Abstract
CotE is a morphogenic protein that controls the assembly of the coat, the proteinaceous structure that surrounds and protects the spore of Bacillus subtilis. CotE has long been thought to interact with several outer coat components, but such interactions were hypothesized from genetic experiment results and have never been directly demonstrated. To study the interaction of CotE with other coat components, we focused our attention on CotC and CotU, two outer coat proteins known to be under CotE control and to form a heterodimer. We report here the results of pull-down experiments that provide the first direct evidence that CotE contacts other coat components. In addition, coexpression experiments demonstrate that CotE is needed and sufficient to allow formation of the CotC-CotU heterodimer in a heterologous host.
The spore of Bacillus subtilis is a dormant cell, resistant to harsh conditions and able to survive extreme environmental conditions (25). Spores are produced in a sporangium that consists of an inner cell, the forespore, that will become the mature spore and an outer cell, the mother cell, that will lyse, liberating the mature spore (18, 26). Resistance of the spore to noxious chemicals, lytic enzymes, and predation by soil protozoans is in part due to the coat, a complex, multilayered structure of more than 50 proteins that encases the spore (5, 8, 13). Proteins that constitute the coat are produced in the mother cell and deposited around the outer membrane surface of the forespore in an ordered manner (8).
A small subset of coat proteins have a regulatory role on the formation of the coat. Those proteins, referred to as morphogenic factors, do not affect the synthesis of the coat components but drive their correct assembly outside of the outer forespore membrane (8). Within this subset of regulatory coat proteins, SpoIVA and CotE play a crucial role. SpoIVA (6, 20, 23) is assembled into the basement layer of the coat and is anchored to the outer membrane of the forespore through its C terminus that contacts SpoVM, a small, amphipathic peptide embedded in the forespore membrane (16, 21, 22). A spoIVA-null mutation impairs the assembly of the coat around the forming spore, and as a consequence, coat material accumulates in the mother cell cytoplasm (23).
CotE (28) assembles into a ring and surrounds the SpoIVA basement structure. The inner layer of the coat is then formed between the SpoIVA basement layer and the CotE ring by coat components produced in the mother cell that infiltrate through the CotE ring, while the outer layer of the coat is formed outside of CotE (6). However, not all CotE molecules are assembled into the ring-like structure, and CotE molecules are also found in the mother cell cytoplasm, at least up to 8 h after the start of sporulation (3). CotE was first identified as a morphogenic factor in a seminal study in which an ultrastructural analysis indicated that a cotE-null mutation prevented formation of the electron-dense outer layer of the coat while it did not affect inner coat formation (28). A subsequent mutagenesis study has revealed that CotE has a modular structure with a C-terminal domain involved in directing the assembly of various coat proteins, an internal domain involved in the targeting of CotE to the forespore, and a N-terminal domain that, together with the internal domain, directs the formation of CotE multimers (17). More recently, formation of CotE multimers has been also confirmed by a yeast two-hybrid approach (14). In a global study of protein interactions in the B. subtilis coat, performed by a fluorescence microscopy analysis of a collection of strains carrying cot-gfp fusions, CotE has been proposed to interact with most outer coat components (12).
From those and other studies, the interactions of CotE with coat structural components have been exclusively inferred on the basis of genetic experiment results, i.e., cotE mutants that failed to assemble one or more coat components. Evidence of a direct interaction between CotE and another coat component has never been provided. We addressed this issue by using as a model two coat components, CotC and CotU, known to be controlled by CotE and to form a heterodimer (10, 28).
CotC is an abundant, 66-amino-acid protein known to assemble in the outer coat in various forms: a monomer of 12 kDa, a homodimer of 21 kDa, and two less abundant forms of 12.5 and 30 kDa, probably due to posttranslational modifications of CotC (9). CotU is a structural homolog of CotC of 86 amino acids. The two proteins, which share an almost identical N terminus and a less conserved C terminus, interact, originating the formation of a heterodimer of 23 kDa (10). Heterodimer formation most likely requires a B. subtilis-specific factor since it does not occur in Escherichia coli or Saccharomyces cerevisiae (10). CotC and CotU are synthesized in the mother cell compartment of the sporulating cell but do not accumulate there since they are immediately assembled around the forming spore (10). In a strain carrying a cotE-null mutation, CotC and CotU, together with all other outer coat components, do not assemble around the forming spore (10). CotC and CotU are also dependent on CotH, an additional morphogenic factor involved in coat formation (9). A cotH-null mutation prevents CotC and CotU assembly in the coat as well as their accumulation in the mother cell cytoplasm (10). Since a mutation causing cotH overexpression allows CotC and CotU accumulation in the mother cell cytoplasm (1), it has been proposed that CotH acts by stabilizing CotC and CotU in the mother cell cytoplasm (1, 10).
