on SpeI fragment | This study |
| pJB100 | pEX100T derivative replacing SmaI site with the SpeI site | |
| pJB109 | pJB100 carrying ΔpilT on SpeI fragment | This study |
| pJB110 | Gmr-tagged ΔpilT in pJB100; pX1918-derived Gmr cassette cloned as HindIII fragment into pJB109 | This study |
| pJB111 | pJB100 carrying ΔpilU on SpeI fragment | This study |
| pJB112 | Gmr-tagged ΔpilU in pJB100; pX1918-derived Gmr-cassette cloned as HindIII fragment into pJB109 | This study |
| pJB113 | pJB100 carrying ΔpilT complementation construct on SpeI fragment | This study |
| pJB114 | pJB100 carrying ΔpilU complementation construct on SpeI fragment | This study |
| pJB116 | pJB100 carrying ΔchpA on SpeI fragment; subcloned from pJB4 | This study |
| pJB118 | pJB100 carrying ΔpilG on SpeI fragment; subcloned from pJB60 | This study |
| pJB119 | pJB100 carrying ΔpilH on SpeI fragment; subcloned from pJB62 | This study |
| pJB122 | pJB100 carrying ΔpilB on SpeI fragment | This study |
| pJB124 | pJB100 carrying pilH-His allelic exchange construct on SpeI fragment | This study |
| pJB126 | pJB100 carrying ΔpilG complementation construct on SpeI fragment | This study |
| pJB127 | pJB100 carrying pilG-His allelic exchange construct on SpeI fragment | This study |
| pJB163 | pJB100 carrying ΔpilTΔpilU on the SpeI fragment | This study |
| pJB202 | FLAG-tagged chpA histidine kinase domain in pOK12; subcloned as KpnI/ClaI fragment from pAL64 | This study |
| pJB203 | pJB100 carrying a pilT truncation construct on SpeI fragment | This study |
| pJB207 | chpA(AAA) histidine kinase domain in pOK12 | This study |
| pJB212 | pilU and 1-kb 5′ sequence cloned as SpeI/HindIII fragment into mCTX2 | This study |
| pJB215 | pJB100 carrying ΔfliC on SpeI fragment | This study |
| pJB219 | Modified pOK12 lacking KpnI and ClaI sites; BglII/SacI fragment removed from pOK12 MCS | This study |
| pJB220 | 3′ chpA fragment including 1-kb 3′ flanking sequence in pJB219 | This study |
| pJB221 | chpA including 1 kb of 5′ and 3′ flanking sequence in pJB219 | This study |
| pJB230 | pJB100 carrying ΔfliC complementation construct on SpeI fragment | This study |
| pJB231 | pJB100 carrying ΔchpA complementation construct on SpeI fragment; subcloned from pJB221 | This study |
| pJB232 | chpA-FLAG, including 1 kb of 5′ and 3′ flanking sequence in pJB219; KpnI/ClaI fragment from pJB202 subcloned into pJB221 from which the corresponding KpnI/ClaI fragment had been removed | This study |
| pJB233 | chpA(AAA)-FLAG including 1 kb of 5′ and 3′ flanking sequence in pJB219; KpnI/ClaI fragment from pJB207 subcloned into pJB221 from which the corresponding KpnI/ClaI fragment had been removed | This study |
| pJB234 | pJB100 carrying chpA-FLAG allelic exchange on SpeI fragment; subcloned from pJB232 | This study |
| pJB235 | pJB100 carrying chpA(AAA)-FLAG allelic exchange on SpeI fragment; subcloned from pJB233 | This study |
| pJB241 | pJB100 carrying ΔpilA complementation construct on SpeI fragment | This study |
| pJB246 | pJB100 carrying ΔpilB complementation construct on SpeI fragment | This study |
| pJB247 | pOK12 carrying pilG-His allelic-exchange construct on SpeI fragment | This study |
| pJB248 | pOK12 carrying pilH-His allelic-exchange construct on SpeI fragment | This study |
| pJB249 | pilA reporter construct cloned as XhoI/BamHI fragment into mCTX-lacZ | This study |
| pJB250 | pJB100 carrying ΔpilR on SpeI fragment | This study |
| pJB251 | pJB100 carrying ΔpilR complementation construct on SpeI fragment | This study |
| pJB252 | pOK12 carrying pilG(D58A)-His allelic-exchange construct on SpeI fragment | This study |
| pJB254 | pOK12 carrying pilH(D582)-His allelic-exchange construct on SpeI fragment | This study |
| pJB256 | pJB100 carrying pilG(D58A)-His allelic-exchange construct on SpeI fragment | This study |
| pJB258 | pJB100 carrying pilH(D582)-His allelic-exchange construct on SpeI fragment | This study |
Cbr, carbenicillin resistance; Tcr, tetracycline resistance; Apr, ampicillin resistance; Gmr, gentamicin resistance; Kmr, kanamycin resistance.
Plasmid construction.
All plasmids were purified by using QIAprep spin miniprep columns (Qiagen). Standard recombinant DNA manipulation techniques were used (45). Enzymes were purchased from New England Biolabs, Inc., and used as recommended by the manufacturer. All oligonucleotides used in the present study were designed based on the PAO1 genome sequence (48) and were synthesized by Qiagen or Elim Biopharmaceuticals, Inc. (Hayward, CA). The oligonucleotide primer sequences used for PCR amplification are listed in Table 2. PCR amplifications were carried out with Pfu Turbo DNA polymerase (Stratagene) or Herculase DNA polymerase (Stratagene), and the products were sequenced to ensure that no mutations were introduced during amplification. Intermediate cloning steps were performed using DH5α (Invitrogen). Plasmids were introduced into chemically competent E. coli strains.
TABLE 2.
Oligonucleotides used in this study
| Primer | Oligonucleotide sequence (5′→3′)a |
|---|---|
| chpA22 | GGACTAGTAGATGGCGAGCGAGATGC |
| chpA25 | GAGCTCCTCGCGCAGCGC |
| chpA26.1 | GGCGCGCAACGGCCATTG |
| chpA29 | GGACTAGTCGCAGCGTGCGGCCAGAT |
| chpA-HisKn-F | CTCCTCACCCTCTCCGCCGCCGCCGCCGGCATCCGCCTC |
| chpA-HisKn-R | GAGGCGGATGCCGGCGGCGGCGGCGGAGAGGGTGAGGAG |
| fliC1 | GGACTAGTACCGTTTCGTGGTCTCGA |
| fliC2 | TTAGCGCAGCAGAAGCTTAAGGGCCATGGTGATTTCCTCCAAAGG |
| fliC3 | ATGGCCCTTAAGCTTCTGCTGCGCTAAGCCCGGGAACGGTCACTC |
| fliC4 | GGACTAGTATTGATGGCGTCACGCAT |
| pilA1 | CCATGGATGCCTAACCTCACCCTTGC |
| pilA1(Spe) | ACTAGTATGCCTAACCTCACCCTTGC |
| pilA2 | GAATTCTAAGGTGATCGAAGGTGGCTTGTTTAG |
| pilA3 | GAATTCCATGAATATCTCCATTGATATG |
| pilA4 | TCTAGATTGATCCGTCCGTCCTGCGG |
| pilA4(Spe) | ACTAGTTTGATCCGTCCGTCCTGCGG |
| pilA-Reporter-F | CGCGGATCCGAATCTCTCCGTTGATTA |
| pilA-Reporter-R | CCGCTCGAGGTGTTGGCGGACCAGCTT |
| pilB1 | GCACTAGTATCAACGAGGGCACCCTGCGC |
| pilB2 | TTAATCCTTGGTAAGCTTGTCGTTCATGGGGAAGGAATCGCAGAAGGG |
| pilB3 | ATGAACGACAAGCTTACCAAGGATTAATCCATGGCGGACAAAGCGTTA |
| pilB4 | GCACTAGTTGGATCGCCATGTTGGGAAAG |
| pilG1 | CGGGGTACCCTCAGCGGCTGGGCCACGCCGCG |
| pilG1(Spe) | GCACTAGTCTCAGCGGCTGGGCCACGCCGCG |
| pilG2 | AAGCTTTTGCTGTTCCATGTTCGCCC |
| pilG3 | AAGCTTAAGGGGCGCATCGTCGGCTC |
| pilG4 | TCTAGAATGAAGGGTTGCAGTGCCGC |
| pilG4(Spe) | GCACTAGTATGAAGGGTTGCAGTGCCGC |
| pilG5 | TCAGTGGTGGTGGTGGTGGTGGGAAACGGCGTCCACCGGGGT |
| pilG6 | ACCCCGGTGGACGCCGTTTCCCACCACCACCACCACCACTGA |
| pilG-D58A-F | AACATCATTTTCGTCGCCATCATGATGCCGCGC |
| pilG-D58A-R | GCGCGGCATCATGATGGCGACGAAAATGATGTT |
| pilH1 | GGTACCCGTTATCGAAGGGCGGGTCC |
| pilH1(Spe) | GCACTAGTCGTTATCGAAGGGCGGGTCC |
| pilH4 | TCTAGAGCTTTCTCGAAGTCGTTGCG |
| pilH4(Spe) | GCACTAGTGCTTTCTCGAAGTCGTTGCG |
| pilH15 | TCAGTGGTGGTGGTGGTGGTGGCCCGCCAGCACCGCATTG |
| pilH16 | CAATGCGGTGCTGGCGGGCCACCACCACCACCACCACTGA |
| pilH-D52A-F | GACGTGGTCCTGATGGCCATCGTCATGCCCGGC |
| pilH-D52A-R | GCCGGGCATGACGATGGCCATCAGGACCACGTC |
| pilR1 | GGACTAGTCTGCAGCGCATGCGCACC |
| pilR2 | TCAGTCGATGCCAAGCTTTCGGCTCATGCGTGCGGCTTCCGTCAG |
| pilR3 | ATGAGCCGAAAGCTTGGCATCGACTGAAAGTGAAAAGGCCTGTCC |
| pilR4 | GGACTAGTATCGTCTCGCCGCTCTAT |
| pilT1 | GGACTAGTTGGCACACGGCTTTCATGG |
| pilT1(new) | GGACTAGTAACTCGCCGGCGATCTTC |
| pilT4 | GGACTAGTTGTAGAGCTGGTACAGCGC |
| pilT5 | TCAGAAGTTTTCAAGCTTAATATCCATGGGACTCCCCAATTACAAGC |
| pilT6 | ATGGATATTAAGCTTGAAAACTTCTGATCCTGGCGCCGATCCGC |
| pilT-Trunc2 | CAGGTAATCGAGTCTAGTCTAGACTAGGGGACTCCCCAATTACAAGCAAGCAGG |
| pilT-Trunc3 | GGGAGTCCCCTAGTCTAGACTAGACTCGATTACCTGAACAACACCAAGTACCAC |
| pilT-Trunc4 | GGACTAGTGAAGTTGCACTCATGGTT |
| pilU1 | GGACTAGTAACACAAGCAGGTGCATGC |
| pilU4 | GGACTAGTAAGGTGAAGAACCAGATCG |
| pilU5 | TCAGCGGAAGCGTCTAGAGAATTCCATGATGTTCTCGCTCACTCAGG |
| pilU6 | ATGGAATTCTCTAGACGCTTCCGCTGAACCGTCTTCAGTAGGCCAGC |
| pilTpilU1 | TCAGCGGAAGCGAATATCCATGGGACTCCCCAATTACAAGC |
| pilTpilU2 | ATGGATATTCGCTTCCGCTGAACCGTCTTCAGTAGGCCAGC |
| pilU-CTX-Spe-F | GGACTAGTGACCGGGTGGTCGACGTGTTC |
| pilU-CTX-Hind-R | CCCAAGCTTTCAGCGGAAGCGCCGGCCGGC |
Restriction endonuclease sequences are underlined.
Construction and complementation of in-frame deletion mutants.
All matings were performed as described previously (55). All mutants were confirmed by Southern or PCR analysis and are listed in Table 1.
(i) ChpA.
