Abstract
The outer membrane of Gram-negative bacteria represents the interface between the bacterium and its external environment. It has a critical role as a protective barrier against harmful substances and is also important in host-bacteria interactions representing the initial physical point of contact between the host cell and bacterial cell. RopB is a previously identified outer membrane protein from Rhizobium leguminosarum bv. viciae that is present in free-living cells but absent in bacteroids (H. P. Roest, I. H. Mulders, C. A. Wijffelman, and B. J. Lugtenberg, Mol. Plant Microbe Interact. 8:576-583, 1995). The functions of RopB and the molecular mechanisms of ropB gene regulation have remained unknown. We identified and cloned ropB and two homologs (ropB2 and ropB3) from the R. leguminosarum VF39SM genome. Reporter gene fusions indicated that the expression of ropB was 8-fold higher when cells were grown in complex media than when they were grown in minimal media, while ropB3 expression was constitutively expressed at low levels in both complex and minimal media. Expression of ropB2 was negligible under all conditions tested. The use of minimal media supplemented with various sources of peptides resulted in a 5-fold increase in ropB expression. An increase in ropB expression in the presence of peptides was not observed in a chvG mutant background, indicating a role for the sensor kinase in regulating ropB expression. Each member of the ropB gene family was mutated using insertional mutagenesis, and the mutants were assayed for susceptibility to antimicrobial agents and symbiotic phenotypes. All mutants formed effective nodules on pea plants, and gene expression for each rop gene in bacteroids was negligible. The functions of ropB2 and ropB3 remain cryptic, while the ropB mutant had an increased sensitivity to detergents, hydrophobic antibiotics, and weak organic acids, suggesting a role for RopB in outer membrane stability.
The outer membrane of Gram-negative bacteria represents the interface between the bacterium and its external environment. Consequently, it plays a critical role as a protective barrier against the entry of harmful substances (39) and is also important in host-bacteria interactions since it is the initial point of contact between the host cell and bacterial cell. For instance, the lipopolysaccharide (LPS) of the outer membrane is important for establishing a successful host infection in numerous Gram-negative bacteria (e.g., review reference 25) and protecting the cell from toxic compounds (e.g., review references 32 and 33). Although extensive research has been conducted on the role of LPS during plant infection by rhizobia (10), other components of the outer membrane such as the outer membrane proteins (OMPs) are relatively uncharacterized in Rhizobium.
A major protein component of the outer membrane of Rhizobium leguminosarum is the group II OMP RopB (11, 37). This OmpA-like protein is present in the outer membrane of free-living bacteria but is not found in membrane fractions of bacteroids isolated from Pisum sativum nodules (11). The specific trigger causing the decrease of RopB in bacteroids has not been found (38), and no further study of ropB gene expression in R. leguminosarum has been published to date. However, genes homologous to ropB have been studied in other members of the Rhizobiales, including aopB in Agrobacterium tumefaciens (20, 26), the omp25-omp31 family in Brucella spp. (7, 28, 42, 48), and ropB1 and ropB2 in Sinorhizobium meliloti (3, 6). Mutations in some of these ropB homologs have affected host-microbe interactions. An aopB mutant of A. tumefaciens is attenuated in tumor formation when inoculated onto plants (20). A Brucella abortus omp25 mutant is equally infective as the wild type, but after being internalized into host cells, the mutant is more sensitive to intracellular destruction (28). A ropB1 mutant of S. meliloti exhibits a Fix+ phenotype but is less competitive in nodulating alfalfa when coinoculated with the wild type (6). Some of these OMPs have also been implicated in outer membrane integrity. For example, a S. meliloti ropB1 mutant is more sensitive to the detergent sodium deoxycholate (DOC) (6).
The regulation of OMP gene expression in the Rhizobiales has been best characterized in A. tumefaciens and B. abortus. Expression of aopB increases under acidic conditions, and this regulation is controlled through the two-component signal transduction system ChvI/ChvG (26). A homologous two-component signal transduction system in B. abortus, named BvrR/BvrS, regulates OMP gene expression, including that of omp25 (16, 42).
It is not uncommon for rhizobial genomes to contain multiple ropB-like genes (3, 6, 7, 52). For example, four ropB homologs are predicted in the genome of R. leguminosarum bv. viciae 3841 (52). Given the importance of OMPs in several of the Rhizobiales species, we were interested in further characterizing the genes in the ropB family. Insertional mutagenesis and gusA reporter gene transcriptional fusions were used to investigate the function and regulation of the ropB gene family members in R. leguminosarum bv. viciae VF39SM.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. R. leguminosarum strains were grown on tryptone-yeast (TY) medium (5 g tryptone, 3 g yeast extract, 3.5 mM CaCl2 per liter H2O) or Vincent's minimal medium (VMM) (47) at 30°C, and Escherichia coli strains were grown on Luria-Bertani (LB) medium (40) at 37°C. Difco products supplied by Becton Dickinson were used for media preparation and included Bacto tryptone, Difco yeast extract, Difco casein digest, Bacto peptone, and Bacto soytone. When required, R. leguminosarum strains were grown in the presence of the following concentrations of antibiotics: gentamicin (Gm), 30 μg/ml; neomycin (Nm), 50 μg/ml; streptomycin (Sm), 500 μg/ml; spectinomycin (Sp), 100 μg/ml; and tetracycline (Tc), 5 μg/ml. E. coli strains were cultured in the following concentrations of antibiotics, where appropriate: ampicillin (Ap), 100 μg/ml; Gm, 15 μg/ml; kanamycin (Km), 50 μg/ml; Sp, 100 μg/ml; and Tc, 10 μg/ml.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristicsa | Reference(s) or source |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | endA1 hsdR17 supE44 thi-1 recA1 gyrA96 relA1 Δ(argF-lacZYA)U169 φ80lacZ ΔM15 | Invitrogen |
| S17-1 | RP4 tra region, mobilizer strain, Spr | 41 |
| TOP10 | F−mcrA Δ(mrr-hsdRMS-mcrBC)φ80lacZΔM15 ΔlacX74 recA1 araD139 galU galK Δ(ara-leu)7697 rpsL (Strr) endA1 nupG | Invitrogen |
| R. leguminosarum | ||
| VF39SM | Biovar viciae, spontaneous streptomycin-resistant mutant of VF39, Smr | 34 |
| 3841 | Biovar viciae, JB300 derivative streptomycin resistant, genome sequenced | 52 |
| 38EV27 | 3841 fabF2 fabF1 mutant, deficient in 27-hydroxyoctacosanoate modified LPS | 45 |
| DF20 | VF39SM, chvGΩKm mutant, unable to grow on complex media, Smr Nmr | This study |
| DF33 | VF39SM, ropBΩKm mutant, Smr Nmr | This study |
| DF15 | VF39SM, ropB2ΩKm mutant, Smr Nmr | This study |
| DF42 | VF39SM, ropB3ΩKm mutant, Smr Nmr | This study |
| Plasmids | ||
| pCRII-TOPO | TOPO TA cloning vector, Kmr Apr | Invitrogen |
| pCRS530 | Vector containing CAS-GNm cassette, Apr Kmr | 36 |
| pDG71 | Broad-host-range vector containing tryptophan promoter, Tcr | 14 |
| pFus1par | Broad-host-range vector with promoterless gusA for transcriptional fusions, par-stabilized, Tcr | 36, 51 |
| pHC41 | IncP broad-host-range vector, Tcr | 9 |
| pHP45ΩKm | Vector containing excisable Kmr gene cassette | 12 |
| pJQ200SK | Suicide vector, mob, sacB, Gmr | 35 |
| pDF4 | pFus1par with ropB::gusA transcriptional fusion, Tcr | This study |
| pDF37 | Broad-host-range vector pHC41 containing functional copy of ropB, Tcr | This study |
| pDF38 | Broad-host-range vector pDG71 containing functional copy of chvG, downstream of tryptophan promoter, Tcr | This study |
| pDF40 | pFus1par with ropB2::gusA transcriptional fusion, Tcr | This study |
| pDF41 | pFus1par with ropB3::gusA transcriptional fusion, Tcr | This study |
Apr, Gmr, Kmr, Nmr, Smr, Spr, and Tcr denote resistance to ampicillin, gentamicin, kanamycin, neomycin, streptomycin, spectinomycin, and tetracycline, respectively.
