Abstract
Magnetotactic bacteria (MTB) synthesize unique organelles termed “magnetosomes,” which are membrane-enclosed structures containing crystals of magnetite or greigite. Magnetosomes form a chain around MamK cytoskeletal filaments and provide the basis for the ability of MTB to navigate along geomagnetic field lines in order to find optimal microaerobic habitats. Genomes of species of the MTB genus Magnetospirillum, in addition to a gene encoding the tubulin-like FtsZ protein (involved in cell division), contain a second gene termed “ftsZ-like,” whose function is unknown. In the present study, we found that the ftsZ-like gene of Magnetospirillum gryphiswaldense strain MSR-1 belongs to a 4.9-kb mamXY polycistronic transcription unit. We then purified the recombinant FtsZ-like protein to homogeneity. The FtsZ-like protein efficiently hydrolyzed ATP and GTP, with ATPase and GTPase activity levels of 2.17 and 5.56 μmol phosphorus per mol protein per min, respectively. The FtsZ-like protein underwent GTP-dependent polymerization into long filamentous bundles in vitro. To determine the role of the ftsZ-like gene, we constructed a ftsZ-like mutant (ΔftsZ-like mutant) and its complementation strain (ΔftsZ-like_C strain). Growth of ΔftsZ-like cells was similar to that of the wild type, indicating that the ΔftsZ-like gene is not involved in cell division. Transmission electron microscopic observations indicated that the ΔftsZ-like cells, in comparison to wild-type cells, produced smaller magnetosomes, with poorly defined morphology and irregular alignment, including large gaps. Magnetic analyses showed that ΔftsZ-like produced mainly superparamagnetic (SP) magnetite particles, whereas wild-type and ΔftsZ-like_C cells produced mainly single-domain (SD) particles. Our findings suggest that the FtsZ-like protein is required for synthesis of SD particles and magnetosomes in M. gryphiswaldense.
Magnetotactic bacteria (MTB) can orient themselves along geomagnetic field lines and search for microaerophilic environments. These capabilities are based on unique prokaryotic organelles termed magnetosomes (3). Magnetosomes are nanometer-size magnetic particles of iron oxide (magnetite; Fe3O4) or iron sulfide (greigite; Fe3S4) (4, 5, 45), enclosed within intracytoplasmic vesicles of the magnetosome membrane (MM) (3, 43). Magnetosome formation is a complex process involving vesicle formation, iron transportation, nucleation and growth of magnetite crystals, and their assembly into chain-like structures. A model for magnetosome formation has been proposed by Komeili (18) and Schüler (44). According to this model, magnetosome vesicles are invaginated from the inner membrane, and protein sorting to the MM occurs concurrently. The protein MamA was suggested to activate magnetosome vesicles for magnetite biomineralization (19). With the help of the MamK and MamJ proteins, the membrane invaginations are then assembled into a chain structure. The bacterial actin-like MamK can form filaments required for maintaining magnetosome organization and function (20, 33). The ΔmamJ mutant shows no change in the synthesis of magnetite crystals but is unable to produce a straight magnetosome chain (42). The acidic MamJ is implicated in the control of magnetosome chain assembly. The final step is magnetite biomineralization, including iron uptake and the nucleation and growth of magnetite crystals. Four small proteins, MamG, MamF, MamD, and MamC, control the grain size of magnetite crystals (41). The acidic Mms6, which is a tightly bound constituent of the MM in Magnetospirillum magneticum AMB-1, had a striking effect on the morphology of growing magnetite crystals in vitro (2, 34).
In MTB, most of the genes involved in magnetosome formation are clustered in a genomic magnetosome island (MAI) (52). In addition to the actin-like mamK gene, MAIs of Magnetospirillum spp. contain a tubulin-like gene termed “ftsZ-like.” Comparisons among five sequenced strains of MTB (Magnetospirillum gryphiswaldense strain MSR-1, M. magneticum strain AMB-1, magnetic coccus strain MC-1, Magnetospirillum magnetotacticum strain MS-1, and Desulfovibrio magneticus strain RS-1) showed that the ftsZ-like gene is conserved only among the Magnetospirillum strains (30, 36). The FtsZ-like protein of M. gryphiswaldense has ∼84% similarity to the FtsZ-like proteins of M. magneticum and M. magnetotacticum. ftsZ-like is the last gene of the mamXY cluster in magnetospirilla. In M. gryphiswaldense MSR-1, the ∼4.9-kb mamXY cluster, which is located ∼28 kb downstream of the mamAB operon, consists of the mamY, mamX, mamH-like, and ftsZ-like genes. The predicted FtsZ-like protein consists of tubulin and tubulin C-terminal domains. These domains are present in prokaryotic homologs of eukaryotic tubulins. The ftsZ gene encodes a GTPase (9, 35). The bacterial FtsZ protein is an essential component of the cell division apparatus, assembling in a GTP-dependent manner a cytokinetic ring structure (Z ring) that mediates cell division (6). The Z ring then recruits members of the cell division complex, including FtsA, ZipA, MinC, and others that promote cytokinesis (53). The function of the FtsZ-like protein is unknown.