The chpA in-frame deletion mutant was constructed by allelic replacement and has been described previously (55). To complement the ΔchpA mutation, a 3′ fragment of chpA, including 1 kb of 3′-flanking sequence was amplified by using the chpA26.1/chpA29(Xba) PCR primer pair and cloned as an MluI/XbaI fragment into pJB219 (a modified version of pOK12 in which the KpnI and ClaI sites have been removed from the MCS) to form pJB220. A 5′ fragment of chpA, including 1 kb of 5′ flanking sequence was amplified by using the chpA22(HindIII)/chpA25 PCR primer pair and cloned as an HindIII/MluI fragment into pJB220 to form pJB221. Full-length chpA and 1 kb of 5′- and 3′-flanking sequence were subcloned as a SpeI fragment from pJB221 to pJB100 to form pJB231. pJB231 was transformed into E. coli S17.1, and transformants were mated to the ΔchpA strain to create a ΔchpA derivative complemented for chpA at its endogenous locus.
(ii) PilA.
5′ and 3′ pilA deletion construct fragments were amplified by using the pilA1/pilA2 and pilA3/pilA4 PCR primer pairs. An adenosine residue was added to the 3′ end of the PCR products and the modified products ligated into pGEM-T. The 5′ deletion construct was excised from pGEM-T as an NcoI/EcoRIII fragment. The 3′ deletion construct was excised from pGEM-T as an EcoRI/XbaI fragment. Excised 5′ and 3′ deletion construct fragments were concatamerized and cloned as an NcoI/XbaI fragment into pOK12. The deletion construct was excised from pOK12 as a SpeI fragment and ligated into the allelic exchange vector pJEN34 to form pJEN34PAO1KO3. pJEN34PAO1KO3 was transformed into E. coli S17.1, and transformants were mated to PAO1 to create an unmarked ΔpilA mutant. This procedure deleted amino acids 2 to 149 of the PilA protein. To complement the ΔpilA mutation, pilA and 1 kb of 5′ and 3′ flanking sequence were amplified by using the pilA1(Spe)/pilA4(Spe) PCR primer pair. The product was cloned as a SpeI fragment into pJB100 to form pJB241. pJB241 was transformed into E. coli S17.1, and transformants were mated to the ΔpilA strain to create a ΔpilA derivative complemented for pilA at its endogenous locus.
(iii) PilB.
5′ and 3′ pilB deletion construct fragments were amplified by using the pilB1/pilB2 and pilB3/pilB4 PCR primer pairs. PCR products were used as templates in a sequence overlap extension (SOE) reaction with the pilB1/pilB4 PCR primer pair. The SOE product was cloned as a SpeI fragment into pJB100 for form pJB122. pJB122 was transformed into E. coli S17.1, and transformants were mated to PAO1 to create an unmarked ΔpilB mutant. This procedure deleted amino acids 4 to 564 of the PilB protein. To complement the ΔpilB mutation, pilB and 1 kb of the 5′- and 3′-flanking sequence were amplified by using the pilB1/pilB4 PCR primer pair. The product was cloned as a SpeI fragment into pJB100 to form pJB246. pJB246 was transformed into E. coli S17.1, and transformants were mated to ΔpilB to create a ΔpilB derivative complemented for pilB at its endogenous locus.
(iv) PilG.
5′ and 3′ pilG deletion construct fragments were amplified by using the pilG1/pilG2 and pilG3/pilG4 PCR primer pairs. An adenosine residue was added to the 3′ end of the PCR products, and the modified products were ligated into pGEM-T to form pJB43 (5′ deletion construct) and pJB44 (3′ deletion construct). The 5′ deletion construct was excised from pJB43 as an EcoRI/HindIII fragment. The 3′ deletion construct was excised from pJB44 as a HindIII/XbaI fragment. Excised 5′ and 3′ deletion construct fragments were concatamerized and cloned as an EcoRI/XbaI fragment into pOK12 to form pJB60. The deletion construct was excised from pJB60 as a SpeI fragment and ligated into the allelic exchange vector pJEN34 to form pJB87. pJB87 was transformed into E. coli S17.1, and transformants were mated to PAO1 to create an unmarked ΔpilG mutant. This procedure deleted amino acids 5 to 87 of the PilG protein. To complement the ΔpilG mutation, pilG and 1 kb of 5′ and 3′ flanking sequence were amplified by using the pilG1(Spe)/pilG4(Spe) PCR primer pair. The product was cloned as a SpeI fragment into pJB100 to form pJB126. pJB126 was transformed into E. coli S17.1 and transformants were mated to the ΔpilG strain to create a ΔpilG derivative complemented for pilG at its endogenous locus. The deletion and complementation of pilH has been described previously (3).
(v) PilR.
5′ and 3′ pilR deletion construct fragments were amplified by using the pilR1/pilR2 and pilR3/pilR4 PCR primer pairs. PCR products were used as templates in an SOE reaction with the pilR1/pilR4 PCR primer pair. The SOE product was cloned as a SpeI fragment into pJB100 for form pJB250. pJB250 was transformed into E. coli S17.1, and transformants were mated to PAO1 to create an unmarked ΔpilR mutant. This procedure deleted amino acids 4 to 443 of the PilR protein. To complement the ΔpilR mutation, pilR and 1 kb of 5′ and 3′ flanking sequence were amplified by using the pilR1/pilR4 PCR primer pair. The product was cloned as a SpeI fragment into pJB100 to form pJB251. pJB251 was transformed into E. coli S17.1, and transformants were mated to the ΔpilR strain to create a ΔpilR derivative complemented for pilR at its endogenous locus.
(vi) PilT.
5′ and 3′ pilT deletion construct fragments were amplified by using the pilT1/pilT5 and pilT6/pilT4 PCR primer pairs. PCR products were used as templates in an SOE reaction with the pilT1/pilT4 PCR primer pair. The SOE product was cloned as a SpeI fragment into pJB100 for form pJB109. pJB109 was transformed into E. coli S17.1, and transformants were mated to PAO1 to create an unmarked ΔpilT mutant. This procedure deleted amino acids 4 to 342 of the PilT protein. To complement the ΔpilT mutation, pilT and 1 kb of 5′ and 3′ flanking sequence were amplified by using the pilT1/pilT4 PCR primer pair. The product was cloned as a SpeI fragment into pJB100 to form pJB113. pJB113 was transformed into E. coli S17.1, and transformants were mated to the ΔpilT mutant to create a ΔpilT derivative complemented for pilT at its endogenous locus.
The pilT truncation mutation (pilTTrunc) deletes 429 bp of the pilT coding sequence including the start codon (amino acids 1 to 143 of the PilT protein) and introduces stop codons in all three reading frames. The mutant retains 783 bp of sequence upstream of pilU. The 5′ and 3′ truncation construct fragments were amplified by using the pilT1(new)/pilT-Trunc2 PilT-Trunc3/pilT-Trunc4 PCR primer pairs. PCR products were used as templates in an SOE reaction with the pilT1(new)/pilT-Trunc4 PCR primer pair. The SOE product was cloned as a SpeI fragment into pJB100 for form pJB203. pJB203 was transformed into E. coli S17.1, and transformants were mated to PAO1 to create the pilTTrunc mutant.
(vii) PilU.
5′ and 3′ pilU deletion construct fragments were amplified by using the pilU1/pilU5 and pilU6/pilU4 PCR primer pairs. PCR products were used as templates in an SOE reaction with the pilU1/pilU4 PCR primer pair. The SOE product was cloned as a SpeI fragment into pJB100 for form pJB111. pJB111 was transformed into E. coli S17.1, and transformants were mated to PAO1 to create an unmarked ΔpilU mutant. This procedure deleted amino acids 4 to 380 of the PilU protein. To complement ΔpilU, pilU and 1 kb of 5′- and 3′-flanking sequence were amplified by using the pilU1/pilU4 PCR primer pair. The product was cloned as a SpeI fragment into pJB100 to form pJB114. pJB114 was transformed into E. coli S17.1, and transformants were mated to the ΔpilU mutant to create a ΔpilU derivative complemented for pilU at its endogenous locus.
(viii) FliC.
5′ and 3′ fliC deletion construct fragments were amplified by using the fliC1/fliC4 and fliC3/fliC4 PCR primer pairs. PCR products were used as templates in an SOE reaction with the fliC1/fliC4 PCR primer pair. The SOE product was cloned as a SpeI fragment into pJB100 for form pJB215. pJB215 was transformed into E. coli S17.1, and transformants were mated to PAO1 to create an unmarked ΔfliC mutant. This procedure deleted amino acids 4 to 486 of the FliC protein. To complement the ΔfliC mutation, fliC and 1 kb of 5′- and 3′-flanking sequence were amplified by using the flic1/flic4 PCR primer pair. The product was cloned as a SpeI fragment into pJB100 to form pJB230. pJB230 was transformed into E. coli S17.1, and transformants were mated to the ΔfliC mutant to create a ΔfliC derivative complemented for fliC at its endogenous locus.
A pEX100T-based chpA deletion construct was made by subcloning the chpA deletion construct as a SpeI fragment from pJB4 to pJB100 to form pJB116. pJB116 was transformed into E. coli S17.1, and transformants were mated to PAO1ΔpilH, PAO1ΔpilT, PAO1ΔpilU, and PAO1ΔpilTΔpilU constructs (see below) to create PAO1ΔchpAΔpilH, PAO1ΔchpAΔpilT, PAO1ΔchpAΔpilU, and PAO1ΔchpAΔpilTΔpilU, respectively.
To construct PAO1ΔpilGΔpilH, the 5′ deletion construct from pJB43 was excised as an XhoI/HindIII fragment, concatamerized with the 3′ deletion construct from pJB48 excised as a HindIII/XbaI fragment, and cloned as an XhoI/XbaI fragment into pOK12 to form pJB96. The deletion construct was excised from pJB96 as a SpeI fragment and ligated into the allelic exchange vector pJEN34 to form pJB97. pJB97 was transformed into E. coli S17.1, and transformants were mated to PAO1 to create an unmarked ΔpilG ΔpilH mutant. Gentamicin-resistant derivatives of the pilT and pilU deletion constructs were made by subcloning a gentamicin resistance encoding cassette as a HindIII fragment from pX1918GT (46) into pJB109 and pJB111 to form pJB110 and pJB112, respectively. pJB110 and pJB112 were transformed into E. coli S17.1, and transformants were mated to PAO1ΔpilG to created PAO1ΔpilGΔpilT-gent and PAO1ΔpilGΔpilU-gent. PAO1ΔpilGΔpilT-gent and PAO1ΔpilGpilU-gent were then mated to S17.1 strains carrying pJB109 and pJB111 to create the PAO1ΔpilGΔpilT and PAO1ΔpilGΔpilU constructs. A pEX100T-based pilG deletion construct was made by subcloning the pilG deletion construct as a SpeI fragment from pJB59 to pJB100 to form pJB118. pJB118 was transformed into E. coli S17.1, and transformants were mated to the ΔpilT ΔpilU strain (see below) to create strain PAO1ΔpilGΔpilTΔpilU.
Strains PAO1ΔpilBΔpilH and PAO1ΔpilBΔpilTΔpilU were constructed by mating the ΔpilH and ΔpilT ΔpilU strains respectively, to E. coli S17.1 carrying pJB122. The PAO1ΔpilBΔpilT and PAO1ΔpilBΔpilU strains were constructed by mating the PAO1ΔpilB strain to E. coli S17.1 carrying pJB109 and pJB111, respectively.
5′ and 3′ PAO1ΔpilTΔpilU deletion construct fragments were amplified by using the pilT1/pilTpilU1 and pilTpilU2/pilU4 PCR primer pairs. PCR products were used as templates in an SOE reaction with the pilT1/pilU4 PCR primer pair. The SOE product was cloned as a SpeI fragment into pJB100 for form pJB163. pJB163 was transformed into E. coli S17.1 and transformants were mated to PAO1 to create PAO1ΔpilTΔpilU. A pEX100T-based pilH deletion construct was made by subcloning the pilH deletion construct as a SpeI fragment from pJB62 to pJB100 to form pJB119. pJB119 was transformed into E. coli S17.1, and transformants were mated to ΔpilT, ΔpilU, and ΔpilT ΔpilU constructs to create strains PAO1ΔpilHΔpilT, PAO1ΔpilHΔpilU, and PAO1ΔpilHΔpilTΔpilU, respectively.
(ix) PAO1ΔpilTCTX-pilU strains.
pilU, along with 1,000 bp of upstream sequence, was introduced in single copy onto the P. aeruginosa chromosome at the phage attachment site attB by using the mini-CTX system (22). The CTX-pilU construct was amplified by using the pilU-CTX-Spe-F(1kb)/pilU-CTX-Hind-R PCR primer pair. The PCR product was cloned as a SpeI/HindIII fragment into mini-CTX2 to form pJB212. pJB212 was transformed into E. coli S17.1, and transformants were mated to all pilT mutant strains except the PAO1ΔpilTTrunc and PAO1ΔpilTΔpilU strains to create PAO1ΔpilTCTX-pilU derivatives. The plasmid backbone was removed by mating strains to E. coli SM10 carrying pFLP2, as described previously (22).
