Abstract
The ability of Ralstonia solanacearum to cause disease in plants depends on its type III secretion system (T3SS). The expression of the T3SS and its effector substrates is coordinately controlled by a regulatory cascade, at the bottom of which is HrpB. Transcription of the hrpB gene is activated by a plant-responsive regulator named HrpG, which is a master regulator of a wide array of pathogenicity functions in R. solanacearum. We have identified in the genome of strain GMI1000 a close paralog of hrpG (83% overall similarity at the protein level) that we have named prhG. Despite this high similarity, the expression pattern of prhG is remarkably different from that of hrpG: prhG expression is activated after growth of bacteria in minimal medium but not in the presence of host cells, while hrpG expression is specifically induced in response to plant cell signals. We provide genetic evidence that prhG is a transcriptional regulator that, like hrpG, controls the expression of hrpB and the hrpB-regulated genes under minimal medium conditions. However, the regulatory functions of prhG and hrpG are distinct: prhG has no influence on hrpB expression when the bacteria are in the presence of plant cells, and transcriptomic profiling analysis of a prhG mutant revealed that the PrhG and HrpG regulons have only one pathogenicity target in common, hrpB. Functional complementation experiments indicated that PrhG and HrpG are individually sufficient to activate hrpB expression in minimal medium. Rather surprisingly, a prhG disruption mutant had little impact on pathogenicity, which may indicate that prhG has a minor role in the activation of T3SS genes when R. solanacearum grows parasitically inside the plant. The cross talk between pathogenicity regulatory proteins and environmental signals described here denotes that an intricate network is at the basis of the bacterial disease program.
In order to successfully colonize plants, bacterial pathogens must deploy a genetic program to express virulence genes, which enable adaptation to living conditions inside the host (25). A set of genes that is highly expressed upon contact with the eukaryotic host is the type III secretion system (T3SS) present in many plant and animal bacterial pathogens. This system is a major genetic determinant of disease development, as it translocates bacterial proteins—called effectors—into host cells to interfere with cellular defense responses and facilitate bacterial colonization (reviewed in references 12, 20, and 39). In bacterial plant pathogens, the T3SS is encoded by a cluster of some 20 hrp genes named after the inability of mutants deficient in them to cause a hypersensitive response in resistant plants or pathogenicity in susceptible hosts (21). In all of the species studied, the transcription of the T3SS and most of its effector substrates is tightly controlled by a succession of consecutively activated regulators.
Ralstonia solanacearum is the agent that causes bacterial wilt in more than 200 plant hosts, including important crops such as potato and tomato, and has long been studied as a model plant vascular pathogen (10, 14). In R. solanacearum GMI1000, the regulatory cascade driving T3SS gene expression has been very well characterized (see references 6, 32, and 38 for a review). In this bacterium, the transcription of type III secretion genes is regulated by the HrpB regulator, which belongs to the AraC family (16). HrpB may directly bind to a specific hrpII box found in the promoters of most of its target genes (8). The transcription of the hrpB gene is activated by the HrpG transcriptional activator, which belongs to the OmpR/PhoB subfamily of two-component response regulators (7, 13). The key role of the HrpB and HrpG regulators is illustrated by the fact that mutant strains deficient in any of them are nonpathogenic (7, 16, 35). Activation of the T3SS regulatory cascade in R. solanacearum is controlled by the outer membrane receptor protein PrhA. It has been shown that PrhA senses an unidentified, nondiffusible, plant-specific signal and transfers the activating signal through PrhR, PrhI, and PrhJ to drive HrpG transcription (1, 6).
Transcription of hrp genes is much higher when bacteria are cultured in defined minimal media rather than complete media (3, 30). This induction pattern seems to be general in all of the plant pathogens studied, and it has been attributed to the fact that minimal media somehow mimic the conditions encountered in the apoplastic environment (30, 32). In R. solanacearum, this metabolic regulation is superimposed on the induction caused by plant cell contact and seems to be integrated into the cascade by HrpG, HrpB, or both (7, 15, 16). This conclusion is based on the observation that only hrpB and hrpG, and not the upstream components of the hrp regulatory cascade, are required for hrp gene induction when the bacteria are grown in minimal medium.
Bacterial plant pathogens of the genus Xanthomonas share part of the hrp regulatory cascade of R. solanacearum, in contrast to other bacterial plant pathogens, where T3SS transcriptional control involves σ54-dependent enhancer-binding regulators (32). Indeed, the HrpG and HrpB (named HrpX) regulators and their functions have been found to be conserved in Xanthomonas campestris pv. vesicatoria, although the upper components of the cascade have not yet been identified (36, 37). More generally, HrpX and HrpG orthologs have been found in all of the genome sequences of Xanthomonas T3SS-harboring species (29). The two different regulatory systems clearly coincide with the two subgroups into which the hrp gene clusters have been classified on the basis of gene conservation (2).