In the present study, we found that the ftsZ-like gene was expressed as a polycistronic mamXY unit under physiological conditions. The purified FtsZ-like protein displayed both ATPase and GTPase activities and polymerized into tubulin filament bundles in a GTP-dependent manner in vitro. Deletion of the ftsZ-like gene had no effect on cell division but resulted in the production of superparamagnetic (SP) crystals at ambient temperature.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids used in this study are described in Table 1. Escherichia coli strains were grown in Luria-Bertani (LB) medium at 37°C. M. gryphiswaldense strains were grown at 30°C in optimized flask medium (OFM) (49). Sterilized ferric citrate was added as the iron source after autoclaving. For conjugation, M. gryphiswaldense strains were cultured in selection medium, in which NH4Cl and yeast extract were replaced by 4 g sodium glutamate (22). For liquid culture, strains were kept in 250-ml serum bottles containing 100 ml medium, with shaking at 100 rpm. Microaerobic conditions occurred at high cell densities as a result of oxygen consumption. The following antibiotics and concentrations (μg ml−1) were used: kanamycin (Km), 50 for E. coli and 5 for M. gryphiswaldense; gentamicin (Gm), 20 for E. coli and 5 for M. gryphiswaldense; and tetracycline (Tc), 12.5 for E. coli and 5 for M. gryphiswaldense.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| E. coli strains | ||
| DH5α | endA1 hsdR17(r− m+) supE44 thi-1 recA1 gyrA (NalR) | 39 |
| relA1 Δ(lacZYA-argF)U169 deoR [φ80dlacZΔM15] | ||
| S17-1 | thi endA recA hsdR with RP4-2-Tc::Mu-Km::Tn7 integrated in the chromosome, Smr | 47 |
| M. gryphiswaldense strains | ||
| MSR-1 | Wild type | DSM6361 |
| ΔftsZ-like | ftsZ-like-deficient mutant, Gmr | Present study |
| ΔftsZ-like_C | ΔftsZ-like-carrying plasmid pCFZL, Gmr Tcr | Present study |
| Plasmids | ||
| pRK415 | Cloning vector, pRK290 derivative, Tcr | 17 |
| pK19mobsacB | sacB (modified from B. subtilis), lacZ, Kmr | 40 |
| pUCGm | pUC1918 carrying the aacC1 gene, Gmr | 46 |
| pET-28a-c(+) | Expression vector, T7 promoter, Kmr | Novagen |
| pDFZ1 | pK19mobsacB derivative for ftsZ-like deletion, Kmr | Present study |
| pDFZ2 | pK19mobsacB derivative for ftsZ-like deletion, Kmr Gmr | Present study |
| pCFZL | pRK415 derivative for ftsZ-like expression, Tcr | Present study |
| pEFZL | pET-28a-c(+) derivative for ftsZ-like expression, Kmr | Present study |
Reverse transcription (RT)-PCR.
M. gryphiswaldense cells were grown until exponential phase, and total RNA was isolated by the SV total RNA isolation system (Promega), according to manufacturer's instructions. RT-PCR analyses were performed with 0.6 μg RNA pretreated with RQ1 RNase-free DNase (Promega), using Moloney murine leukemia virus (MMLV) reverse transcriptase (Tiangen). A negative-control reaction for RT-PCR was performed using total RNA without MMLV reverse transcriptase to confirm the lack of genomic DNA contamination in each reaction mixture. Primers used to amplify the cDNA from total RNA were random primers. Primers used to amplify the intergenic regions of the mamXY cluster were as follows: 5′mamY-mamY_f/5′mamY-mamY_r (5′-AAGCGACCATCAATGTCCACT-3′; 5′-CCTCCGTTAATCGTCTTTGATAC-3′). mamY-mamX_f/mamY-mamX_r (5′-ATTGGTGCCGAGAATGTGCG-3′; 5′-TGTGATCTTCCCACGGATTGG-3′), mamX-mamH-like_f/mamX-mamH-like_r (5′-CCCATGTGGATCGACCTTGC-3′; 5′-CCCACGCAGACGATGGAGAC-3′), mamH-like-ftsZ-like_f/mamH-like-ftsZ-like_r (5′-GCGGCCTTCCTCCTGCTGAT-3′; 5′-AGGGCTTGGGCGTCCGTATT-3′), and ftsZ-like-3′ftsZ-like_f/ftsZ-like-3′ftsZ-like_r (5′-GTTATGAGCGAGATGGGCAAGG-3′; 5′-GGCCGTCTTCATGGTGTTGG-3′).
Construction of the ftsZ-like deletion mutant.
Gene replacement was performed by homologous recombination of flanking sequence regions of the ftsZ-like gene between the M. gryphiswaldense MSR-1 chromosome and pDFZ2. A 1,385-bp upstream region and a 1,396-bp downstream region of ftsZ-like were amplified using the primers DCftsZ-likeu_f/DCftsZ-likeu_r (5′-CGCGGATCCGGTGTGTTCATCATCATGTGCCTG-3′; 5′-AAAACTGCAGAATACCATTTAGCCGATTCGCAGGG-3′) and DCftsZ-liked_f/DCftsZ-liked_r (5′-AAAACTGCAGTGAGCCGCTCCGGAAGAATCAAGC-3′; 5′-CCCAAGCTTGGGATACTGACTAGAGCGGTTTCACG-3′), respectively (restriction sites are underlined). These two fragments were cloned into the pK19mobsacB vector at the BamHI and HindIII sites, yielding plasmid pDFZ1. Finally, the aacC1 gene (encoding Gmr) from pUCGm was inserted into pDFZ1 at the PstI site, resulting in suicide plasmid pDFZ2. pDFZ2 was transferred from E. coli S17-1 into M. gryphiswaldense MSR-1 by biparental conjugation as described previously (38). Gmr colonies were isolated after 7 days and replica printed to identify Kms colonies. Gmr Kms colonies resulting from a double-crossover recombination event were confirmed by PCR and Southern blot analysis. To complement the ftsZ-like mutant (ΔftsZ-like mutant), the M. gryphiswaldense ftsZ-like gene was amplified using primer CLftsZ-like_f/CLftsZ-like_r (5′-CTAGTCTAGACACCACGGGAGCAGCCCTTATTTTTGGCGTTGAACCCATTAGCTTCTG-3′; 5′-CGGGGTACCGCTTGATTCTTCCGGAGCGGC-3′) (restriction sites are underlined). The DNA fragment encoding the ftsZ-like gene was cloned into the XbaI and KpnI sites of the pRK415 vector, resulting in pCFZL.
Expression and purification of the FtsZ-like protein.
The DNA fragment encoding the ftsZ-like gene was amplified from chromosomal DNA of M. gryphiswaldense MSR-1 by PCR and cloned into the SacI and XhoI sites of the pET28a(+) (Novagen) vector. The resultant plasmid pEFZL expressed the FtsZ-like protein with a His6 tag fused at its N terminus. The primer used for FtsZ-like cloning was EXftsZ-like_f/EXftsZ-like_r (5′-CCCGAGCTCATTTTTGGCGTTGAACCCATTAGC-3′ and 5′-CCGCTCGAGTCAGGCGATACCGGTGGC-3′) (restriction sites are underlined). E. coli BL21(DE3) containing the pEFZL plasmid was cultured in LB medium supplemented with 50 μg/ml kanamycin and grown at 37°C. Protein expression was induced at A600 of 0.6 by 0.5 mM isopropyl-1-thio-β-d-galactopyranoside. Cells were harvested after 6 h, pelleted at 8,000 rpm and 4°C for 10 min, and resuspended in lysis buffer (50 mM NaH2PO4, 300 mM NaCl, and 10 mM imidazole at pH 8.0). Cells were disrupted by sonication, and the solution was centrifuged at 12,000 × g for 10 min at 4°C. The supernatant was loaded onto a nickel-nitrilotriacetic acid spin column (Qiagen) equilibrated with lysis buffer. The column was washed four times with wash buffer (50 mM NaH2PO4, 1 M NaCl, 50 mM imidazole, 0.5% [vol/vol] Triton X-100, and 5% [vol/vol] glycerol at pH 8.0). Proteins were eluted by stepwise increases of imidazole concentrations (150, 200, 250 mM) in elution buffer (50 mM NaH2PO4, 300 mM NaCl, and 250 mM imidazole at pH 8.0). The N-terminal His6 tag was removed by digestion with thrombin (15). The eluate was dialyzed against 25 mM HEPES-NaOH (pH 7.2), 1 mM dithiothreitol (DTT), 0.1 mM EDTA, and 10% glycerol and stored at −80°C until use. The proteins were dialyzed against a specific buffer before being used for assaying a specific property. Proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) with Coomassie blue R-250 staining and quantified by the Bradford method, using bovine serum albumin (BSA) as standard. Matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) was performed at Beijing Genomics Institute (BGI).