(x) chpA-FLAG and chpA point mutants.
The region of chpA encoding the histidine kinase domain and the 2X-FLAG tag was subcloned as a ClaI/KpnI fragment from pAL64 (37) into pOK12 to form pJB202. The KpnI/ClaI fragment was subcloned from pJB202 into pJB221 from which the corresponding ClaI/KpnI fragment had been removed to form pJB232. Full-length chpA-FLAG, including 1 kb 5′ and 3′ flanking sequence, was subcloned into pJB100 to form pJB234. pJB234 was transformed into E. coli S17.1, and transformants were mated to the ΔchpA mutant to create PAO1ΔchpA-FLAG.
pJB202 was the substrate for a QuickChange (Stratagene) mutagenesis reaction using the chpA-HisKn-F/chpA-HisKn-R PCR primer pair. The mutation replaces each of the three residues in the signature DXG motif of the G1 box of ChpA with alanines (D2091A, D2092A, and G2093A), chpA(AAA)-FLAG. The mutated histidine kinase domain, including the 2X-FLAG tag, was subcloned as a KpnI/ClaI fragment from pJB207 into pJB221 from which the corresponding ClaI/KpnI fragment had been removed to form pJB233. Full-length chpA(AAA)-FLAG, including 1 kb of 5′ and 3′ flanking sequence, was subcloned into pJB100 to form pJB235. pJB235 was transformed into E. coli S17.1, and transformants were mated to the ΔchpA mutant to create PAO1ΔchpA(AAA)-FLAG.
(xi) pilG-His, pilH-His, and pilG and pilH point mutants.
5′ and 3′ allelic-exchange constructs to generate C-terminal His6-tagged derivatives of the pilG and pilH mutants were amplified by using the pilG1(Spe)/pilG5 and pilG6/pilG4(Spe) and the pilH1/pilH15 and pilH16/pilH4 PCR primer pairs, respectively. PCR products were used as templates in SOE reactions with the pilG1(Spe)/pilG4(Spe) and pilH1/pilH4 primer pairs. The SOE products were cloned blunt into SmaI-digested pEX100T to form pJB124 (pilH-His) and pJB127 (pilG-His). pJB124 and pJB127 were transformed into E. coli S17.1, and transformants were mated to ΔpilH and ΔpilG mutants to create the pilH-His and pilG-His constructs, respectively.
pilG-His and pilH-His allelic-exchange constructs were amplified from pJB127 and pJB124, respectively, by using pilG1(Spe)/pilG4(Spe) and pilH1(Spe)/pilH4(Spe) PCR primer pairs cloned as Spe fragments into pOK12 to form pJB247 and pJB248. pJB247 and pJB248 were substrates for QuickChange (Stratagene) mutagenesis reactions by using pilG-D58A-F/pilG-D58A-R and pilH-D52A-F/pilH-D52A-R PCR primer pairs to form pJB252 and pJB254, respectively. The pilG(D58A)-His and pilH(D52A)-His allelic-exchange constructs were subcloned as SpeI fragments from pJB252 and pJB254 into pJB100 to form pJB256 and pJB258. pJB256 and pJB258 were transformed into E. coli S17.1 and transformants were mated to ΔpilG and ΔpilH strains to create PAO1ΔpilG(D58A)-His and PAO1ΔpilH(D52A)-His.
PpilA-lacZ reporter strains.
A transcriptional fusion of the pilA promoter to lacZ was introduced in single copy onto the P. aeruginosa chromosome at the phage attachment site attB using the mini-CTX system (4). A 500-bp portion of sequence upstream of the pilA start codon was amplified by using the pilA-Reporter-F/pilA-Reporter-R PCR primer pair. The PCR product was cloned as an XhoI/BamHI fragment into mCTX2-lacZ to form pJB249. pJB249 was transformed into E. coli S17.1, and transformants were mated to all single mutant strains (except the ΔpilT strain) and pilTTrunc to create CTX-PpilA derivatives. The plasmid backbone was removed by mating strains to E. coli SM10 carrying pFLP2, as described previously (4).
TM assay.
Subsurface twitching motility (TM) stab assays were performed as previously described (1). TM was quantified by taking three separate measurements of twitching zone diameter from five independent stabs.
Generation of the α-PilA antibody.
Polyclonal α-PilA antibodies were raised commercially in rabbits against the synthetic peptide WACKSTQDPMFTPKGCD (Invitrogen). The peptide represents amino acids 132 to 148 of PilA. For the present study, serum collected 8 weeks postinoculation with the antigen was used.
Immunoblotting.
Isolation of intracellular pilin for cells cultured on solid media was performed as follows. A portion (100 μl) of a 2-ml LB culture grown shaking overnight at 37°C was plated onto 1.5% LB agar and grown at 37°C for 16 h. After growth, bacteria were scraped from the agar surface and resuspended in 5 ml of phosphate-buffered saline (PBS). A volume of cells equivalent to an optical density at 600 nm (OD600) of 5.0 was brought up to 1 ml in PBS. Cells were vortexed for 30 min to remove surface pili and harvested by centrifugation (20,000 × g for 10 min). The supernatant was removed, and cells were resuspended in 100 μl of BugBuster protein extraction reagent (Novagen). Then, 1 μl of Benzonase nuclease (Novagen) was added, and the resuspended cells were incubated by gentle mixing at room temperature for 30 min. Cellular debris was removed by centrifugation (20,000 × g for 10 min), and 90 μl of supernatant was added to 90 μl of 2× NuPage LDS protein sample buffer (Invitrogen) and 20 μl of 10× NuPage sample reducing agent (Invitrogen). Samples were boiled for 5 min. Protein concentration was determined by using a Bio-Rad protein assay. To isolate surface pili, bacteria were grown as described above at 37°C for 16 h on 1.5% LB agar, scraped from the agar surface, and resuspended in 5 ml of PBS. A volume of cells equivalent to an OD600 of 20.0 was brought up to 1 ml in PBS. Cells were vortexed at room temperature for 30 min to remove surface pili. The suspension was centrifuged at room temperature (20,000 × g for 10 min), and the supernatant was collected and centrifuged a second time (20,000 × g for 10 min) to remove all cellular debris. The resulting supernatant was incubated overnight at 4°C in 100 mM MgCl2 to precipitate pili, as described previously (1). The precipitate was collected by centrifugation at 4°C (20,000 × g for 20 min), the supernatant was removed, and the pellet was resuspended in 100 μl of PBS and 100 μl of 2× NuPage LDS protein sample buffer (Invitrogen), including 1× NuPage sample reducing agent (Invitrogen). The samples were boiled for 5 min.
Isolation of intracellular pilin for cells cultured in liquid medium was performed as follows. Bacteria were grown shaking at 37°C for 16 h in 10 ml of LB medium. A volume of cells equivalent to an OD600 of 5.0 was brought up to 1.5 ml in PBS and processed as described above. To isolate surface pili, bacteria were grown shaking at 37°C for 16 h in 10 ml of LB broth. A volume of cells equivalent to an OD600 of 20.0 was brought up to 6 ml in PBS. Cells were harvested by centrifugation (6,000 × g for 10 min), resuspended in 1 ml of PBS and processed as described above.
To detect cell-associated and surface pilin for cells cultured on both solid medium and in liquid medium, 15 μg of total cell-associated protein or 10 μl of surface pilin samples (1/20 of the total sample) was separated on Novex-NuPAGE 12% Bis-Tris SDS-PAGE gels (Invitrogen) and electroblotted onto Immun-Blot PVDF membrane (Bio-Rad) in a previously described Tris-glycine system (50). Membranes were blocked in 5% skim milk and probed with a 1:100,000 dilution of primary α-PilA antibody in 5% skim milk powder (0.05% Tween 20) in PBS. Membranes were then incubated with a 1:25,000 dilution of goat anti-rabbit immunoglobulin G conjugated with horseradish peroxidase (Jackson Immunoresearch Laboratories) in 5% skim milk powder (0.05% Tween 20) in PBS, followed by detection by enhanced chemiluminescence using the Amersham ECL Western blotting detection kit (GE Healthcare).
Cell-associated and surface flagellin samples for both plate and broth cultured cells were prepared as described for pilin samples. Detection of cell-associated and surface flagellin for both plate- and broth-cultured cells was carried out as described for pilin immunoblots, with the exception that membranes were probed with a 1:200,000 dilution of anti-FlaB antibody (kindly provided by Daniel Wozniak, Ohio State University [18]).
To quantify the levels of surface pili, samples were run in triplicate and immunoblotted for PilA and FliC as described above. Images of immunoblots were acquired on the ChemiDoc XRS (Bio-Rad) and quantified by using QuantityOne image analysis software. Surface pili levels were normalized to FliC. Isolation and detection of PAO1ΔchpA-FLAG and PAO1ΔchpA(AAA)-FLAG was performed as described previously (37). Briefly, to prepare protein samples, cells were harvested from 2 ml of culture grown shaking for 16 h at 37°C, resuspended in 400 μl of SDS-PAGE sample buffer, passaged through a 27.5 gauge needle three times, and boiled for 5 min. Then, 40 μl (1/10 of total sample volumes) was separated by SDS-PAGE and immunoblotted with 2.5 μg of α-FLAG M2 monoclonal antibody/ml using 2.5 μg of α-FLAG M2 monoclonal antibody (Sigma)/ml.
Phage sensitivity assays.
Phage sensitivity was assayed using the Tfp-specific P. aeruginosa bacteriophage PO4 (kindly provided by Lori Burrows, McMaster University), as previously described (52). Briefly, the phage stock was titered by adding 10 μl of serial diluted phage to 100 μl of stationary-phase PAO1, followed by incubation at room temperature for 30 min, and then added to 3 ml of 0.8% LB top agar, poured onto 1.5% LB agar, and incubated overnight at 37°C. To determine the phage sensitivity of mutant strains, triplicate cultures started from single colonies were grown shaking for 16 h at 37°C. Next, 100 μl of culture was infected with 10 μl of diluted phage, followed by incubation at room temperature for 30 min. After incubation, samples were added to 3 ml of 0.8% LB top agar, followed by incubation for 16 h at 37°C. To quantify the data, the plaques were counted, and the PFU of phage/ml were determined.
β-Galactosidase assays.
β-Galactosidase assays were performed as previously described (52) with the following modifications. For assays performed on strains cultured in liquid media, cultures started from single colonies were grown in triplicate with shaking for 16 h at 37°C in 5 ml of LB medium. For assays performed on strains cultured on solid media, cultures started from single colonies were grown in triplicate shaking overnight at 37°C in 2 ml of LB medium. Then, 100 μl of overnight culture was plated to 1.5% LB agar and grown for 16 h. After growth, cells were scraped from the agar surface, and resuspended in 5 ml of PBS and diluted to an OD600 of 4.0 before being assayed as described previously (52). Optical densities (OD600 and OD420) were recorded by using a SPECTRAmax microplate spectrophotometer (Molecular devices) and SoftMaxPro 4.3.1. Specific activities were calculated relative to the OD600.
Statistical analysis.
Statistical significance was determined by analysis of variance using Instat software. Differences were considered to be significant at a P of <0.05.
RESULTS
chpA, pilG, and pilB regulate pilus extension.
