Abstract
Proteins that bind σ factors typically attenuate the function of the σ factor by restricting its access to the RNA polymerase (RNAP) core enzyme. An exception to this general rule is the Crl protein that binds the stationary-phase sigma factor σS (RpoS) and enhances its affinity for the RNAP core enzyme, thereby increasing expression of σS-dependent genes. Analyses of sequenced bacterial genomes revealed that crl is less widespread and less conserved at the sequence level than rpoS. Seventeen residues are conserved in all members of the Crl family. Site-directed mutagenesis of the crl gene from Salmonella enterica serovar Typhimurium and complementation of a Δcrl mutant of Salmonella indicated that substitution of the conserved residues Y22, F53, W56, and W82 decreased Crl activity. This conclusion was further confirmed by promoter binding and abortive transcription assays. We also used a bacterial two-hybrid system (BACTH) to show that the four substitutions in Crl abolish Crl-σS interaction and that residues 1 to 71 in σS are dispensable for Crl binding. In Escherichia coli, it has been reported that Crl also interacts with the ferric uptake regulator Fur and that Fur represses crl transcription. However, the Salmonella Crl and Fur proteins did not interact in the BACTH system. In addition, a fur mutation did not have any significant effect on the expression level of Crl in Salmonella. These results suggest that the relationship between Crl and Fur is different in Salmonella and E. coli.
In bacteria, transcription depends on a multisubunit RNA polymerase (RNAP) consisting of a catalytically active core enzyme (E) with a subunit structure α2ββ′ω that associates with any one of several σ factors to form different holoenzyme (Eσ) species. The σ subunit is required for specific promoter binding, and different σ factors direct RNAP to different classes of promoters, thereby modulating the gene expression patterns (17). The RNA polymerase holoenzyme containing the σ70 subunit is responsible for the transcription of most genes during exponential growth (17). When cells enter stationary phase or are under specific stress conditions (high osmolarity, low pH, or high and low temperatures) during exponential growth, σS, which is encoded by the rpoS gene, becomes more abundant, associates with the core enzyme, and directs the transcription of genes essential for the general stress response and for stationary phase survival (17, 20, 25).
Sigma factors compete for binding to a limited amount of the core polymerase (16, 17, 20, 34). σ70 is abundant throughout the growth cycle and has the highest affinity of all sigma factors for E in vitro (20). In contrast, levels of σS reach only about one-third of the σ70 levels upon entry into stationary phase, and σS exhibits the lowest affinity for E of all sigma factors in vitro (20). The cell uses at least two strategies to ensure the switch between σ70- and σS-associated RNA polymerases and to allow gene expression to be reprogrammed upon entry into stationary phase. Several factors (Rsd, 6S RNA, ppGpp, and DksA) indirectly increase σS competitiveness by decreasing the ability of σ70 to bind to E (25). In addition, the unconventional regulatory protein Crl increases the performance of σS.
The crl gene product is a regulator of σS activity in Escherichia coli (2, 14, 36, 50) and Salmonella (38, 40, 41). In both species, the Crl protein binds σS (2, 12) and facilitates RNA polymerase holoenzyme EσS formation (12, 50), thereby enhancing σS effectiveness (10, 14, 27, 38, 40, 41, 50). The Crl protein of Salmonella binds σS with a stoichiometry of 1:1 and increases the affinity of σS for the core enzyme 7-fold (12). In contrast, Crl does not bind σ70 and does not modify the affinity of σ70 for the core enzyme (12). Lelong et al. (28) reported that in E. coli W3110, Crl can interact with the ferric uptake regulator Fur, a key protein for the control of intracellular iron concentration (see reference 8 and references therein) and that Fur represses crl transcription (28).
In an attempt to gain further insight into Crl function, we searched for homologues of Crl in protein databases. Residues conserved in all members of the Crl family were replaced by using site-directed mutagenesis experiments to identify residues important for Crl activity in Salmonella. We used the bacterial adenylate cyclase two-hybrid system (BACTH) (22) to determine whether these residues and specific regions of Crl are required for efficient interaction with σS. We also used the BACTH system to assess the effect of the Crl mutations on the interaction of Crl with Fur.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Strains and plasmids are listed in Table 1. Bacteriophage P22HT105/1int was used to transfer mutations between Salmonella strains by transduction (44). Green plates, for screening for P22-infected cells or lysogens, were prepared as described previously (48). Strains were routinely cultured in Luria Bertani (LB) medium (43). Minimal medium was M9 (43) containing 20 mM glucose. For routine monitoring of multicellular behavior (rdar morphotype), Salmonella strains were grown at 28°C on LB agar plates without NaCl (LB0) and supplemented with Congo red (40 μg ml−1; CR plates) as described previously (42). Antibiotics were used at the following concentrations: ampicillin (Ap), 100 μg ml−1; carbenicillin (Cb), 100 μg ml−1; chloramphenicol (Cm), 15 μg ml−1 for the chromosomal resistance gene and 30 μg ml−1 for the plasmid resistance gene; kanamycin (Km), 50 μg ml−1; and tetracycline (Tet) 20 μg ml−1.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Characteristic(s) | Source or referencea |
|---|---|---|
| Strains | ||
| E. coli | ||
| MC1061K | araD139 Δ(ara-leu)767 Δ(lacIPOZY) X74 rpsL galU galK rpoS::kan | 26 |
| JM109 | recA1 supE44 endA1 hsdR17 gyrA96 relA1 thi Δ(lac-proAB) F′(traD36 proAB+lacIqlacZΔM15) | 43 |
| BTH101 | F−cya-99 araD139 galE15 galK16 rpsL1 (Strr) hsdR2 mcrA1 mcrB1 | D. Ladant |
| S. serovar Typhi | ||
| T2 | Wild type | CISb |
| S. serovar Typhimurium | ||
| C52 | Wild type | 26 |
| C52K | C52 ΔrpoS::kan | 26 |
| ATCC 14028 | Wild type | ATCC |
| ATCCrpoS | ATCC 14028 ΔrpoS | 41 |
| ATCCcrl | ATCC 14028 Δcrl::Cm | 40 |
| JS402 | Δfur-41::cat | 19 |
| ATCCfur | ATCC 14028 Δfur-41::cat | |
| ATCCrpoSfur | ATCC 14028 ΔrpoS Δfur-41::cat | |
| ATCCcrl csgD-lacZ | ATCC 14028 Δcrl::Cm csgD-lacZY | 40 |
| ATCCsdhA-lacZ-k | ATCC 14028 sdhA-lacZY-Km | 41 |
| ATCCfur sdhA-lacZ-k | ATCC 14028 Δfur-4l::cat sdhA-lacZY-Km | |
| ATCCcrl-lacZ | ATCC 14028 crl-lacZY-Km | |
| ATCCrpoS crl-lacZ | ATCC 14028 ΔrpoS::Cm crl-lacZY-Km | |
| ATCCfur crl-lacZ | ATCC 14028 Δfur-41::cat crl-lacZY-Km | |
| ATCCrpoSfur crl-lacZ | ATCC 14028 ΔrpoS Δfur-41::cat crl-lacZY-Km | |
| 2922K | ATCC 14028 ΔSTM2922::Km | 41 |
| 2922Kcrl | ATCC 14028 ΔSTM2922::Km Δcrl::Cm | 41 |
| 2922KrpoS | ATCC 14028 ΔSTM2922::Km ΔrpoS::Cm | 41 |
| 2922KrpoST2 | ATCC 14028 ΔSTM2922::Km rpoST2 | |
| 2922KrpoST2crl | 2922K rpoST2 Δcrl::Cm | |
| 2922KkatE-lacZ | 2922K Tn5B21-2.4 | 41 |
| 2922KrpoS katE-lacZ | 2922KrpoS Tn5B21-2.4 | 41 |
| 2922Kcrl katE-lacZ | 2922Kcrl Tn5B21-2.4 | 41 |
| 2922KrpoST2katE-lacZ | 2922KrpoST2 Tn5B21-2.4 | |
| 2922KrpoST2crl katE-lacZ | 2922K rpoST2 Δcrl::Cm Tn5B21-2.4 | |
| Plasmids | ||
| pACYC184 | Cloning vector; Cmr Tetr | 6 |
| pACcrl-1 | pACYC184 with the promoterless crl gene cloned into the cat gene (crl is transcribed from the cat promoter); Tetr | 40 |
| pACcrlY22A | pACcrl-1 with mutation Y22A in Crl | |
| pACcrlF35A | pACcrl-1 with mutation F35A in Crl | |
| pACcrlC28A | pACcrl-1 with mutation C28A in Crl | |
| pACcrlC37A | pACcrl-1 with mutation C37A in Crl | |
| pACcrlC41A | pACcrl-1 with mutation C41A in Crl | |
| pACcrlF53A | pACcrl-1 with mutation F53A in Crl | |
| pACcrlW56A | pACcrl-1 with mutation W56A in Crl | |
| pACcrlG74A | pACcrl-1 with mutation G74A in Crl | |
| pACcrlG80A | pACcrl-1 with mutation G80A in Crl | |
| pACcrlW82A | pACcrl-1 with mutation W82A in Crl | |
| pACcrlF103A | pACcrl-1 with mutation F103A in Crl | |
| pACcrlC28-37 | pACcrl-1 with mutations C28A and C37A in Crl | |
| pACcrlC28-41 | pACcrl-1 with mutations C28A and C41A in Crl | |
| pACcrlC37-41 | pACcrl-1 with mutations C37A and C41A in Crl | |
| pACcrlC28-37-41 | pACcrl-1 with mutations C28A, C37A, and C41A in Crl | |
| pUC4K | Source of Km resistance cartridge | Pharmacia |
| pSTF4 | spvRAB-lacZ fusion in pQF50; Cbr | 26 |
| pACrpoST2 | pACYC184 with a 6-kb BglII fragment carrying rpoST2, Cmr | |
| pUCrpoST2 | pUC19 with a 3.3 kb SphI-ScaI fragment carrying rpoST2, Cbr | |
| pUCrpoST2K | pUCrpoST2 with STY3047::Km, Cbr Kmr | |
| pQE60 | Vector for expression of His-tagged proteins, Cbr | Qiagen |
| pQE30 | Vector for expression of His-tagged proteins, Cbr | Qiagen |
| pQErpoST2 | pQE60::rpoST2 expresses a σST2-His6 protein, Cbr | |
| pQE30rpoS | pQE30::rpoS expresses a His6-σS protein, Cbr | |
| pQEcrl | pQE30::crl expresses a His6-Crl protein, Cbr | 40 |
| pQEcrlY22A | pQEcrl with mutation Y22A in Crl | |
| pQEcrlF53A | pQEcrl with mutation F53A in Crl | |
| pQEcrlW56A | pQEcrl with mutation W56A in Crl | |
| pQEcrlW82A | pQEcrl with mutation W82A in Crl | |
| pKT25 | BACTH vector designed to express a given polypeptide fused in frame at its N-terminal end with T25 fragment; p15 ori; Kmr | 23 |
| pKNT25 | BACTH vector designed to express a given polypeptide fused in frame at its C-terminal end with T25 fragment; p15 ori; Kmr | 23 |
| pUT18 | BACTH vector designed to express a given polypeptide fused in frame at its C-terminal end with T18 fragment; ColE1 ori; Apr | 23 |
| pUTCrl | pUT18 expressing Crl-T18 | |
| pUTCrlY22A | pUT18 expressing Crl-T18 with mutation Y22A in Crl | |
| pUTCrlR51A | pUT18 expressing Crl-T18 with mutation R51A in Crl | |
| pUTCrlE52A | pUT18 expressing Crl-T18 with mutation E52A in Crl | |
| pUTCrlF53A | pUT18 expressing Crl-T18 with mutation F53A in Crl | |
| pUTCrlG55A | pUT18 expressing Crl-T18 with mutation G55A in Crl | |
| pUTCrlW56A | pUT18 expressing Crl-T18 with mutation W56A in Crl | |
| pUTCrlW57A | pUT18 expressing Crl-T18 with mutation W57A in Crl | |
| pUTCrlG80A | pUT18 expressing Crl-T18 with mutation G80A in Crl | |
| pUTCrlW82A | pUT18 expressing Crl-T18 with mutation W82A in Crl | |
| pKNTCrl | pKNT25 expressing Crl-T25 | |
| pKTRpoS | pKT25 expressing T25-RpoS | |
| pKTRpoSΔ12-71 | pKTRpoS with in frame deletion of residues 12-71 in RpoS | |
| pKTRpoSΔ1-38 | pKTRpoS with deletion of residues 1-38 in RpoS | |
| pKTRpoSΔ1-71 | pKTRpoS with deletion of residues 1-71 in RpoS | |
| pKTFur | pKT25 expressing T25-Fur | |
| pUTFur | pUT18 expressing Fur-T18 |
This study, unless otherwise noted.