Here we provide the first direct evidence that CotE interacts with two other coat components, CotC and CotU, and show that CotE is essential and sufficient to mediate CotC-CotU interaction to form a heterodimer.
MATERIALS AND METHODS
Bacterial strains and transformation.
B. subtilis strains used in this study are listed in Table 1. Plasmid amplification for subcloning experiments, nucleotide sequence analysis, and transformation of Escherichia coli competent cells were performed with E. coli strain DH5α (24). E. coli strain BL21(DE3) (Novagene) was used for protein overexpression. Bacterial strains were transformed by previously described procedures: CaCl2-mediated transformation of E. coli competent cells (24) and two-step transformation of B. subtilis (4).
TABLE 1.
Bacillus subtilis and Escherichia coli strains used
| Strain name | Relevant genotype | Reference or source |
|---|---|---|
| B. subtilis | ||
| PY79 | Prototrophic | 27 |
| BZ213 | cotE::cat | 28 |
| 67 | spoIVA | 7 |
| RH263 | spoIVA cotE::cat | This work |
| E. colia | ||
| RH52 | cotC::his | 9 |
| RH59 | cotU::his | 10 |
| RH62 | cotC::his | 10 |
| RH63 | cotC::his cotU::his | 10 |
| RH125 | cotC::his cotU::his cotE | This work |
| RH136 | cotE | This work |
| AP19 | cotE::his | This work |
All E. coli strains are derivatives of strain BL21(D3) transformed with various plasmids. The relevant genotypes shown for E. coli strains are those of the contained plasmid.
Genetic and molecular procedures.
Isolation of plasmids, restriction digestion, and ligation of DNA were carried out by standard methods (24). Chromosomal DNA from B. subtilis was isolated as described elsewhere (4). A spoIVA cotE double mutant was obtained by transforming a competent cell of strain 67 (spoIVA) (Table 1) with chromosomal DNA extracted from strain BZ213 (cotE::cat), generating strain RH263.
Sporulating cells lysates and immunoblot analysis.
Sporulation of all B. subtilis strains was induced by the exhaustion method (4). Sporulating cells were harvested after 8 and 10 h from the onset of sporulation, and mother cells and forespore fractions were isolated as described before (10). Whole-cell lysates of sporulating cells were prepared by sonication (10) followed by detergent treatment (62.5 mM Tris-HCl [pH 6.8], 4% SDS, 5% glycerol, 2% β-mercaptoethanol, 0.003% bromophenol blue) at 100°C for 7 min. Fifty micrograms (mother cell extract or whole-cell lysates) or 15 μg (forespore extract) of total proteins was subjected to immunoblot analysis with the anti-CotC or anti-CotU antibodies as described previously (10), except that polyvinylidene difluoride membranes were used instead of nitrocellulose.
Overproduction of six-His-tagged and untagged CotE.
To overexpress CotE in E. coli, the coding region of the cotE gene was PCR amplified using the B. subtilis chromosomal DNA as a template and oligonucleotides E-rbs-PstI-F (CTGCAGTTTAAGAAGGAGATATACATATGTCTGAATACAGGGAAT [PstI and NdeI restriction sites are underlined, and the ribosome binding site is italicized]) and E-STOP (CCAAGCTTATTCTTCAGGATCTCCCAC [HindIII restriction site is underlined]) as primers. The amplification product of 582 bp was digested with NdeI and HindIII and ligated to the same sites of the expression vector pRSETB (Invitrogen), to obtain pRH134. By digesting pRSETB with NdeI the His tag present on the vector was removed.