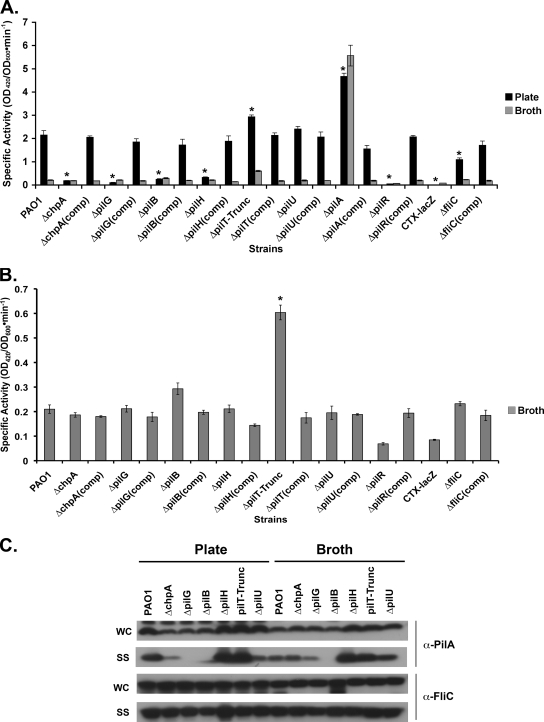
We constructed in-frame chpA (55), pilG, pilH (3), pilB, pilT, and pilU deletion mutants in strain PAO1 and assayed them for Tfp function, including TM, intracellular and surface pilin levels, and phage sensitivity. The present study represents the first in-depth phenotypic analysis of in-frame deletions of these genes in the same strain background and is the first instance in which all mutations have been complemented by reintroduction of the genes onto the chromosome at their endogenous loci. TM was measured by the subsurface stab assay. To determine surface and intracellular pilin levels, plate-grown bacteria were harvested after 16 h. Surface pili were sheared by vigorous vortexing, the bacteria were removed by centrifugation, and the sheared pili were precipitated from the supernatant. Pilin in the pelleted bacteria constituted the intracellular pilin fraction. Infection with phage PO4, which requires Tfp for adsorption, is a sensitive measure of surface pili. For example, some mutants which lack surface pili by Western analysis remain sensitive to infection by phage PO4 (56).
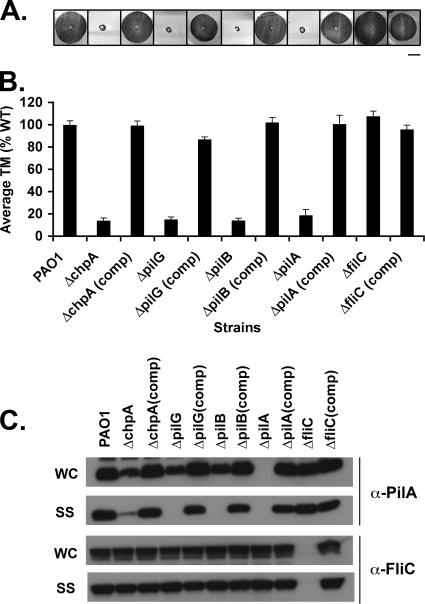
Consistent with previous results (12, 13, 42, 54-56), all mutants had defects in TM (Fig. 2A and B and Fig. 3A and B). As expected (42), the ΔpilB mutant lacked surface pili (Fig. 2C) and was resistant to infection with phage PO4 (Table 3). The ΔpilG mutant also lacked detectable surface pili, and the ΔchpA mutant had greatly reduced surface pili as determined by Western blot analysis (Fig. 2C). Both mutants, however, remained susceptible to phage infection (Table 3), suggesting that they assembled small amounts of functional surface pili. With the exception of pilB and pilT single, double, and triple mutants, infection of all strains with phage PO4 produced nearly as many plaques as when wild-type (WT) PAO1 was infected. However, in all cases plaques formed on mutant lawns were turbid compared to plaques formed on WT lawns (data not shown). All mutants had wild-type levels of flagellin on the cell surface, indicating that presentation of surface structures other than Tfp is not affected in these mutant backgrounds (Fig. 2C). Although Western blot analysis of cell lysates showed a slight decrease in pilin production in the ΔpilB, ΔpilG, and ΔchpA mutants compared to the wild type (Fig. 2C), this decrease is not sufficient to account for their profound defect in TM and surface presentation. TM, pilin production, surface piliation, and phage sensitivity could be complemented by reintroduction of each of the genes at their endogenous chromosomal loci (Fig. 2). Taken together, and consistent with previous observations, these data suggest that chpA, pilG, and pilB are involved in the regulation of pilus extension.
FIG. 2.
Assays of pilus function for mutants defective in pilus extension. (A) Subsurface TM assay of PAO1, the indicated in-frame deletion mutants, and complemented (comp) strains in which the wild-type gene was reintroduced at its endogenous locus. TM assays were performed by the subsurface stab method, followed by Coomassie blue staining, as described previously (1). Bar, 1 cm. (B) Graph depicting TM zone diameters for the indicated strains. The diameter is expressed as a percentage of PAO1 TM. Shown are the means ± the standard deviation (SD, n = 5). The residual zones observed in ΔpilA, ΔchpA, ΔpilG, and ΔpilB strains represent colony growth. (C) Intracellular and surface pilin and flagellin levels. For the indicated strains, surface structures (SS) including pili and flagella were sheared by vigorous vortexing of bacteria cultured on solid media and separated from cells (WC) by centrifugation. The sheared pili and flagella were precipitated. WC (15 μg of total protein) and SS (5% of the total resuspended volume of the precipitate) samples were separated by SDS-PAGE and immunoblotted with a polyclonal antibody to PilA (α-pilA) or to FliC (α-FliC).
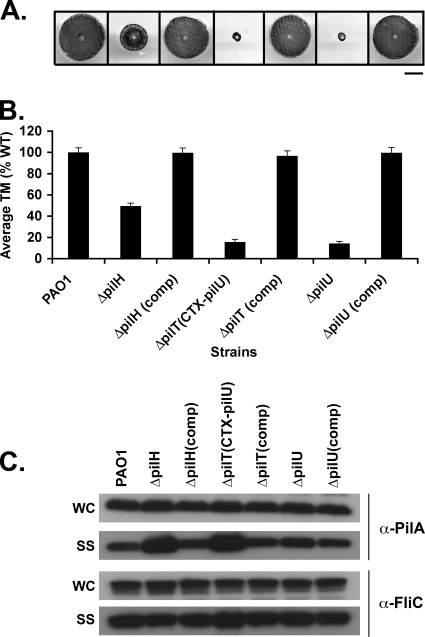
FIG. 3.
Assays of pilus function for mutants defective in pilus retraction. (A) Subsurface TM assays were performed as described in Fig. 1. The bar represents 1 cm. (B) Graph depicting average twitching zone diameters for the indicated strains. Shown are the means ± the SD (n = 5) The residual zone observed in the ΔpilT and ΔpilB strains represents colony growth and is similar to that of the ΔpilA strain. (C) Surface structures (SS) and intracellular (WC) preparations of the indicated strains were immunoblotted with a polyclonal antibody to PilA (α-pilA) or to FliC (α-FliC), as described in Fig. 2.
TABLE 3.
Sensitivity of various strains to infection by the pilus-specific phage PO4
| Strain | Strain sensitivity (avg PFU/ml ± SD [109])a |
|---|---|
| PAO1 | 3.96 ± 0.53 |
| PAO1ΔchpA | 3.30 ± 0.70 |
| PAO1ΔchpA(comp) | 3.75 ± 0.79 |
| PAO1ΔpilG | 2.85 ± 0.72 |
| PAO1ΔpilG(comp) | 4.52 ± 0.66 |
| PAO1ΔpilB | 0.00 ± 0.00* |
| PAO1ΔpilB(comp) | 3.92 ± 0.32 |
| PAO1ΔpilA | 0.00 ± 0.00* |
| PAO1ΔpilA(comp) | 4.12 ± 0.56 |
| PAO1ΔfliC | 3.92 ± 0.56 |
| PAO1ΔfliC(comp) | 5.41 ± 0.14 |
| PAO1ΔpilH | 2.96 ± 0.58† |
| PAO1ΔpilH(comp) | 3.48 ± 0.45 |
| PAO1ΔpilTCTX-pilU | 0.00 ± 0.00* |
| PAO1ΔpilT(comp) | 3.39 ± 0.17 |
| PAO1ΔpilU | 2.88 ± 0.33 |
| PAO1ΔpilU(comp) | 4.11 ± 0.48 |
| PAO1ΔchpAΔpilH | 4.00 ± 0.45† |
| PAO1ΔchpAΔpilTCTX-pilU | 0.00 ± 0.00* |
| PAO1ΔchpAΔpilU | 1.85 ± 0.30 |
| PAO1ΔchpAΔpilTΔpilU | 0.00 ± 0.00* |
| PAO1ΔpilGΔpilH | 3.93 ± 0.72† |
| PAO1ΔpilGΔpilTCTX-pilU | 0.00 ± 0.00* |
| PAO1ΔpilGΔpilU | 3.07 ± 0.78 |
| PAO1ΔpilGΔpilTΔpilU | 0.00 ± 0.00* |
| PAO1ΔpilBΔpilH | 0.00 ± 0.00* |
| PAO1ΔpilBΔpilTCTX-pilU | 0.00 ± 0.00* |
| PAO1ΔpilBΔpilU | 0.00 ± 0.00* |
| PAO1ΔpilBΔpilTΔpilU | 0.00 ± 0.00* |
| PAO1ΔpilHΔpilTCTX-pilU | 0.00 ± 0.00* |
| PAO1ΔpilTΔpilU | 0.00 ± 0.00* |
| PAO1ΔpilHΔpilU | 2.87 ± 0.83† |
| PAO1ΔpilHΔpilTΔpilU | 0.00 ± 0.00* |
| PAO1chpA-FLAG | 3.45 ± 0.63 |
| PAO1ΔchpA(AAA)-FLAG | 3.28 ± 0.63 |
| PAO1pilG-His | 3.17 ± 0.29 |
| PAO1pilG(D58A)-His | 2.99 ± 0.39 |
| PAO1pilH-His | 3.31 ± 0.34 |
| PAO1pilH(D52A)-His | 4.23 ± 0.23† |
*, No plaques were detectable even when strains were infected with 109 phage. †, The ΔpilH plaque morphology was distinguished by large zones of seemingly directional lysis and poorly defined plaques.
pilH and pilT regulate pilus retraction.
We next examined ΔpilH, ΔpilT, and ΔpilU mutants to confirm their role in the regulation of pilus retraction. All three of these mutants, in contrast to the ΔchpA, ΔpilG, and ΔpilB mutants, had WT levels of intracellular pilin (Fig. 3C). The ΔpilT mutant was completely defective for TM, having TM zones comparable to those of a ΔpilA mutant (Fig. 3A and B). It had significantly increased levels of surface pili relative to WT (P < 0.05, Fig. 5D) and was resistant to phage PO4 (Table 3). The ΔpilU mutant showed greatly decreased TM (Fig. 3A and B) but was phage sensitive (Table 3) and had levels of surface pili statistically indistinguishable from the WT (Fig. 3C and 5D). The ΔpilH mutant had a TM zone intermediate to wild type and ΔpilA (Fig. 3A and B) and exhibited increased levels of surface pili relative to the WT (P < 0.05, Fig. 3C and 5D) but, in contrast to the ΔpilT mutant, it was phage PO4 sensitive (Table 3). The morphology of plaques formed by PO4 phage on the ΔpilH mutant was distinguished by a tear-drop-shaped zone of lysis and poorly defined plaques, even at low titers of phage (data not shown). This phenotype is unique among the Tfp regulatory factors addressed in the current study and does not appear to have been described for any other factor involved in the biogenesis or function of Tfp. For each of these strains, WT TM, surface piliation, and phage sensitivity could be restored by reintroduction of each of the genes at their endogenous chromosomal loci (Fig. 3).
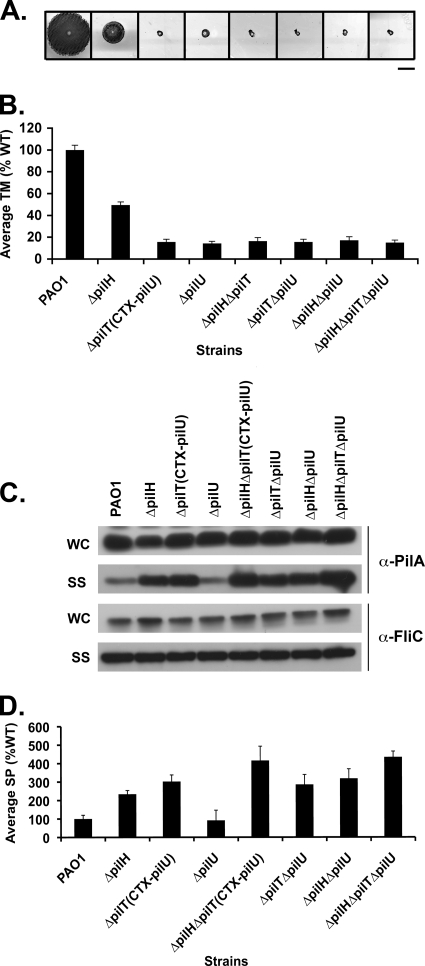
FIG. 5.