Nucleoside triphosphate (NTP) hydrolysis.
The nucleoside triphosphatase (NTPase) activity of FtsZ-like was measured with the EnzChek phosphate assay kit (catalog no. E6646; Molecular Probes, Invitrogen), which utilizes the spectrophotometric shift of maximal absorbance from 330 to 360 nm when the substrate 2-amino-6-mercapto-7-methylpurine riboside (MESG) is converted enzymatically by purine nucleoside phosphorylase (PNP) into ribose 1-phosphate and 2-amino-6-mercapto-7-methylpurine. For accurate measurement of NTPase activity, 1 mM GTP was preincubated with the kit reagent for 10 min at 22°C to deplete free phosphate before mixing with the protein sample, which included FtsZ-like, in 50 mM HEPES at pH 7.5, 200 mM KCl, 175 mM NaCl, and 2.5 mM MgCl2. The concentration of FtsZ-like protein in each experiment was 5 μM. Absorbance at 360 nm of the sample was recorded every 10 s at room temperature using a UVmini-1240 UV-visible (UV-VIS) spectrophotometer (Shimadzu). For control experiments in which GTP was added to the regenerating system in the absence of FtsZ-like, the A360 readout was flat, indicating that all inorganic phosphate present in GTP was converted to a signal by the PNP. The rate of NTP hydrolysis was calculated based on the regression line of phosphate standards, and the value for preexisting inorganic phosphate in GTP was subtracted. Experiments were performed in triplicate, and mean values are presented.
Transmission electron microscopy (TEM).
M. gryphiswaldense cells (wild-type, ΔftsZ-like, and the ΔftsZ-like complementation strain [ΔftsZ-like_C strain] cells) were grown in OFM at 30°C until stationary phase and concentrated from suspension by centrifugation (12,000 rpm, 1 min). Cells and isolated magnetosomes were adsorbed onto carbon-coated copper grids and rinsed twice with double-distilled H2O. Samples were observed with a conventional TEM (Philips Tecnai F30) at 120 kV and a high-resolution TEM (JEOL 2010) at 200 kV.
An electron microscopic assay for FtsZ-like polymerization was performed as described previously (54). FtsZ-like protein (6 mM) was incubated in assembly buffer (50 mM Tris at pH 7.5, 1 mM MgCl2, 1 mM GTP, 10 mM CaCl2) at 37°C for 5 min. Samples (5 μl each) were placed on a carbon-coated copper grid, blotted, negatively stained with 1% uranyl acetate, blotted again, and observed with the conventional TEM.
Magnetic analyses.
To characterize the magnetic properties of magnetosomes, freeze-dried cell samples (washed with double-distilled H2O) were analyzed. Low-temperature magnetic measurements were taken using a Quantum Design MPMS XL-5 magnetometer (sensitivity, 5.0 × 10−10 Am2). Thermal demagnetization of remanence acquired in a 2.5-T field at 5 K (hereafter named SIRM5K_2.5T) after two pretreatments was measured from 5 K to 300 K. The first pretreatment was cooling the sample from 300 K to 5 K in a zero field (zero field cooled [ZFC]), and the second was cooling it from 300 K to 5 K in a 2.5-T field (field cooled [FC]). The Verwey transition temperature (Tv) was defined as the temperature corresponding to the maximal first-order derivative dM/dT of the FC curve. Room temperature first-order reversal curves (FORCs) were measured on an alternating gradient magnetometer (sensitivity, 1.0 × 10−11 Am2; MicroMag model 2900) using the protocol of Roberts et al. (37). FORC diagrams were calculated using FORCinel version 1.05 software, with a smoothing factor (SF) of 2 (14). FORC diagrams provide information on the domain state, coercivity, and magnetostatic interaction of magnetic crystals. In a FORC diagram, coercive field (Hc) and magnetostatic interaction fields (Hb) are indicated on the horizontal and vertical axes, respectively.
RESULTS
Transcriptional organization of the mamXY gene cluster.
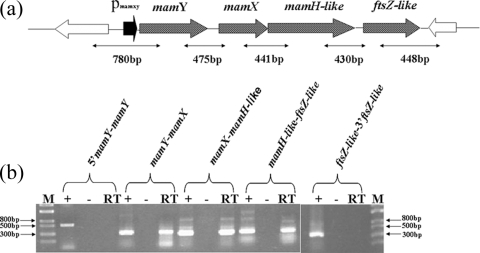
The ftsZ-like gene is located at the last position of the operon-like, colinear organization of the mamXY cluster (Fig. 1a). We applied RT-PCR to determine whether ftsZ-like is transcribed under physiological conditions and whether adjacent genes are cotranscribed, using primers that amplify intergenic regions of the mamXY cluster. Transcripts were detected for all tested intergenic junctions but not for regions located at the 5′ and 3′ ends of the first and last genes of this cluster (Fig. 1b). Amplicons obtained from the cDNAs were the same size as amplicons obtained from genomic DNA. Negative-control experiments, performed by omitting reverse transcriptase during RT reactions, revealed the complete absence of DNA in the RNA samples. These results indicate that the ftsZ-like gene is expressed from a single, long, polycistronic mamXY transcript unit under physiological conditions.
FIG. 1.