Bioinformatic analysis.
The genome sequence of R. leguminosarum 3841 (52) was used as a template to design PCR primers for the amplification of ropB1, ropB2, and ropB3 genes from R. leguminosarum VF39SM. The Oligo program was used to select PCR primers. Previous genetic analysis indicated that the two genomes are very similar (15, 51). DNA sequencing to confirm the proper identity of the PCR amplicons was used. Amino acid sequences for proteins from rhizobial species were obtained from RhizoBase (http://bacteria.kazusa.or.jp/rhizobase/), and those for Agrobacterium tumefaciens, Brucella melitensis, and Brucella abortus were obtained from GenBank. Homologous sequences were analyzed using BLASTP (1). Protein sequences were aligned using the ClustalW program (24). NJplot was used to construct tree diagrams.
Conjugation.
Insertional mutagenesis and reporter gene fusion constructs were mobilized from E. coli into R. leguminosarum using conjugation. Donor and recipient cells were cultured overnight. Conjugation was performed by mixing 0.5 ml of donor S17-1 cells (optical density at 600 nm [OD600] of ∼1.0) with 0.5 ml of recipient R. leguminosarum cells (OD600 of ∼0.5) in a microcentrifuge tube. The mixture was pelleted by centrifugation at 9,632 × g for 5 min, the supernatant was removed, and the pellet was resuspended in 50 μl of sterile water. The cell suspension was spotted onto TY agar or VMM agar containing 0.5 mM proline and incubated at 30°C for 24 to 48 h, after which the cells were scraped off and resuspended in 1 ml of sterile water. A total of 100 μl of cells was plated on the appropriate selective media.
Insertional mutagenesis of ropB, ropB2, ropB3, and chvG.
Insertional mutagenesis via allelic exchange was performed as described by Quandt and Hynes using the suicide vector pJQ200SK (35). Each gene to be mutated was amplified using PCR and cloned into the cloning vector pCRII-TOPO, according to the manufacturer's instructions (Invitrogen). The gene was inactivated by the insertion of an antibiotic cassette (12) into an internal restriction site. The inactivated gene was subsequently cloned into pJQ200SK and transformed into E. coli S17-1 cells for conjugation into R. leguminosarum. Putative double-crossover mutants were selected for growth on either TY or VMM containing the appropriate antibiotics and 5% (wt/vol) sucrose. Arising colonies were further screened for gentamicin sensitivity. Putative mutants found to be gentamicin sensitive were confirmed using Southern blotting and PCR analysis. The cloning details for construction of each individual construct are described below.
The primers RopBFor (GACGCCCTTGTAATGAGC) and RopBRev (CGCCAATCCCCCGAAAAC) were used to clone the ropB gene into the vector pCRII-TOPO. The construct pCRII::ropB was digested with BamHI/XhoI, and the fragment containing ropB was ligated into pJQ200SK. An internal SalI site was used to interrupt ropB by inserting the ΩKm cassette from p1918ΩKm using SalI to create construct pDF33.
The ropB2 mutant was created by cloning a 1.8-kb region containing the ropB2 gene using the primers RopB2ProFor (5′-TCAAGACGCAGCAACTCAAGCA-3′) and RopB2R (5′-GCTATTATTTCATCGTGCCGACCC-3′) into pCRII-TOPO. The construct was digested with BamHI/XhoI, and the fragment containing ropB2 was ligated into pJQ200SK. An internal SalI site was used to interrupt ropB2 by inserting a ΩKm cassette excised from pHP45ΩKm to create the construct pDF15.
The construct used to mutate ropB3 was created by amplifying the 5′ end of the ropB3 gene using the PCR primers RopB3F5 (5′-CGGGAGCTCATGGCGAAGTATCAATCC-3′) and RopB3R5 (5′-CGGGATCCCGGTGCGTCATAAGAGGG-3′) and the 3′ end of the ropB3 gene using the PCR primers RopB3F3 (5′-CGGGATCCCACGAGTGGAGGCATCAG-3′) and RopB3R3 (5′-CGGCTCGAGTGAGTGTCCAGTTCCGTG-3′). The 5′ fragment was digested with SstI/BamHI and ligated into similarly digested pJQ200SK. The resulting plasmid and the amplified 3′ fragment of the ropB3 gene were then digested with BamHI/XhoI and ligated together. The ropB3 gene was then interrupted with the ΩKm cassette from pHP45ΩKm at the internal BamHI site to create the construct pDF42.
The construct used to mutate chvG was created by cloning a 1.8-kb region containing the chvG gene into pCRII-TOPO using the PCR primers ChvGF (5′-TGTCGGAAATGTTGAGGGCT-3′) and ChvGR (5′-CGGTGATGTTTTCGGCTCTC-3′). The chvG region was removed by digesting it with BamHI/SphI and ligated into pBBRKG1 (15). The ΩKm cassette from pHP45ΩKm was inserted at an internal SstI site of chvG to interrupt it. pBBRKG1::chvGΩKm was then digested with XbaI/SpeI, with the fragment containing chvGΩKm ligated into pJQ200SK cut with XbaI to create pDF20.
Construction of ropB, ropB2, and ropB3 promoter fusions.
A transcriptional fusion for ropB was constructed by cloning the ropB promoter region into pFus1par. The primers RopBProFor (TCGAATTTCAGCCATGCGGC) and RopBProRev (AACGCCGTAAACGATCTGGC) were used to amplify a 442-bp fragment from VF39SM. This fragment included 169 bp upstream of the ropB start codon. This fragment was cloned into pCRII, according to the supplier's instructions, to create pCRII::ropBPro. An EcoRI-digested fragment was then cloned into pFus1par in the proper orientation (verified by DNA sequencing) to create the transcriptional fusion pDF4.
A transcriptional fusion for ropB2 was constructed by cloning the ropB2 promoter region into pFus1par. The primers RopB2ProFor (TCAAGACGCAGCAACTCAAGCA) and RopB2ProRev (CGATGCCGTAGACGATATTTCCAG) were used to amplify a 1,194-bp fragment, including 924 bp upstream of the ropB2 start codon. This fragment was cloned into pCRII, according to supplier's instructions. A 1.2-kb XhoI/KpnI fragment was cloned from this plasmid into XhoI/KpnI-digested pFus1par to create the transcriptional fusion pDF40, which was verified by DNA sequencing.
A transcriptional fusion for ropB3 was created by using PCR primers RopB3ProF (5′-CGGGGTACCATGGCGAAGTATCAATCC-3′) and RopB3ProR (5′-CGGTGCGTCATAAGAGGG-3′) and by amplifying a 529-bp fragment containing the ropB3 promoter. This fragment was cloned into pCRII-TOPO, according to manufacturer's instructions. The ropB3 promoter region was liberated by digesting it with EcoRI/KpnI and ligated into a similarly digested pFus1par vector to create the ropB3 transcriptional fusion pDF41, which was verified by DNA sequencing.