PilH enhances the function of PilT but not PilU. (A) Subsurface TM assays were performed as described in Fig. 1. Bar, 1 cm. (B) Graph depicting average TM zone diameters for the indicated strains. Shown are the means ± the SD (n = 5). The residual zone of TM in all of the mutants represents colony growth and is similar to that of the ΔpilA mutant. (C) A 1:5 dilution of surface structures (SS) and cell intracellular (WC) preparations of the indicated strains were immunoblotted with a polyclonal antibody to PilA (α-pilA) or to FliC (α-FliC), as described in Fig. 2. (D) Quantification of surface piliation levels relative to a FliC loading control. Surface piliation levels are expressed as a percentage of PAO1. Shown are the means ± the SD (n = 3).
Whereas all of the other mutants (i.e., the ΔchpA, ΔpilG, ΔpilB, ΔpilH, and ΔpilU mutants) could be complemented for TM in trans, the ΔpilT mutant could only be complemented by the reintroduction of pilT onto the chromosome at the pilT locus (data not shown). This finding suggests that the in-frame deletion of pilT might affect transcription of the adjacent pilU gene by disrupting cis-regulatory sequences required for pilU transcription located within the pilT coding sequence. Indeed, previous studies suggest that pilT and pilU are transcribed separately (56). We constructed a derivative of the ΔpilT mutant (PAO1ΔpilTCTX-pilU) in which the pilU gene, including 1,000 bp of upstream sequence, was introduced at an exogenous chromosomal locus using the mini-CTX system (22). This mutant was TM defective, had increased levels of surface pili, was phage resistant, and had WT levels of intracellular pilin and could be complemented for TM by expression of pilT in cis or in trans (Fig. 3, Table 3, and data not shown). This strain was used in the remainder of our studies except where indicated.
The phenotype of ΔpilT mutants differed from that of ΔpilH mutants in several important ways. PAO1ΔpilTCTX-pilU had increased surface pili relative to the ΔpilH mutant by Western blot analysis (Fig. 3C and 5C), although the difference was not statistically significant (Fig. 5D). PAO1ΔpilTCTX-pilU was phage PO4 resistant, whereas the ΔpilH mutant was phage PO4 sensitive. Together, these data are consistent with the idea that PilT functions as a motor protein which serves to drive pilus retraction and whose activity is enhanced but is not completely dependent upon PilH.
chpA, pilG, and pilB function upstream of pilH, pilT, and pilU.
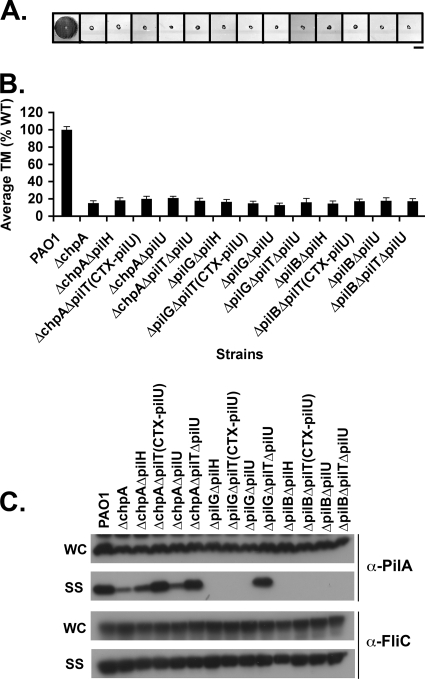
We hypothesize that ChpA acts as a histidine kinase to phosphorylate PilG and/or PilH. Phosphorylated PilG, in turn modulates PilB-dependent pilus extension, while phosphorylated PilH is required for PilT- and/or PilU-dependent pilus retraction. This model leads to the testable genetic prediction that double mutants in any factor involved in regulating pilus extension (chpA, pilG, or pilB) and any factor involved in regulating pilus retraction (pilH, pilT, and pilU) should have reduced surface pili. We constructed informative double mutants and assayed them for TM, intracellular and surface pilin levels, and phage sensitivity. All double mutants were defective for TM (Fig. 4A and B). This finding is most notable in the case of pilH double mutants, which no longer exhibit any TM, indicating that ChpA, PilG, and PilB all function upstream of PilH.
FIG. 4.
chpA, pilG, and pilB function upstream of pilH, pilT, and pilU. (A) Subsurface TM assays were performed as described in Fig. 1. Bar, 1 cm. (B) Graph depicting average TM zone diameters for the indicated strains. Shown are the means ± the SD (n = 5). The residual zone of TM in all of the mutants represents colony growth and is similar to of the ΔpilA mutant. (C) Surface structures (SS) and intracellular (WC) preparations of the indicated strains were immunoblotted with a polyclonal antibody to PilA (α-pilA) or to FliC (α-FliC), as described in Fig. 2.
chpA double mutants (PAO1ΔchpAΔpilH, PAO1ΔchpAΔpilTCTX-pilU, and PAO1ΔchpAΔpilU) retained some surface piliation (Fig. 4C), although less than seen in the single pilH, pilT, or pilU mutants (Fig. 3C) but more than seen in the single chpA mutant (Fig. 2C). Furthermore, PAO1ΔchpAΔpilH and PAO1ΔchpAΔpilTCTX-pilU exhibited phage sensitivity similar to PAO1ΔpilH and PAO1ΔpilTCTX-pilU (Table 3), respectively, indicating that PilH and PilT function downstream of ChpA. The observation that PAO1ΔchpAΔpilU shows reduced surface piliation relative to WT (Fig. 4C), where the ΔpilU mutant has surface pilin levels similar to WT (Fig. 3C) indicates that, whatever role PilU plays in the regulation of pilus dynamics, it functions downstream of ChpA. Thus, we conclude that ChpA functions upstream of PilH, PilT, and PilU.
We next compared various combinations of pilG or pilB double mutants. PAO1ΔpilGΔpilH and PAO1ΔpilGΔpilTCTX-pilU had the same phage sensitivity as PAO1ΔpilH and PAO1ΔpilTCTX-pilU, respectively (Table 3). Likewise, PAO1ΔpilBΔpilH and PAO1ΔpilBΔpilU were resistant to phage infection. PAO1ΔpilGΔpilH, PAO1ΔpilGΔpilTCTX-pilU, PAO1ΔpilGΔpilU, PAO1ΔpilBΔpilH, PAO1ΔpilBΔpilTCTX-pilU, and PAO1pilBΔpilU all lacked surface pili (Fig. 4C). These data are consistent with the model that both PilG and PilB function upstream of PilH, PilT, and PilU.
Our work (Table 3) and others (3, 12) have shown that chpA and pilG mutants remain susceptible to infection by phage PO4, whereas pilB mutants are immune to this pilus-specific phage (42). These observations suggest that some PilB-dependent pilus extension must occur in the absence of upstream signaling input from either ChpA or PilG. To test this hypothesis, we constructed PAO1ΔchpAΔpilTΔpilU, PAO1ΔpilGΔpilTΔpilU, and PAO1ΔpilBΔpilTΔpilU and assayed them for TM, pilin production, and surface pili. We reasoned that in these sensitized genetic backgrounds, in which pilus retraction has been disabled, we would be able to detect any residual surface pili resulting from basal PilB activity. All triple mutants were defective for TM (Fig. 4A and B). Detectable levels of surface pili were found for PAO1ΔchpAΔpilTΔpilU and PAO1ΔpilGΔpilTΔpilU, but not PAO1ΔpilBΔpilTΔpilU (Fig. 4C). These data indicate that PilB-dependent extension occurs in the absence of signaling input from either ChpA or PilG and that PilB is absolutely required for pilus extension.
PilH enhances the function of PilT but not that of PilU.
PAO1ΔpilTCTX-pilU is defective for TM and has increased levels of surface piliation relative to the WT, ΔpilH, and ΔpilU strains (Fig. 3C). If PilH is involved in mediating PilT or PilU activity, we would predict that PAO1ΔpilHΔpilTCTX-pilU and PAO1ΔpilHΔpilU should recapitulate the phenotypes of PAO1ΔpilTCTX-pilU and PAO1ΔpilU, respectively. To test this prediction, we constructed PAO1ΔpilHΔpilTCTX-pilU and PAO1ΔpilHΔpilU and assayed them for TM, pilin production, surface pili, and phage sensitivity.
PAO1ΔpilHΔpilTCTX-pilU was defective in TM, similar to PAO1ΔpilTCTX-pilU, and was clearly distinguishable from the intermediate TM of PAO1ΔpilH (Fig. 5A and B). PAO1ΔpilHΔpilTCTX-pilU was resistant to phage PO4 infection (Table 3), suggesting that PilH and PilT function in the same pathway. PAO1ΔpilHΔpilTCTX-pilU reproducibly showed significantly increased levels of surface pili relative to PAO1ΔpilH and PAO1ΔpilTCTX-pilU (P < 0.05, Fig. 5C and D). These data suggest that PilH enhances the retractile activity of PilT and that some additional retractile activity, enhanced perhaps by PilH, must be present.
PAO1ΔpilHΔpilU had a TM phenotype indistinguishable from that of the ΔpilU strain (Fig. 5A and B), resembled the ΔpilH strain in its unusual plaque morphology when infected with phage PO4 (Table 3), and had increased levels of surface pili relative to the ΔpilU strain (Fig. 5C and D). The simplest interpretation of these results is that both PilH and PilU regulate TM, but that they do not operate in the same pathway to mediate retraction. The conclusion that PilU does not appear to function in pilus retraction is further supported by examination of PAO1ΔpilTΔpilU and PAO1ΔpilHΔpilTΔpilU. ΔpilTΔpilU had surface piliation levels similar to those of PAO1ΔpilTCTX-pilU (Fig. 5C and D). Likewise, PAO1ΔpilHΔpilTΔpilU had surface piliation levels similar to those of PAO1ΔpilHΔpilTCTX-pilU (Fig. 5C and D). Thus, the loss of PilU function did not further increase the amount of surface pili, suggesting that the additional retractile activity is not encoded by pilU.
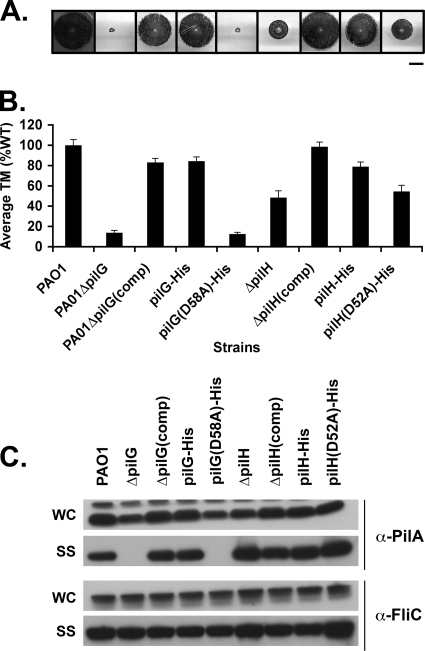
The phosphoacceptor sites of PilG and PilH are required for their function.
Our results thus far support a model in which the CheY-like response regulator proteins PilG and/or PilH are targets of ChpA kinase activity, suggesting that their ability to be phosphorylated is required for function. To test this hypothesis, we constructed strains in which we replaced the WT allele on the chromosome with either His-tagged pilG or pilH (pilG-His, pilH-His) or His-tagged pilG and pilH derivatives in which the putative phosphoacceptor site aspartate residues were replaced with alanine residues [pilG(D58A)-His and pilH(D52A)-His]. The various strains were assayed for TM, pilin production, and surface pili. The pilG-His strain showed no defect in TM, intracellular pilin levels, or surface piliation (Fig. 6), suggesting that the His tag did not interfere with function of PilG. In the case of the pilH-His strain, the epitope tag slightly decreased its function, as the strain carrying the His-tagged protein showed slightly decreased TM relative to WT (Fig. 6) and increased surface piliation, although not nearly to the same extent as ΔpilH.
FIG. 6.
The phosphoacceptor sites of PilG and PilH are required for function. (A) Subsurface TM assays of PAO1 (WT) and the ΔpilG, ΔpilG(comp), ΔpilH, ΔpilH(comp), His-tagged pilG (pilG-His), and pilH (pilH-His) mutants, as well as the His-tagged pilG and pilH point mutant strains PAO1ΔpilG(D58A)-His and PAO1ΔpilH(D52A)-His. Bar, 1 cm. (B) Graph depicting average TM zone diameters for the indicated strains. Shown are the means ± the SD (n = 5). The residual zone of TM in the pilG mutants represents colony growth and is similar to that of the ΔpilA mutant. (C) Surface structures (SS) and intracellular (WC) preparations of the indicated strains were immunoblotted with a polyclonal antibody to PilA (α-pilA) or to FliC (α-FliC), as described in Fig. 2.