WHO Reference Center for Salmonella (Institut Pasteur, Paris).
Oxidative shock survival assay.
Cells were grown to stationary phase in LB medium (optical density at 600 nM [OD600] of 3.5 to 4), washed, resuspended in phosphate-buffered saline (PBS), and mixed with H2O2 at a final concentration of 15 mM. Aliquots of bacteria were removed at time intervals, and numbers of colony forming cells were determined on LB plates.
DNA manipulations and sequence analysis.
Standard molecular biology techniques were used (43). Oligonucleotides were obtained from Sigma-Aldrich (France). DNA sequencing was performed by Cogenics (France). DNA and amino acid sequence analyses were conducted using the BLAST programs at the NCBI (National Center for Biotechnology Information).
Cloning of the rpoST2 allele.
We demonstrated that the nucleotide sequence of a PCR-amplified rpoS gene from Salmonella enterica serovar Typhi strain T2 (rpoST2) is identical to that in Salmonella ATCC 14028 (41) except for a G→A mutation at position 318, which does not change the amino acid sequence, and a deletion of 8 nucleotides starting at position 109, which results in a frameshift and appearance of a premature stop codon in rpoST2. Total DNA from S. Typhi T2 was cleaved with BglII, and the resulting fragments were cloned into the BamHI site of pACYC184. Recombinant plasmids were then transformed into the E. coli rpoS mutant strain MC1061K carrying an rpoS-dependent spvRAB-lacZ fusion on pSTF4 (26). Selection of Cbr Cmr lactose-positive (Lac+) clones and subsequent examination of their plasmid DNA by restriction analysis and DNA sequencing of rpoS produced pACrpoST2 containing a 6-kb BglII fragment carrying rpoST2. A 3.3-kb SphI-ScaI fragment carrying rpoST2 was subsequently cloned into the SphI and SmaI sites of pUC19 to yield pUCrpoST2.
Construction of S. Typhimurium 2922KrpoST2.
The rpoS allele in S. enterica serovar Typhimurium ATCC 14028 (rpoSSTM) was replaced by the rpoST2 allele using the following strategy (41). pUCrpoST2 contains the rpoST2 allele of S. Typhi strain T2 and the downstream genes, including a gene encoding a putative decarboxylase (named STY3047 in S. Typhi and STM2922 in S. Typhimurium) that is not regulated by rpoS (1). In pUCrpoST2K, a kanamycin resistance (Km) cartridge is located in the SmaI site of STY3047. pUCrpoST2K was introduced by electroporation into ATCCrpoS, where it was unstable. Recombination of the Km cartridge into the host genome with simultaneous loss of pUCrpoST2K resulted in recombinants that were resistant to kanamycin and sensitive to carbenicillin. A Kmr Cbs Cms recombinant was selected and checked by PCR for the presence of the STM2922::Km mutation and simultaneous replacement of the ΔrpoS::Cm mutation by the rpoST2 allele. The rpoST2 and STM2922::Km alleles were then cotransduced to ATCCrpoS. Transductants that were Kmr but Cms were selected, and transduction of the STM2922::Km mutation and simultaneous replacement of the ΔrpoS::Cm mutation by the rpoST2 allele were confirmed by PCR. One Kmr Cms strain, designated 2922KrpoST2, was also checked by DNA sequencing for the presence of the rpoST2 allele.
Determination of the N-terminal sequence of σST2.
pQErpoST2, which produces the rpoST2 gene with a C-terminal His6 extension under the control of the pQE60 isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible promoter, was constructed as follows. Primers specific to the ends of rpoS (5′-CCGTCAAGAATTCACGGGTAGGAGCCACCTTATGA-3′ and 5′-TGATCGAGATCTCTCGCGGAACAGCGCTTCGATATTCA-3′; restriction sites are underlined) were used to amplify a 1-kb fragment of S. Typhi T2 total DNA by PCR. EcoRI and BglII restriction sites were incorporated at its 5′ and 3′ ends, respectively. After digestion with EcoRI and BglII, the PCR-amplified fragment was ligated into the EcoRI and BglII sites of pQE60. The nucleotide sequence of the rpoS insert in pQErpoST2 was verified. In this construct, rpoST2 is expressed from its own translation sequences. E. coli JM109 carrying pQErpoST2 was grown in LB medium containing ampicillin at 37°C to an optical density of 0.9, when 0.7 mM IPTG was added. The cells were harvested after 5 h, resuspended in buffer A (50 mM Na2HPO4, pH 8, 300 mM NaCl) and then disrupted in by a French press. The crude cell extract was supplemented with benzonase (1,250 U) and imidazole (10 mM) and centrifuged at 27,000 × g for 30 min. The supernatant was added to a 1.5-ml Ni-nitrilotriacetic acid (NTA) column and washed with buffer A containing 20 mM imidazole. His6-tagged σS from S. Typhi strain T2 (σST2-His6) was eluted with buffer A containing 300 mM imidazole. After electrophoresis in a 10% polyacrylamide-SDS gel, the σST2-His6 protein was transferred on a polyvinylidene difluoride (PVDF) membrane, stained with amido black, and excised from the membrane for N-terminal sequence analysis. Amino-terminal Edman degradation was carried out on an Applied Biosystems sequencer.
Construction of a chromosomal crl-lac transcriptional fusion.
We previously created a chromosomal mutation in the crl gene of Salmonella ATCC 14028 using PCR-generated linear DNA fragments and the λRed recombination method (yielding strain ATCCcrl) (40). A single-copy crl-lacZY transcriptional gene fusion was constructed in ATCCcrl using conditional plasmids containing promoterless lacZY genes and the FLP recognition target (FRT) site, as described previously (11). PCR assays were used to ensure integration of the plasmids in the correct location and to exclude the presence of multiple plasmid integrants (using standard test primers, such as those described in reference 11). We also used flanking locus-specific primers to amplify junction fragments that were subsequently analyzed by DNA sequencing. Isogenic strains were constructed by phage P22 HT int-mediated transduction of the mutations into the appropriate strains.
Enzymatic assays.
β-Galactosidase activity was measured as described by Miller (31) and is expressed in Miller units.
BACTH assays.
The BACTH assay is dependent upon the functional reconstitution of the Bordetella pertussis adenylate cyclase T18 and T25 subdomains by two interacting partners (22). The resulting cyclic AMP (cAMP) binds to and activates the transcription activator cAMP receptor protein (CRP), a positive regulator of the lac and mal operons involved in lactose and maltose (Mal) catabolism. Derivatives of pUT18, pKT25, and pKNT25, used in the BACTH assays, were constructed by cloning PCR-amplified DNA fragments encoding the protein of interest from S. Typhimurium between the PstI and EcoRI sites of pUT18 and pKNT25 and the XbaI and KpnI sites of pKT25. The DNA encoding Crl was amplified using primers Crl-Pst (5′-CA TGC CTG CAG AAG GAG ATC GCA ATG ACG TTA CCG AGT-3′) and Crl-Eco (5′-T CGA TGA ATT CGA TGC CGA CAG TTT TAC CGG CTC GTC GT-3′), restricted by PstI and EcoRI (sites are underlined), and cloned into the PstI and EcoRI sites of both pUT18 and pKNT25 to yield pUTCrl and pKNTCrl, respectively. The DNA encoding RpoS was amplified using primers RpoS-Xba (5′-TCG ACT CTA GAT ATG AGT CAG AAT ACG CTG AAA GTT CAT-3′) and RpoS-Kpn (5′-AC TTA GGT ACC TTA CTC GCG GAA CAG CGC TTC GAT-3′), restricted by XbaI and KpnI (sites are underlined), and cloned into the XbaI and KpnI sites of pKT25 to yield pKTRpoS. The DNA encoding Fur was amplified using primers Fur-Pst (5′-CAT GCC TGC AGC ATG ACT GAC AAC AAT ACC GCA-3′) and Fur-Eco (5′-T CGA TGA ATT CGA TTT AGT CGC GTC ATC GTG CGC GT-3′) for cloning into the PstI and EcoRI sites (underlined) of pUT18 (yielding pUTFur) and using Fur-Xba (5′-TCG ACT CTA GAC ATG ACT GAC AAC AAT ACC GCA TTA AAG A-3′) and Fur-Kpn (5′-AC TTA GGT ACC TTA TTT AGT CGC GTC ATC GTG CGC GT-3′) for cloning into the XbaI and KpnI sites (underlined) of pKT25 (yielding pKTFur). pKTRpoSΔ12-71 is a derivative of pKTRpoS with an in-frame deletion of an internal HpaI-DraI fragment (residues 12 to 71 of RpoS). pKTRpoSΔ1-38 contains a DNA fragment amplified with primers RpoST2-Xba (5′-TCG ACT CTA GAC CTG GCT GAA GAA GAG CTG TTA TCG CAA-3′; restriction site is underlined) and RpoS-Kpn (see above) and cloned into the XbaI and KpnI sites of pKT25. pKTRpoSΔ1-71 contains a DNA fragment amplified with primers RpoS-Xba5 (5′-TCG ACT CTA GAA ACA GCC GAA GAA GAA GTC TAT TTT GCG CGT-3′; restriction site is underlined) and RpoS-Kpn (see above) and cloned into the XbaI and KpnI sites of pKT25. All plasmids were confirmed to be correct by DNA sequencing. The E. coli BTH101 cya strain was transformed with derivatives of plasmids pKT25, pKNT25, and pUT18 encoding proteins fused to the T25 and T18 fragments of the B. pertussis adenylate cyclase. Cotransformants were plated onto MacConkey maltose plates supplemented with carbenicillin, kanamycin, and 0.5 mM IPTG to assess the Mal+ phenotype and on LB plates supplemented with 5-bromo-4-chloro-indolyl-β-d-galactoside (X-Gal; 40 μg ml−1), carbenicillin, kanamycin, and 0.5 mM IPTG to assess the Lac+ phenotype and β-galactosidase activity. Plates were incubated at 30°C for 3 days; colonies were then collected, and their β-galactosidase activities were measured as described by Miller (31). For immunodetection of the Crl-T18 and T25-σS chimeras, BTH101 derivatives were grown to stationary phase (OD600 of 3.5) in LB medium in the presence of 2 mM cAMP and 0.5 mM IPTG to induce fully the lac promoter on pKT25, pKNT25, and pUT18.