Assays for β-glucuronidase (gusA) activity.
β-Glucuronidase was selected as the reporter gene to assay for rop gene expression (19). Assays for β-glucuronidase activity were carried out using a modified procedure originally described by Miller (30), with 3 mg/ml 4-nitrophenyl β-d-glucuronide replacing 4 mg/ml 2-nitrophenyl β-d-galactopyranoside. Reporter gene expression from free-living cells was measured using R. leguminosarum strains carrying the transcriptional fusions grown in the appropriate media at 30°C and harvested in late exponential phase. These data were calculated as Miller units using the following formula: (A420 × 1,000)/(OD600 × time × dilution). Reporter gene expression was also measured in the bacteroid state as described by Wang et al. (49). In brief, 10 to 15 nodules were harvested at 4 weeks postinoculation and crushed in a buffer containing 1 ml of 0.25 M mannitol and 0.05 M Tris-HCl (pH 7.5). After allowing the plant debris to settle, 100 μl of this bacteroid suspension was used to measure gusA activity. To confirm that the fusion plasmid was maintained in the nodule environment, 10 to 20 individual nodules were surface sterilized (as described below), crushed, and plated in duplicate on media containing Sm or Sm-Tc to confirm that the bacteria isolated from the nodule remain Tc resistant and therefore carry the fusion plasmid. In all cases, >95% plasmid retention was observed.
To determine if the mutation of a particular ropB gene family member altered gene expression of the other members of the ropB gene family, each gene fusion plasmid was mobilized into the corresponding ropB1, ropB2, or ropB3 mutants via conjugation. Subsequent assays for β-glucuronidase were performed as described above.
Nodulation experiments.
Pea seeds (Pisum sativum cv. Trapper) were surface sterilized in 2.625% sodium hypochlorite for 5 min and then in 70% ethanol for 5 min, followed by three washes in sterile water. The sterile seeds were then placed on 1.25% water agar plates and allowed to germinate in the dark at room temperature. After approximately 3 days, they were planted in modified magenta jars that were assembled to resemble Leonard jars (47). The magenta jars contained sterile vermiculite as the solid support medium and approximately 200 ml of Hoagland's plant medium (18). Two pea seeds were planted into each jar, and each seed was inoculated with 500 μl of an overnight culture (OD600 of ∼1) of the appropriate R. leguminosarum strain resuspended in sterile water. The nodules were harvested after 4 weeks. The protocol described by Yost et al. (51) was used to assess the ability of the ropB mutant to compete against the wild type during nodulation.
Antimicrobial sensitivity assays.
Sensitivity to detergents was evaluated by comparing the growth of strains on agar media, both with and without detergent. Serial dilutions of each R. leguminosarum strain being evaluated were plated on TY, TY containing 1.5 mM DOC, and TY containing 0.35 mM sodium dodecyl sulfate (SDS) plates and incubated for at least 4 days at 30°C. After this time, the numbers of CFU were counted, and the log10 reduction in growth due to the detergent was calculated. MIC assays were conducted for erythromycin and l-pyroglutamic acid (PGA) in TY and VMM broth. Growth was determined by measuring absorbance at OD600.
Isolation of cell envelope fractions and SDS-Tricine-PAGE analysis.
Cells being used for cell envelope preparation were grown in either TY or VMM media using conditions described above. Cells were harvested by centrifugation at 4,000 × g for 10 min at 4°C and stored frozen at −70°C in membrane buffer (50 mM sodium phosphate buffer at pH 7.0, 7.5% [wt/vol] glycerol, 50 mM NaCl). The frozen cell slurry was thawed on ice, and phenylmethylsulfonyl fluoride (PMSF) was added to a final concentration of 0.1 mM. The cell slurry was subjected to 3 rounds of 45 s of sonication (0.14 to 0.15 W) on ice using a Misonix Microson ultrasonic cell disruptor. The sonicated cell slurry was centrifuged at 11,000 × g for 10 min at 4°C to remove unbroken cells and subsequently centrifuged in a Beckman Coulter Optima L-90 K ultracentrifuge at 40,000 rpm for 90 min at 4°C in a Ti70 rotor to separate the membrane fraction from the cytosolic fraction. The membrane fraction was resuspended in membrane buffer, and total membrane protein amounts of each sample were determined using a modified Lowry assay (43) and frozen for storage at −70°C. SDS-Tricine-polyacrylamide gel electrophoresis (PAGE) was performed on isolated membranes from each sample. Membrane samples were resolved on either 12% or 16.5% SDS-Tricine-PAGE to resolve protein bands of <40 kilodaltons. All membrane protein samples were loaded onto the gel to final amounts of 2.5 μg or 5 μg. Membrane protein samples were loaded at either room temperature (22 to 25°C) or heated to 100°C before loading to resolve proteins of lower molecular weight. Proteins separated by SDS-Tricine-PAGE gels were visualized by conventional Coomassie blue staining and by trichloroethanol (TCE) staining. TCE was added to the gels during casting, at a final concentration of 0.5% (vol/vol) TCE, to visualize proteins with tryptophan residues by UV irradiation at 300 nm, according to the method described by Ladner et al. (23). The TCE staining technique increased the ability to visualize membrane proteins within the gel by ≥60% in comparison to conventional Coomassie blue staining, with no difference in protein migration. Coomassie blue staining was performed on all TCE-stained gels.
Nucleotide sequence accession numbers.
The DNA sequences for ropB1, ropB2, and ropB3 from R. leguminosarum VF39SM have been deposited in GenBank under the following respective accession numbers: FJ561446, FJ561447, and FJ561448.
RESULTS
The R. leguminosarum VF39SM genome has three genes encoding RopB paralogs.
The genome of R. leguminosarum 3841 (52) has four ropB-like genes, based on a BLASTP search using the amino acid sequence of RopB from R. leguminosarum bv. viciae 248 (37). The gene RL1589, found on chromosome 3841, is annotated as ropB and encodes a protein which is 98% identical to the characterized RopB from R. leguminosarum 248 (37). Of the other three ropB-like genes, one is found on the chromosome (RL1307) and two are found on the plasmids pRl10 and pRl7 (pRL100173 and pRL70177). The DNA sequences of these genes were used to create PCR primers with the intention of amplifying homologous genes in the VF39SM genome. Genes homologous to RL1589, RL1307, and pRL100173 were amplified from the VF39SM genome, while a homolog of pRL70177 was not amplified. This is in agreement with data from a recently completed genome sequencing project of VF39SM (M. F. Hynes et al., unpublished data). Analysis of the genome indicates that the genetic region of pRL7 containing pRL70177 is absent from the VF39SM genome (Hynes et al., unpublished). Furthermore, primers that bind within the pRL70177 coding sequence did not amplify a fragment from VF39SM genomic DNA, even under low stringency conditions. The PCR amplicons containing the three ropB-like genes in VF39SM were subsequently sequenced to ensure they contained ropB homologs.