Recent transcriptomic profiling analyses have determined the HrpB and HrpG regulons and provided evidence that, besides activating HrpB transcription (and thus, T3SS expression), HrpG also controls the expression of other virulence determinants (33). From these studies, control of T3SS gene expression is now viewed as a regulatory network connected with other virulence determinants, rather than a linear cascade. HrpG is located at a central node in this network, integrating diverse environmental signals and controlling most of the bacterial functions that promote disease (33). In this work, we have identified in the genome sequence of R. solanacearum GMI1000 a new regulatory gene encoding a protein highly similar to HrpG. We have characterized this novel regulatory gene, named prhG, and we provide evidence that it is a new player in the complex network controlling T3SS gene expression. PrhG appears to be specifically involved in the control of the hrpB gene in response to environmental signals encountered by the bacteria when they are grown under minimal medium conditions.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The relevant characteristics of the plasmids and bacterial strains used in this work are listed in Table 1. Escherichia coli strains were grown at 37°C in Luria-Bertani medium (5). R. solanacearum strains were grown in complete BG medium or in MP minimal medium supplemented with glutamate at a 20 mM final concentration. The composition of BG medium is as follows (g liter−1): Bacto peptone, 10; Casamino Acids, 1; yeast extract, 1. For agar plates, BG medium was supplemented with glucose (5 g liter−1) and triphenyltetrazolium chloride (0.05 g liter−1). The composition of MP medium is as follows (g liter−1): FeSO4 · 7H2O, 1.25 × 10−4; (NH4)2SO4, 0.5; MgSO4 · 7H2O, 0.05; KH2PO4, 3.4. The pH was adjusted to 7 with KOH. When needed, antibiotics were added to the media at the following final concentrations (mg liter−1): kanamycin, 50; spectinomycin, 40 for R. solanacearum; gentamicin, 10; tetracycline, 10; ampicillin, 100 for E. coli.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Reference or source |
|---|---|---|
| E. coli DH5α | F−recA lacZΔM15 | Bethesda Research Laboratories |
| R. solanacearum | ||
| GMI1000 | Wild-type strain | 31 |
| GMI1425 | hrpG::Tn5-B20 mutant | 7 |
| GMI1421 | hrpY::Tn5-B20 mutant | 33 |
| GMI1475 | hrpB::Tn5-B20 mutant | 16 |
| GMI1525 | hrpB::Ω mutant | 16 |
| GMI1613 | popA::Tn5-B20 mutant | 4 |
| GMI1755 | GMI1000 ΔhrpG mutant | 34 |
| GRS404 | prhG::lacZ mutant | This work |
| GRS445 | GMI1000 ΔprhG mutant | This work |
| GRS491 | GMI1000 ΔhrpG ΔprhG mutant | This work |
| GRS497 | RSp0201::lacZ mutant | This work |
| Plasmids | ||
| pBluescript KS(+) | Cloning vector, Ampr | Stratagene |
| pLAFR6 | pLAFR1 with trp terminators, Tcr | 18 |
| pBBL12 | pLAFR6 with a 1.5-kb PstI fragment carrying hrpG | 7 |
| pCZ367 | pUC18-derived vector used for insertional mutagenesis, Apr Gmr | 9 |
| pCM184 | Allelic exchange vector, Kmr | 23 |
| pGG15 | pLAFR6 carrying the cre recombinase gene | This work |
| pSG371 | pCZ367 carrying a 0.5-kb HindIII-XbaI prhG fragment | This work |
| pLP2 | PCR fragment containing the prhG gene and 500 bp upstream the predicted coding sequence cloned in pLAFR6, Tcr | This work |
| pLP5 | PCR fragment containing the prhG gene and 500 bp upstream the predicted coding sequence cloned in pBBL12, Tcr | This work |
DNA manipulations and genetic constructs.
Standard recombinant DNA techniques were performed as described previously (5). Restriction enzymes, DNA ligase, and other DNA enzymes were used according to the manufacturers' recommendations. PCR amplification of prhG with 500 bp of its promoter region was performed with Pfx polymerase (Invitrogen) using genomic DNA of R. solanacearum from strain GMI1000 and primers Amo2360 (5′-TCCCACATCGTGTCGACTT-3′) and Aval2360H3 (5′-AAGCTTTGCCTGCGCCGTCCCG-3′).