(a) Diagram of the mamXY cluster of M. gryphiswaldense strain MSR-1; (b) transcriptional analysis of mamXY cluster by RT-PCR. Expected sizes of PCR products are indicated below the arrows. Agarose gel electrophoresis of PCR products is shown at the bottom of each panel. Lanes: RT, RT-PCR; −, negative control with reverse transcriptase omitted; +, positive control with genomic DNA as the template; M, DNA size marker.
The FtsZ-like protein is an ATPase and GTPase.
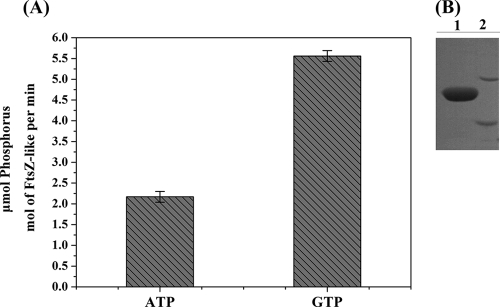
The FtsZ-like protein consists of 323 amino acids and has a predicted molecular mass of 33.7 kDa. The molecular mass of His-FtsZ-like, determined by SDS-PAGE, was ∼40 kDa (Fig. 2B). The His-FtsZ-like protein showed somewhat higher migration than that expected from the calculated molecular weight but was identified by MALDI-TOF MS.
FIG. 2.
Detection of ATP and GTP hydrolytic activities of the purified FtsZ-like protein. (A) ATP and GTP hydrolytic activities of FtsZ-like. (B) SDS-PAGE of purified His-FtsZ-like protein. Lane 1, purified His-FtsZ-like protein; lane 2, molecular mass markers of 45 kDa (upper) and 35 kDa.
The presence of the tubulin signature motif and FtsZ signature sequences within FtsZ-like suggested that the protein might be a GTPase. We therefore tested the ability of FtsZ-like to hydrolyze GTP and other NTPs, using a spectrophotometric method that measures the phosphate released from NTP hydrolysis. Native FtsZ-like protein hydrolyzed GTP efficiently (Fig. 2A). Purified FtsZ-like displayed GTPase activity, converting 5.56 ± 0.13 μmol phosphorus per mol FtsZ-like per min. GTPase activity of M. gryphiswaldense FtsZ-like was higher than those reported for E. coli FtsZ (48) and Bacillus subtilis FtsZ (8). Unexpectedly, in a test of ATPase activity, FtsZ-like converted 2.17 ± 0.13 μmol phosphorus per mol FtsZ-like per min, an efficiency that is ∼2.6 times lower than that for GTP (Fig. 2A). FtsZ-like did not hydrolyze TTP or CTP (data not shown). These results indicate that FtsZ-like protein has both ATPase and GTPase functions.
FtsZ-like undergoes GTP-dependent polymerization.
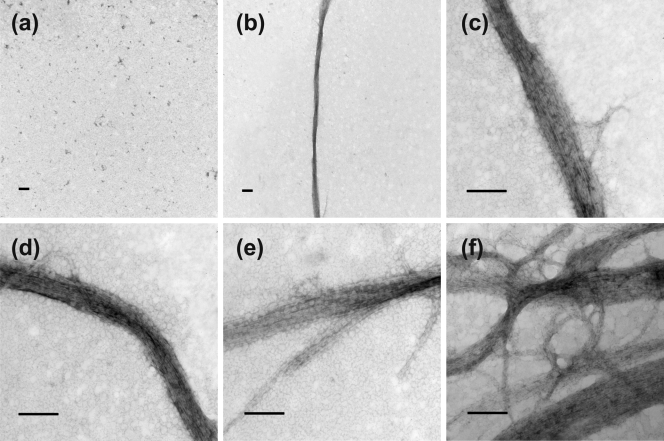
The capacity of the FtsZ-like protein to undergo GTP-dependent polymerization was evaluated using electron microscopy. No polymerization was observed in the absence of GTP (Fig. 3a). In the presence of GTP and 6 mM protein, FtsZ-like formed long, straight, polymeric structures (Fig. 3b), which actually consisted of bundles of protofilaments (Fig. 3c and d). Measurement of individual bundles showed lengths of up to 13.2 μm and widths of up to 195.8 nm. Macromolecular crowding of the protofilaments prevented us from observing single FtsZ-like filaments, and it was therefore difficult to estimate the number of protofilaments per bundle. Occasional bifurcation of polymers was observed at the ends of bundles (Fig. 3e). TEM observations suggested that interconnection between bundles to form a network occurs by lateral alignment of protofilaments (Fig. 3f).
FIG. 3.
TEM images of FtsZ-like polymers. (a) FtsZ-like in the absence of GTP; (b) well-developed bundle of FtsZ-like in the presence of GTP; (c to f) filamentous bundles of FtsZ-like. Scale bars, 1 μm in panels a and b and 200 nm in panels c to f.
Characterization of the ΔftsZ-like mutant.
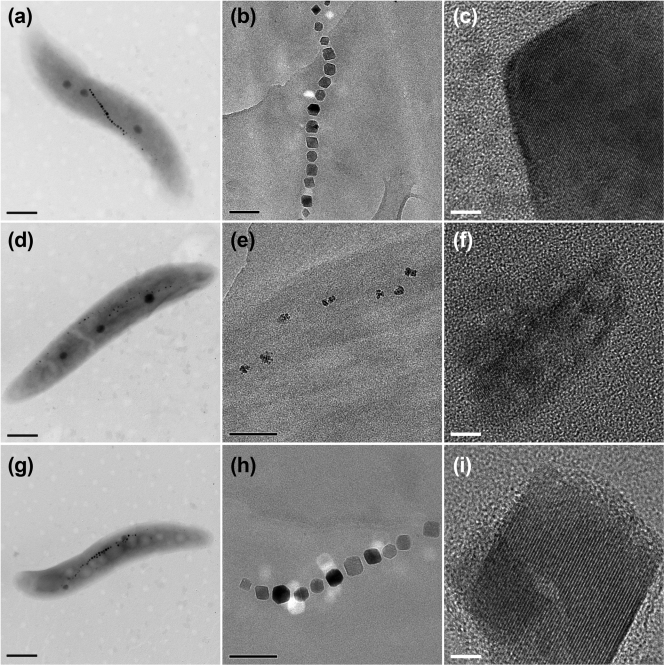
To determine the role of the ftsZ-like gene in the formation of functional magnetosomes, a ΔftsZ-like deletion mutant was constructed. Although the highest optical density at 565 nm (OD565) value of ΔftsZ-like cells was slightly lower than that of the wild type, they both reached exponential stage after 21 h of culture in medium supplemented with 60 μM iron, and deletion of the ftsZ-like gene had no major effect on growth (see Fig. S1 in the supplemental material). TEM micrographs of the mutant cells showed distinctively small magnetosome crystals (Fig. 4). The average magnetosome size for the ΔftsZ-like mutant was 24.0 ± 4.6 nm, compared to that of 37.9 ± 5.3 nm for wild-type cells (Table 2); this difference was statistically significant (P < 0.001; t test). High-resolution TEM observation showed that magnetosomes produced by ΔftsZ-like cells had poorly defined morphology, poor crystallization, and irregular distribution with large gaps (Fig. 4d to f). Magnetosomes produced by wild-type cells were clearly distinct, with symmetrical cuboid crystals, perfect crystal structure, and linear chain assembly (Fig. 4a to c). Formation of the aberrant magnetosome crystals in the ΔftsZ-like mutant was not compensated by their high iron concentration (500 μM), as indicated by TEM observations (data not shown). The difference in the number of magnetosomes per cell between the mutant and the wild type (Table 2) was not statistically significant (P > 0.05). Gaps between adjacent magnetosome particles in ΔftsZ-like were larger, and the magnetosome chain was longer than that of the wild type.