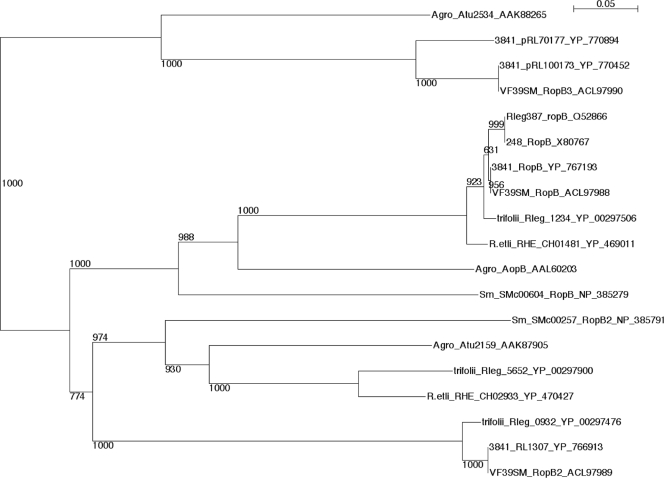
The predicted RopB-like proteins from VF39SM were identical to those found in strain 3841. The VF39SM homolog of RL1589 was named ropB, as it encodes a protein identical to RopB from strain 3841. The VF39SM homolog of RL1307 was named ropB2 based on its corresponding amino acid sequence, sharing homology with RopB2 from S. meliloti 1021 (Fig. 1). As there has been no annotation of pRL100173 or related homologs, and since it encodes a predicted outer membrane protein that is homologous to RopB (26% identical and 43% similarity), we suggest that it be named ropB3. Figure 1 provides a phylogenetic tree of RopB homologs in the rhizobia.
FIG. 1.
A neighbor-joining tree derived from a ClustalW alignment of the amino acid sequences deduced from the ropB gene family is shown, with bootstrap values. Amino acid sequences were obtained from GenBank. Abbreviations: VF39SM, R. leguminosarum bv. viciae VF39SM; 3841, R. leguminosarum bv. viciae 3841; Agro, Agrobacterium tumefaciens; Sm, Sinorhizobium meliloti; R.etli, Rhizobium etli CFN42; trifolii, R. leguminosarum bv. trifolii WSM1325; 248, R. leguminosarum bv. viciae 248; Rleg387, R. leguminosarum bv. viciae 387. Protein names were included if they were available; otherwise, the GenBank locus tag was used to identify each protein. The GenBank accession numbers are listed following the protein name or locus tag.
Insertional mutagenesis of ropB, ropB2, and ropB3.
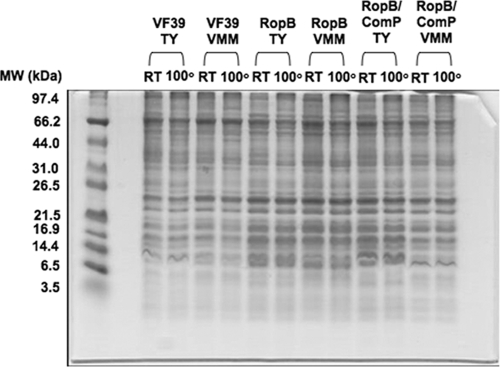
Individual colonies of mutant strains DF33, DF15, and DF42 were isolated on TY agar media to evaluate any changes in colony morphology as a result of the mutations in ropB, ropB2, and ropB3, respectively. After approximately 4 days of incubation, wild-type VF39SM showed convex, circular colonies with perfect borders, while DF33 colonies had irregular edges and were flat. Complementation of DF33 with a plasmid-localized functional copy of ropB (pDF37) produced an intermediate phenotype where the colonies did not have perfectly round edges but were convex like the wild type. In addition, DF33 had a mucoid-like layer around the colony that was not present in VF39SM or the pDF37-complemented mutant. No changes in the colony morphology of the DF15 or DF42 mutant strains were observed. Furthermore, changes in colony morphology for the ropB mutant were not observed on VMM. The effect of a ropB mutation on the profile of membrane proteins was determined by fractionating cell envelope proteins and observing the protein profile on SDS-Tricine-PAGE gels. Mutation of ropB did not appear to cause any dramatic changes in the membrane protein profile of VF39SM (Fig. 2).
FIG. 2.
Tricine-SDS-PAGE (16.5%) of isolated membranes from strains grown in TY and VMM. The strains tested were VF39SM (VF39), the ropB mutant (RopB), and the ropB mutant complemented with plasmid pDF37 (RopB/ComP). The lanes for each strain are indicated on the figure. A total of 5 μg of membrane protein was loaded into each lane of the gel and heated at either room temperature (RT) or to 100°C for 5 min. Coomassie blue gel staining was performed to visualize protein bands.
As stated previously, some rhizobial strains with outer membrane defects are impaired in the symbioses with their host plant (6). To evaluate whether the RopB family of OMPs participate in the plant-host relationship, pea plants were inoculated with the ropB family mutants, and their ability to form functional nodules was assessed. Plants inoculated with DF33, DF15, or DF42 all possessed large pink nodules that contained differentiated bacteroids when examined under a light microscope. In addition, the plants inoculated with these mutants exhibited healthy shoots similar to those exhibited by the control inoculated with the wild type. A ropB mutant in S. meliloti is less competitive for nodulation sites than the wild type when coinoculated on alfalfa (6). Therefore, we also examined the competitive abilities of the ropB mutant for competing against the wild type during nodulation. Three trials were completed using pea plants and a near to 1:1 ratio of the mutant and wild type. The subsequent ratio of mutant nodules to wild-type nodules recovered after 4 weeks of growth was only significantly (chi-square test; P < 0.05) reduced in one of the three trials, and this reduction was slight (data not shown). These results suggest that the ropB mutant in R. leguminosarum may not be substantially impaired in its ability to compete with the wild type during nodulation.
Defects in the outer membrane are also known to cause increased sensitivity to antimicrobial compounds, such as detergents, cationic peptides, and antibiotics (17). The detergents sodium dodecyl sulfate (SDS) and sodium deoxycholate (DOC) were added to TY agar plates at concentrations of 0.35 mM and 1.5 mM, respectively. The ropB2 and ropB3 mutants did not have an increased sensitivity to either of the detergents (Table 2). Mutation of ropB, on the other hand, increased sensitivity to both of the detergents, resulting in a >3-log reduction in growth in the presence of the detergents (Table 2). Complementation of the ropB mutant with the pDF37 plasmid restored the mutant's ability to grow in the presence of detergents. MIC assays revealed that DF33 is also more sensitive to the hydrophobic antibiotic erythromycin and the weak organic acid l-pyroglutamic acid, while mutants DF15 and DF42 showed no difference compared to the wild type.
TABLE 2.
Effect of antimicrobial compounds on growth of the ropB family mutants
| Strain | Log growth reductiona |
MICb |
||||
|---|---|---|---|---|---|---|
| PCA |
Erythromycin |
|||||
| 1.5 mM DOC | 0.35 mM SDS | TY | VMM | TY | VMM | |
| VF39SM | 0.191 ± 0.057 | 0.263 ± 0.112 | 5.5 | 6.0 | 8.83 × 10−3 | 0.027 |
| DF33 (ropB mutant) | 4.85 ± 1.22* | 3.86 ± 1.21* | 4.5 | 5.5 | 1.36 × 10−3 | 0.010 |
| DF33 (ropB mutant) + pDF37 | 0.351 ± 0.163 | 0.708 ± 0.386 | 5.5 | 6.0 | 8.83 × 10−3 | 0.027 |
| DF15 (ropB2 mutant) | 0.103 ± 0.179 | 0.101 ± 0.355 | 5.5 | 6.0 | 8.83 × 10−3 | 0.027 |
| DF42 (ropB3 mutant) | 0.123 ± 0.251 | 0.026 ± 0.131 | 5.5 | 6.0 | 8.83 × 10−3 | 0.027 |
Growth reduction values were calculated by subtracting the number of log CFU/ml grown on TY plus detergent from the number of log CFU/ml grown on TY. Data are presented as the average results ± standard deviations from at least three trials. An asterisk indicates a significant difference in growth between the wild type and the mutant (t test; P of <0.01).
Strains were grown in either TY or VMM broth with various concentrations of PCA or erythromycin. MICs are presented in mM and are representative values from a minimum of three separate replicates.