An RSp0201 disruption mutant was created as previously described (9), using integrative plasmid pCZ367, into which an internal fragment of the gene was cloned after PCR amplification using primers 0201H3 (5′-GAAGCTTCGTTATCGAGCAAC-3′) and 0201Xba (5′-ATCTAGATCCGCCGGCTGAAG-3′). Plasmids (Table 1) were introduced into R. solanacearum strains by electroporation (2.5 kV, 200 W, 25 mF, 0.2-cm cuvette gap).
Creation of an unmarked ΔprhG mutant strain.
A prhG deletion mutant was generated by using allelic exchange vector pCM184 for antibiotic marker recycling (23). Two DNA fragments flanking the prhG sequence were amplified by using primers 2360R1 (5′-GAATTCTCTCGCCCGGTCGGC-3′) and 2360NotI (5′-GCGGCCGCAAAGCTGGAGGGAGAGG-3′) for the left flanking fragment and 2360Sc2 (5′-CCGCGGCGCATGTCGAATGGATGAA-3′) and 2360Sc1 (5′-GAGCTCTTACCCGCGTTGCAACG-3′) for the right flanking fragment.
Both fragments were cloned into pCM184, and the resulting construct was used to transform GMI1000 to generate a ΔprhG::nptII mutant. The kanamycin resistance cassette was then excised in vivo using plasmid pGG15, which carries a copy of the cre recombinase in a pLAFR6 backbone vector. The deletion of prhG in resulting strain GRS445 was then checked by PCR.
Transcriptome analysis.
RNA extraction and probe labeling were performed as already described (26). The R. solanacearum whole-genome DNA microarray with 65/70-mer oligonucleotides specific to the annotated open reading frames (ORFs) of GMI1000 was used for all of the transcriptomic profiling experiments. Hybridizations were conducted manually under the following conditions. Microarray slides were prehybridized for 1 h at 42°C in DIG Easy buffer (Roche) containing 10 mM salmon sperm DNA. Hybridizations were carried out for 13 h under the same conditions, except that the heat-denatured and labeled probes were added to the reaction mixture. Following hybridization, slides were washed once in 1× SSC (0.15 mM NaCl plus 0.015 M sodium citrate)-0.1% sodium dodecyl sulfate at 60°C for 10 min and once in 0.1× SSC for 1 min at room temperature. Slides were finally rinsed in isopropanol at 37°C and dried by 3 min of centrifugation at 1,000 × g.
Hybridized slides were scanned using a GenePix 4000B dual-channel confocal laser scanner (Axon Instruments, Union City, CA).
Signal quantification and data analysis were achieved using GenePix Pro (version 3.0; Molecular Devices, Sunnyvale, CA) and GeneSight software (version 4.1.6; Biodiscovery Inc., El Segundo, CA). For each array, following local background subtraction, the signal for each spot was normalized on the basis of the mean value of the intensity of all of the spots corresponding to R. solanacearum ORFs. The ratio of expression of the wild-type strain to the prhG deletion mutant or of the wild-type strain to the prhG-overexpressing mutant was calculated. The two ratios corresponding to the two replicates present on the same slide were averaged, and the averaged ratio was converted to its log2 value.
For each experimental condition (wild-type strain versus prhG deletion mutant and wild-type strain versus prhG-overexpressing mutant), at least two independent RNA extractions from independent cultures were used and a minimum of six hybridizations were performed with dye swap labeling. In order to minimize false positives, only genes with high levels of significance (P < 0.05 by Student's t test) and an absolute log2 value ratio of greater than 1.3 were considered in this study.
All of the primary data from transcriptome experiments, as well as the experimental protocols used, are available from the ArrayExpress depository (accession number E-MEXP-2456).
Pathogenicity tests.
Pathogenicity tests by soil inoculation were performed by watering 4-week-old tomato plants (Lycopersicum esculentum cv. Marmande) with 50 ml of a suspension containing 107 or 108 CFU ml−1. Pathogenicity tests by stem injection were performed by injecting 20 μl of a 106 CFU ml−1 bacterial suspension into the stems of 4-week-old plants. Disease tests on Arabidopsis thaliana accession Col-0 were performed by immersing plants with cut or uncut roots into bacterial suspensions of 107 and 108 CFU ml−1.
Disease development was scored daily by using a disease index scale ranging from 0 for no symptoms to 4 for completely wilted plants (27). Within-group Kaplan-Meier survival estimates (the total number of plants surviving [i.e., with disease index scores below 3] out of the total number inoculated for each group) were computed for each time interval in order to build Kaplan-Meier survival curves for each group. The log rank test was used to perform between-group comparisons, testing the equivalence of the Kaplan-Meier survival curves for a pair of groups.
Creation of lacZ reporter fusion strains.