To overexpress a six-His-tagged copy of CotE in E. coli, the coding region of the cotE gene was amplified by PCR using B. subtilis chromosomal DNA as a template and oligonucleotides E-NdeI-F (TAGGAATTCCATATGTCTGAATACAGGGAAT [underlined is the EcoRI restriction site]) and E-STOP (CCAAGCTTATTCTTCAGGATCTCCCAC [underlined is the HindIII restriction site]) as primers. The amplified fragment of 564 bp was digested with EcoRI and HindIII and ligated to plasmid pRSETB (Invitrogen), previously digested with the same enzymes. The recombinant plasmid, pAP18, carried the cotE coding region fused to a six-His tag under the transcriptional control of a T7 inducible promoter.
Plasmids pRH134 and pAP18 were checked by nucleotide sequence analysis and used to transform competent cells of E. coli BL21(DE3) (Novagen) to create RH136 and AP19, respectively (Table 1). CotE and CotE-His were produced by autoinduction by growing cells at 37°C for 18 h with orbital shaking (150 rpm) and by using the Overnight Express autoinduction system 1 according to the manufacturer's instructions (Novagen). CotE-His protein was purified under denaturing conditions via Ni-nitrilotriacetic acid (NTA) affinity chromatography as recommended by the manufacturer (Qiagen, Inc.) and used to raise specific antibodies in mice by PriMM Srl (Italy).
Coexpression of cotC, cotU, and cotE in E. coli.
The coding part of cotE, amplified by PCR using oligonucleotides E-rbs-PstI-F and E-STOP as described above, was digested with PstI and HindIII and inserted in the same sites of pRH62 (10), immediately downstream of cotC::his, to create pRH122. In this plasmid, both genes were under the control of the same T7 promoter and formed a single transcriptional unit. The new recombinant plasmid was used to transform the competent cells of E. coli strain RH59 (10), harboring cotU::his under the control of T7 promoter, to create strain RH125. Proteins were produced from these E. coli strains by the autoinduction procedure, as described above.
Pull-down experiments.
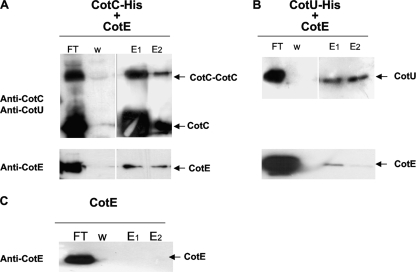
Strains RH52, RH59, and RH136 (Table 1) were grown for 18 h at 37°C in autoinduction medium (see above). Samples (14 ml) were collected by centrifugation and resuspended in 1.5 ml of lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole, 2 mg/ml lysozyme, and 0.01 mg/ml RNase). After 30 min at 4°C, the lysates were sonicated (20-min pulses at 20 Hz with a Sonicator Ultrasonic liquid processor; Heat System Ultrasonic Inc., NY). The suspension was clarified by centrifugation at 13,000 × g at 4°C for 20 min, and protein concentration was determined by a Bio-Rad assay. Three hundred micrograms of extract from strain RH52 or RH59 was applied to Ni-NTA magnetic agarose beads (Qiagen), separately. After 1 h of incubation at room temperature with shaking, the beads were washed with 2.5 ml of wash buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole), and 300 μg of extract from strain RH136 was added to both samples and incubated for 1 h at room temperature with shaking. Unbound proteins were removed by washing with wash buffer at three different concentrations of imidazole (40 mM, 100 mM, and 250 mM [indicated as W in Fig. 2]). Bound proteins were eluted using the wash buffer at increasing concentrations of imidazole (500 mM and 1 M [indicated as E1 and E2 in Fig. 2, respectively]). Eluted proteins were resolved on SDS-12.5% PAGE gels and subjected to immunoblot analysis.
FIG. 2.
Results of a pull-down experiment performed by binding CotC-His (A) or CotU-His (B) to Ni-NTA magnetic beads. Untagged CotE was then added, and flowthrough (FT), washed (W), and eluted (E1 and E2) proteins collected as described in Materials and Methods. Proteins were fractionated on 12.5% polyacrylamide gels, electrotransferred to membranes, and reacted with anti-CotC, anti-CotU, and anti-CotE antibodies. The same experiment was also performed without CotC-His or CotU-His (C).