FIG. 4.
TEM images of cells and magnetosomes of M. gryphiswaldense strains. Cells (left column) were viewed by conventional TEM (scale bar, 500 nm; Philips Tecnai F30). Magnetosome chains of whole cells (middle column; scale bar, 100 nm) and crystal lattice of isolated magnetosomes (right column; scale bar, 5 nm) were viewed by high-resolution TEM (JEOL 2010). (a to c) Wild type; (d to f) ΔftsZ-like; (g to i) ΔftsZ-like_C.
TABLE 2.
Comparative data on magnetosome diameters and numbers in three M. gryphiswaldense strains grown in OFM until stationary phasea
| Strain | Avg diam of magnetosomes (nm) | Avg no. of magnetosomes per cell |
|---|---|---|
| Wild type | 37.9 (5.3) | 25.2 (5.4) |
| ΔftsZ-like | 24.0 (4.6) | 26.1 (4.4) |
| ΔftsZ-like_C | 39.2 (6.7) | 26.5 (3.8) |
Standard deviations are given in parentheses.
To test whether the above-described phenotypes were really caused by deletion of the ftsZ-like gene, the mutant was complemented with ftsZ-like expressed from the lac promoter. Complementation of the ΔftsZ-like strain with plasmid pCFZL containing the entire ftsZ-like gene produced cells (ΔftsZ-like_C) having magnetosomes with physical properties very similar to those of the wild type, i.e., cuboid shape, linear chain arrangement, and well-defined lattice fringes (Fig. 4g to i; Table 2). Magnetosomes produced by ΔftsZ-like_C had grain size slightly larger than that of the wild type, possibly because of multiple copy expression of ftsZ-like from plasmid pCFZL (Table 2). The diameters of magnetosomes of ΔftsZ-like_C were not statistically different from those of the wild type (P > 0.05), although the OD565 values from the ΔftsZ-like_C culture were slightly lower than those from the wild-type culture (See Fig. S1 in the supplemental material). A likely explanation for this is expression of ftsZ-like from a heterologous promoter carried on the plasmid. Taken together, these findings support the idea that deletion of the ftsZ-like gene was the cause of phenotypes observed in ΔftsZ-like cells.
Magnetic properties of wild-type cells and their ΔftsZ-like deletion mutants.
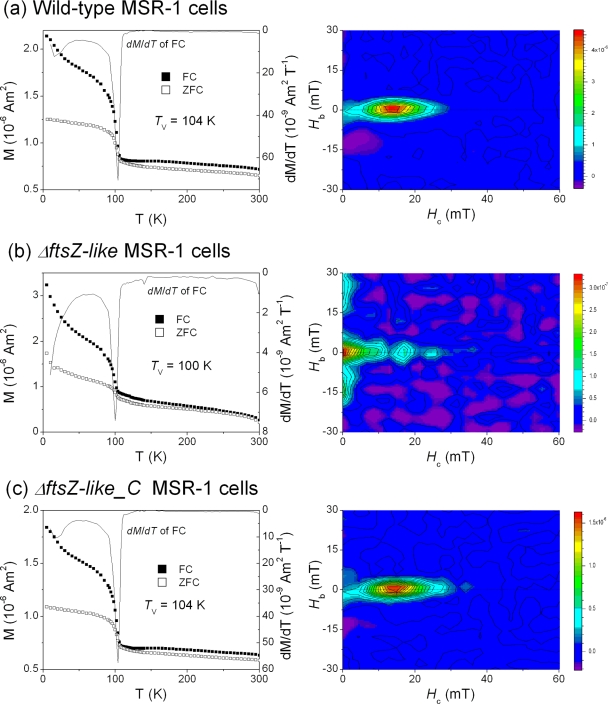
Thermal demagnetization curves and FORC diagrams for whole-cell samples of M. gryphiswaldense MSR-1 wild-type, ΔftsZ-like, and ΔftsZ-like_C cells, as described above, are shown in Fig. 5. Both ZFC and FC curves for all three strains showed sharp drops in remanence between 90 and 110 K, which corresponds to the Verwey transition of magnetosome magnetite (23, 27, 32). The presence of Verwey transition confirms that the magnetosomes synthesized by ΔftsZ-like cells are magnetite rather than other minerals, e.g., maghemite or ferrihydrite (11). However, Verwey transition behaviors differed between ΔftsZ-like and the wild type. ΔftsZ-like_C showed thermal demagnetization behavior that was nearly identical to that of the wild type, and both had sharp Verwey transitions, with Tv of 104 K (Fig. 5a and c). In contrast, ΔftsZ-like showed a suppressed Verwey transition, with Tv of 100 K (Fig. 5b). The remanence carried by ΔftsZ-like decayed dramatically below 50 K and fell to 8.1% of the FC_SIRM5K_2.5T value at 300 K, which is much lower than the corresponding percentages for the wild type (33.5%) and ΔftsZ-like_C (34.5%). These findings indicate a significant contribution of superparamagnetic (SP) magnetite particles (11, 23), probably at the threshold between SP and single-domain (SD) particles, in ΔftsZ-like cells, which is consistent with the TEM observations described above.