The prhG::lacZ fusion was created by using integration plasmid pCZ367 (9). An internal fragment of the prhG gene was PCR amplified by using primers 2360FW (5′-GTAAGCTTCCACACGGCCGGTCAGC-3′) and 2360REV2 (5′-GTCTAGATGTGCTGCTCGATG-3′) and cloned as a 0.5-kb HindIII-XbaI fragment into pCZ367 to generate plasmid pSG371. This plasmid was used to transform R. solanacearum GMI1000, and a single integrative event was selected by using pCZ367 gentamicin resistance (9). Recombinant clones were then checked by PCR.
lacZ reporter fusions with the hrpG, prhG, hrpB, popA, and hrpY genes were introduced into the different genetic backgrounds (ΔhrpG, ΔprhG, and ΔhrpG ΔprhG) by transformation with the genomic DNA of the donor strains. Recombinant strains were selected on media with adequate antibiotics, and the correct genomic insertion of the lacZ fusion was checked by PCR.
Plant cell cultures and bacterium-plant cell cocultures.
The A. thaliana At-202 (accession Col-0) and tomato Msk8 cell suspension lines were grown in Gamborg B5 (Flow Laboratories) and T-MSMO (Sigma) media, respectively (22). For the bacterium-plant cell cocultures, 10-ml samples of Arabidopsis and tomato cell suspensions were inoculated with R. solanacearum. After 16 h of incubation at 28°C, the coculture was filtered and the bacterial cells were recovered for β-galactosidase assays.
β-Galactosidase assays.
β-Galactosidase assays were performed as described previously (4). β-Galactosidase activity was measured according to Miller (24), and values were expressed in Miller units or as a percentage of the activity of the strain carrying a lacZ fusion in a wild-type genetic background.
Protein sequence analysis.
The analysis was performed with the MEGA versus 3.1 software (http://www.megasoftware.net/) using a neighbor-joining algorithm with 1,000 replications to build the tree and calculate the bootstrap values. The accession numbers of the 10 protein sequences compared in this analysis are as follows: PrhG from R. solanacearum GMI1000, NP_522584; PrhG from R. solanacearum Molk2, YP_002255075; PrhG from R. solanacearum UW551, ZP_00945868; HrpG from R. solanacearum GMI1000, CAA07190; HrpG from R. solanacearum Molk2, YP_002254969; HrpG from R. solanacearum UW551, ZP_00944901; HrpG from Xanthomonas oryzae pv. oryzae PXO99A, AAK92203; HrpG from X. campestris pv. vesicatoria 85-10, YP_363045; HrpG from X. campestris pv. campestris ATCC 33913, NP_636540; OmpR from E. coli K-12, AAA58202.
RESULTS
Identification of prhG, a close paralog of hrpG, the master regulator of pathogenicity functions in R. solanacearum.
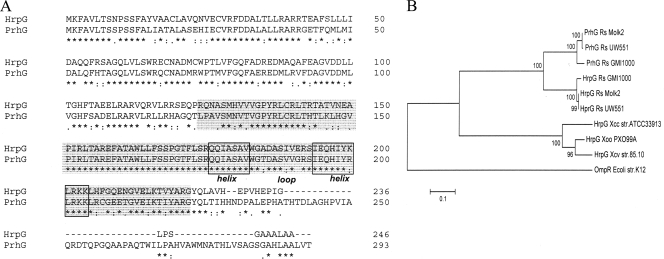
Annotation of the genome of strain GMI1000 revealed the existence of a gene, RSp1023 (hereafter named prhG), homologous to the T3SS regulatory gene hrpG. A comparison of the gene products reveals that the two proteins are 72% identical (83% similar). The main difference between the two proteins is that PrhG is slightly longer, as it contains an ∼50-amino-acid C-terminal extension which is absent in HrpG (Fig. 1A). Like HrpG, the PrhG protein belongs to the OmpR/PhoB family of two-component response regulators (13) and possesses a highly conserved helix-turn-helix motif DNA-binding domain (residues 173 to 205). The relatedness between PrhG and HrpG in this predicted DNA-binding domain reaches 96% amino acid identity. Also similarly to hrpG, no predicted sensor histidine kinase-encoding gene was found in the vicinity of prhG. Although prhG is located 200 kb away from the hrp gene cluster, the two immediate flanking genes (RSp1022 and RSp1024) are predicted to encode type III-dependent effectors (28), which could be suggestive of a functional link of this genomic region with the T3SS.
FIG. 1.