Increased expression of ropB in EDTA.
Addition of EDTA to the growth media chelates the calcium present, and removal of calcium decreases the stability of the outer membrane (32). We hypothesized that the addition of EDTA to VMM might increase expression of ropB, since phenotypic data suggested a role for RopB in outer membrane stability. Gene expression of strains grown in VMM containing 1 mM EDTA was compared to expression of those in VMM alone. The presence of EDTA caused a >2-fold increase in ropB expression (Table 3). The ropB2 and ropB3 fusions did not exhibit an increase in expression of a similar magnitude (Table 3).
TABLE 3.
Promoter activity of ropB, ropB2, and ropB3 in VF39SM under various growth conditions
| Plasmid | gusA reporter fusion |
gusA activitya |
|||
|---|---|---|---|---|---|
| Strains grown in TY | Strains grown in VMM | Strains grown in VMM + 1 mM EDTA | Bacteroids | ||
| pDF4 | ropB::gusA | 5.37 × 103 ± 1.40 × 103 | 690 ± 279* | 1.61 × 103 ± 47.8 | 233 ± 62.4* |
| pDF40 | ropB2::gusA | 240 ± 27.8 | 194 ± 32.3 | 139 ± 62.6 | 116 ± 30.0 |
| pDF41 | ropB3::gusA | 417 ± 61.9 | 199 ± 30.0 | 307 ± 53.9 | 114 ± 11.9 |
Data are expressed as the means ± standard deviations from at least three assays, and values are expressed as Miller units (27). The average background level of gusA activity for TY-grown cells carrying pFus1 is approximately 250 ± 27.2 Miller units. An asterisk indicates a significantly different mean expression compared to expression in TY for each respective gene (t test; P of <0.005).
Expression of the ropB gene family in free-living and bacteroid cells.
The absence of RopB in the outer membranes of bacteroids has been observed previously using Western blotting (38). To determine if RopB disappearance is correlated to transcriptional downregulation of ropB, the ropB::gusA transcriptional fusion pDF4 was created. The promoter activity of ropB was measured in free-living cells grown in complex TY media and bacteroids isolated from pea plants. There was a 23-fold decrease in ropB expression in the nodule compared to that in TY-grown cells (Table 3). This is consistent with the findings of Roest et al. (38) and suggests that diminished RopB in bacteroids is due to decreased rates of gene transcription. A growth condition in TY that reduced ropB expression to the level of expression observed in bacteroid cells could not be identified, but during these investigations, an 8-fold reduction in ropB expression was observed when the fusion strain was grown in VMM (Table 3).
Both the ropB2 and ropB3 fusions were close to background levels in bacteroid cells, suggesting negligible transcriptional expression in bacteroids. Free-living expression of ropB2 and ropB3 were observed at levels below ropB expression in both TY medium and VMM (Table 3). The expression of the ropB2 fusion was close to background levels, indicating that it is unlikely that this gene is expressed under the conditions assayed. In contrast, expression of ropB2 in S. meliloti is increased in bacteroids compared to that in free-living cells grown in complex media (3). The ropB3 fusion had modest expression in TY and expression close to background levels in VMM.
Increased expression of ropB in peptide-rich media.
The increased expression of ropB in TY relative to VMM was investigated further. Addition of either tryptone or yeast extract to VMM resulted in an increase of ropB expression, while additional CaCl2 had no effect (Table 4). Inclusion of both tryptone and yeast extract in VMM had an additive effect on ropB expression (Table 4). The components of VMM did not cause a decrease in ropB expression when added to TY (data not shown). Tryptone and yeast extract are rich in peptides; therefore, other peptide sources were tested to determine if they caused an increase in ropB expression. The addition of hydrolyzed casein, meat peptone, or soytone to VMM all produced a similar increase in ropB expression (Table 4). Amino acids do not increase ropB expression, since the addition of individual or groups of amino acids to VMM did not increase ropB expression (data not shown).
TABLE 4.
Promoter activity of ropB when grown in the presence of peptides
| Medium | GusA activity assayed using pDF4a |
|---|---|
| TY | 5.37 × 103 ± 1.40 × 103* |
| VMM | 690 ± 279 |
| VMM + CaCl2b | 789 ± 135 |
| VMM + yeast extractb | 2.66 × 103 ± 128* |
| VMM + tryptoneb | 2.74 × 103 ± 94.9* |
| VMM + tryptone + yeast extractb | 4.50 × 103 ± 278* |
| VMM + 1% hydrolyzed casein | 3.48 × 103 ± 570* |
| VMM + 0.5% meat peptone | 3.03 × 103 ± 132* |
| VMM + 0.5% soytone | 3.43 × 103 ± 94.2* |
| VMM + 2.5 mM IQYc | 436 ± 109 |
| VMM + 2.5 mM YVLc | 445 ± 11.3 |
| VMM + 2.5 mM FSDKIAKc | 467 ± 28.8 |
ropB promoter activity was assayed using gusA fusion plasmid pDF4 in a VF39SM wild-type background. Data are expressed as the means ± standard deviations from at least three assays, and values are expressed as Miller units (27). The background level of gusA activity for TY grown cells is approximately 250 Miller units. *, growth in TY or in VMM amended with peptide-rich medium components resulted in a statistically significant increase in ropB expression compared to that in growth in VMM (t test; P of <0.001).
Consists of 0.5% tryptone, 0.3% yeast extract, and 3.4 mM CaCl2 used at the same concentration as TY.
Synthesized peptides with sequences specified by the single-letter amino acid code were added to VMM.
Regulation of ropB by the sensor kinase ChvG.
Expression of ropB homologs in other Alphaproteobacteria is regulated by a two-component signal transduction system. For example, ChvI/ChvG regulates aopB expression in response to acidic pH (20, 26) in A. tumefaciens. A homologous system in B. abortus named BvrR/BvrS regulates OMP gene expression, including omp25-omp31 (16), and mutation of the bvrR-bvrS genes results in decreased virulence (42). R. leguminosarum has a two-component signal transduction system that is homologous to those found in A. tumefaciens and B. abortus. The sensor kinase of this system, also denoted chvG in the R. leguminosarum 3841 genome, was cloned and mutated in VF39SM in order to determine if it plays a role in ropB regulation.
The VF39SM chvG mutant DF20 was unable to grow on complex media, similar to the chvG mutant of A. tumefaciens (8). The growth of the chvG mutant was substantially delayed compared to the growth of the wild type, and the mutant does not form effective nodules on pea plants. DF20 was cultured routinely on VMM agar because the mutant grew very poorly in VMM broth. The poor growth of this mutant echoes the difficulty in obtaining loss-of-function chvG (exoS) mutants in S. meliloti (9), and the chvI and chvG mutants were obtained only recently in S. meliloti (4). In order to determine if ChvG plays a role in regulating expression of the ropB family, the transcriptional fusions pDF4, pDF40, and pDF41 were introduced into the chvG mutant background. We wanted to compare ropB expression in VMM and TY between the chvG mutant and wild type. However, this comparison was not possible due to the inability of the chvG mutant to grow in TY broth. Further investigation into growth phenotypes of the chvG mutant led to the discovery that the chvG mutant can grow on VMM agar supplemented with 0.5% tryptone when 3.4 mM CaCl2 is present. However, the mutant was incapable of growing under similar conditions in broth culture. In light of this, VMM agar with 3.4 mM CaCl2 and VMM with 0.5% tryptone and 3.4 mM CaCl2 were used to study the role that ChvG plays in the expression of the genes in the ropB family.