FIG. 5.
Low-temperature magnetic (remanence) measurements (left) and room temperature FORC diagrams (right) for wild-type (a), ΔftsZ-like (b), and ΔftsZ-like_C (c) cells.
FORC diagrams for wild-type and ΔftsZ-like_C cells showed smoothly closed contours around 13 mT and 14 mT, respectively, with slight overlap at the Hb axis (Fig. 5a and c), indicating predominantly fine-grained SD magnetosome particles and small amounts of SP magnetosome particles. Both strains showed a narrow vertical spread (<5 mT) on the Hc axis. In contrast to the wild type and ΔftsZ-like_C, ΔftsZ-like cells showed much lower coercivity (<5 mT) and a considerably wider vertical spread along the Hb axis (>20 to 30 mT) (Fig. 5b). ΔftsZ-like cells also had small closed contours around an Hc of 16.8 mT, indicating the presence of SD magnetosome particles. Taken together, these findings suggest that ΔftsZ-like cells have a much higher proportion of SP particles, or particles at the SP/SD threshold, compared to that of the wild type or ΔftsZ-like_C.
DISCUSSION
The function of the FtsZ-like protein in M. gryphiswaldense strain MSR-1 was studied by deletion mutation and complementation analysis. TEM observations indicated that FtsZ-like affects magnetosome formation. ΔftsZ-like deletion mutant cells produced small magnetosomes (average size, 24.0 ± 4.6 nm) with aberrant shapes, irregular alignment, and poor crystallization. This was confirmed by magnetic analyses at low and room temperatures, which indicated that ΔftsZ-like cells produce a higher proportion of SP magnetosome magnetite with less stoichiometry in comparison to ΔftsZ-like_C and wild-type cells.
Although the FtsZ-like protein evidently contributes to magnetosome formation, the mechanism remains to be elucidated. Alignment analyses of FtsZ-like using BLAST (1) and ClustalW programs showed high similarity with the FtsZ proteins of several bacterial species, i.e., ∼42% similarity with FtsZ of E. coli, Bacillus subtilis, and Bacillus anthracis and 26% similarity with that of Pyrococcus abyssi. FtsZ-like consists of 323 amino acids, while the FtsZ proteins of the other bacteria range from 350 to 400 amino acids (21). Several regions of homology with the FtsZ proteins are located between amino acids 1 and 320 of FtsZ-like. Since FtsZ-like is not essential for growth of M. gryphiswaldense MSR-1 (see Fig. S1 in the supplemental material), it presumably does not share the cell division function of the FtsZ proteins. The presence of a heptapeptide known as the “tubulin signature motif” (GGGTGTG) is crucial for the binding of GTP (9, 28, 35) and polymerization of the FtsZ proteins (10, 26). Like the FtsZ proteins, FtsZ-like displays GTP hydrolysis activity (Fig. 2A). FtsZ-like also hydrolyzed ATP, with an efficiency that was ∼2.6 times lower than that for GTP (Fig. 2A); such ATP hydrolysis function has not been reported for other FtsZ proteins. No significant hydrolysis activity was detected for other nucleotides.
GTP-dependent polymerization is essential for the functioning of tubulin family proteins, including FtsZ. In vitro, purified FtsZ proteins display assembly into a wide variety of polymer morphologies (including tubules, sheets, asters, straight and curved protofilaments, and minirings) (7, 12, 24, 25, 29, 31, 54). FtsZ-like was also found to undergo GTP-dependent polymerization (Fig. 3). Under TEM, the polymers appeared as straight, long, filamentous bundles, similar to those of the MamK polymer (51). The MamK cytoskeletal filament has been suggested to contribute to the maintenance of magnetosome organization and function (20, 33). Both the tubulin-like protein FtsZ-like and actin-like protein MamK in MTB belong to the group of bacterial cytoskeletal proteins. Interestingly, the chimeric protein MamKI, identified from noncultured MTB, comprises an N-terminal FtsZ-like domain fused to a C-terminal MamK-like domain (16). Such fusion of tubulin-like and actin-like domains within a single polypeptide in MTB suggests possible interaction of these proteins in strain MSR-1. On the other hand, bacterial two-hybrid assay analyses indicated that FtsZ-like does not interact directly with MamK protein (see Fig. S2 in the supplemental material). The relationships between FtsZ-like and MamK, and with other magnetosome-associated proteins, await further study.
Biomineralization and the assembly of bacterial magnetosomes into chains are intracellular processes under strict genetic control (13). There are additional components and factors that contribute to magnetosome chains, e.g., interparticle connections, membrane vesicles, cytoskeletal filaments, and the magnetosomal matrix (50). These components are highly ordered with various magnetosome-associated proteins. Detailed reconstruction of magnetosomal components in vitro will provide new insights into the functions of magnetosome-associated proteins and the mechanism of magnetosome formation. FtsZ-like cytoskeletal filaments may play a regulatory or accessory role in magnetosome formation, by undergoing GTP-dependent dynamic polymerization to regulate magnetosome assembly and synthesis. Extensive future studies will clarify this role.
The finding that a FtsZ-like protein is involved in magnetosome synthesis in M. gryphiswaldense MSR-1 has important implications for the mechanism of magnetosome biomineralization and functional concepts of the bacterial cytoskeleton.
Supplementary Material
Acknowledgments
We thank Tao Song (Institute of Electrical Engineering, Chinese Academy of Sciences) for the Cmag determination. We thank Xuhui Gao (Testing Center, the University of Science and Technology of China) and Junzhen Jia and Haihong Liu (Electron Microscopy Laboratory, China Agricultural University) for the high-resolution TEM observations. We also thank Yaoping Zhang (Department of Bacteriology, University of Wisconsin—Madison) for helpful comments on the manuscript.
This study was supported by the Chinese High Technology Research and Development Program (grant 2007AA021805) and the Chinese National Natural Science Foundation (grants 30570023 and 30870043).