Construction of plasmids with mutated crl genes.
Site-directed mutagenesis was performed using a QuikChange II Site-directed mutagenesis kit (Stratagene) as recommended by the manufacturer. Site-directed mutagenesis of plasmid pACcrl-1 yielded plasmids pACcrlY22A, pACcrlF35A, pACcrlC28A, pACcrlC37A, pACcrlC41A, pACcrlF53A, pACcrlW56A, pACcrlG74A, pACcrlG80A, pACcrlW82A, pACcrlF103A, pACcrlC28-37, pACcrlC28-41, pACcrlC37-C41, and pACcrlC28-37-41 (where subscripts indicate the mutations in crl) (Table 1). Plasmids pUTCrlY22A, pUTCrlF53A, pUTCrlW56A, pUTCrlW82A, and pUTCrlG80A (Table 1) were obtained by cloning PCR-amplified DNA fragments from plasmids pACcrlY22A, pACcrlF53A, pACcrlW56A, pACcrlW82A, and pACcrlG80A, respectively, between the PstI and EcoRI sites of pUT18 using primers Crl-Pst and Crl-Eco (as described above). Plasmids pUTCrlR51A, pUTCrlE52A, pUTCrlG55A, and pUTCrlW57A were obtained by site-directed mutagenesis of plasmid pUTCrl (Table 1). All plasmids were confirmed to be correct by DNA sequencing.
Electrophoresis and immunoblot analysis of proteins.
Whole-cell extracts were prepared, and SDS-polyacrylamide gel electrophoresis was carried out as described by Silhavy et al. (46). The amount of protein in whole-cell lysates was determined using a detergent-compatible protein assay kit (Bio-Rad). Equal amounts of protein were loaded in each slot. The molecular sizes of the proteins were estimated using molecular size standards (Fermentas, France). Rabbit antibodies against the Crl protein of Salmonella were from Robbe-Saule et al. (40). Rabbit antibodies against the σS protein of S. enterica serovar Typhimurium were from Coynault et al. (7). Proteins were transferred to Amersham Hybond P membranes (GE Healthcare) and incubated with the polyclonal rabbit antibody serum as previously described (7). Bound antibodies were detected using a secondary anti-rabbit antibody linked to peroxidase and an Amersham ECL Plus Western blotting detection system kit (GE Healthcare).
Overproduction and purification of His6-Crl variants.
A total of 250 ml of JM109 carrying pQEcrl wild-type or mutant was grown in LB medium containing carbenicillin at 30°C to an optical density of 0.6, and then 1 mM IPTG was added. After 30 min, the temperature was lowered to 23°C. The cells were harvested after 4 h, washed, resuspended in 10 ml of buffer A (25 mM Tris, pH 8.0, 1 M NaCl, 0.01% NP-40) supplemented with complete EDTA-free antiprotease (Roche), as described by the manufacturer, and then disrupted in a French press (Aminco). The crude cell extract was centrifuged at 15,000 × g for 30 min. The supernatant was adjusted to 3 mM imidazole, added to 1.5 ml of Ni-nitrilotriacetic acid agarose (Qiagen), and gently mixed for 1 h. The slurry was packed onto an Econo-Pac column (Bio-Rad) and washed with 15 ml of buffer A containing 20 mM imidazole. His6-Crl was eluted with buffer A containing 250 mM imidazole, dialyzed against buffer B (20 mM Tris-HCl, pH 8, 50 mM NaCl, 0.01% NP-40), and added to DNA-cellulose equilibrated in the same buffer (0.3 ml; Sigma D-8515). The DNA-cellulose was removed by centrifugation, and the concentrations of the proteins were determined by a Bradford assay.
Gel retardation assays.
The DNA encoding rpoS was amplified using primers HK1 (5′-AGGCTCGGATCCATGAGTCAGAATACGCTGAAAGTTCAT-3′) and HK2 (5′-TTCCGAAAGCTTTTACTCGCGGAACAGCGCTTCGATATT-3′; restriction sites are underlined) and cloned into the BamHI and HindIII sites of pQE30 to yield pQE30rpoS. The nucleotide sequence of the rpoS insert in pQE30rpoS was checked by DNA sequencing. S. Typhimurium His6-σS produced from pQE30rpoS was purified using Ni-affinity chromatography as described for His6-σST2. E. coli core (Epicentre) was used to reconstitute the σS-holoenzyme, which was incubated with variant Crl proteins (wild type or mutants) or buffer. Nine microliters of the protein complexes was then added to 3 μl of 32P-labeled katN fragment in buffer Glu (40 mM HEPES pH 8.0, 10 mM MgCl2, 100 mM K-glutamate, 2 mM dithiothreitol [DTT]) containing 500 μg ml−1 bovine serum albumin (BSA) and incubated at 28°C for 20 min. The final concentrations of core, His6-σS, Crl, and katN fragments were, respectively, 8 nM, 32 nM, 3 μM, and 1 nM. After the addition of 3 μl of loading buffer (buffer Glu containing 50% sucrose, 0.025% xylene cyanol blue, and 150 μg ml−1 of heparin), the mixture was loaded onto a 6% native polyacrylamide gel run in TG buffer (25 mM Tris, 192 mM glycine, pH 8.5) at 8 V/cm. The gel was dried before being autoradiographed and quantified using a PhosphorImager (Molecular Dynamics).
Abortive initiation assays.
A mixture of the 207-bp lacUV5 fragment (10 nM), ApA (2 mM), and 80 μM [α-32P]UTP was added to an equal volume of the holoenzyme (60 nM; ratio of His6-σS to core of 2.5: 1) preincubated with buffer or Crl (wild type or variants) at a 6 μM concentration at 28°C. Aliquots were removed at various times. The aliquots were spotted onto Whatman 3MM paper prespotted with 100 mM EDTA, and chromatograms were developed as previously described (40).
RESULTS
Distribution and sequence conservation of Crl in sequenced microbial genomes.
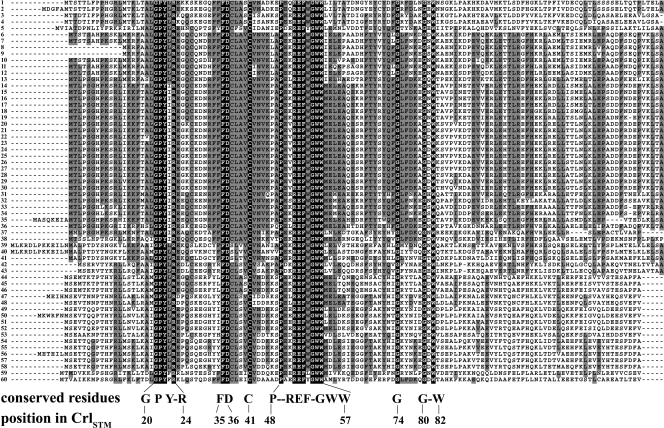
A BLAST search of 1,264 sequenced bacterial genomes (www.ncbi.nlm.nih.gov) revealed 143 genomes containing a crl gene. They are all Gammaproteobacteria and contain an rpoS gene (Table 2). Crl was absent from many rpoS-containing bacteria, for example, Pseudomonas, in which RpoS plays an important role (45). Thus, crl is not as widely distributed as rpoS, and it is also less conserved at the sequence level (Table 2). Sequence comparison of the 143 Crl proteins identified 60 Crl sequences that differed by at least one amino acid. Alignment of these 60 Crl sequences by ClustalW revealed 17 residues that are conserved in all members of the Crl family (Fig. 1).
TABLE 2.
Distribution and sequence conservation of Crl in sequenced bacterial genomes
| Family and genusa | Identity with CrlSTM (%)b | Identity with RpoSSTM (%)c |
|---|---|---|
| Enterobacteriaceae | ||
| Salmonella | 96-100 | 99-100 |
| Citrobacter | 90 | 100 |
| Escherichia | 83-84 | 98-99 |
| Shigella | 83-84 | 99 |
| Enterobacter | 74-77 | 97 |
| Klebsiella | 75-76 | 98 |
| Serratia | 69 | 94 |
| Erwinia | 66 | 95 |
| Yersinia | 63-66 | 84-97 |
| Pectobacterium | 64 | 91 |
| Proteus | 49 | 79 |
| Providencia | 40-45 | 78 |
| Other families | ||
| Aeromonas | 43-44 | 85-84 |
| Vibrio | 38-50 | 72-82 |
| Unclassified Vibrionales | 46 | 81 |
| Photobacterium | 42-49 | 79-80 |
| Moritella | 38 | 76 |
| Psychromonas | 35-44 | 77 |
Sequenced bacterial genomes containing a crl gene.
Range of percentage of identity between the amino acid sequence of the Crl proteins in strains belonging to the indicated genus and that CrlSTM (133 residues; NP-459316, shown in Fig. 1, line 16).
FIG. 1.