Preparation of samples for matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) analysis.
Strain RH125 was grown for 18 h at 37°C in autoinduction medium (see above). Cells (100 ml) were washed with 10 ml of 1× phosphate-buffered saline (PBS), suspended with 5 ml of 1× PBS and 5 ml of 2× cracking buffer (120 mM Tris-HCl [pH 6.8], 2% SDS, 20% glycerol, 2% β-mercaptoethanol), and heated at 100°C for 10 min. The suspension was clarified by centrifugation at 13,000 × g for 20 min, and the supernatant was diluted 1:10 by using binding buffer (20 mM sodium phosphate, 500 mM NaCl, 10 mM β-mercaptoethanol, 10% glycerol, 25 mM imidazole [pH 7.4]). The diluted sample was applied to a HisTrap HP column (GE Healthcare Europe GmbH, Milan, Italy) equilibrated with 10 ml of binding buffer. The column was washed with 10 ml of binding buffer, and proteins were eluted with the same buffer, supplemented with increasing concentrations of imidazole (50 mM, 100 mM, 250 mM, and 500 mM). Purified proteins were resolved on SDS-15% PAGE gels, and bands were sent for MALDI-mass spectrometry analysis.
RESULTS
cotE expression is required to allow the interaction between CotC and CotU.
It has previously been reported that in a strain carrying a cotE-null mutation, CotC and CotU are not assembled around the forming spore and they accumulate in the mother cell compartment but fail to interact and do not form the CotC-CotU heterodimer (10). Possible explanations for the lack of interaction in the cotE mutant are that the interaction either is CotE dependent or can occur only when CotC and CotU are already assembled on the forming coat.
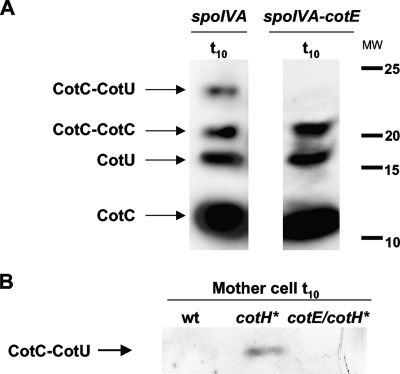
To discriminate between these two possibilities, we decided to verify whether the heterodimer could form in the mother cell cytoplasm. Since CotC and CotU do not accumulate in the mother cell of a wild-type strain but immediately assemble around the spore (9, 10), we used strains either carrying a null mutation in the spoIVA (23) or overexpressing the cotH gene (1). In a spoIVA mutant, the coat is known to be dispersed in the mother cell cytoplasm and not assembled around the spore (23), while overexpression of cotH is known to allow accumulation of CotC and CotU in the mother cell cytoplasm (1). As shown in Fig. 1A, in a spoIVA-null mutant, the heterodimer was formed but only in the presence of a wild-type allele of cotE. An identical result was obtained with the strain overexpressing cotH (Fig. 1B). Taken together, these data support the hypothesis that the CotC-CotU interaction is CotE dependent and that it can occur independently from the assembly of the two proteins on the spore coat.
FIG. 1.
Western blots of proteins extracted 10 h after the onset of sporulation (t10) from sporulating cells of a spoIVA mutant or a spoIVA cotE double mutant (A) and from the mother cell fraction of a strain overexpressing cotH (indicated as cotH*) (B). Proteins were fractionated on 15% polyacrylamide gels, electrotransferred to membranes, and reacted with anti-CotU antibody. Identical results were obtained with anti-CotC antibody. MW, molecular weights in thousands; wt, wild type.
Formation of CotC homodimers and CotC-CotU heterodimers does not involve cysteine residues.
The CotC-CotU heterodimer as well as the CotC-CotC homodimer were observed on a Western blot under denaturing conditions (Fig. 1A) after extraction from sporulating cells by treatment with 0.1 N NaOH at 4°C (19), indicating that both dimers are resistant to reducing and denaturing conditions.