Consistent with the idea that signaling through PilG and PilH is required for their function, the phenotypes of the PAO1ΔpilG(D58A)-His or PAO1ΔpilH(D52A)-His strains were indistinguishable from those of the ΔpilG and ΔpilH strains, respectively. PAO1ΔpilG(D58A)-His was deficient for TM and lacked surface pili (Fig. 6A). PAO1ΔpilH(D52A)-His had an intermediate TM phenotype, increased surface pili, and formed aberrant plaques (Fig. 6 and Table 3). We were unable to detect His-tagged wild-type or mutant PilG or PilH by immunoblot analysis in either soluble or total cell lysates. Therefore, it is not possible to formally rule out that the defects observed in TM and surface piliation for PAO1ΔpilG(D58A)-His and PAO1ΔpilH(D52A)-His were secondary to reduced protein levels. We consider this extremely unlikely, however, since the equivalent point mutation has been made in many other CheY homologs without affecting protein levels or protein stability (23, 24, 38).
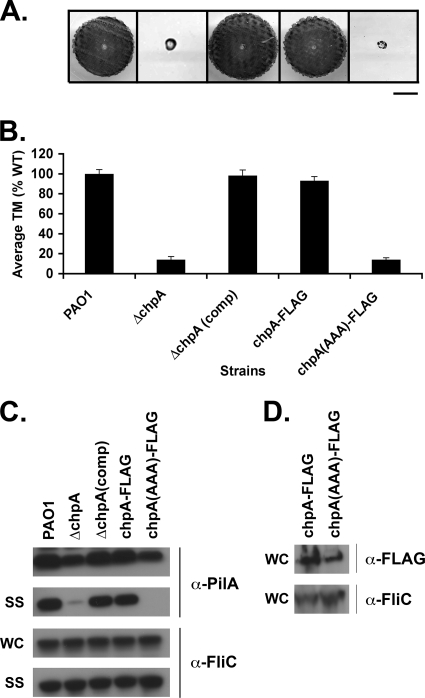
The histidine kinase domain of ChpA is required for function.
To test whether the histidine kinase activity of ChpA is required for its function, we mutated the histidine kinase domain. Although histidine kinase mutants are typically constructed by altering the histidine residue that is the target of autophosphorylation (20), this approach is not possible for ChpA, since it encodes eight potential sites of phosphorylation (six histidine-containing phosphotransfer domains and two novel serine- and threonine-containing phosphotransfer domains [55]). Instead, we mutated the signature DXG motif of the G1 box of a chromosomal FLAG-tagged chpA derivative to AAA. The G1 box, along with the G2 box, is implicated in the binding and hydrolysis of ATP (21). E. coli cheA in which the DXG motif of the G1 box has been altered fails to autophosphorylate, shows decreased affinity for ATP in vitro, and is deficient for chemotaxis in vivo (15, 21). ChpA was C-terminally fused to FLAG so that levels of the wild-type and mutant protein could be compared.
We assayed the PAO1ΔchpA-FLAG and PAO1ΔchpA(AAA)-FLAG strains for TM, intracellular pilin levels, and surface piliation. PAO1ΔchpA-FLAG was indistinguishable from the WT in all assays (Fig. 7A, B, and C), suggesting that the FLAG tag does not interfere with ChpA function. In contrast, the PAO1ΔchpA(AAA)-FLAG histidine kinase mutant exhibited a phenotype similar to that of PAO1ΔchpA. It was defective for TM, had slightly reduced levels of intracellular pilin, and had greatly reduced surface pili (Fig. 7A, B, and C). Immunoblots with α-FLAG showed equal production of ChpA-FLAG and ChpA(AAA)-FLAG, indicating that the defects in pilin function in the ChpA histidine kinase mutant was not due decreased protein levels (Fig. 7D). ChpA-FLAG and ChpA(AAA)-FLAG proteins were only detectable in the insoluble cell lysate fraction (Fig. 7D and data not shown), suggesting that ChpA may be membrane bound. In support of this idea, the TMpred program, which can be used to identify potential transmembrane regions and orientations, strongly predicts three transmembrane domains for ChpA (data not shown). These data provide genetic evidence that the histidine kinase domain of ChpA is required for its function.
FIG. 7.
The histidine kinase domain of ChpA is required for function. (A) Subsurface TM assay of PAO1 (WT) and the ΔchpA, ΔchpA(comp), and FLAG-tagged chpA (chpA-FLAG) mutants, as well as the FLAG-tagged chpA histidine kinase mutant [chpA(AAA)-FLAG]. Bar, 1 cm. (B) Graph depicting average TM zone diameters for the indicated strains. Shown are the means ± the SD (n = 5). The residual zone of TM in the ChpA mutants represents colony growth and is similar to that of the ΔpilA mutant. (C) Surface structures (SS) and intracellular (WC) preparations of the indicated strains were immunoblotted with a polyclonal antibody to PilA (α-pilA) or to FliC (α-FliC), as described in Fig. 2. (D) Levels of ChpA-FLAG and ChpA(AAA)-FLAG. To prepare protein samples, cells were harvested from 2 ml of culture grown shaking for 16 h at 37°C, resuspended in 400 μl of SDS-PAGE sample buffer, passaged through a 27.5 gauge needle three times, and boiled for 5 min. Then, 10% of the total sample volumes were separated by SDS-PAGE and immunoblotted with 2.5 μg of α-FLAG M2 monoclonal antibody/ml.
The Chp system may functionally regulate pilA transcription by controlling pilus extension and retraction.
Immunoblot analysis of the ΔchpA, ΔpilG, and ΔpilB cell lysates shows an ∼2-fold decrease in intracellular pilin levels relative to WT (Fig. 2C). To determine whether this defect occurred at the level of pilA transcription, a PpilA-lacZ transcriptional fusion reporter was constructed. The construct contained 500 bp of sequence upstream of the pilA start codon, which should be sufficient to include all cis-regulatory sequences required for pilA transcription (25, 26, 30, 32). This reporter fusion was integrated at the CTX site (4) in various mutant strain backgrounds. For bacteria cultured on solid media, pilA transcription is increased ∼2-fold in the ΔpilA strain compared to the WT (Fig. 8A; P < 0.05), suggesting that PilA may serve as a negative regulator of pilA transcription. Unexpectedly, β-galactosidase activity was reduced by ca. 90 to 95% relative to the WT in ΔchpA, ΔpilG, and ΔpilB::CTX-PpilA-lacZ reporter strains when assayed from bacteria cultured on solid media (Fig. 8A; P < 0.05).
FIG. 8.
The Chp system regulates pilA transcription by mediating intracellular levels of PilA. (A) The indicated strains harboring the PpilA-lacZ fusion and PAO1 harboring a promoterless lacZ gene (CTX-lacZ) integrated into the chromosome at the attB site were cultured in liquid (A and B) or on solid (A) medium for 16 h. The β-galactosidase activity of samples was measured, and the specific activities relative to the OD600 were calculated. Shown are the means ± the SD (n = 3). *, P < 0.05. (C) Surface structures (SS) and intracellular (WC) preparations of the indicated strains cultured in liquid (broth) or on solid (plate) medium were immunoblotted with a polyclonal antibody to PilA (α-pilA) or to FliC (α-FliC), as described in Fig. 2. The higher-molecular-weight band above pilin in the whole-cell lysates represents a cross-reactive band present in all samples.
As a negative control, we constructed an in-frame deletion in pilR, the response regulator in the PilR/PilS two-component regulatory system which, along with σ54, is required for pilA transcription (25, 26, 30, 32). Although the defects in pilA transcription in the ΔchpA, ΔpilG, and ΔpilB strains do not appear as severe as in the ΔpilR strain, where the levels of transcription are similar to a promoterless lacZ gene incorporated at the CTX site (CTX-lacZ, Fig. 8A), they are not statistically different from the ΔpilR strain. pilA transcription was slightly decreased in a ΔfliC background (Fig. 8A). Our finding that intracellular pilin levels are only decreased twofold in the ΔchpA, ΔpilG, and ΔpilB strains, whereas pilA transcription is decreased 90 to 95%, suggests the possibility that intracellular levels of PilA may also be regulated posttranscriptionally.
We next measured pilA transcription in the ΔpilH, ΔpilT, and ΔpilU strains. Since the CTX site of PAO1ΔpilTCTX-pilU already contained an exogenous gene (pilU), we constructed a PilT mutant (PAO1pilTTrunc) missing the first 143 amino acids of the protein but which retained the 783 bp upstream of PilU, including the presumptive upstream regulatory sequences necessary for pilU production. We introduced the PpilA-lacZ reporter construct into the CTX site of pilTTrunc. This mutant is defective for TM, has increased surface pili, and could be complemented for TM by expression of pilT in cis or in trans (data not shown). pilTTrunc reproducibly showed a 40% increase in β-galactosidase activity relative to WT when assayed from bacteria cultured on solid media (Fig. 8A; P < 0.05), a finding consistent with the notion that pilin disassembly or intracellular pilin levels may regulate pilin transcription. pilA promoter activity was unchanged relative to the WT in the ΔpilU strain, further supporting the idea that PilU does not function in retraction. pilA transcription in the PAO1ΔpilH::CTX-PpilA-lacZ reporter strain, which is hyperpiliated, was reduced to 15% of WT, similar to the levels observed for the ΔchpA, ΔpilG, and ΔpilB strains (Fig. 8A). In addition to its role in mediating pilus retraction, it is possible that PilH may have a positive role in regulating pilA transcription.
We investigated and compared pilA transcription in WT and mutant strains cultured in liquid media. For WT PAO1, the levels of pilA transcription were decreased by ca. 95% for cells grown in liquid culture to stationary phase compared to cells cultured on solid media (Fig. 8A; P < 0.0001). This finding suggests that pilA transcription is highly repressed in WT cells cultured in liquid media. Furthermore, given that the intracellular and surface pilin levels were similar under the two different growth conditions (Fig. 8C), these results suggest that the turnover of pilin is much slower in cells cultured in liquid compared to on solid media. Nonetheless, even in liquid culture, pilA transcription was increased 27-fold relative to WT in the ΔpilA background (Fig. 8B, P < 0.05). The absolute levels of pilA transcription for broth- and plate-cultured bacteria in the ΔpilA background were similar, suggesting that pilin functions as a negative regulator under both culture conditions and that pilA transcription was maximally derepressed in the absence of PilA.
Similar to PAO1pilTTrunc bacteria cultured on solid media, the PAO1pilTTrunc strain cultured in broth had increased levels of surface pili and showed a threefold increase in pilA transcription compared to the WT (Fig. 8A and B; P < 0.05). The observation that PAO1pilTTrunc shows a moderate increase in β-galactosidase activity suggests that pili remain functional under liquid culture conditions and, as such, PilT continues to act as a functional regulator of pilA transcription by mediating intracellular levels of PilA. Similarly, the hyperpiliated phenotype of ΔpilH suggests a role in regulating pilus retraction in liquid as on solid media (Fig. 8C).
We did not detect decreases in pilA transcription for mutants defective in pilin export (i.e., the ΔchpA, ΔpilG, and ΔpilB mutants) under liquid culture conditions (Fig. 8B). Of interest, the levels of intracellular and surface pilin in ΔchpA and ΔpilG mutants were similar to those of the WT, suggesting that the role they play in the regulation of pilus dynamics may be culture condition specific (Fig. 8B). The ΔpilB mutant, however, lacks detectable surface pili when assayed from plates or out of broth, underscoring its essential role in controlling pilus extension (Fig. 8C). Unlike on solid media, however, the ΔpilB mutant export defect does not correspond to a decrease in intracellular pilin levels or in pilA transcription (Fig. 8B). Similarly, an apparent increase in pilin production in the ΔpilH strain does not correspond to increased pilA transcription (Fig. 8B). Some of these inconsistencies may be due, in part, to the global decrease in pilA transcription in cells cultured in liquid compared to on solid media, a finding that may be indicative of an overall decrease in pilus activity under liquid culture condition and which may make subtle changes in levels of pilA transcription difficult to detect.