Alignment of the proteins from the Crl family. Alignment of the Crl protein sequences was performed with ClustalW. Residues that are identical in all of the Crl proteins are shown in black boxes, and positions of these conserved residues in the Crl sequence from S. enterica serovar Typhimurium ([CrlSTM] 133 residues; NP-459316, line 16) are shown below the alignment. The accession numbers for the Crl sequences are as follows (line numbers are in parentheses): for Salmonella, ZP-02346845 (14), ZP-02663633 (15)NP-459316 (16), YP-215307 (17), and YP-001571691 (18); for Citrobacter, YP-001454497 (19); for Escherichia, NP-285957 (22), NP-414775 (23), YP-001742400 (25), YP-002383844 (26), YP-002327819 (27), NP-752324 (28), YP-539315 (29), and ZP-02902139 (30); for Shigella, YP-309300 (20), YP-402173 (21), and NP-706240 (24); for Enterobacter, YP-001175503 (31), ZP-03281271 (32), and YP-001439177 (35); for Klebsiella, YP-001333935 (33) and YP-002240279 (34); for Serratia, YP-001477200 (13); for Erwinia, YP-001908518 (36); for Yersinia, YP-001007379 (6), ZP-00829910 (7), ZP-00821198 (8), ZP-00826726 (9), ZP-00833806 (10), YP-001402111 (11), and NP-668295 (12); for Pectobacterium, YP-051554 (37); for Proteus, YP-002150140 (38); for Providencia, ZP-03313695 (39), ZP-03317513 (40), and ZP-02958311 (41); for Aeromonas, YP-857911 (42) and YP-001140774 (43); for Vibrio, ZP-01235672 (1), NP-231906 (44), ZP-01957597 (45), ZP-01980856 (46), NP-759329 (47), NP-933650 (48), ZP-01065753 (49), YP-002418001 (50), ZP-00993088 (51), ZP-01870715 (53), NP-797054 (54), ZP-01260295 (55), YP-001444378 (56), ZP-01987497 (57), and ZP-02197346 (58); for unclassified Vibrionales, ZP-01813308 (52); for Photobacterium, ZP-01159718 (2), YP-129053 (3), and ZP-01218949 (4); for Moritella, ZP-01898329 (5); for Psychromonas, YP-944255 (59) and ZP-01216084 (60).
Analysis of the 16 completed genomic sequences of Salmonella (www.ncbi.nlm.nih.gov) indicated that the Crl sequence is highly conserved (Fig. 1 and Table 2). Interestingly, the crl gene in S. enterica serovar Paratyphi B SPB7 lacks a G at position 73 in the open reading frame, causing the appearance of a premature stop codon and the likely production of a truncated, nonfunctional Crl protein.
The first 71 amino acids of σS are nonessential for Crl binding.
Crl is less conserved than the full-length RpoS protein (Table 2). However, the first 55 residues of RpoS are poorly conserved, whereas the remaining 275 residues of RpoS are highly conserved (data not shown). For instance, the level of sequence identity between RpoSSTM and the RpoS proteins from the enterobacteria Proteus mirabilis and Providencia stuartii is 43% and 40% for amino acids 1 to 55 compared to 87% and 86% for amino acids 56 to 330, respectively. These first 55 residues define a specific domain of σS that is not well conserved in other sigma factors besides σ70 (corresponding roughly to region σ1.1) (see also Discussion section and references 32 and 51) and might thus play a role in the specificity of recognition of σS by Crl. In addition, σ1.1 and Crl might play complementary roles in the formation of EσS. Crl increases the affinity of σS for the core enzyme (12). σ1.1 stabilizes the interaction between σ70 and core RNAP (18) and might play a similar role in the σS-core interaction. These possibilities prompted us to determine whether this domain of σS is required for σS activity and for Crl activation.
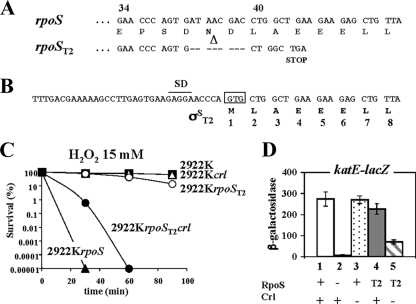
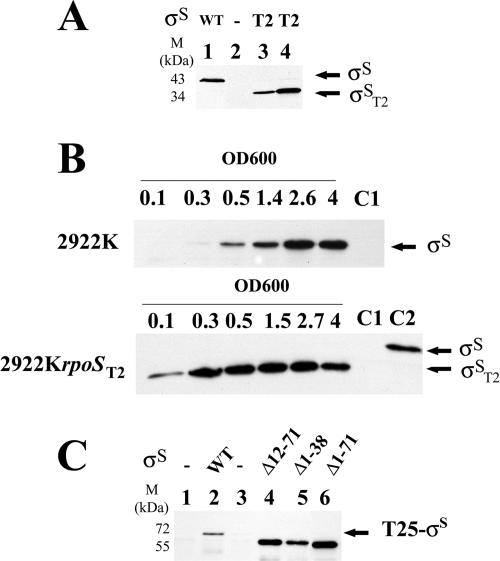
We first examined the sensitivity to Crl activation of a σS mutant lacking the first 39 residues of σS (σST2) (Fig. 2 and 3) and thus lacking most of region σ1.1 (32, 51). rpoST2 is a natural rpoS allele from S. Typhi strain T2 that contains a deletion of 8 nucleotides in the 5′ end of the coding sequence, resulting in the appearance of a premature stop codon (Fig. 2A). Surprisingly, rpoST2 was able to direct the synthesis of a 35-kDa σS protein (Fig. 3A, σST2). N-terminal sequence analysis indicated that σST2 has an N-terminal Met residue followed by a sequence that is identical to that downstream of residue 39 in native σS (Fig. 2B). We inferred that σST2 is encoded from the mutant gene in which translation initiates at a GTG start codon preceding codon 40 of σS in rpoST2 and upstream of which is a putative ribosome binding site (Fig. 2B, Shine-Dalgarno sequence [SD]). Translational reinitiation within the E. coli rpoS gene has also been reported previously (15). To facilitate characterization of the rpoST2 allele in isogenic backgrounds, the rpoS gene from S. Typhimurium ATCC 14028 was replaced by rpoST2, yielding strain 2922KrpoST2. During exponential growth, σST2 was produced in larger amounts than the wild-type σS protein (OD600 of 0.1 to 1.4) (Fig. 3B), likely because the N-terminal 11 residues of σS involved in ClpX recognition and σS proteolysis by ClpP (13, 49) were not present in σST2. σS is required for bacterial resistance to various stresses during stationary phase (the so-called general stress resistance) (20, 25). The σS-dependent gene katE encodes a catalase, an enzyme that detoxifies hydrogen peroxide (H2O2) and contributes to the resistance of Salmonella to oxidative stress in stationary phase (38). Despite the large amount of σST2 produced, strain 2922KrpoST2 was slightly less resistant to H2O2 than the wild-type strain 2922K and exhibited lower expression of the katE-lacZ fusion (Fig. 2C and D). This suggested that σST2 is less active than wild-type σS. In vitro transcription experiments with σST2 confirmed this hypothesis (data not shown). Interestingly, the levels of H2O2 resistance and the expression of the catalase gene katE were more dependent on Crl activation in 2922KrpoST2 than in the wild-type strain (Fig. 2C and D). This indicated that the σST2 protein is able to interact with Crl and that its activity is highly dependent on Crl.
FIG. 2.
Characterization of the rpoS mutant allele rpoST2. (A) Relevant portion of the rpoS sequence in the wild-type rpoS allele and the rpoST2 mutant allele. The deletion of 8 nucleotides in rpoST2, compared to the rpoS wild type, is shown. This mutation results in the appearance of a premature stop codon in rpoST2. (B) Determination of the N-terminal sequence of σST2. Production of σST2 likely results from translational reinitiation in rpoST2 at the GTG codon that is preceded by a putative ribosome binding site (Shine-Dalgarno sequence [SD]). (C) Resistance to hydrogen peroxide of the indicated S. Typhimurium strains. Cells were grown to stationary phase in LB medium at 37°C, washed, and resuspended in PBS to an OD600 of 0.1, and H2O2 15 mM was added. A representative experiment is shown. Similar results were obtained in repeat experiments. (D) Expression of a katE-lacZ gene fusion in Salmonella carrying the wild-type rpoS and mutant rpoST2 alleles. Lane 1, 2922KkatE-lacZ; lane, 2, 2922KrpoS katE-lacZ; lane 3, 2922Kcrl katE-lacZ; lane 4, 2922KrpoST2 katE-lacZ; lane 5, 2922KrpoST2crl katE-lacZ. β-Galactosidase activity was measured in overnight LB cultures at 37°C according to the method of Miller (31).
FIG. 3.
Expression of σS wild-type (WT) and mutant proteins. (A) Detection of σST2 produced from the rpoST2 allele. Overnight LB cultures at 37°C were analyzed by Western blotting with anti-σS antibodies. Five micrograms of total protein was loaded in each slot. Lane 1, S. Typhimurium wild-type strain C52; lane 2, S. Typhimurium ΔrpoS mutant C52K; lane 3, S. Typhi strain T2; lane 4, E. coli MC1061K harboring plasmid pACrpoST2. (B) Expression of σS and σST2 as a function of bacterial cell growth. Salmonella strains 2922K and 2922KrpoST2 were grown in LB medium at 37°C. Exponential-phase cultures (optical density at 600 nm of 0.5) were diluted into LB medium prewarmed at 37°C to prolong the exponential phase. Aliquots were removed at various time intervals and analyzed by Western blotting with anti-σS antibodies. Ten micrograms of total protein was loaded in each slot. The growth phase was determined by measuring culture turbidity at the OD600. Lane C1, 2922KrpoS (OD600 of 4); lane C2, 2922K (OD600 of 1.4). (C) Expression of wild-type and truncated T25-σS hybrid proteins. The E. coli BTH101 cya strain harboring pKT25 and its derivatives were grown to stationary phase (OD600 of 3.5) in LB medium in the presence of 2 mM cAMP and 0.5 mM IPTG to fully induce the lac promoter on pKT25. Ten micrograms of total protein was loaded in each slot and analyzed by Western blotting with anti-σS antibodies. Lane 1, no plasmid; lane 2, pKTRpoS; lane 3, pKT25; lane 4, pKTRpoSΔ12-71; lane 5, pKTRpoSΔ1-38; lane 6, pKTRpoSΔ1-71.