(A) ClustalW analysis of homologous proteins HrpG and PrhG (72% global identity and 96% in the helix-loop-helix domain). The predicted DNA-binding and transactivation domain is highlighted in gray, and the helix-loop-helix domain is boxed. (B) Neighbor-joining analysis showing the relationships among the PrhG and HrpG protein sequences of three R. solanacearum strains (GMI1000, Molk2, and UW551) and three Xanthomonas sp. strains (X. oryzae pv. oryzae strain PXO99A, X. campestris pv. vesicatoria strain 85-10, and X. campestris pv. campestris strain ATCC 33913). The E. coli K-12 OmpR protein sequence was added as an outgroup. The tree was constructed and the bootstrap values were calculated by using the neighbor-joining algorithm with 1,000 replications. The asterisks, periods, and colons below the sequences in panel A indicate identical residues, conserved substitutions, and semiconserved substitutions, respectively.
Analysis of the phylogenetic relationships with homologous sequences in the databases revealed that prhG is conserved in R. solanacearum strains taxonomically distant from GMI1000, such as Molk2 or UW551 (Fig. 1B). Secondly, this cladogram showed that the PrhG sequence group is closer to the R. solanacearum HrpG group than to the Xanthomonas sp. HrpG clade. This observation and the fact that the amino acid sequence identity between HrpG and PrhG is significantly higher than that between HrpG and its Xanthomonas orthologues (37 to 40%) support the view that prhG might be the result of an ancient hrpG duplication event that took place in R. solanacearum before the divergence of its various strains. Interestingly, none of the sequenced Xanthomonas sp. genomes was found to harbor a close paralog of HrpG, as observed in all of the R. solanacearum genomes investigated.
A prhG mutant is slightly reduced in pathogenicity on tomato plants.
To evaluate the role of prhG in R. solanacearum pathogenicity, an unmarked deletion mutant derivative was generated from strain GMI1000. The resulting strain, GRS445, was used for inoculation assays on tomato, the natural host of GMI1000. Routine infection tests were performed either by injection of bacteria into the stem or by drenching the plants with a bacterial suspension, an inoculation method which better mimics natural infection conditions.
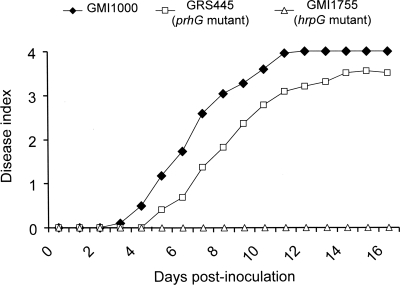
The pathogenicity assays done by drenching tomato plants showed no significant difference in the appearance of symptoms due to the mutant compared to the wild-type strain (data not shown). A difference in virulence was observed when a bacterial suspension of 104 CFU was injected directly into the stems. Under these conditions, symptoms appeared with a slight delay when the plants were inoculated with the prhG mutant strain compared to the wild-type strain (Fig. 2). We applied a log rank test to determine whether the disease curves obtained with the wild-type strain and the prhG mutant derivative were significantly different under these conditions. A score of 3 was used to discriminate between live plants (score below 3) and dead plants (score of 3 or above). With these criteria, the P value is 0.00997, showing that the difference between the two disease curves is indeed significant and that prhG is required for full pathogenicity on tomato.
FIG. 2.
Disease progress curves of R. solanacearum wild-type strain GMI1000 and the prhG and hrpG deletion mutant strains on tomato plants. An equivalent number of bacterial cells (104 CFU) was injected into the stems of susceptible tomato seedlings, which were monitored for 18 days and rated on a disease index scale where 0 indicates healthy plants and 4 indicates 100% wilted. All of the assays had a minimum of 24 plants per treatment per assay. Results are representative of three independent experiments.
Pathogenicity assays were also performed on another host plant, A. thaliana ecotype Columbia, which is susceptible to GMI1000. No significant difference in pathogenicity was observed between the prhG mutant and the wild-type strain when A. thaliana plants with uncut or cut roots were inoculated (data not shown).
prhG and hrpG respond to different environmental signals.