DISCUSSION
P. aeruginosa uses Tfp for locomotion, adherence, and biofilm formation during growth in diverse environments. We have previously described the initial characterization of the Chp signaling system, a putative chemosensory system that regulates Tfp function and biofilm formation (3, 55). In the present study, we used genetic analysis to further understand the mechanism by which the Chp system controls Tfp function. We present data consistent with the hypothesis that PilA is a negative regulator of pilA transcription and that the Chp system, along with the ATPases PilB and PilT, serves to functionally regulate pilA transcription by modulating intracellular levels of PilA.
Through the construction and complementation of in-frame deletion mutants, we have confirmed the role of ChpA, PilG, and PilB in the regulation of pilus extension and the role of PilH and PilT in retraction. Our extensive analysis of Tfp phenotypes of double and triple mutants in factors involved in extension (chpA, pilG, and pilB) and those involved in retraction (pilH and pilT) suggest that factors involved in extension function upstream of those involved in retraction. Furthermore, our data indicate that while PilB is absolutely required for pilus extension, it retains some extension activity in the absence of upstream signaling input from either ChpA or PilG. Our data also indicate that PilH enhances PilT activity and that PilT retains some retractile activity in the absence of PilH.
It is intriguing, although not without precedence (51), that the Chp system uses two CheY-like proteins, PilG and PilH, to influence ATPase function, though the exact mechanism remains to be elucidated. These response regulators could affect association of PilB or PilT with the inner membrane components of the pilus system. Alternatively, they could promote PilB or PilT hexamer formation, or they could affect PilB or PilT turnover or polar localization. Whether these two response regulators compete for phosphorylation or have distinct subcellular localization will also be of interest to investigate in the future.
Our data present conflicting evidence for the role of PilU in mediating pilus retraction. PilU is a member of the family of hexameric secretion ATPases and demonstrates ATPase activity in vitro (8), has a high degree of homology to PilT (39% identical and 61% similar), and likely arose by gene duplication (56). Like PAO1ΔpilTCTX-pilU, PAO1ΔpilU is severely defective in TM, but it is phage sensitive and thus retains some retractile function (56; the present study). The most straightforward model is that PilU functions as an accessory retractile motor to PilT. Indeed, older published electron microscopy data show hyperpiliation of a pilU insertion mutant (56), suggesting a role for PilU in retraction, though surface pili levels were not measured.
In support of an accessory role for PilU in pilus retraction, we observed (i) that PAO1ΔpilHΔpilT exhibited increased surface piliation relative to PAO1ΔpilH and PAO1ΔpilTCTX-pilU; (ii) that residual surface piliation due to basal PilB activity is only apparent in a ΔpilG background when both pilT and pilU are deleted (i.e., PAO1ΔpilGΔpilTΔpilU); and (iii) that the levels of surface piliation in strain PAO1ΔchpAΔpilTpilU are clearly increased relative to strains PAO1ΔchpA and PAO1ΔchpAΔpilTCTX-pilU. However, if PilU has retractile activity, even if minor, we would predict that the PAO1ΔpilTΔpilU strain would have increased surface piliation relative to PAO1ΔpilTCTX-pilU. What we observed, however, is that the degree of surface piliation for PAO1ΔpilTΔpilU is the same as that for PAO1ΔpilTCTX-pilU and that the surface piliation of PAO1ΔpilHΔpilTΔpilU is similar to that of PAO1ΔpilHΔpilTCTX-pilU. It remains to be determined whether the number or the length of the pili differ or whether their spatial distribution is altered in these mutants. Nonetheless, these observations suggest an additional PilH-dependent retraction factor that is not PilU.
We show for the first time that the histidine kinase domain of ChpA is essential for Tfp function. ChpA is one of the most complex CheA homologs yet to be described. In addition to its histidine kinase domain, it possesses eight potential sites of phosphorylation and a CheY-like receiver domain at its C terminus. Previous studies have demonstrated that the second and third Hpt domains contribute to TM, while the CheY domain is absolutely essential; a potential role in the regulation of swarming motility has also been shown for Hpt4 and Hpt5 (37). It is possible that the ChpA histidine kinase domain catalyzes the autophosphorylation of Hpt2 and/or Hpt3, which then transfers phosphate to the C-terminal CheY domain. Alternatively, the phosphates may be transferred to PilG and/or PilH, which may compete with the CheY domain for phosphorylation. A combination of biochemical and genetic analyses will be required to further elucidate this complex signaling pathway. ChpA may have an additional role in the regulation of type III secretion. β-Galactosidase assays show decreased transcription of the type III secreted effector exoT, and microarray analysis suggests that ChpA plays some role in the regulation of expression of the type III secretion system (our unpublished data). How this regulation occurs and whether it is dependent upon the N-terminal domain of ChpA, which is homologous to FimL, a known regulator of type III secretion (53), or upon ChpA signaling, remains to be determined.
Our studies suggest a role for Tfp dynamics in regulating pilA transcription. First, we found that pilA transcription is much lower in WT bacteria cultured in liquid compared to bacteria grown on solid media, despite the fact that the amount of intracellular and surface pili is the same under both conditions. This finding suggests that pilin turnover is decreased in liquid-grown cultures relative to plate-grown cultures and may correlate with a decreased need for pilus pilus retraction or extension in liquid culture conditions. Second, in the absence of PilA or in the absence of retraction, pilA transcription is increased, suggesting that unassembled pilin subunits may function as a negative regulator of pilA transcription. Specifically, we observed a twofold increase in pilA transcription in ΔpilA and a reproducible but smaller increase in transcription in a pilT mutant. When we attempted to overproduce PilA by expression from a plasmid-borne copy of pilA under an inducible promoter, we were unable to increase pilin levels or to affect transcription from the native pilA promoter driving lacZ (unpublished data). Our data are consistent with previous reports that showed negative-feedback regulation of pilA transcription by PilA in P. aeruginosa (16) and in M. xanthus (29, 58), where it is postulated that unassembled pilin subunits are sensed by the PilR/PilS two-component regulatory system (29). Whitchurch and Mattick (56), in contrast, reported Northern blot analysis demonstrating decreased pilA mRNA in pilT or pilU insertion mutants. Third, our data suggest that the Chp system, along with the ATPases PilB and PilT, serves to functionally regulate pilA transcription by modulating intracellular levels of PilA. Specifically, we show that mutants with defects in PilA export (i.e., ΔchpA, ΔpilB, and ΔpilG mutants) have decreased pilA transcription, whereas a mutant defective in pilin disassembly (PAO1ΔpilTTrunc) demonstrated increased pilA transcription. Similarly, Johnson et al. (31) showed that pilA transcription by both Northern analysis and lacZ assay was upregulated in hyperpiliated mutants and downregulated in hypopiliated mutants. Our data are consistent with observations made in other organisms, such as the cyanobacterium Synechocystis sp. strain PCC6803 and Neisseria gonorrhea, where hyperpiliated pilT mutants have been shown to accumulate higher than normal levels of pilA1 transcript (5, 14). One attractive model is that as the pilus is dissembled, PilS senses pilin levels associated with the inner membrane and regulates PilR activity and pilA transcription.
We note that transposon insertion mutations in pilG and other Chp system components (pilH, pilI, and pilJ) have been reported to have WT levels of pilA transcription in strain PAK (12, 13), although at least one chpA insertion mutant has been shown to have a defect in pilA transcription (55). Point mutations to an Hpt domain (H1088Q) and the cheY domain (D2406N) of chpA resulted in increased pilA transcription by Northern analysis (37). The reason for the discrepancy between the insertion mutant, PAO1ΔchpA, and the point mutants remains to be determined; it may reflect strain differences or differences in culture conditions. It is also unclear if the increase in pilA transcription in the chpA point mutants corresponds to increased surface piliation. Mutations in components of other putative chemosensory systems proposed to regulate Tfp-driven motility have been shown to have defects in pilA transcription (57).
Our studies suggest that PilH regulates pathways in addition to pilin retraction, suggesting that this CheY homolog interfaces with multiple signal transduction circuits. The ΔpilH mutant has aberrant TM and increased surface pili. Based on its hyperpiliation phenotype, we would predict that the pilH mutant would show increased levels of pilA transcription; however, it is actually reduced to levels near those of the ΔchpA, ΔpilG, and ΔpilB mutants, suggesting that pilH may play some additional, positive role in regulating pilA transcription. Unique among the Chp system mutants, PAO1ΔpilH also shows altered pyocyanin and rhamnolipid production that correspond to changes in phzA1 and rhlA transcription and increased virulence in a Drosophila infection model (our unpublished data). Site-directed pilH mutants, in which the aspartate residue in the putative phosphoacceptor site is replaced with alanine, recapitulate the virulence factor production phenotypes of the null mutant (our unpublished data). Further studies will need to be carried out to determine the full range of PilH function.
The data presented here provides genetic evidence consistent with a model in which ChpA acts as a histidine kinase to phosphorylate PilG and/or PilH. PilG-P, in turn, modulates PilB to drive pilus extension, and PilH-P modulates PilT to drive pilus retraction (Fig. 1). Future studies will focus on complementing these genetic studies with biochemical analysis. The ability of ChpA to autophosphorylate, and on which of its eight potential phosphorylation sites, and to participate in phospho-transfer reactions with PilG, PilH, or its own CheY domain has yet to be demonstrated in vitro. Moreover, direct interactions between ChpA and PilG, ChpA and PilH, PilG and PilB, and PilH, and PilT/PilU remain to be shown. These interactions may be more complex than we previously imagined. A recent study demonstrated direct binding of PilZ and FimX, two proteins previously shown to have roles in the regulation of Tfp function, to PilB (19). Whether and how PilG interacts with this complex remains to be determined. It is also unclear whether a similar or completely novel complex exists for PilT and whether and how PilH might interact with those components of the retraction apparatus.
Acknowledgments
We thank Carol Gross, Cynthia Whitchurch, and current and past members of the Engel lab for discussion and comments. We thank Lori Burrows, Daniel Wozniak, and Rebecca Wilson for the generous donation of reagents.
This study was supported by NIH grant R01 AI42806 (J.N.E.).
Footnotes
Published ahead of print on 11 December 2009.
REFERENCES
- 1.Alm, R., and J. S. Mattick. 1995. Identification of a gene, pilV, required for type 4 fimbrial biogenesis in Pseudomonas aeruginosa, whose product possesses a pre-pilin-like leader sequence. Mol. Microbiol. 16:485-496. [DOI] [PubMed] [Google Scholar]
- 2.Baker, M. D., P. M. Wolanin, and J. B. Stock. 2006. Signal transduction in bacterial chemotaxis. Bioessays 28:9-22. [DOI] [PubMed] [Google Scholar]
- 3.Barken, K. B., S. J. Pamp, L. Yang, M. Gjermansen, J. J. Bertrand, M. Klausen, M. Givskov, C. B. Whitchurch, J. N. Engel, and T. Tolker-Nielsen. 2008. Roles of type IV pili, flagellum-mediated motility and extracellular DNA in the formation of mature multicellular structures in Pseudomonas aeruginosa biofilms. Environ. Microbiol. 10:2331-2343. [DOI] [PubMed] [Google Scholar]
- 4.Becher, A., and H. P. Schweizer. 2000. Integration-proficient Pseudomonas aeruginosa vectors for isolation of single-copy chromosomal lacZ and lux gene fusions. Biotechniques 29:948-952. [DOI] [PubMed] [Google Scholar]
- 5.Bhaya, D., N. R. Bianco, D. Bryant, and A. Grossman. 2000. Type IV pilus biogenesis and motility in the cyanobacterium Synechocystis sp. PCC6803. Mol. Microbiol. 37:941-951. [DOI] [PubMed] [Google Scholar]
- 6.Bradley, D. E., and T. L. Pitt. 1974. Pilus-dependence of four Pseudomonas aeruginosa bacteriophages with non-contractile tails. J. Gen. Virol. 23:1-15. [DOI] [PubMed] [Google Scholar]
- 7.Chiang, P., and L. L. Burrows. 2003. Biofilm formation by hyperpiliated mutants of Pseudomonas aeruginosa. J. Bacteriol. 185:2374-2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chiang, P., L. M. Sampaleanu, M. Ayers, M. Pahuta, P. L. Howell, and L. L. Burrows. 2008. Functional role of conserved residues in the characteristic secretion NTPase motifs of the Pseudomonas aeruginosa type IV pilus motor proteins PilB, PilT, and PilU. Microbiology 154:114-126. [DOI] [PubMed] [Google Scholar]
- 9.Cosson, P., L. Zulianello, O. Join-Lambert, F. Faurisson, L. Bebbie, M. Benghezal, C. van Delden, L. K. Curty, and T. Kohler. 2002. Pseudomonas aeruginosa virulence analyzed in a Dictyostelium discoideum host system. J. Bacteriol. 184:3027-3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318-1322. [DOI] [PubMed] [Google Scholar]
- 11.D'Argenio, D. A., L. A. Gallagher, C. A. Berg, and C. Manoil. 2001. Drosophila as a model host for Pseudomonas aeruginosa infection. J. Bacteriol. 183:1466-1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Darzins. 1993. The pilG gene product, required for Pseudomonas aeruginosa pilus production and twitching motility, is homologous to the enteric, single domain response regulator CheY. J. Bacteriol. 175:5934-5944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Darzins, A. 1994. Characterization of a Pseudomonas aeruginosa gene cluster involved in pilus biosynthesis and twitching motility: sequence similarity to the chemotaxis proteins of enterics and the gliding bacterium Myxococcus xanthus. Mol. Microbiol. 11:137-153. [DOI] [PubMed] [Google Scholar]
- 14.Dietrich, M., H. Mollenkopf, M. So, and A. Friedrich. 2009. Pilin regulation in the pilT mutant of Neisseria gonorrhoeae strain MS11. FEMS Microbiol. Lett. 296:248-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ellefson, D. D., U. Weber, and A. J. Wolfe. 1997. Genetic analysis of the catalytic domain of the chemotaxis-associated histidine kinase CheA. J. Bacteriol. 179:825-830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elleman, T. C., P. A. Hoyne, D. J. Stewart, N. M. McKern, and J. E. Peterson. 1986. Expression of pili from Bacteroides nodosus in Pseudomonas aeruginosa. J. Bacteriol. 168:574-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Engel, J. N. 2003. Molecular pathogenesis of acute Pseudomonas aeruginosa infections, p. 201-230. In A. Hauser and J. Rello (ed.), Severe infections caused by Pseudomonas aeruginosa. Kluwer Academic/Plenum Press, New York, NY.