The 5-fold increase in expression of ropB in the wild-type background in the presence of tryptone was abolished in the chvG mutant background (Table 5). The mutation of chvG had no effect on the regulation of ropB2, as the expression of ropB2 remained very low when grown on either type of media and regardless of the background. The expression patterns of ropB3 were similar in both the wild-type and chvG mutant backgrounds. The inability of the chvG mutant to grow in broth culture precluded us from determining the role of ChvG in upregulating ropB under Ca2+-limiting conditions.
TABLE 5.
Effect of a chvG mutation on ropB expression in the presence of tryptone
| Strain | Expressionb |
||
|---|---|---|---|
| VMM + CaCl2a | VMM + tryptone + CaCl2a | Fold induction | |
| VF39SM | 310 ± 58.7 | 1.59 × 103 ± 380 | 5.11 ± 0.260 |
| chvG mutant | 430 ± 9.50 | 665 ± 100 | 1.54 ± 0.210* |
Strains were cultured on VMM agar with 3.4 mM CaCl2 added and 0.5% tryptone, as indicated.
Expression data is expressed as the means ± standard deviations from at least three assays, and values are expressed as Miller units (27). Fold induction is measured by comparing the expression in the presence and absence of 0.5% tryptone. An asterisk indicates that a significant difference in the effect of tryptone on the increase in ropB expression between the chvG mutant and the wild type was observed (t test; P of <0.0001).
TY-dependent upregulation of ropB is abolished in an LPS mutant lacking 27-hydroxyoctacosanoate-modified lipid A.
The LPS of R. leguminosarum, and of other members of Rhizobiaceae, is modified by the addition of 27-hydroxyoctacosanoate (also referred to as very long chain fatty acid) to the 2′ position of lipid A (13, 46). We recently isolated a mutant of R. leguminosarum 3841 where the lipid A is lacking the 27-hydroxyoctacosanoate modification (45). The ropB, ropB2, and ropB3 transcriptional fusion plasmids were mated into this mutant as well as into the 3841 wild-type strain. gusA gene expression from pDF4 was measured in both wild-type and mutant strains grown in TY or VMM. A 4-fold upregulation of ropB expression in TY broth relative to VMM broth was observed in wild-type 3841 carrying pDF4, similar to the upregulation observed in VF39SM (data not shown). Notably, the increased expression in the ropB fusion that is typical when the fusion strain is grown in TY broth was not evident in the LPS mutant (Table 6). The expression patterns in the ropB2 and ropB3 fusions were unchanged between the wild type and the LPS mutant (Table 6).
TABLE 6.
Expression of the ropB gene family fusions in a mutant deficient in 27-hydroxyoctacosonate-modified LPS
| Plasmid | Fusion | GusA activity for straina: |
|
|---|---|---|---|
| 3841 | 38EV27 | ||
| pDF4 | ropB::gusA | 6.25 × 103 ± 1.74 × 103 | 235 ± 35.7* |
| pDF40 | ropB2::gusA | 201 ± 21.7 | 196 ± 8.11 |
| pDF41 | ropB3::gusA | 381 ± 44.6 | 480 ± 139 |
Strains were grown in TY broth and assayed for GusA activity using the modified Miller assay described in Materials and Methods. Data are expressed as the means ± standard deviations from at least three assays, and values are expressed as Miller units (27). An asterisk indicates a significant difference in ropB gene expression between the wild type and lipid A mutant (t test; P of <0.0001).
DISCUSSION
Phenotypic and gene expression data from this study support the hypothesis that RopB, like OmpA (32) of E. coli, is an important structural component of the outer membrane, while the roles of RopB2 and RopB3 remain cryptic. Mutations that lead to structural defects in the outer membrane often increase outer membrane permeability and increase a cell's sensitivity to hydrophobic antimicrobials (17, 44). Of the rop mutants, only the ropB mutant was sensitive to detergents, the hydrophobic weak organic acid pyroglutamic acid (50), and the hydrophobic antibiotic erythromycin. The contribution of RopB to outer membrane stability may involve an interaction with LPS. Other eight-stranded β-barrel OMPs with secondary structure similar to that of RopB have been proposed to interact with LPS, including Omp21 from Comamonas acidovorans (2), OprH from Pseudomonas aeruginosa (5), and Omp25 from Brucella (7). A possible interaction between RopB and LPS is supported by the fact that ropB expression is significantly reduced in a LPS lipid A mutant of R. leguminosarum 3841 (Table 6). The reduction of ropB expression in the lipid A mutant may contribute to the increased sensitivity to detergents and other membrane-associated stressors observed in LPS mutants lacking 27-hydroxyoctacosanoate-modified lipid A (13, 45, 46). Outer membrane protein interaction with LPS at the divalent cation binding sites has been suggested for the Pseudomonas aeruginosa OMP OprH. Increased oprH expression is thought to stabilize the outer membrane under Mg2+-limiting growth conditions by compensating for the decrease in divalent cations (5, 31, 53). The possibility that RopB functions similarly to OprH is supported by the increased expression of ropB when VF39SM is grown in media containing EDTA (Table 3), which chelates 98% of the Ca2+ present in VMM (H. Weger, personal communication). A role of calcium in rhizobial OMP gene regulation has been noted previously. Kim and colleagues (22) have observed that in Sinorhizobium fredii USDA257, expression of Omp22, an OMP with homology to RopB, increases under calcium-limiting growth conditions. They also noted that the majority of Omp22 was found in the extracellular fraction when cells are calcium limited, suggesting that calcium is required for the structural integrity necessary to maintain its localization in the outer membrane (22). de Maagd and colleagues (11) have noted that LPS is also released into the growth media under calcium-limiting growth conditions. It would be interesting to ascertain if the RopB is also found to be associated with the LPS excreted into the growth medium.
The functions of ropB2 and ropB3 in VF39SM remain elusive, as we could not identify any phenotypic changes in these mutants, and we were unable to identify conditions under which gene expression was increased. Expression of ropB2 did not increase in a ropB or ropB3 mutant background, suggesting it does not compensate for the loss of ropB or ropB3. Furthermore, ropB3 expression was not increased in the ropB mutant background, suggesting that RopB3 does not compensate for loss of RopB.
The outer membrane of Rhizobium leguminosarum undergoes structural changes during bacteroid maturation, including a transition in the LPS from a hydrophilic to a predominantly hydrophobic nature (21), and the downregulation of the ropB genes in mature bacteroids emphasizes the changes that occur to the cell envelope during bacteroid maturation. The data also confirm the earlier Western blot data of Roest et al. (38), indicating that the absence of these OMPs in mature bacteroids is due to downregulation of gene expression. It is notable that the difference in ropB gene expression between cells grown in VMM and bacteroids is not large, especially compared to ropB expression in TY (Table 3). Therefore, interpretation of the physiological significance of a change in gene expression from a free-living to bacteroid state should be considered in the context of the growth parameters used for the free-living cells.
A role for ChvG in the regulation of ropB expression is consistent with the ChvG-dependent OMP gene regulation observed in other Rhizobiales (16, 26, 29, 42). However, ChvG-dependent induction of OMP gene expression due to the presence of growth components rich in peptides has not been previously reported. Regulation of OMPs in response to growth in complex media has been reported for Omp21 from C. acidovorans, which is present in cells grown in complex media and absent in cells grown in minimal media (2). Given the direct correlation between peptide-rich media and ropB expression, we attempted to identify specific peptides that may be involved in regulation using synthetic peptides derived from the peptide sequences resulting from an enzymatic hydrolysis of casein. These sequences were selected based on their reported high antimicrobial activity in E. coli (27). However, these peptides did not induce ropB gene expression (Table 4). Identification of a particular peptide can be daunting, given the number of diverse peptides generated from the enzymatic hydrolysis of casein. It is also possible that peptides indirectly induce expression of ropB through ChvG, and this requires a mixture of several peptides. It is interesting to note that a chvG mutant is incapable of growing on TY and that ropB, which is regulated by chvG, is strongly induced in TY. Although a ropB mutant is still able to grow in TY, it does exhibit colony morphology that is different from that of the wild type (data not shown). The role of ChvG in inducing ropB expression under Ca2+-limiting growth conditions remains to be elucidated. A major hurdle in this experiment is the poor growth of the chvG mutant in the presence of EDTA and also in broth media.