Footnotes
Published ahead of print on 18 December 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Arakaki, A., J. Webb, and T. Matsunaga. 2003. A novel protein tightly bound to bacterial magnetic particles in Magnetospirillum magneticum strain AMB-1. J. Biol. Chem. 278:8745-8750. [DOI] [PubMed] [Google Scholar]
- 3.Bazylinski, D. A., and R. B. Frankel. 2004. Magnetosome formation in prokaryotes. Nat. Rev. Microbiol. 2:217-230. [DOI] [PubMed] [Google Scholar]
- 4.Blakemore, R. P. 1975. Magnetotactic bacteria. Science 190:377-379. [DOI] [PubMed] [Google Scholar]
- 5.Blakemore, R. P. 1982. Magnetotactic bacteria. Annu. Rev. Microbiol. 36:217-238. [DOI] [PubMed] [Google Scholar]
- 6.Bramhill, D., and C. M. Thompson. 1994. GTP-dependent polymerization of Escherichia coli FtsZ protein to form tubules. Proc. Natl. Acad. Sci. U. S. A. 91:5813-5817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carballido-Lopez, R., and J. Errington. 2003. A dynamic bacterial cytoskeleton. Trends Cell. Biol. 13:577-583. [DOI] [PubMed] [Google Scholar]
- 8.Chung, K. M., H. H. Hsu, H. Y. Yeh, and B. Y. Chang. 2007. Mechanism of regulation of prokaryotic tubulin-like GTPase FtsZ by membrane protein EzrA. J. Biol. Chem. 282:14891-14897. [DOI] [PubMed] [Google Scholar]
- 9.de Boer, P., R. Crossley, and L. Rothfield. 1992. The essential bacterial cell-division protein FtsZ is a GTPase. Nature 359:254-256. [DOI] [PubMed] [Google Scholar]
- 10.Dougherty, C. A., C. R. Sage, A. Davis, and K. W. Farrell. 2001. Mutation in the beta-tubulin signature motif suppresses microtubule GTPase activity and dynamics, and slows mitosis. Biochemistry 40:15725-15732. [DOI] [PubMed] [Google Scholar]
- 11.Dunlop, D. J., and Ö. Özdemir (ed.). 1997. Rock magnetism: fundamentals and frontiers. Cambridge University Press, Cambridge, United Kingdom.
- 12.Erickson, H. P., D. W. Taylor, K. A. Taylor, and D. Bramhill. 1996. Bacterial cell division protein FtsZ assembles into protofilament sheets and minirings, structural homologs of tubulin polymers. Proc. Natl. Acad. Sci. U. S. A. 93:519-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Faivre, D., and D. Schüler. 2008. Magnetotactic bacteria and magnetosomes. Chem. Rev. 108:4875-4898. [DOI] [PubMed] [Google Scholar]
- 14.Fischer, H., G. Mastrogiacomo, J. F. Löffler, R. J. Warthmann, P. G. Weidler, and A. U. Gehring. 2008. Ferromagnetic resonance and magnetic characteristics of intact magnetosome chains in Magnetospirillum gryphiswaldense. Earth Planet. Sci. Lett. 270:200-208. [Google Scholar]
- 15.Hefti, M. H., C. J. G. Van Vugt-Van der Toorn, R. Dixon, and J. Vervoort. 2001. A novel purification method for histidine-tagged proteins containing a thrombin cleavage site. Anal. Biochem. 295:180-185. [DOI] [PubMed] [Google Scholar]
- 16.Jogler, C., W. Lin, A. Meyerdierks, M. Kube, E. Katzmann, C. Flies, Y. X. Pan, R. Amann, R. Reinhardt, and D. Schüler. 2009. Toward cloning of the magnetotactic metagenome: identification of magnetosome island gene clusters in uncultivated magnetotactic bacteria from different aquatic sediments. Appl. Environ. Microbiol. 75:3972-3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keen, N. T., S. Tamaki, D. Kobayashi, and D. Trollinger. 1988. Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene 70:191-197. [DOI] [PubMed] [Google Scholar]
- 18.Komeili, A. 2007. Molecular mechanisms of magnetosome formation. Annu. Rev. Biochem. 76:351-366. [DOI] [PubMed] [Google Scholar]
- 19.Komeili, A., H. Vali, T. J. Beveridge, and D. K. Newman. 2004. Magnetosome vesicles are present before magnetite formation, and MamA is required for their activation. Proc. Natl. Acad. Sci. U. S. A. 101:3839-3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Komeili, A., Z. Li, D. K. Newman, and G. J. Jensen. 2006. Magnetosomes are cell membrane invaginations organized by the actin-like protein MamK. Science 311:242-245. [DOI] [PubMed] [Google Scholar]
- 21.Lee, K. N., I. Padmalayam, B. Baumstark, S. L. Baker, and R. F. Massung. 2003. Characterization of the ftsZ gene from Ehrlichia chaffeensis, Anaplasma phagocytophilum, and Rickettsia rickettsii, and use as a differential PCR target. DNA Cell Biol. 22:179-186. [DOI] [PubMed] [Google Scholar]
- 22.Li, F., Y. Li, W. Jiang, Z. F. Wang, and J. L. Li. 2004. Development of a genetic manipulation system and screening of magnetosome deleted mutants for Magnetospirillum gryphiswaldense. Acta Microbiol. Sin. 44:440-444. (In Chinese.) [Google Scholar]
- 23.Li, J. H., Y. X. Pan, G. J. Chen, Q. S. Liu, L. X. Tian, and W. Lin. 2009. Magnetite magnetosome and fragmental chain formation of Magnetospirillum magneticum AMB-1: transmission electron microscopy and magnetic observations. Geophys. J. Int. 177:33-42. [Google Scholar]
- 24.Löwe, J., and L. A. Amos. 1999. Tubulin-like protofilaments in Ca2+-induced FtsZ sheets. EMBO J. 18:2364-2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Löwe, J., and L. A. Amos. 2000. Helical tubes of FtsZ from Methanococcus jannaschii. Biol. Chem. 381:993-999. [DOI] [PubMed] [Google Scholar]
- 26.Lu, C., J. Stricker, and H. P. Erickson. 2001. Site-specific mutations of FtsZ—effects on GTPase and in vitro assembly. BMC Microbiol. 1:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moskowitz, B. M., R. B. Frankel, and D. A. Bazylinski. 1993. Rock magnetic criteria for the detection of biogenic magnetite. Earth Planet. Sci. Lett. 120:283-300. [Google Scholar]