Since both CotC and CotU have a cysteine residue at position 32 (10), we decided to verify whether those cysteines were involved in the formation of the homo- and/or heterodimer. To this aim, we exposed proteins extracted from a wild-type strain to extreme reducing conditions (10 mM or 80 mM dithiothreitol or 1% β-mercaptoethanol, for either 60 min at 80°C or overnight at 37°C) and to subsequent alkylation with iodoacetic acid (IAA) (15). IAA binds to reduced cysteines and prevents them from reforming sulfur bridges. None of the tested conditions impaired formation of the dimers after either 1 h of incubation at 60°C or an overnight incubation at 37°C (not shown), thus excluding the possibility that they were dependent on cysteine-mediated sulfur bridges.
CotE binds to CotC and CotU.
To verify whether CotE directly interacts with CotC and/or CotU, we overexpressed in E. coli a His-tagged version of cotC (CotC-His), a His-tagged version of cotU (CotU-His), or an untagged version of cotE and performed an in vitro His tag pull-down assay. After autoinduction, E. coli cells were lysed by sonication, and Ni-NTA magnetic beads were incubated with extracts of cells expressing CotC-His or CotU-His. Beads were then washed and incubated with the extract of cells expressing untagged CotE. After additional washes, proteins were eluted and used for Western blot experiments with anti-CotC, anti-CotU, anti-His, or anti-CotE antibodies. As shown in Fig. 2, untagged CotE bound Ni-NTA beads when CotC-His (panel A) or CotU-His (panel B) was present. In the absence of CotC-His or CotU-His, untagged CotE was not able to bind to the Ni-NTA beads (Fig. 2C). Similarly, purified bovine serum albumin (BSA; 0.1 mg; New England Biolabs) was also not able to bind Ni-NTA beads in the absence of CotC-His or CotU-His (data not shown).
Although only a small fraction of CotE was able to bind the beads carrying either CotC-His (Fig. 2A) or CotU-His (Fig. 2B) and the majority of the protein remained in the flowthrough, these in vitro results demonstrated that CotE directly interacts with CotC and CotU and are the first proof of a direct interaction between CotE and other components of the B. subtilis spore coat.
CotE mediates the interaction of CotC and CotU.
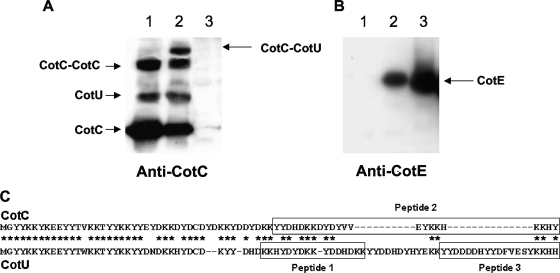
To investigate whether the interaction with CotE was sufficient to induce the formation of the CotC-CotU heterodimer, we overexpressed all three proteins together in the same E. coli strain and used the extracted proteins for Western blot analysis with anti-CotC, anti-CotU, anti-His, and anti-CotE antibodies. As previously reported (10), when only cotC and cotU were overexpressed together in E. coli, three proteins were recognized by anti-CotC and anti-CotU antibodies (Fig. 3A, lane 1). The three bands corresponded in size to CotC, CotU, and CotC homodimers (10). When cotE was also expressed in the same cells, a new protein complex was formed (Fig. 3A, lane 2). The additional protein did not contain CotE, since when the same gel was reacted against anti-CotE antibody, a CotE-specific signal was observed at a different position on the gel (Fig. 3B). It is unlikely that the additional protein is an unspecific signal, since it was not present in lane 1 (CotC-CotU) or lane 3 (CotE) of Fig. 3A and identical results were obtained with anti-CotU and anti-His antibodies (not shown).
FIG. 3.
Coexpression experiment results. Extracts from E. coli cells overproducing CotC and CotU (lane 1), CotC, CotU, and CotE (lane 2) and CotE (lane 3) were fractionated on 15% polyacrylamide gels, electrotransferred to membranes, and reacted with anti-CotC (A) or anti-CotE (B) antibody. Results identical to those of panel A were obtained with anti-CotU antibody (not shown). Panel C shows the position of the three peptides identified as diagnostic of CotC or CotU by the MALDI-TOF experiment results shown in Table 2.