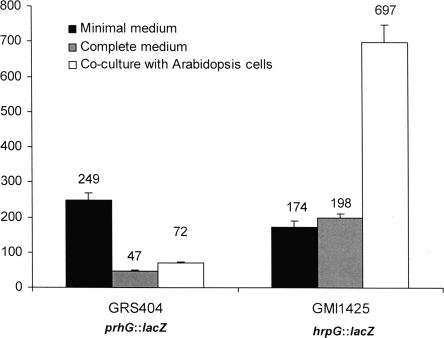
Since R. solanacearum hrpG was known to display a distinct expression pattern, depending on the environmental conditions used (7), we determined the expression profile of prhG under the same conditions. In these assays, bacteria carrying a chromosomal lacZ fusion to hrpG (strain GMI1425) or prhG (strain GRS404) were grown in complete medium, in minimal medium supplemented with glutamate as the sole carbon source, or in the presence of A. thaliana or tomato host cell suspensions. Figure 3 shows the levels of β-galactosidase activity monitored for each strain under each set of conditions. It appears that the prhG::lacZ fusion was induced in minimal medium (fivefold with respect to complete medium). However, in contrast to the specific and strong expression of the hrpG::lacZ fusion (strain GMI1425) detected in the presence of plant cells, the prhG::lacZ fusion was not induced under this condition. In the presence of Arabidopsis plant cells, prhG expression levels were even lower than those detected in minimal medium and ∼10 times lower than those of hrpG. Similar observations were made when both strains were grown in tomato cell suspensions (data not shown). These data show that the two regulatory genes respond differently to environmental signals: transcription of hrpG is specifically enhanced in response to plant signals, while transcription of prhG appears to be mainly induced under minimal medium conditions.
FIG. 3.
Expression of hrpG and prhG in different media and coculture with plant cells. Strains GRS404 (prhG::lacZ) and GMI1425 (hrpG::lacZ) were grown for 16 h in minimal medium supplemented with glutamate at a 20 mM final concentration in B medium (complete medium) and in coculture with Arabidopsis At-202 cells in Gamborg medium. β-Galactosidase activity is expressed in Miller units. Each measurement corresponds to the average of four replicates, and bars indicate standard deviations.
Transcriptomic profiling of a prhG mutant reveals that the PrhG regulon significantly differs from the HrpG regulon.
In order to determine the complete PrhG regulon, a GMI1000 whole-genome microarray (26) was used to compare the transcriptomes of R. solanacearum GMI1000 and its prhG deletion mutant derivative. Both strains were grown to late exponential phase in minimal medium, which has been shown to induce the expression of hrp genes (3), as well as prhG. Using >1.3 as the threshold of the absolute log2 ratio (an approximately threefold difference from the wild-type RNA level), we identified 95 genes as activated by PrhG and 1 gene (RSc1863) as repressed. The PrhG regulon largely overlaps the previously described HrpB regulon: nearly 75% of the genes which are activated by PrhG have already been proven to be activated by HrpB, and the gene repressed by PrhG is also repressed by HrpB (26) (see Table S1 in the supplemental material).
The comparison of the transcriptomes of R. solanacearum GMI1000 and a prhG-overexpressing strain (GMI1000/pLP2) enabled us to identify 21 additional genes upregulated by PrhG, 17 of which also belong to the HrpB regulon, and 22 additional genes repressed by PrhG, 15 of which are also repressed by HrpB. Finally, in order to avoid possible heterologous complementation of PrhG by HrpG, we also compared gene expression in the ΔhrpG ΔprhG double mutant with that in the ΔhrpG ΔprhG mutant expressing a prhG copy on a plasmid (pLP2). Here again, almost all of the differentially expressed genes were found to be part of the hrpB regulon (data not shown).
In conclusion, the transcriptomic profiling analyses performed showed that PrhG mainly controls the expression of hrpB and thus of the genes regulated by HrpB. Besides the genes belonging to the HrpB regulon, 39 genes appeared to have a transcription level specifically altered in the prhG mutant, 32 of which are under positive control and only 7 of which are under negative control (see Table S1 in the supplemental material). Except for one gene, none of the genes known to be regulated by HrpG independently of HrpB (33) was found to be regulated by PrhG. The exception is the RSp0201 gene, which encodes a conserved hypothetical protein of unknown function. We generated a disruption mutant of RSp0201 in GMI1000 and found that its pathogenicity was not altered on tomato plants (data not shown). Therefore, rather surprisingly, the overlap between the PrhG and HrpG regulons is essentially reduced to the HrpB regulon.
PrhG controls the expression of hrpB and HrpB-regulated genes.
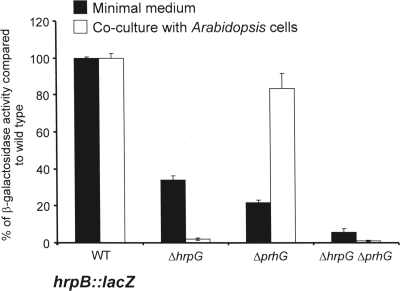
To confirm the control of hrpB expression by PrhG, the hrpB::lacZ fusion from strain GMI1475 (16) was introduced into the prhG deletion mutant. The level of transcription of hrpB in this strain was measured by β-galactosidase activity and compared to that measured in a wild-type genetic background. The results showed a threefold decrease in hrpB expression in the prhG deletion mutant after growth of the strains in minimal medium compared to that in the wild-type genetic background (Fig. 4). In contrast, no decrease in hrpB expression was measured in the prhG deletion mutant compared to its expression in the wild-type strain upon coculture of these strains with tomato or A. thaliana cell suspensions. The comparison of hrpB expression in the mutant strains indicates that both HrpG and PrhG are required for full induction of hrpB in minimal medium but that in the presence of plant cells, only HrpG is involved in the induction of hrpB expression.