We previously used the bacterial two-hybrid system (BACTH system) (22) to detect an interaction between Crl and σS (12). C-terminal fusions of S. Typhimurium σS to T25 (T25-σS) and N-terminal fusions of S. Typhimurium Crl to T18 (Crl-T18) were produced in the E. coli BTH101 cya lac+ strain from pKTRpoS and pUTCrl, respectively, and the resulting β-galactosidase activities were measured (Table 3). As expected (12), T25-σS interacted with Crl-T18 (Table 3). In this system, the first 38 residues of σS were dispensable for efficient binding of Crl (Table 3, compare T25-σS and T25-σSΔ1-38 in the presence of Crl-T18). Moreover, deletion of residues 1 to 38 from σS resulted in increased levels of the chimera and β-galactosidase activity in the BACTH assay (Fig. 3C lanes 2 and 5; Table 3). This is probably again due to the absence of the ClpX recognition site in T25-σSΔ1-38. Interestingly, deletion of residues 1 to 71 in T25-σSΔ1-71 and deletion of residues 12 to 71, which does not remove the ClpX recognition site (13, 49) in T25-σSΔ12-71, had a more pronounced effect on σS levels and β-galactosidase activity than deletion of residues 1 to 38 (in T25-σSΔ1-38) (Fig. 3C, lanes 4 to 6; Table 3). This indicated that the first 71 residues in σS are nonessential for Crl binding and further suggested that the deletion of this region induces a conformational change in σS that favors its stability. Altogether, these results suggested that Crl interacts with the conserved amino acid region (72 to 330) of σS.
TABLE 3.
BACTH analysis of Crl interactions with σS and Fur
| Coexpressed proteins | β-Galactosidase activity (Miller units)a |
|---|---|
| T18 + T25 | 47 ± 6 |
| Crl-T18 + T25 | 47 ± 6 |
| T18 + T25-σS | 43 ± 3 |
| Crl-T18 + T25-σS | 263 ± 38 |
| Crl-T18 + T25-σSΔ1-38 | 455 ± 33 |
| Crl-T18 + T25-σSΔ1-71 | 1830 ± 186 |
| Crl-T18 + T25-σSΔ12-71 | 2352 ± 146 |
| CrlY22A-T18 + T25-σS | 44 ± 5 |
| CrlR51A-T18 + T25-σS | 59 ± 3 |
| CrlE52A-T18 + T25-σS | 102 ± 12 |
| CrlF53A-T18 + T25-σS | 48 ± 5 |
| CrlG55A-T18 + T25-σS | 138 ± 19 |
| CrlW56A-T18 + T25-σS | 44 ± 4 |
| CrlW57A-T18 + T25-σS | 74 ± 4 |
| CrlG80A-T18 + T25-σS | 205 ± 27 |
| CrlW82A-T18 + T25-σS | 55 ± 6 |
| Crl-T18 + T25-Fur | 43 ± 1 |
| Fur-T18 + Crl-T25 | 58 ± 6 |
| Fur-T18 + T25-Fur | 243 ± 32 |
| Fur-T18 +T25 | 55 ± 3 |
| T18 + T25-Fur | 42 ± 2 |
| T18 + Crl-T25 | 52 ± 5 |
The efficiencies of functional complementation between the indicated proteins were quantified by measuring β-galactosidase activities in E. coli BTH101 cells harboring the corresponding plasmids as described in Materials and Methods. β-Galactosidase activity was measured according to the method of Miller (31).
Identification of conserved residues important for Crl activity.
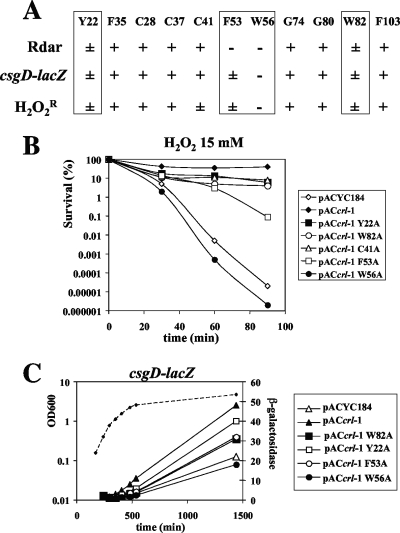
Alanine substitution mutagenesis was performed to investigate the functional relevance of the conserved residues in Crl. The conserved residues are located in four regions of Crl (one-third of them are aromatic amino acids) (Fig. 1). As a first step, one or two residues in each conserved region were mutated. We also mutated the three cysteine residues in Crl. pACcrl-1 expresses the crl gene from S. Typhimurium under the control of the vector cat promoter and is able to complement a Δcrl mutant of Salmonella (40). Site-directed mutagenesis of pACcrl-1 yielded 11 derivatives expressing altered Crl proteins (Table 1 and Fig. 4A). The ability of the mutated crl genes to complement the Salmonella Δcrl mutant was assessed using three different tests (Fig. 4 and 5). The development of the rdar morphotype depends on Crl (40) and was used as a qualitative test (Fig. 5). This colony morphology is caused by production of curli and cellulose and is correlated with biofilm formation and expression of the regulatory gene csgD (42). In ATCC 14028, the expression level of a csgD-lacZ fusion and the H2O2 resistance level at 28°C both depend on Crl (38, 40) and were used as quantitative assays (Fig. 4B and C).
FIG. 4.
Site-directed mutagenesis of the Salmonella crl gene and characterization of the crl mutant alleles. Site-directed mutagenesis of pACcrl-1 was performed, yielding 11 derivatives expressing Crl proteins in which residues Y22, F35, C28, C37, C41, F53, W56, G74, G80, W82, and F103 are replaced by alanines. The ability of the mutated crl genes to complement the Salmonella Δcrl mutant was assessed using three different tests: the H2O2 resistance level at 28°C, the expression level of a csgD-lacZ fusion, and development of the rdar morphotype. (A) Summary of the results obtained: +, complementation level similar to that obtained with pACcrl-1 expressing the wild-type Crl protein; −, complementation level similar to that obtained with the negative control pACYC184; ±, partial complementation level. Complementation experiments for the rdar morphotype are shown in Fig. 5. (B) Complementation of the Δcrl mutant for resistance to H2O2. ATCCcrl derivatives containing the indicated plasmids were grown to stationary phase in LB medium at 28°C, washed, and resuspended in PBS to an OD600 of 1, and 15 mM H2O2 was added. A representative experiment is shown. Similar results were obtained in repeat experiments. (C) Complementation of the Δcrl mutant for expression of a csgD-lacZ fusion. ATCCcrl csgD-lacZ derivatives containing the indicated plasmids were grown in LB0 (LB without NaCl) medium at 28°C. Exponential-phase cultures (optical density at 600 nm of 0.5) of Salmonella were diluted into LB0 medium prewarmed at 28°C to prolong the exponential phase. Aliquots were removed at various time intervals, and β-galactosidase activity measured (lines) according to the method of Miller (31). The growth phase was determined by measuring the culture turbidity at an optical density of 600 nm (dashed line; the growth curve was similar for all the strains). The measurements were repeated twice, and a representative experiment is shown.
FIG. 5.
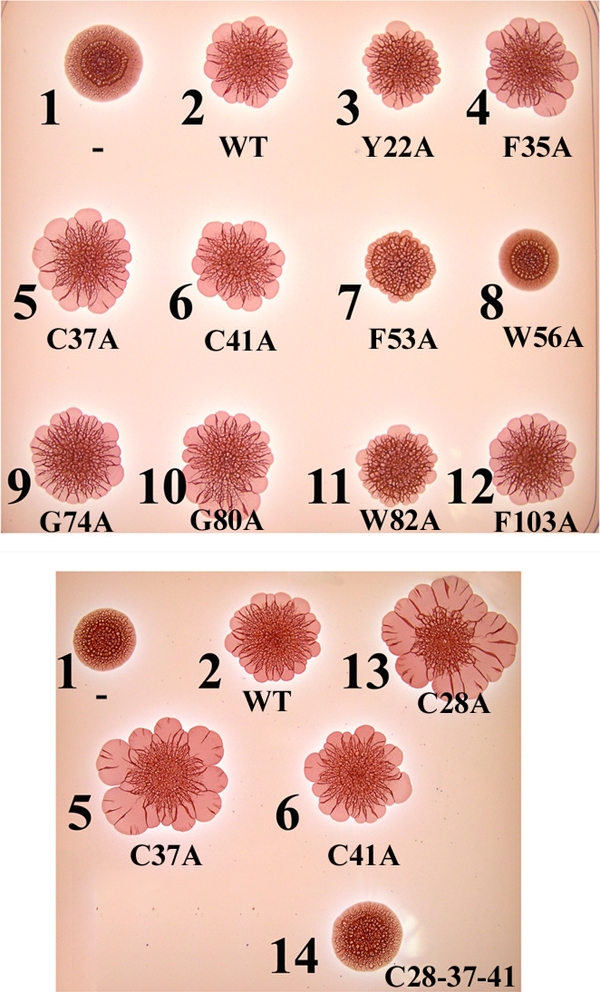
The rdar morphotypes of Salmonella ATCCcrl harboring different plasmids. The Δcrl mutant harboring the vector pACYC184, the wild-type crl gene on pACcrl-1, and mutated pACcrl-1 derivatives (Table 1) were grown for 5 days on CR plates at 28°C. Plasmids are as follows: spot 1, pACYC184; spot 2, pACcrl-1; spot 3, pACcrlY22A; spot 4, pACcrlF35A; spot 5, pACcrlC37A; spot 6, pACcrlC41A; spot 7, pACcrlF53A; spot 8, pACcrlW56A; spot 9, pACcrlG74A; spot 10, pACcrlG80A; spot 11, pACcrlW82A; spot 12, pACcrlF103A; spot 13, pACcrlC28A; spot 14, pACcrlC28-37-41. Complementation of the crl mutation was observed with pACcrl-1 and its derivatives containing mutations F35A, C28A, C37A, C41A, G74A, G80A, and F103A.
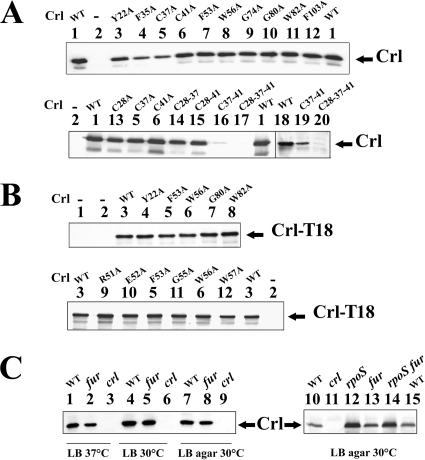
Derivatives of pACcrl-1 expressing the Crl proteins with F35A, C28A, C37A, G74A, G80A, and F103A substitutions complemented Δcrl as well as pACcrl-1 did (Fig. 4 and 5 and data not shown). Two substitutions, W56A and F53A, located in a conserved motif of Crl (residues 48 to 57) (Fig. 1), substantially diminished complementation in all three tests (Fig. 4 and 5, spots 7 and 8). Two additional substitutions, Y22A and W82A, also decreased the complementation levels but to a lesser extent (Fig. 4 and 5, spots 3 and 11). These four substitutions did not significantly decrease the cellular level of Crl (Fig. 6A) and, thus, likely affected its activity.
FIG. 6.