To determine the nature of the proteins formed when CotC, CotU, and CotE were overexpressed together in E. coli, we performed a MALDI-TOF mass spectrometry analysis. The four proteins shown in Fig. 3A (lane 2) were excised from the SDS-polyacrylamide gel and subjected to MALDI-TOF by using a MALDI-TOF micro MX (Waters Co., Manchester, United Kingdom), as described previously (2). Due to the high percentage of identity between CotC and CotU amino acid sequences (10), it was necessary to identify diagnostic peptides to discriminate between the two coat proteins. In particular, three peptides at the C-terminal end of the two proteins were found to be diagnostic: peptides 1 and 3 of CotU and peptide 2 of CotC (Fig. 3C). As reported in Table 2, peptides 1 and 3 (diagnostic of CotU) were found in the protein tentatively assigned as CotU, while only peptide 2 (diagnostic of CotC) was found in proteins indicated as CotC monomers and CotC-CotC homodimers. All three peptides were found in the additional protein of Fig. 3A (lane2), confirming that it was the CotC-CotU heterodimer (Table 2).
TABLE 2.
Molecular masses obtained by MALDI-TOF mass spectrometry of tryptic digests of bands CotC, CotU, and CotC-CotUb
| Peptide | Sequence positionsa | Theoretical Mr (M+H)+ | Experimental Mr (M+H)+ of band: |
|||
|---|---|---|---|---|---|---|
| CotC | CotU | CotC-CotC | CotC-CotU | |||
| 1 | 40-54 | 1,869.83 | 1,869.78 | 1,869.78 | ||
| 2 | 45-66 | 2,964.42 | 2,964.43 | 2,964.46 | 2,964.46 | |
| 3 | 68-86 | 2,539.04 | 2,539.01 | 2,538.91 | ||
Refers to the amino acid sequence of CotC (peptide 2) and CotU (peptides 1 and 3) as reported in Fig. 3C.
Theoretical and experimental masses together with peptide sequence positions are reported (Da).
These results confirm the nature of the proteins expressed in E. coli and indicate that no additional factors other than CotE are needed to mediate CotC-CotU heterodimer formation.
DISCUSSION
The main outcome of this work is that it provides the first direct evidence that CotE interacts with a coat component other than CotE itself. CotE of B. subtilis is a morphogenic factor required for outer coat formation (28), known to form multimers organized in a ring-like structure assembled around the forming spore (6, 14, 17). CotE has been proposed as a major regulatory factor of outer coat assembly, and its interaction with several components of the coat has been inferred on the basis of genetic experiment results (12). We provide here direct evidence that CotE interacts with CotC and with CotU, two outer coat components whose assembly around the spore has previously been indicated as CotE dependent (10, 28). Assembly of both CotC and CotU is also dependent on CotH and CotG (9, 10, 12). However, while the effect of CotG on CotC/CotU has not been studied in detail, their dependency on the expression of cotH is likely to reflect a stability problem. It has previously been proposed that CotH either protects CotC and CotU from a proteolytic cleavage or acts on them in a chaperone-like way (1, 9). Therefore, CotH probably controls the assembly of CotC and CotU only indirectly, by regulating their presence in the mother cell cytoplasm. The pull-down experiment results shown in Fig. 2 demonstrate that CotE is retained on Ni-NTA magnetic beads only when CotC-His or CotU-His has previously been bound to the beads, indicating that CotE directly binds CotC and CotU. A future challenge will be to define these interactions by identifying the amino acid residues involved in CotE-CotC and CotE-CotU contacts.
A second important result of this work is that CotE is sufficient to mediate CotC-CotU interaction in E. coli cells. The coexpression experiment results shown in Fig. 3 and the MALDI-TOF analysis of the proteins produced in E. coli clearly indicate that only in the presence of CotE do the two coat components interact, forming the heterodimer previously observed in the coat protein fraction of B. subtilis spores (10). However, at this stage, we cannot explain how CotE mediates CotC-CotU interaction and can hypothesize only that CotE either recognizes CotC and CotU as substrates and catalyzes their interaction or acts as a platform to which CotC and CotU bind. In the latter case, the CotE role would be to hold the two coat components close to each other, allowing their spontaneous or self-catalyzed interaction. Although an enzymatic activity associated with CotE has never been demonstrated, we observed that CotE contains a region of homology with the consensus of the Walker A domain of ATPases (consensus, A/GXXXXGKT [11]; CotE, A47 - - - G51K52T53). However, so far we have been unable to detect any ATPase activity associated with E. coli-purified CotE (data not shown). Additional experiments will be needed to clarify the CotE role in CotC-CotU interaction and to discriminate between the two hypotheses.