FIG. 4.
Expression of hrpB in different genetic backgrounds after 16 h of growth in minimal medium supplemented with glutamate at a 20 mM final concentration and in coculture with Arabidopsis At-202 cells in Gamborg medium. β-Galactosidase activity is expressed as a percentage of the expression level measured in the wild-type (WT) genetic background. Each measurement corresponds to the average of four replicates, and bars indicate standard deviations.
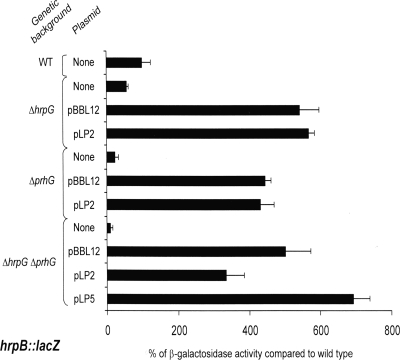
We also monitored the expression of two hrpB-regulated genes, popA, which codes for an R. solanacearum harpin (4), and hrpY, which encodes the structural component of the Hrp pilus (34). As shown in Table 2, expression of a popA::lacZ fusion decreased 23-fold in a prhG deletion mutant compared to that in the wild-type genetic background after the growth of these strains in minimal medium. For the hrpY::lacZ fusion, only a fourfold decrease was observed under the same conditions, but the impact was more important than that observed in the hrpG mutant. The transcription of both popA and hrpY was almost completely abolished in the double-deletion mutant.
TABLE 2.
β-Galactosidase activities obtained with hrpY::lacZ and popA::lacZ fusions in different genetic backgrounds
| Genetic background | Avg (SD) β-galactosidase activity (Miller units)a with reporter fusion: |
|
|---|---|---|
| hrpY::lacZ | popA::lacZ | |
| Wild type | 741.0 (14.10) | 1,677.0 (181.00) |
| ΔhrpG | 590.5 (4.90) | 44.0 (2.80) |
| ΔprhG | 193.0 (15.60) | 73.0 (2.80) |
| ΔhrpG ΔprhG | 9.0 (1.40) | 11.0 (5.70) |
Measurements of four replicates were made after 16 h of bacterial growth in minimal medium supplemented with glutamate at a 20 mM final concentration.
Taken together, these results show that both HrpG and PrhG are necessary for full induction of hrpB and its targets in minimal medium.
Evidence that PrhG and HrpG can control hrpB expression independently.
To get a better understanding of the regulation mechanism involving HrpG and PrhG in minimal medium, we conducted a complementation analysis of R. solanacearum mutants with hrpG, prhG, or both regulatory genes deleted, all bearing an hrpB::lacZ fusion. Three plasmid constructions were used: a plasmid carrying a copy of prhG (pLP2) under the control of its promoter, a plasmid with a copy of hrpG (pBBL12) under the control of its promoter, and a plasmid containing both regulators (pLP5), each under the control of its own promoter. Plasmids were introduced into the different hrpB::lacZ strains (the wild type and the ΔhrpG, ΔprhG, and ΔhrpG ΔprhG mutants), and the expression of hrpB was measured by β-galactosidase assays.
The results presented in Fig. 5 indicate that both HrpG and PrhG are able to induce hrpB expression on their own. In the absence of the chromosomal copy of hrpG and prhG, plasmid pBBL12 or pLP2 carrying a copy of hrpG or prhG, respectively, is able to restore a significant and comparable expression level of the hrpB::lacZ reporter fusion. These results also show that under the same growth conditions and expressed on the same backbone vector, prhG and hrpG have similar hrpB promoter induction strengths. The complementation of the double mutant strain with pLP5, carrying both regulators, also restored hrpB expression, but not to a significantly higher level than the plasmids carrying a single regulatory gene. In conclusion, these complementation experiments show that HrpG and PrhG act independently to control the expression of their common target, hrpB, and suggest that no cooperation mechanism is involved in this control.
FIG. 5.
Expression of hrpB in different genetic backgrounds after 16 h of growth in minimal medium supplemented with glutamate at a 20 mM final concentration. pBBL12, pLP2, and pLP5 are low-copy-number plasmids carrying, respectively, a copy of hrpG, a copy of prhG, and both hrpG and prhG, all under the control of their own promoters. β-Galactosidase activity is expressed as a percentage of the level of expression measured in the wild-type (WT) genetic background devoid of any plasmids. Each measurement corresponds to the average of three replicates, and bars indicate standard deviations.