Expression of Crl wild-type (WT) and mutant proteins. (A) Detection of Crl produced from pACcrl-1 and its mutant derivatives in Salmonella ATCCcrl. ATCCcrl strains, harboring the vector pACYC184 and the pACcrl-1 derivatives as indicated below, were grown to stationary phase (OD600 of 4) in LB medium at 37°C and analyzed by Western blotting with anti-Crl antibodies. One microgram of total protein was loaded in slots 1 to 17. Lane 1, pACcrl-1; lane 2, pACYC184; lane 3, pACcrlY22A; lane 4, pACcrlF35A; lane 5, pACcrlC37A; lane 6, pACcrlC41A; lane 7, pACcrlF53A; lane 8, pACcrlW56A; lane 9, pACcrlG74A; lane 10, pACcrlG80A; lane 11, pACcrlW82A; lane 12, pACcrlF103A; lane 13, pACcrlC28A; lane 14, pACcrlC28-37; lane 15, pACcrlC28-41; lane 16, pACcrlC37-41; lane 17, pACcrlC28-37-41. Five micrograms of total protein was loaded in slots 18 to 20. Lane 19, pACcrlC37-41; lane 20, pACcrlC28-37-41; lane 18, a stationary phase culture of ATCC 14028 in LB medium at 37°C was used as control. (B) Expression of wild-type and altered Crl-T18 hybrid proteins. The E. coli BTH101 cya strain harboring pUT18 and its derivatives were grown to stationary phase (OD600 of 3) in LB medium in the presence of 2 mM cAMP and 0.5 mM IPTG to fully induce the lac promoter on pUT18. Five micrograms of total protein was loaded in each slot and analyzed by Western blotting with anti-Crl antibodies. Lane 1, no plasmid; lane 2, pUT18; lane 3, pUTCrl; lane 4, pUTCrlY22A; lane 5, pUTCrlF53A; lane 6, pUTCrlW56A; lane 7, pUTCrlG80A; lane 8, pUTCrlW82A; lane 9, pUTCrlR51A; lane 10, pUTCrlE52A; lane 11, pUTCrlG55A; lane 12, pUTCrlW57A. (C) Detection of Crl in Salmonella ATCC 14028 (lanes 1, 4, 7, 10, and 15) and its mutant derivatives ATCCfur (lanes 2, 5, 8, and 13), ATCCcrl (lanes 3, 6, 9, and 11), ATCCrpoS (lane 12), and ATCCrpoSfur (lane 14). Cultures were grown in LB medium (LB) overnight at 30°C or 37°C or on LB agar at 30°C, as described previously (28), and analyzed by Western blotting with anti-Crl antibodies. One microgram of total protein was loaded in each slot.
The Salmonella Crl protein contains three cysteines, (C28, C37, and C41), one of which (C41) is conserved in all Crl members. Alanine substitutions for the cysteine residues (C28A, C37A, and C41A) did not significantly affect the levels of Crl protein or its activity (Fig. 4, 5, and 6A). Only a slight effect of C41A on H2O2 resistance was observed (Fig. 4B). Interestingly, replacement of all three cysteine residues substantially decreased Crl levels (Fig. 6A, lanes 17 and 20). Examination of the Crl family members indicated that they all contain at least two cysteine residues. Altogether, these results suggested that the presence of at least one, and probably two, cysteine residue(s) is required for Crl stability. Double mutations in the cysteine residues in Crl (Crl with the mutations C28A and C37A [CrlC28-37], CrlC28-41, and CrlC37-41) were constructed to determine what combination of cysteine substitution affected Crl stability. The combinations of C28A and C37A substitutions and C28A and C41A substitutions did not significantly affect the levels of Crl protein (Fig. 6A, lanes 14 and 15). In contrast, alanine substitution for both C37 and C41 substantially decreased Crl levels (Fig. 6A, lane 16), suggesting that these two cysteine residues are important for Crl stability. Levels of the triple CrlC28-37-41 mutant protein were lower than the level of CrlC37-41 (Fig. 6A, lanes 19 and 20). This suggests that the C28A substitution further decreased the stability of CrlC37-41.
Screening of the crl mutations in the BACTH system and in vitro assays.
We next determined whether the mutations selected above affected the Crl-σS interaction. DNA encoding proteins with Y22A, F53A, W56A, or W82A substitutions or the neutral G80A substitution were introduced into pUTCrl, and the ability of the Crl-T18 derivatives to interact with σS was assessed in the presence of T25-σS (Table 3). Interestingly, the Y22A, F53A, W56A, and W82A substitutions, but not the G80A substitution, all abolished the interaction between Crl-T18 and T25-σS (Table 3). These substitutions did not affect levels of Crl-T18 (Fig. 6B). The altered Crl-T18 hybrid proteins were also unable to interact with the T25-σS derivative lacking amino acids 12 to 71 (data not shown). In conclusion, the Y22A, F53A, W56A, and W82A substitutions all abolished the interaction between Crl and σS, either directly or indirectly, through conformational changes in Crl.
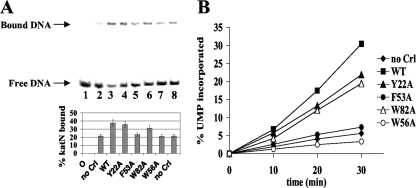
The activities of the variant proteins were also probed in vitro by gel retardation and abortive initiation assays, based on the ability of Crl to increase the amount of promoter complexes formed by the σS-RNA polymerase (EσS) and activate transcription initiation (12, 40). As shown on the autoradiogram of the native polyacrylamide gel (Fig. 7A, lane 3), EσS binding to the 32P-labeled katN promoter was stimulated 2-fold by wild-type Crl. The Y22A and W82A variants were still able to increase the formation of EσS-katN complexes by about 1.8- and 1.6-fold, respectively (Fig. 7A, lanes 4 and 6). In contrast, the F53A and W56A variants of Crl (Fig. 7A, lanes 5 and 7) had hardly any effect, in agreement with their highly reduced ability to stimulate csgD-lacZ expression or hydrogen peroxide resistance. The gel retardation methodology is only semiquantitative, however, and the addition of heparin to disrupt nonspecific core-DNA binding before electrophoresis might have disturbed weak interactions between Crl variants and σS. Therefore, abortive initiation assays, which do not require heparin addition and can be exploited for more quantitative studies, were conducted using the well-characterized lacUV5 promoter, which can be transcribed in vitro by Eσ70 and EσS and, unlike katN, has the advantage of producing large amounts of abortive products (29). As expected, EσS produced less ApApUpU tetranucleotide from lacUV5 in the presence of the Crl variants than with the wild-type protein (Fig. 7B). The results were more obvious than in gel retardation assays since wild-type Crl stimulated EσS-lacUV5 abortive transcription by 4-fold under our conditions. After a 30-min incubation at 28°C, the observed decrease in ApApUpU synthesis was 35% for Y22A, 40% for W82A, and 90% for F53A. The addition of W56A Crl resulted in an even lower activity than in the absence of Crl, suggesting a slight inhibitory effect of this variant on σS-dependent transcription. Altogether, these results emphasize the role of the REF-GWW motif (Fig. 1) in Crl activation of σS-dependent promoters. Consistent with this hypothesis, the R51A and W57A substitutions and, to a lesser extent, the E52A and G55A substitutions affected the interaction between Crl and σS in the BACTH assay (Table 3) but did not affect levels of Crl-T18 (Fig. 6B).
FIG. 7.
Effects of the Crl variants on EσS promoter binding and abortive transcription. (A) Band shift analysis of EσS binding to the katN-labeled fragment in the absence of Crl (lanes 2 and 8) and in the presence of wild-type (WT) Crl (lane 3) or its variants Y22A (lane 4), F53A (lane 5), W82A (lane 6), and W56A (lane 7). Lane 1, no protein. A typical autoradiogram is shown. The bands corresponding to free and bound DNAs are indicated by arrows, and the percentage of bound DNA indicated below each lane is the average of two experiments. (B) Abortive initiation assays. EσS was preincubated in the absence of Crl or in the presence of wild-type Crl or its variants Y22A, F53A, W82A, and W56A. After addition of a mixture containing the lacUV5 fragment, the ApA dinucleotide, and [α-32P]UTP, the incorporation of labeled [α-32P]UMP into abortive transcripts was monitored as a function of time. The measurements were repeated twice, and a representative experiment is shown.
Fur-Crl interactions in Salmonella.
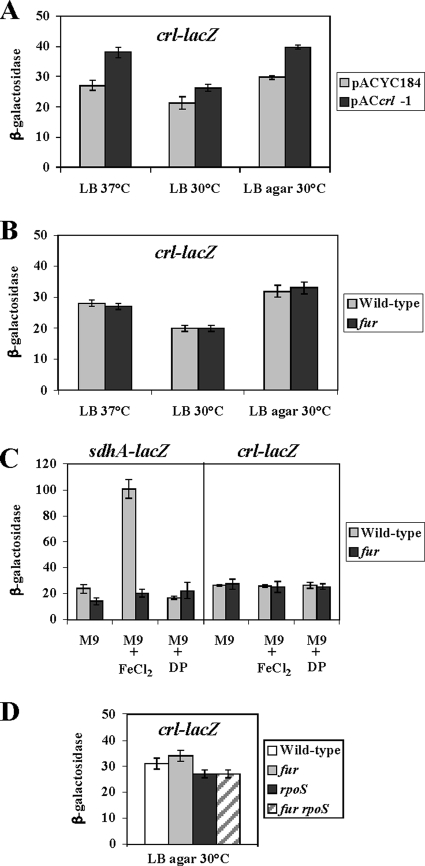
To determine whether the four substitutions studied above affected Crl-σS interaction specifically, we assessed their effect on the interaction of Crl with another partner. In a previous study using E. coli W3110, an interaction between Crl and the ferric uptake regulator Fur was detected by copurification (28). To investigate the interaction between the Salmonella Crl and Fur proteins in the BACTH assay, we constructed plasmids pUTFur encoding a Fur-T18 protein and pKTFur encoding a T25-Fur protein. Significant interactions between the Crl and the Fur hybrid proteins were not detected (Table 3). The Fur hybrid proteins were stable, as indicated by the fact that the expected dimerization of Fur-T18 and T25-Fur (reference 8 and references therein) gave a positive signal (Table 3).
crl transcription in E. coli W3110 is repressed by Fur (by 100-fold) and by Crl (by more than 10-fold) (28). To assess the possible relationship between Crl and Fur in Salmonella ATCC 14028, we examined the effects of Fur and Crl on crl expression. We first compared Crl levels in the wild-type and fur strains by immunodetection of the Crl protein (Fig. 6C). Cells were grown to stationary phase in LB medium at 30°C and 37°C and also on LB agar at 30°C, a growth condition used to study the regulation of crl expression by Fur in E. coli W3110 (28). The amount of Crl detected in the fur strain was the same as in the wild-type strain under all three conditions (Fig. 6C, compare lanes 1 and 2, 4 and 5, and 7 and 8). In addition, the levels of crl-lacZ-encoded β-galactosidase were not significantly affected either by the fur mutation or by the production of Crl in trans from plasmid pACcrl-1 (Fig. 8A and B). These results suggested that crl transcription is not repressed by Fur and Crl in ATCC 14028.