- 28.Mukherjee, A., K. Dai, and J. Lutkenhaus. 1993. Escherichia coli cell division protein FtsZ is a guanine nucleotide binding protein. Proc. Natl. Acad. Sci. U. S. A. 90:1053-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mukherjee, A., and J. Lutkenhaus. 1994. Guanine nucleotide-dependent assembly of FtsZ into filaments. J. Bacteriol. 176:2754-2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakazawa, H., A. Arakaki, S. Narita-Yamada, et al. 2009. Whole genome sequence of Desulfovibrio magneticus strain RS-1 revealed common gene clusters in magnetotactic bacteria. Genome Res. 19:1801-1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oliva, M. A., S. Huecas, J. M. Palacios, J. Martin-Benito, J. M. Valpuesta, and J. M. Andreu. 2003. Assembly of archaeal cell division protein FtsZ and a GTPase-inactive mutant into double-stranded Filaments. J. Biol. Chem. 278:33562-33570. [DOI] [PubMed] [Google Scholar]
- 32.Pan, Y. X., N. Petersen, M. Winklhofer, A. F. Davila, Q. S. Liu, T. Frederichs, M. Hanzlik, and R. X. Zhu. 2005. Rock magnetic properties of uncultured magnetotactic bacteria. Earth Planet. Sci. Lett. 237:311-325. [Google Scholar]
- 33.Pradel, N., C. L. Santini, A. Bernadac, Y. Fukumori, and L. F. Wu. 2006. Biogenesis of actin-like bacterial cytoskeletal filaments destined for positioning prokaryotic magnetic organelles. Proc. Natl. Acad. Sci. U. S. A. 103:17485-17489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prozorov, T., S. K. Mallapragada, B. Narasimhan, L. Wang, P. Palo, M. Nilsen-Hamilton, T. J. Williams, D. A. Bazylinski, R. Prozorov, and P. C. Canfield. 2007. Protein-mediated synthesis of uniform superparamagnetic magnetite nanocrystals. Adv. Funct. Mater. 17:951-957. [Google Scholar]
- 35.RayChaudhuri, D., and J. T. Park. 1992. Escherichia coli cell-division gene ftsZ encodes a novel GTP-binding protein. Nature 359:251-254. [DOI] [PubMed] [Google Scholar]
- 36.Richter, M., M. Kube, D. A. Bazylinski, T. Lombardot, F. O. Glöckner, R. Reinhardt, and D. Schüler. 2007. Comparative genome analysis of four magnetotactic bacteria reveals a complex set of group-specific genes implicated in magnetosome biomineralization and function. J. Bacteriol. 189:4899-4910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roberts, A. P., C. R. Pike, and K. L. Verosub. 2000. First-order reversal curve diagrams: a new tool for characterizing the magnetic properties of natural samples. Geophys. Res. 105:28461-28475. [Google Scholar]
- 38.Rong, C. B., Y. J. Huang, W. J. Zhang, W. Jiang, and Y. Li. 2008. Ferrous iron transport protein B gene (feoB1) plays an accessory role in magnetosome formation in Magnetospirillum gryphiswaldense strain MSR-1. Res. Microbiol. 159:530-536. [DOI] [PubMed] [Google Scholar]
- 39.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 40.Schäfer, A., A. Tauch, W. Jäger, J. Kalinowski, G. Thierbach, and A. Pühler. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69-73. [DOI] [PubMed] [Google Scholar]
- 41.Scheffel, A., A. Gardes, K. Grunberg, G. Wanner, and D. Schüler. 2008. The major magnetosome proteins MamGFDC are not essential for magnetite biomineralization in Magnetospirillum gryphiswaldense but regulate the size of magnetosome crystals. J. Bacteriol. 190:377-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scheffel, A., M. Gruska, D. Faivre, A. Linaroudis, J. M. Plitzko, and D. Schüler. 2006. An acidic protein aligns magnetosomes along a filamentous structure in magnetotactic bacteria. Nature 440:110-114. [DOI] [PubMed] [Google Scholar]
- 43.Schüler, D. 2004. Molecular analysis of a subcellular compartment: the magnetosome membrane in Magnetospirillum gryphiswaldense. Arch. Microbiol. 181:1-7. [DOI] [PubMed] [Google Scholar]
- 44.Schüler, D. 2008. Genetics and cell biology of magnetosome formation in magnetotactic bacteria. FEMS Microbiol. Rev. 32:654-672. [DOI] [PubMed] [Google Scholar]
- 45.Schüler, D., and R. B. Frankel. 1999. Bacterial magnetosomes: microbiology, biomineralization and biotechnological applications. Appl. Microbiol. Biotechnol. 52:464-473. [DOI] [PubMed] [Google Scholar]
- 46.Schweizer, H. P. 1993. Small broad-host-range gentamycin resistance gene cassettes for site specific insertion and deletion mutagenesis. Biotechniques 15:831-834. [PubMed] [Google Scholar]
- 47.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Biotechnology 1:784-791. [Google Scholar]
- 48.Sossong, T. M., Jr., M. R. Brigham-Burke, P. Hensley, and K. H. Pearce, Jr. 1999. Self-activation of guanosine triphosphatase activity by oligomerization of the bacterial cell division protein FtsZ. Biochemistry 38:14843-14850. [DOI] [PubMed] [Google Scholar]
- 49.Sun, J. B., F. Zhao, T. Tang, W. Jiang, J. S. Tian, Y. Li, and J. L. Li. 2008. High-yield growth and magnetosome formation by Magnetospirillum gryphiswaldense MSR-1 in an oxygen-controlled fermentor supplied solely with air. Appl. Microbiol. Biotechnol. 79:389-397. [DOI] [PubMed] [Google Scholar]
- 50.Taoka, A., R. Asada, H. Sasaki, K. Anzawa, L. F. Wu, and Y. Fukumori. 2006. Spatial localizations of Mam22 and Mam12 in the magnetosomes of Magnetospirillum magnetotacticum. J. Bacteriol. 188:3805-3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Taoka, A., R. Asada, L. F. Wu, and Y. Fukumori. 2007. Polymerization of the actin-like protein MamK, which is associated with magnetosomes. J. Bacteriol. 189:8737-8740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ullrich, S., M. Kube, S. Schubbe, R. Reinhardt, and D. Schüler. 2005. A hypervariable 130-kilobase genomic region of Magnetospirillum gryphiswaldense comprises a magnetosome island which undergoes frequent rearrangements during stationary growth. J. Bacteriol. 187:7176-7184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weiss, D. S. 2004. Bacterial cell division and the septal ring. Mol. Microbiol. 54:588-597. [DOI] [PubMed] [Google Scholar]
- 54.Yu, X. C., and W. Margolin. 1997. Ca2+-mediated GTP-dependent dynamic assembly of bacterial cell division protein FtsZ into asters and polymer networks in vitro. EMBO. J. 16:5455-5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.