An additional result of this work is that CotC-CotC homodimers and CotC-CotU heterodimers are resistant to denaturing and reducing conditions and are not dependent on the single cysteine residue that both proteins have at position 32 (Fig. 3C). While CotC-CotC homodimers form spontaneously in E. coli (9), CotC-CotU heterodimers require CotE to form. Preliminary results indicate that amino acids at the C terminus of CotC are involved in the formation of both the homo- and the heterodimers (R. Isticato and E. Ricca, unpublished results), but in this case also, additional experiments will be needed to clarify the nature of those interactions.
Acknowledgments
We thank A. Parente, Mass Spectrometry and Protein Sequencing Research Group (Department of Life Sciences, Second University of Naples, Italy), for the MALDI-TOF analysis, A. De Maro for helpful discussions, and L. Di Iorio for technical assistance.
This work was supported by a grant (KBBE-2007-207948) from the EU 7th Framework to E.R.
Footnotes
Published ahead of print on 18 December 2009.
REFERENCES
- 1.Baccigalupi, L., G. Castaldo, G. Cangiano, R. Isticato, R. Marasco, M. De Felice, and E. Ricca. 2004. GerE-independent expression of cotH leads to CotC accumulation in the mother cell compartment during Bacillus subtilis sporulation. Microbiology 150:3441-3449. [DOI] [PubMed] [Google Scholar]
- 2.Chambery, A., A. Farina, A. Di Maro, M. Rossi, C. Abbondanza, B. Moncharmont, L. Malorni, G. Cacace, G. Pocsfalvi, A. Malorni, and A. Parente. 2006. Proteomic analysis of MCF-7 cell lines expressing the zinc-finger or the proline-rich domain of retinoblastoma-interacting-zinc-finger protein. J. Proteome Res. 5:1176-1185. [DOI] [PubMed] [Google Scholar]
- 3.Costa, T., M. Serrano, L. Steil, U. Völker, C. P. Moran, Jr., and A. O. Henriques. 2007. The timing of cotE expression affects Bacillus subtilis spore coat morphology but not lysozyme resistance. J. Bacteriol. 189:2401-2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cutting, S., and P. B. Vander Horn. 1990. Genetic analysis, p. 27-74. In C. Harwood and S. Cutting (ed.), Molecular biological methods for Bacillus. John Wiley and Sons, Chichester, United Kingdom.
- 5.Driks, A. 2002. Maximum shields: the assembly and function of the bacterial spore coat. Trends Microbiol. 10:251-254. [DOI] [PubMed] [Google Scholar]
- 6.Driks, A., S. Roels, B. Beall, C. P. Moran, Jr., and R. Losick. 1994. Subcellular localization of proteins involved in the assembly of the spore coat of Bacillus subtilis. Genes Dev. 8:234-244. [DOI] [PubMed] [Google Scholar]
- 7.Errington, J., and J. Mandelstam. 1986. Use of a lacZ gene fusion to determine the dependence pattern and the spore compartment expression of sporulation operon spoVA in spo mutants of Bacillus subtilis. J. Gen. Microbiol. 132:2977-2985. [DOI] [PubMed] [Google Scholar]
- 8.Henriques, A. O., and C. P. Moran, Jr. 2007. Structure, assembly and function of the spore surface layers. Annu. Rev. Microbiol. 61:555-588. [DOI] [PubMed] [Google Scholar]
- 9.Isticato, R., G. Esposito, R. Zilhão, S. Nolasco, G. Cangiano, M. De Felice, A. O. Henriques, and E. Ricca. 2004. Assembly of multiple CotC forms into the Bacillus subtilis spore coat. J. Bacteriol. 186:1129-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Isticato, R., A. Pelosi, R. Zilhão, L. Baccigalupi, A. O. Henriques, M. De Felice, and E. Ricca. 2008. CotC-CotU heterodimerization during assembly of the Bacillus subtilis spore coat. J. Bacteriol. 190:1267-1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jonathan, C., M. Swaffield, and D. Purugganan. 1997. The evolution of the conserved ATPase. History of an ancient protein module. J. Mol. Evol. 45:549-563. [DOI] [PubMed] [Google Scholar]