FIG. 8.
Expression of crl-lacZ and sdhA-lacZ gene fusions in Salmonella. Expression of the gene fusions in the Salmonella strains indicated was determined in cultures grown under different conditions. Cultures were grown overnight in LB medium (LB) at 37°C or 30°C or on LB agar at 30°C, as described previously (28). M9, minimal M9 medium with 20 mM glucose; M9+FeCl2, M9 with 100 μM ferrous chloride; M9+DP, M9 with the iron chelator 2,2′-dipyridyl (100 μM). β-Galactosidase activity was measured according to the method of Miller (31). (A) Expression of the crl-lacZ gene fusion in ATCCcrl-lacZ strains harboring pACYC184 and pACcrl-1 that expresses the crl gene under the control of the cat promoter of pACYC184 (40). (B) Expression of the crl-lacZ gene fusion in ATCCcrl-lacZ and ATCCfur crl-lacZ. (C) Expression of the sdhA-lacZ gene fusion in ATCCsdhA-lacZ-k and ATCCfur sdhA-lacZ-k and expression of the crl-lacZ gene fusion in ATCCcrl-lacZ and ATCCfur crl-lacZ. (D) Expression of the crl-lacZ gene fusion in ATCCcrl-lacZ, ATCCrpoS crl-lacZ, ATCCfur crl-lacZ and ATCCrpoSfur crl-lacZ.
In the presence of iron, Fur negatively controls transcription of a small RNA, RyhB, which facilitates degradation of the sdhCDBA mRNA encoding succinate dehydrogenase (30). Consistent with this finding, sdhA-lacZ-encoded β-galactosidase in ATCC 14028 was dependent on both iron and Fur (Fig. 8C). This demonstrated that regulation by Fur is indeed functional in ATCC 14028. Fur did not exert any effect on the crl-lacZ fusion (Fig. 8C). Also, no effect of Fur on crl-lacZ expression was detected in LB medium supplemented with iron (data not shown).
E. coli W3110 has many genes that are differentially expressed in E. coli MG1655 (52), and many laboratory stocks of W3110 contain mutations in rpoS (21, 52; also www.ncbi.nlm.nih.gov). These findings prompted us to evaluate the effect of the fur mutation on crl expression in an rpoS background. Crl production and crl-lacZ-encoded β-galactosidase levels were not affected by the fur mutation in the Salmonella rpoS mutant (Fig. 6C, lanes 12 and 14, and 8D). As previously shown (38), Crl production was increased by the rpoS mutation (Fig. 6C, lanes 10, 12, and 15) by a mechanism that likely operates at the posttranscriptional level (Fig. 8D).
Altogether, these results suggest that, in contrast to E. coli W3110, Crl and Fur do not interact in Salmonella.
DISCUSSION
Analysis of the protein sequence databases revealed the narrow distribution of Crl homologues in bacteria. In contrast, RpoS homologues are found in many Gram-negative bacteria of the gamma, delta, and beta subdivisions (www.ncbi.nlm.nih.gov). Several hypotheses might explain the absence of Crl in bacteria containing an RpoS homologue. Crl compensates for a low affinity of σS for the E and might be dispensable in bacteria expressing a σS protein with a high affinity for E. Moreover, the physiological impact of Crl is greatest at low concentrations of σS (40, 41), and the environment can modulate the impact of Crl by affecting the σS level (38). Thus, depending on the bacterial lifestyles and environment encountered, cellular physiology, and the mechanisms of regulation of rpoS expression, Crl might be dispensable. Alternatively, functional homologues of Crl might exist in some rpoS-containing bacteria, or some species might use alternative strategies to favor σS interaction with E. Interestingly, we did not find Crl homologues in bacteria that do not contain rpoS. This is consistent with previous work suggesting that the main function of Crl is to favor σS interaction with RNA polymerase (41, 50).
Of the 16 Salmonella strains whose genome sequence is completed, one, S. Paratyphi SB7, contains a frameshift mutation in crl that results in the appearance of a premature stop codon. This observation is reminiscent of previous findings that rpoS null mutants can be found in natural isolates of S. Typhi (7, 37, 39) and is consistent with our previous observation that a Δcrl mutation increases the competitive fitness of Salmonella by attenuating σS activity (41). The selection of rpoS mutants in bacterial populations likely results from a growth advantage of rpoS mutants in the absence of environmental stress (24, 33, 34). However, in environments where bacteria encounter mild stress, ΔrpoS mutants are outcompeted by the wild-type strain, whereas rpoS attenuated mutants exhibit increased fitness (33, 41, 53). We can predict that the beneficial effect of a crl mutation is significant when the survival strategy requires “enough but not too much” σS activity.
We showed that four conserved residues (Y22, F53, W56, and W82) are important for Crl activity and for Crl-σS interaction but not for Crl stability. In contrast, a Crl protein in which each of its three cysteine residues was replaced was unstable. The Y22A and W82A substitutions had a more dramatic effect in the BACTH system than in the complementation tests and the in vitro assays (Table 3 and Fig. 4, 5, and 7). The IPTG-inducible lac promoter on pKT25 and pUT18 is induced by cAMP, and, thus, a mutation that decreases the efficiency of the interaction between T25-σS and Crl-T18 also lowers the expression level of the hybrid proteins, thereby amplifying the effect of the mutation (24; also D. Ladant, personal communication).
The F53A and W56A substitutions are located in a conserved motif in the middle part of the protein (Fig. 1, REF-GWW). Analysis of four additional substitutions (R51A, E52A, G55A, and W57A) in the BACTH assay confirmed the role of the REF-GWW motif in Crl-σS interactions (Table 3 and Fig. 6B). The two other substitutions, Y22A and W82A, are outside of this motif. At least some of these mutations likely affect Crl binding to σS indirectly through conformational changes. The three-dimensional structure of Crl would provide new insight into this issue. Unfortunately, our attempts to crystallize Crl have been unsuccessful. In addition, deletion experiments in the BACTH system did not allow us to delineate a subregion of Crl involved in σS binding. Crl might spread along σS, or, more likely, a specific conformation of Crl might be required for efficient interaction with σS.
The primary determinants of σS involved in the binding of Crl are unknown, but the lack of interaction between Crl and σ70 (12) suggests that the interaction involves sequence determinants or structural features that are specific to σS. The vast majority of σ factors, including σS, belong to the so-called σ70 family, reflecting their relationship to the principal σ factor of E. coli, σ70. Sequence alignments of σ70 family members of groups 1 and 2, to which σS belongs, reveal that they have four conserved regions (regions 1 to 4) (32, 35). Among these, region 2 (subregions 2.1 to 2.4) and region 4 (subregions 4.1 and 4.2) contain DNA-binding domains that mediate recognition of the conserved −10 and −35 elements of σ70-dependent promoters, respectively. The linear division of σ70 factors into functionally distinct regions is largely confirmed by structural data, which revealed that primary sigma factors have four flexibly linked domains, σ1.1, σ2, σ3, and σ4, containing regions 1.1, 1.2 to 2.4, 3.0 to 3.1, and 4.1 to 4.2, respectively (5, 32). Free σ factors generally do not specifically bind promoter DNA, and the N-terminal σ1.1 region is autoinhibitory (4, 9). σ1.1 might act indirectly to inhibit promoter binding by stabilizing a compact conformation of σ that is incompatible with promoter recognition (47). Binding to core RNAP induces large movements of the σ domains (3), converting σ into an active conformation in which the DNA binding determinants in σ2 and σ4 are exposed (32).
σ1.1 shows little conservation between σ70 and σS and is not found in the alternative sigma factors of the σ70 family (32, 35). Therefore, the specificity of recognition between Crl and σS might involve σ1.1. However, our findings that (i) σST2, a protein lacking residues 1 to 39, including most of region σ1.1 in native σS (32, 51) is dependent on Crl activation (Fig. 2), and (ii) deletion of residues 1 to 71 of σS, which removes region σ1.1 and part of subregion 1.2 (32, 51) does not abolish Crl-σS interaction (Table 3), ruled out this possibility. Experiments are in progress to determine which region of σS is involved in Crl binding. In addition to its autoinhibitory role, σ1.1 stabilizes the interaction between σ70 and core RNAP (18). Our finding that σST2 is less active but more dependent on Crl activation than wild-type σS (Fig. 2C and D and data not shown) is consistent with a similar role for σ1.1 in σS-core interaction. Crl increases the affinity of σS for core RNAP (12) and might partially compensate for the absence of a full region σ1.1 in σST2.
Acknowledgments
We are grateful to Anthony Pugsley for critical reading of the manuscript. We thank James Slauch for providing the Salmonella fur strain. We are grateful to Gouzel Karimova and Daniel Ladant for providing the cya strain and BACTH vectors and for helpful discussions.
Footnotes
Published ahead of print on 11 December 2009.