Future experiments are planned to define in greater detail how ChvG induces expression of ropB and the possible mechanism for reduced ropB expression in a lipid A mutant. We will also continue our characterization of ropB2 and ropB3 in an effort to assign functions to these genes. Understanding the regulation and function of outer membrane proteins will further define the important role of the outer membrane in both Gram-negative symbioses and pathogenesis.
Acknowledgments
This research has been supported by a Natural Sciences and Engineering Research Council (NSERC) grant to C.K.Y., and D.L.F. and E.M.V. were supported by Postgraduate Scholarships from NSERC.
We thank M. F. Hynes for providing unpublished VF39SM genome data and R. J. Turner for helpful discussions.
Footnotes
Published ahead of print on 18 December 2009.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baldermann, C., A. Lupas, J. Lubieniecki, and H. Engelhardt. 1998. The regulated outer membrane protein Omp21 from Comamonas acidovorans is identified as a member of a new family of eight-stranded β-sheet proteins by its sequence and properties. J. Bacteriol. 180:3741-3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnett, M. J., C. J. Toman, R. F. Fisher, and S. R. Long. 2004. A dual-genome symbiosis chip for coordinate study of signal exchange and development in a prokaryote-host interaction. Proc. Natl. Acad. Sci. U. S. A. 101:16636-16641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bélanger, L., K. A. Dimmick, J. S. Fleming, and T. C. Charles. 2009. Null mutations in Sinorhizobium meliloti exoS and chvI demonstrate the importance of this two-component regulatory system for symbiosis. Mol. Microbiol. 74:1223-1237. [DOI] [PubMed] [Google Scholar]
- 5.Bell, A., M. Bains, and R. E. Hancock. 1991. Pseudomonas aeruginosa outer membrane protein OprH: expression from the cloned gene and function in EDTA and gentamicin resistance. J. Bacteriol. 173:6657-6664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campbell, G. R., L. A. Sharypova, H. Scheidle, K. M. Jones, K. Niehaus, A. Becker, and G. C. Walker. 2003. Striking complexity of lipopolysaccharide defects in a collection of Sinorhizobium meliloti mutants. J. Bacteriol. 185:3853-3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caro-Hernández, P., L. Fernandez-Lago, M. J. De Miguel, A. I. Martin-Martin, A. Cloeckaert, M. J. Grillo, and N. Vizcaino. 2007. Role of the Omp25/Omp31 family in outer membrane properties and virulence of Brucella ovis. Infect. Immun. 75:4050-4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Charles, T. C., and E. W. Nester. 1993. A chromosomally encoded two-component sensory transduction system is required for virulence of Agrobacterium tumefaciens. J. Bacteriol. 175:6614-6625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng, H. P., and G. C. Walker. 1998. Succinoglycan production by Rhizobium meliloti is regulated through the ExoS-ChvI two-component regulatory system. J. Bacteriol. 180:20-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Castro, C., A. Molinaro, R. Lanzetta, A. Silipo, and M. Parrilli. 2008. Lipopolysaccharide structures from Agrobacterium and Rhizobiaceae species. Carbohydr. Res. 343:1924-1933. [DOI] [PubMed] [Google Scholar]
- 11.de Maagd, R., R. de Rijk, I. Mulders, and B. Lugtenberg. 1989a. Immunological characterization of Rhizobium leguminosarum outer membrane antigens by use of polyclonal and monoclonal antibodies. J. Bacteriol. 171:1136-1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fellay, R., J. Frey, and H. Krisch. 1987. Interposon mutagenesis of soil and water bacteria: a family of DNA fragments designed for in vitro insertional mutagenesis of Gram-negative bacteria. Gene 52:147-154. [DOI] [PubMed] [Google Scholar]
- 13.Ferguson, G. P., R. M. Roop, and G. C. Walker. 2002. Deficiency of a Sinorhizobium meliloti bacA mutant in alfalfa symbiosis correlates with alteration of the cell envelope. J. Bacteriol. 184:5625-5632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gage, D. J. 2002. Analysis of infection thread development using gfp- and DsRed-expressing Sinorhizobium meliloti. J. Bacteriol. 184:7042-7046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gilbert, K. B., E. M. Vanderlinde, and C. K. Yost. 2007. Mutagenesis of the carboxy terminal protease CtpA decreases desiccation tolerance in Rhizobium leguminosarum. FEMS Microbiol. Lett. 272:65-74. [DOI] [PubMed] [Google Scholar]
- 16.Guzmán-Verri, C., L. Manterola, A. Sola-Landa, A. Parra, A. Cloeckaert, J. Garin, J. P. Gorvel, I. Moriyón, E. Moreno, and I. López-Goñi. 2002. The two-component system BvrR/BvrS essential for Brucella abortus virulence regulates the expression of outer membrane proteins with counterparts in members of the Rhizobiaceae. Proc. Natl. Acad. Sci. U. S. A. 99:12375-12380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hancock, R. E. 1984. Alterations in outer membrane permeability. Annu. Rev. Microbiol. 38:237-264. [DOI] [PubMed] [Google Scholar]
- 18.Hoagland, D. R., and D. I. Arnon. 1938. The water-culture method for growing plants without soil. University of California Agricultural Experiment Station, circulars 347-353. University of California, Berkeley, CA.