- 12.Kim, H., M. Hahn, P. Grabowski, D. McPherson, M. M. Otte, R. Wang, C. C. Ferguson, P. Eichenberger, and A. Driks. 2006. The Bacillus subtilis spore coat protein interaction network. Mol. Microbiol. 59:487-502. [DOI] [PubMed] [Google Scholar]
- 13.Klobutcher, L. A., K. Ragkousi, and P. Setlow 2006. The Bacillus subtilis spore coat provides “eat resistance” during phagocytic predation by the protozoan Tetrahymena thermophila. Proc. Natl. Acad. Sci. U. S. A. 103:165-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krajcíková, D., M. Lukácová, D. Müllerová, S. M. Cutting, and I. Barák. 2009. Searching for protein-protein interactions within the Bacillus subtilis spore coat. J. Bacteriol. 191:3212-3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lane, L. C. 1978. A simple method for stabilizing protein-sulfhydryl groups during SDS-gel electrophoresis. Anal. Biochem. 86:655-664. [DOI] [PubMed] [Google Scholar]
- 16.Levin, P. A., N. Fan, E. Ricca, A. Driks, R. Losick, and S. Cutting. 1993. An unusually small gene required for sporulation by Bacillus subtilis. Mol. Microbiol. 9:761-771. [DOI] [PubMed] [Google Scholar]
- 17.Little, S., and A. Driks. 2001. Functional analysis of the Bacillus subtilis morphogenetic spore coat protein CotE. Mol. Microbiol. 42:1107-1120. [DOI] [PubMed] [Google Scholar]
- 18.Losick, R., P. Youngman, and P. J. Piggot. 1986. Genetics of endospore formation in Bacillus subtilis. Annu. Rev. Genet. 20:625-669. [DOI] [PubMed] [Google Scholar]
- 19.Nicholson, W. L., and P. Setlow. 1990. Sporulation, germination and outgrowth, p. 391-450. In C. Harwood and S. Cutting (ed.), Molecular biological methods for Bacillus. John Wiley and Sons, Chichester, United Kingdom.
- 20.Piggot, P. J., and J. G. Coote. 1976. Genetic aspects of bacterial endospore formation. Bacteriol. Rev. 40:908-962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramamurthi, K. S., K. S. Clapham, and R. Losick. 2006. Peptide anchoring spore coat assembly to the outer forespore membrane in Bacillus subtilis. Mol. Microbiol. 62:1547-1557. [DOI] [PubMed] [Google Scholar]
- 22.Ramamurthi, K. S., and R. Losick. 2008. ATP-driven self-assembly of a morphogenetic protein in Bacillus subtilis. Mol. Cell 31:406-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roels, S., A. Driks, and R. Losick. 1992. Characterization of spoIVA, a sporulation gene involved in coat morphogenesis in Bacillus subtilis. J. Bacteriol. 174:575-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 25.Setlow, P. 2006. Spores of Bacillus subtilis: their resistance to and killing by radiation, heat and chemicals. J. Appl. Microbiol. 101:514-525. [DOI] [PubMed] [Google Scholar]
- 26.Stragier, P., and R. Losick 1996. Molecular genetics of sporulation in Bacillus subtilis. Annu. Rev. Genet. 30:297-341. [DOI] [PubMed] [Google Scholar]
- 27.Youngman, P., J. B. Perkins, and R. Losick. 1984. A novel method for the rapid cloning in Escherichia coli of Bacillus subtilis chromosomal DNA adjacent to Tn917 insertion. Mol. Gen. Genet. 195:424-433. [DOI] [PubMed] [Google Scholar]
- 28.Zheng, L., W. P. Donovan, P. C. Fitz-James, and R. Losick. 1988. Gene encoding a morphogenic protein required in the assembly of the outer coat of the Bacillus subtilis endospore. Genes Dev. 2:1047-1054. [DOI] [PubMed] [Google Scholar]