REFERENCES
- 1.Bang, I.-S., J. G. Frye, M. McClelland, J. Velayudhan, and F. C. Fang. 2005. Alternative sigma factor interactions in Salmonella: σE and σH promote antioxidant defences by enhancing σS levels. Mol. Microbiol. 56:811-823. [DOI] [PubMed] [Google Scholar]
- 2.Bougdour, A., C. Lelong, and J. Geiselmann. 2004. Crl, a low temperature-induced protein in Escherichia coli that binds directly to the stationary phase sigma subunit of RNA polymerase. J. Biol. Chem. 279:19540-19550. [DOI] [PubMed] [Google Scholar]
- 3.Callaci, S., E. Heyduk, and T. Heyduk. 1999. Core RNA polymerase from E. coli induces a major change in the domain arrangement of the sigma 70 subunit. Mol. Cell 3:229-238. [DOI] [PubMed] [Google Scholar]
- 4.Camarero, J. A., A. Shekhtman, E. A. Campbell, M. Chlenov, T. M. Gruber, D. A. Bryant, S. A. Darst, D. Cowburn, and T. W. Muir. 2002. Autoregulation of a bacterial σ factor explored by using segmental isotopic labeling and NMR. Proc. Natl. Acad. Sci. U. S. A. 99:8536-8541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campbell, E. A., L. F. Westblade, and S. A. Darst. 2008. Regulation of bacterial RNA polymerase σ factor activity: a structural perspective. Curr. Opin. Microbiol. 11:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang, A. C., and S. N. Cohen. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 134:1141-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coynault, C., V. Robbe-Saule, and F. Norel. 1996. Virulence and vaccine potential of Salmonella typhimurium mutants deficient in the expression of the RpoS (sigma S) regulon. Mol. Microbiol. 22:149-160. [DOI] [PubMed] [Google Scholar]
- 8.D'Autréaux, B., L. Pecqueur, A. Gonzalez de Peredo, R. E. M. Diederix, C. Caux-Thang, L. Tabet, B. Bersch, E. Forest, and I. Michaud-Soret. 2007. Reversible redox- and zinc-dependent dimerization of the Escherichia coli Fur protein. Biochemistry 46:1329-1342. [DOI] [PubMed] [Google Scholar]
- 9.Dombroski, A. J., W. A. Walter, M. T. Jr. Record, D. A. Siegele, and C. A. Gross. 1992. Polypeptides containing highly conserved regions of transcription initiation factor sigma 70 exhibit specificity of binding to promoter DNA. Cell 70:501-512. [DOI] [PubMed] [Google Scholar]
- 10.Dong, T., M. G. Kirchhof, and H. E. Schellhorn. 2008. RpoS regulation of gene expression during exponential growth of Escherichia coli K12. Mol. Genet. Genomics 279:267-277. [DOI] [PubMed] [Google Scholar]
- 11.Ellermeier, C. D., A. Janakiraman, and J. M. Slauch. 2002. Construction of targeted single copy lac fusions using lambda Red and FLP-mediated site-specific recombination in bacteria. Gene 290:153-161. [DOI] [PubMed] [Google Scholar]
- 12.England, P., L. F. Westblade, G. Karimova, V. Robbe-Saule, F. Norel, and A. Kolb. 2008. Binding of the unorthodox transcription activator, Crl, to the components of the transcription machinery. J. Biol. Chem. 283:33455-33464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flynn, J. M., S. B. Neher, Y-I. Kim, R. T. Sauer, and T. A. Baker. 2003. Proteomic discovery of cellular substrates of the ClpXP protease reveals five classes of ClpX-recognition signals. Mol. Cell 11:671-683. [DOI] [PubMed] [Google Scholar]
- 14.Gaal, T., M. J. Mandel, T. J. Silhavy, and R. L. Gourse. 2006. Crl facilitates RNA polymerase holoenzyme formation. J. Bacteriol. 188:7966-7970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gowrishankar, J., K. Yamamoto, P. R. Subbarayan, and A. Ishihama. 2003. In vitro properties of RpoS (σS) mutants of Escherichia coli with postulated N-terminal subregion 1.1 or C-terminal region 4 deleted. J. Bacteriol. 185:2673-2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grigorova, I. L., N. J. Phleger, V. K. Mutalik, and C. A. Gross. 2006. Insights into transcriptional regulation and σ competition from an equilibrum model of RNA polymerase binding to DNA. Proc. Natl. Acad. Sci. U. S. A. 103:5332-5337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gruber, T. M., and C. A. Gross. 2003. Multiple sigma subunits and the partitioning of bacterial transcription space. Annu. Rev. Microbiol. 57:441-466. [DOI] [PubMed] [Google Scholar]
- 18.Hinton, D. M., S. Vuthoori, and R. Mulamba. 2006. The bacteriophage T4 inhibitor and coactivator AsiA inhibits Escherichia coli RNA polymerase more rapidly in the absence of σ70 region 1.1: evidence that region 1.1 stabilizes the interaction between σ70 and core. J. Bacteriol. 188:1279-1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ikeda, J. S., A. Janakiraman, D. G. Kehres, M. E. Maguire, and J. M. Slauch. 2005. Transcriptional regulation of sitABCD of Salmonella enterica serovar Typhimurium by MntR and Fur. J. Bacteriol. 187:912-922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ishihama, A. 2000. Functional modulation of Escherichia coli RNA polymerase. Annu. Rev. Microbiol. 54:499-518. [DOI] [PubMed] [Google Scholar]
- 21.Jishage, M., and A. Ishihama. 1997. Variation in RNA polymerase sigma subunit composition within different stocks of Escherichia coli W3110. J. Bacteriol. 179:959-963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karimova, G., J. Pidoux, A. Ullmann, and D. Ladant. 1998. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc. Natl. Acad. Sci. U. S. A. 95:5752-5756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karimova, G., C. Robichon, and D. Ladant. 2009. Characterization of YmgF, a 72-residue inner membrane protein that associates with the Escherichia coli cell division machinery. J. Bacteriol. 191:333-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.King, T., A. Ishihama, A. Kori, and T. Ferenci. 2004. A regulatory trade-off as a source of strain variation in the species Escherichia coli. J. Bacteriol. 186:5614-5620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klauck, E., A. Typas, and R. Hengge. 2007. The σS subunit of RNA polymerase as a signal integrator and network master regulator in the general stress response in Escherichia coli. Sci. Prog. 90:103-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kowarz, L., C. Coynault, V. Robbe-Saule, and F. Norel. 1994. The Salmonella typhimurium katF (rpoS) gene: cloning, nucleotide sequence, and regulation of spvR and spvABCD virulence plasmid genes. J. Bacteriol. 176:6852-6860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lelong, C., K. Aguiluz, S. Luche, L. Kuhn, J. Garin, T. Rabilloud, and J. Geiselmann. 2007. The Crl-RpoS regulon of Escherichia coli. Mol. Cell Proteomics 6:648-659. [DOI] [PubMed] [Google Scholar]
- 28.Lelong, C., M. Rolland, M. Louwagie, J. Garin, and J. Geiselmann. 2007. Mutual regulation of Crl and Fur in Escherichia coli W3110. Mol. Cell Proteomics 6:660-668. [DOI] [PubMed] [Google Scholar]
- 29.Malan, T. P., A. Kolb, H. Buc, and W. R. McClure. 1984. Mechanism of CRP-cAMP activation of lac operon transcription initiation activation of the P1 promoter. J. Mol. Biol. 180:881-909. [DOI] [PubMed] [Google Scholar]
- 30.Massé, E., and S. Gottesman. 2002. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 99:4620-4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 32.Murakami, K. S., and S. A. Darst. 2003. Bacterial RNA polymerase: the wholo story. Curr. Opin. Struct. Biol. 13:31-39. [DOI] [PubMed] [Google Scholar]
- 33.Notley-McRobb, L., T. King, and T. Ferenci. 2002. rpoS mutations and loss of general stress resistance in Escherichia coli populations as a consequence of conflict between competing stress responses. J. Bacteriol. 184:806-811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nyström, T. 2004. Growth versus maintenance: a trade-off dictated by RNA polymerase availability and sigma factor competition? Mol. Microbiol. 54:855-862. [DOI] [PubMed] [Google Scholar]
- 35.Paget, M. S. B., and J. Helmann. 2003. The σ70 family of sigma factors. Genome Biol. 4:203. http://genomebiology.com/2003/4/1/203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pratt, L. A., and T. J. Silhavy. 1998. Crl stimulates RpoS activity during stationary phase. Mol. Microbiol. 29:1225-1236. [DOI] [PubMed] [Google Scholar]
- 37.Robbe-Saule, V., G. Algorta, I. Rouilhac, and F. Norel. 2003. Characterization of the RpoS status of clinical isolates of Salmonella enterica. Appl. Environ. Microbiol. 69:4352-4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robbe-Saule, V., I. Carreira, A. Kolb, and F. Norel. 2008. Effect of growth temperature on Crl-dependent regulation of σS activity in Salmonella enterica serovar Typhimurium. J. Bacteriol. 190:4453-4459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Robbe-Saule, V., C. Coynault, and F. Norel. 1995. The live oral typhoid vaccine Ty21a is a rpoS mutant and is susceptible to various environmental stresses. FEMS Microbiol. Lett. 126:171-176. [DOI] [PubMed] [Google Scholar]
- 40.Robbe-Saule, V., V. Jaumouille, M. C. Prevost, S. Guadagnini, C. Talhouarne, H. Mathout, A. Kolb, and F. Norel. 2006. Crl activates transcription initiation of RpoS-regulated genes involved in the multicellular behavior of Salmonella enterica serovar Typhimurium. J. Bacteriol. 188:3983-3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robbe-Saule, V., M. D. Lopes, A. Kolb, and F. Norel. 2007. Physiological effects of Crl in Salmonella are modulated by σS level and promoter specificity. J. Bacteriol. 189:2976-2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Römling, U. 2005. Characterization of the rdar morphotype, a multicellular behaviour in Enterobacteriaceae. Cell. Mol. Life Sci. 62:1234-1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 44.Schmieger, H. 1972. Phage P22-mutants with increased or decreased transduction abilities. Mol. Gen. Genet. 119:75-88. [DOI] [PubMed] [Google Scholar]
- 45.Schuster, M., A. C. Hawkins, C. S. Harwood, and E. P. Greenberg. 2004. The Pseudomonas aeruginosa RpoS regulon and its relationship to quorum sensing. Mol. Microbiol. 51:973-985. [DOI] [PubMed] [Google Scholar]
- 46.Silhavy, T. J., M. L. Bernman, and L. W. Ernqvist. 1984. Experiments with gene fusions. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 47.Sorenson, M. K., and S. A. Darst. 2006. Disulfide cross-linking indicates that FlgM-bound and free σ28 adopt similar conformations. Proc. Natl. Acad. Sci. U. S. A. 103:16722-16727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sternberg, N. L., and R. Maurer. 1991. Bacteriophage-mediated generalized transduction in Escherichia coli and Salmonella typhimurium. Methods Enzymol. 204:18-43. [DOI] [PubMed] [Google Scholar]
- 49.Stüdemann, A., M. Noirclerc-Savoye, E. Klauck, G. Becker, D. Schneider, and R. Hengge. 2003. Sequential recognition of two distinct sites in σS by the proteolytic targeting factor RssB and ClpX. EMBO J. 22:4111-4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Typas, A., C. Barembruch, A. Possling, and R. Hengge. 2007. Stationary phase reorganisation of the Escherichia coli transcription machinery by Crl protein, a fine-tuner of σS activity and levels. The EMBO J. 26:1569-1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vassylyev, D. G., S.-I. Sekine, O. Laptenko, J. Lee, M. N. Vassylyeva, S. Borukhov, and S. Yokoyama. 2002. Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6Å resolution. Nature 417:712-719. [DOI] [PubMed] [Google Scholar]
- 52.Vijayendran, C., T. Polen, V. F. Wendisch, K. Friehs, K. Niehaus, and E. Flaschel. 2007. The plasticity of global proteome and genome expression analysed in closely related W3110 and MG1655 strains of a well-studied model organism, Escherichia coli K12. J. Biotechnol. 128:747-761. [DOI] [PubMed] [Google Scholar]
- 53.Zambrano, M. M., and R. Kolter. 1996. GASPing for life in stationary phase. Cell 86:181-184. [DOI] [PubMed] [Google Scholar]