- 18.Garrett, E. S., D. Perlegas, and D. J. Wozniak. 1999. Negative control of flagellum synthesis in Pseudomonas aeruginosa is modulated by the alternative sigma factor AlgT (AlgU). J. Bacteriol. 181:7401-7404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guzzo, C. R., R. K. Salinas, M. O. Andrade, and C. S. Farah. 2009. PILZ protein structure and interactions with PILB and the FIMX EAL domain: implications for control of type IV pilus biogenesis. J. Mol. Biol. 393:848-866. [DOI] [PubMed] [Google Scholar]
- 20.Hess, J. F., K. Oosawa, N. Kaplan, and M. I. Simon. 1988. Phosphorylation of three proteins in the signaling pathway of bacterial chemotaxis. Cell 53:79-87. [DOI] [PubMed] [Google Scholar]
- 21.Hirschman, A., M. Boukhvalova, R. VanBruggen, A. J. Wolfe, and R. C. Stewart. 2001. Active site mutations in CheA, the signal-transducing protein kinase of the chemotaxis system in Escherichia coli. Biochemistry 40:13876-13887. [DOI] [PubMed] [Google Scholar]
- 22.Hoang, T. T., A. J. Kutchma, A. Becher, and H. P. Schweizer. 2000. Integration-proficient plasmids for Pseudomonas aeruginosa: site-specific integration and use for engineering of reporter and expression strains. Plasmid 43:59-72. [DOI] [PubMed] [Google Scholar]
- 23.Inclan, Y. F., S. Laurent, and D. R. Zusman. 2008. The receiver domain of FrzE, a CheA-CheY fusion protein, regulates the CheA histidine kinase activity and downstream signaling to the A- and S-motility systems of Myxococcus xanthus. Mol. Microbiol. 68:1328-1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Inclan, Y. F., H. C. Vlamakis, and D. R. Zusman. 2007. FrzZ, a dual CheY-like response regulator, functions as an output for the Frz chemosensory pathway of Myxococcus xanthus. Mol. Microbiol. 65:90-102. [DOI] [PubMed] [Google Scholar]
- 25.Ishimoto, K. S., and S. Lory. 1989. Formation of pilin in Pseudomonas aeruginosa requires the alternative σ factor (RpoN) of RNA polymerase. Proc. Natl. Acad. Sci. U. S. A. 86:1954-1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ishimoto, K. S., and S. Lory. 1992. Identification of pilR, which encodes a transcriptional activator of the Pseudomonas aeruginosa pilin gene. J. Bacteriol. 174:3514-3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jakovljevic, V., S. Leonardy, M. Hoppert, and L. Sogaard-Andersen. 2008. PilB and PilT are ATPases acting antagonistically in type IV pilus function in Myxococcus xanthus. J. Bacteriol. 190:2411-2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jander, G., L. G. Rahme, and F. M. Ausubel. 2000. Positive correlation between virulence of Pseudomonas aeruginosa mutants in mice and insects. J. Bacteriol. 182:3843-3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jelsbak, L., and D. Kaiser. 2005. Regulating pilin expression reveals a threshold for S motility in Myxococcus xanthus. J. Bacteriol. 187:2105-2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jin, S., K. S. Ishimoto, and S. Lory. 1994. PilR, a transcriptional regulator of piliation in Pseudomonas aeruginosa, binds to a cis-acting sequence upstream of the pilin gene promoter. Mol. Microbiol. 14:1049-1057. [DOI] [PubMed] [Google Scholar]
- 31.Johnson, K., and S. Lory. 1987. Characterization of Pseudomonas aeruginosa mutants with altered piliation. J. Bacteriol. 169:5663-5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson, K., M. L. Parker, and S. Lory. 1986. Nucleotide sequence and transcriptional initiation site of two Pseudomonas aeruginosa pilin genes. J. Biol. Chem. 261:15703-15708. [PubMed] [Google Scholar]
- 33.Kaiser, D. 2006. A microbial genetic journey. Annu. Rev. Microbiol. 60:1-25. [DOI] [PubMed] [Google Scholar]
- 34.Kaiser, D. 2007. Bacterial swarming: a re-examination of cell-movement patterns. Curr. Biol. 17:R561-R570. [DOI] [PubMed] [Google Scholar]
- 35.Klausen, M., A. Aaes-Jorgensen, S. Molin, and T. Tolker-Nielsen. 2003. Involvement of bacterial migration in the development of complex multicellular structures in Pseudomonas aeruginosa biofilms. Mol. Microbiol. 50:61-68. [DOI] [PubMed] [Google Scholar]
- 36.Klausen, M., A. Heydorn, P. Ragas, L. Lambertsen, A. Aaes-Jorgensen, S. Molin, and T. Tolker-Nielsen. 2003. Biofilm formation by Pseudomonas aeruginosa wild type, flagella and type IV pili mutants. Mol. Microbiol. 48:1511-1524. [DOI] [PubMed] [Google Scholar]
- 37.Leech, A. J., and J. S. Mattick. 2006. Effect of site-specific mutations in different phosphotransfer domains of the chemosensory protein ChpA on Pseudomonas aeruginosa motility. J. Bacteriol. 188:8479-8486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li, Y., V. H. Bustamante, R. Lux, D. Zusman, and W. Shi. 2005. Divergent regulatory pathways control A and S motility in Myxococcus xanthus through FrzE, a CheA-CheY fusion protein. J. Bacteriol. 187:1716-1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mandell, G. L., J. E. Bennett, and R. Dolin. 2005. Principles and practice of infectious diseases, 6th ed. Churchill Livingstone, Inc., New York, NY.
- 40.Mattick, J. S. 2002. Type iv pili and twitching motility. Annu. Rev. Microbiol. 56:289-314. [DOI] [PubMed] [Google Scholar]
- 41.Merz, A. J., M. So, and M. P. Sheetz. 2000. Pilus retraction powers bacterial twitching motility. Nature 407:98-102. [DOI] [PubMed] [Google Scholar]
- 42.Nunn, D., S. Bergman, and S. Lory. 1990. Products of three accessory genes, pilB, pilC, and pilD, are required for biogenesis of Pseudomonas aeruginosa pili. J. Bacteriol. 172:2911-2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O'Toole, G. A., and R. Kolter. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30:295-304. [DOI] [PubMed] [Google Scholar]
- 44.Porter, S. L., G. H. Wadhams, and J. P. Armitage. 2008. Rhodobacter sphaeroides: complexity in chemotactic signaling. Trends Microbiol. 16:251-260. [DOI] [PubMed] [Google Scholar]
- 45.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 46.Schweizer, H. P., and T. T. Hoang. 1995. An improved system for gene replacement and xylE fusion analysis in Pseudomonas aeruginosa. Gene 158:15-22. [DOI] [PubMed] [Google Scholar]
- 47.Skerker, J. M., and H. C. Berg. 2001. Direct observation of extension and retraction of type IV pili. Proc. Natl. Acad. Sci. U. S. A. 98:6901-6904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stover, C., X. Pham, A. Erwin, S. Mizoguchi, P. Warrener, M. Hickey, F. Brinkman, W. Hufnagle, D. Kowalik, M. Lagrou, R. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. Brody, S. Coulter, K. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. Wong, Z. Wu, and I. Paulsen. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 49.Tan, M. W., L. G. Rahme, J. A. Sternberg, R. G. Tompkins, and F. M. Ausubel. 1999. Pseudomonas aeruginosa killing of Caenorhabditis elegans used to identify P. aeruginosa virulence factors. Proc. Natl. Acad. Sci. U. S. A. 96:2408-2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. U. S. A. 76:4350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wadhams, G. H., and J. P. Armitage. 2004. Making sense of it all: bacterial chemotaxis. Nat. Rev. Mol. Cell. Biol. 5:1024-1037. [DOI] [PubMed] [Google Scholar]
- 52.Whitchurch, C., S. Beatson, J. Comolli, T. Jakobsen, J. Sargent, J. Bertrand, M. Klausen, L. Waite, T. Tolker-Nielsen, P. J. Kang, J. Mattick, and J. N. Engel. 2005. FimL, a novel Pseudomonas aeruginosa gene product involved in type IV fimbrial function and twitching motility. Mol. Microbiol. 55:1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Whitchurch, C. B., S. A. Beatson, J. C. Comolli, T. Jakobsen, J. L. Sargent, J. J. Bertrand, J. West, M. Klausen, L. L. Waite, P. J. Kang, T. Tolker-Nielsen, J. S. Mattick, and J. N. Engel. 2005. Pseudomonas aeruginosa fimL regulates multiple virulence functions by intersecting with Vfr-modulated pathways. Mol. Microbiol. 55:1357-1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Whitchurch, C. B., M. Hobbs, S. P. Livingston, V. Krishnapillai, and J. S. Mattick. 1991. Characterization of a Pseudomonas aeruginosa twitching motility gene and evidence for a specialized protein export system widespread in eubacteria. Gene 101:33-44. [DOI] [PubMed] [Google Scholar]
- 55.Whitchurch, C. B., A. J. Leech, M. D. Young, D. Kennedy, J. L. Sargent, J. J. Bertrand, A. B. Semmler, A. S. Mellick, P. R. Martin, R. A. Alm, M. Hobbs, S. A. Beatson, B. Huang, L. Nguyen, J. C. Commolli, J. N. Engel, A. Darzins, and J. S. Mattick. 2004. Characterization of a complex chemosensory signal transduction system which controls twitching motility in Pseudomonas aeruginosa. Mol. Microbiol. 52:873-893. [DOI] [PubMed] [Google Scholar]
- 56.Whitchurch, C. B., and J. S. Mattick. 1994. Characterization of a gene, pilU, required for twitching motility but not phage sensitivity in Pseudomonas aeruginosa. Mol. Microbiol. 13:1079-1081. [DOI] [PubMed] [Google Scholar]
- 57.Winther-Larsen, H. C., and M. Koomey. 2002. Transcriptional, chemosensory and cell-contact-dependent regulation of type IV pilus expression. Curr. Opin. Microbiol. 5:173-178. [DOI] [PubMed] [Google Scholar]
- 58.Wu, S. S., and D. Kaiser. 1997. Regulation of expression of the pilA gene in Myxococcus xanthus. J. Bacteriol. 179:7748-7758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zusman, D. R., A. E. Scott, Z. Yang, and J. R. Kirby. 2007. Chemosensory pathways, motility and development in Myxococcus xanthus. Nat. Rev. Microbiol. 5:862-872. [DOI] [PubMed] [Google Scholar]