- 19.Jefferson, R. A., S. M. Burgess, and D. Hirsch. 1986. Beta-glucuronidase from Escherichia coli as a gene-fusion marker. Proc. Natl. Acad. Sci. U. S. A. 83:8447-8451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jia, Y. H., L. P. Li, Q. M. Hou, and S. Q. Pan. 2002. An Agrobacterium gene involved in tumorigenesis encodes an outer membrane protein exposed on the bacterial cell surface. Gene 284:113-124. [DOI] [PubMed] [Google Scholar]
- 21.Kannenberg, E. L., and R. W. Carlson. 2001. Lipid A and O-chain modifications cause Rhizobium lipopolysaccharides to become hydrophobic during bacteroid development. Mol. Microbiol. 39:379-391. [DOI] [PubMed] [Google Scholar]
- 22.Kim, W.-S., J. Sun-Hyung, R.-D. Park, K.-Y. Kim, and H. B. Krishnan. 2005. Sinorhizobium fredii USDA257 releases a 22-kDa outer membrane protein (Omp22) to the extracellular milieu when grown in calcium-limiting conditions. Mol. Plant Microbe Interact. 18:808-818. [DOI] [PubMed] [Google Scholar]
- 23.Ladner, C. L., J. Yang, R. J. Turner, and R. A. Edwards. 2004. Visible fluorescent detection of proteins in polyacrylamide gels without staining. Anal. Biochem. 326:13-20. [DOI] [PubMed] [Google Scholar]
- 24.Larkin, M. A., G. Blackshields, N. P. Brown, R. Chenna, P. A. McGettigan, H. McWilliam, F. Valentin, I. M. Wallace, A. Wilm, R. Lopez, J. D. Thompson, T. J. Gibson, and D. G. Higgins. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947-2948. [DOI] [PubMed] [Google Scholar]
- 25.Lerouge, I., and J. Vanderleyden. 2002. O-antigen structural variation: mechanisms and possible roles in animal/plant-microbe interactions. FEMS Microbiol. Rev. 26:17-47. [DOI] [PubMed] [Google Scholar]
- 26.Li, L., Y. Jia, Q. Hou, T. C. Charles, E. W. Nester, and S. Q. Pan. 2002. A global pH sensor: Agrobacterium sensor protein ChvG regulates acid-inducible genes on its two chromosomes and Ti plasmid. Proc. Natl. Acad. Sci. U. S. A. 99:12369-12374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.López-Expósito, I., F. Minervini, L. Amigo, and I. Recio. 2006. Identification of antibacterial peptides from bovine kappa-casein. J. Food Prot. 69:2992-2997. [DOI] [PubMed] [Google Scholar]
- 28.Manterola, L., C. Guzmán-Verri, E. Chaves-Olarte, E. Barquero-Calvo, M. J. de Miguel, I. Moriyón, M. J. Grilló, I. López-Goñi, and E. Moreno. 2007. BvrR/BvrS-controlled outer membrane proteins Omp3a and Omp3b are not essential for Brucella abortus virulence. Infect. Immun. 75:4867-4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mantis, N. J., and S. C. Winans. 1993. The chromosomal response regulatory gene chvI of Agrobacterium tumefaciens complements an Escherichia coli phoB mutation and is required for virulence. J. Bacteriol. 175:6626-6636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 31.Nicas, T. I., and R. E. Hancock. 1980. Outer membrane protein H1 of Pseudomonas aeruginosa: involvement in adaptive and mutational resistance to ethylenediaminetetraacetate, polymyxin B, and gentamicin. J. Bacteriol. 143:872-878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nikaido, H. 2003. Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 67:593-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nikaido, H., and M. Vaara. 1985. Molecular basis of bacterial outer membrane permeability. Microbiol. Rev. 49:1-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Priefer, U. B. 1989. Genes involved in lipopolysaccharide production and symbiosis are clustered on the chromosome of Rhizobium leguminosarum biovar viciae VF39. J. Bacteriol. 171:6161-6168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Quandt, J., and M. F. Hynes. 1993. Versatile suicide vectors which allow direct selection for gene replacement in Gram-negative bacteria. Gene 127:15-21. [DOI] [PubMed] [Google Scholar]
- 36.Reeve, W. G., R. P. Tiwari, P. S. Worsley, M. J. Dilworth, A. R. Glenn, and J. G. Howieson. 1999. Constructs for insertional mutagenesis, transcriptional signal localization and gene regulation studies in root nodule and other bacteria. Microbiology 145:1307-1316. [DOI] [PubMed] [Google Scholar]
- 37.Roest, H. P., I. H. Mulders, C. A. Wijffelman, and B. J. Lugtenberg. 1995. Isolation of ropB, a gene encoding a 22-kDa Rhizobium leguminosarum outer membrane protein. Mol. Plant Microbe Interact. 8:576-583. [PubMed] [Google Scholar]
- 38.Roest, H. P., L. Goosenderoo, C. A. Wijffelman, R. A. de Maagd, and B. J. Lugtenberg. 1995. Outer-membrane protein changes during bacteroid development are independent of nitrogen-fixation and differ between indeterminate and determinate nodulating host plants of Rhizobium leguminosarum. Mol. Plant Microbe Interact. 8:14-22. [Google Scholar]
- 39.Ruiz, N., T. Wu, D. Kahne, and T. J. Silhavy. 2006. Probing the barrier function of the outer membrane with chemical conditionality. ACS Chem. Biol. 1:385-395. [DOI] [PubMed] [Google Scholar]
- 40.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 41.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Biotechnology (NY) 1:784-791. [Google Scholar]
- 42.Sola-Landa, A., J. Pizarro-Cerdá, M. J. Grilló, E. Moreno, I. Moriyón, J. M. Blasco, J. P. Gorvel, and I. López-Goñi. 1998. A two-component regulatory system playing a critical role in plant pathogens and endosymbionts is present in Brucella abortus and controls cell invasion and virulence. Mol. Microbiol. 29:125-138. [DOI] [PubMed] [Google Scholar]
- 43.Stoscheck, C. M. 1990. Quantitation of protein. Methods Enzymol. 182:50-68. [DOI] [PubMed] [Google Scholar]
- 44.Vaara, M. 1993. Outer membrane permeability barrier to azithromycin, clarithromycin, and roxithromycin in Gram-negative enteric bacteria. Antimicrob. Agents Chemother. 37:354-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vanderlinde, E. M., A. Muszynski, J. J. Harrison, S. F. Koval, D. L. Foreman, H. Ceri, E. L. Kannenberg, R. W. Carlson, and C. K. Yost. A Rhizobium leguminosarum biovar viciae 3841 mutant, deficient in 27-hydroxyoctacosanoate-modified lipopolysaccharide is impaired in desiccation tolerance, biofilm formation, and motility. Microbiology, in press. [DOI] [PMC free article] [PubMed]
- 46.Vedam, V., E. L. Kannenberg, J. G. Haynes, D. J. Sherrier, A. Datta, and R. W. Carlson. 2003. A Rhizobium leguminosarum AcpXL mutant produces lipopolysaccharide lacking 27-hydroxyoctacosanoic acid. J. Bacteriol. 185:1841-1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vincent, J. M. 1970. A manual for the practical study of root-nodule bacteria (IBP handbook no. 15). Blackwell Scientific Publications, Oxford, United Kingdom.
- 48.Vizcaíno, N., P. Caro-Hernandez, and L. Fernandez-Lago. 2004. DNA polymorphism in the omp25/omp31 family of Brucella spp.: identification of a 1.7-kb inversion in Brucella cetaceae and of a 15.1-kb genomic island, absent from Brucella ovis, related to the synthesis of smooth lipopolysaccharide. Microbes Infect. 6:821-834. [DOI] [PubMed] [Google Scholar]
- 49.Wang, Y. P., K. Birkenhead, B. Boesten, S. Manian, and F. O'Gara. 1989. Genetic analysis and regulation of the Rhizobium meliloti genes controlling C4-dicarboxylic acid transport. Gene 85:135-144. [DOI] [PubMed] [Google Scholar]
- 50.Yang, Z. N., T. Suomalainen, A. MayraMakinen, and E. Huttunen. 1997. Antimicrobial activity of 2-pyrrolidone-5-carboxylic acid produced by lactic acid bacteria. J. Food Prot. 60:786-790. [DOI] [PubMed] [Google Scholar]
- 51.Yost, C. K., A. M. Rath, T. C. Noel, and M. F. Hynes. 2006. Characterization of genes involved in erythritol catabolism in Rhizobium leguminosarum bv. viciae. Microbiology 152:2061-2074. [DOI] [PubMed] [Google Scholar]
- 52.Young, J. P., L. C. Crossman, A. W. Johnston, N. R. Thomson, Z. F. Ghazoui, K. H. Hull, M. Wexler, A. R. Curson, J. D. Todd, et al. 2006. The genome of Rhizobium leguminosarum has recognizable core and accessory components. Genome Biol. 7:R34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Young, M. L., M. Bains, A. Bell, and R. E. Hancock. 1992. Role of Pseudomonas aeruginosa outer membrane protein OprH in polymyxin and gentamicin resistance: isolation of an OprH-deficient mutant by gene replacement techniques. Antimicrob. Agents Chemother. 36:2566-2568. [DOI] [PMC free article] [PubMed] [Google Scholar]