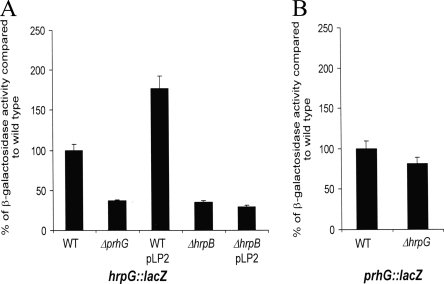
prhG does not control the expression of hrpG and vice-versa.
We then evaluated whether prhG could influence the transcription of its paralog hrpG or if, on the other hand, it could depend on hrpG for its own expression. The interdependency of each regulator was studied by using R. solanacearum strains carrying either a hrpG::lacZ or a prhG::lacZ fusion in a prhG or hrpG deletion background, respectively. β-Galactosidase assays proved that prhG transcription is independent of HrpG under minimal medium conditions, as the activity measured in the hrpG mutant is similar to that measured in the wild-type genetic background (Fig. 6B). Concerning the transcription of hrpG, the results showed a twofold decrease in β-galactosidase activity in the prhG deletion mutant and a twofold increase when an extra copy of prhG is introduced on a plasmid into the hrpG::lacZ strain. Because, it is known that HrpB exerts a positive but relatively modest impact on hrpG expression (16, 26), we investigated whether this slight effect of PrhG on hrpG expression is indirectly mediated through hrpB. As shown in Fig. 6A, this influence of PrhG on hrpG expression is abolished in an hrpB mutant, thus showing that the observed twofold factor maybe due to a positive feedback effect of HrpB on hrpG. Altogether, these data support the view that HrpG and PrhG do not exert direct transcriptional control on each other and belong to different signaling pathways.
FIG. 6.
Expression of hrpG (A) and prhG (B) in different genetic backgrounds after 16 h of growth in minimal medium supplemented with glutamate at a 20 mM final concentration. β-Galactosidase activity is expressed as a percentage of the level of expression measured in the wild-type (WT) genetic background. Each measurement corresponds to the average of four replicates, and bars indicate standard deviations.
DISCUSSION
In this study, we have identified PrhG, a novel component of the regulatory network controlling the expression of pathogenicity genes in R. solanacearum. The determination of the prhG regulon through transcriptomic profiling after growth of the bacteria in minimal medium revealed that a large proportion (75%) of the prhG-dependent genes are part of the previously described hrpB regulon (26). We confirmed by gene reporter fusion assays that expression of hrpB and some hrpB-controlled genes is indeed under the transcriptional control of prhG specifically under minimal medium conditions. PrhG is encoded by an OmpR/PhoB family regulatory gene which is the closest paralog to hrpG, a master regulator of pathogenicity genes in R. solanacearum (33).
Both PrhG and HrpG control the expression of T3SS and type III effector genes through hrpB. It is therefore rather surprising that a prhG mutant is only slightly affected in its pathogenicity and is not completely avirulent, as hrpG (or hrpB) deletion mutants have been shown to be (7). This marked difference in the prhG/hrpG mutant phenotypes on plants clearly reveals that the corresponding proteins have distinct roles, despite their strong relatedness and their having in common a major pathogenicity target (hrpB). The most probable hypothesis to explain the low impact of a prhG mutation on pathogenicity is based on the striking difference observed in the transcription pattern of the prhG/hrpG genes (Fig. 3). In the presence of plant signals, hrpG is much more strongly expressed in the bacterial cell and this low level of prhG expression in a hrpG mutant is probably not sufficient to restore pathogenicity. It is also possible that, in a plant environment, HrpG integrates specific signals or posttranslational modifications that do not impact PrhG so that specific activation of HrpG would bypass any requirement for PrhG. In this scenario, prhG seems to be dispensable for the expression of T3SS genes in plants as hrpB activation is mainly achieved through hrpG. This is supported by the data presented in Fig. 4 showing that the disruption of prhG has nearly no impact on hrpB expression after growth of bacteria in the presence of plant cells while disruption of hrpG abolishes hrpB expression.
The comparison of the defined PrhG and HrpG regulons reveals that they overlap only for the hrpB regulon. HrpG was shown to control the expression of 184 genes independently of hrpB in minimal medium (33), and—with a single exception (RSp0201)—none of these genes appears to be a regulatory target of prhG, thus illustrating the specificity of these regulatory proteins. This specificity raises intriguing questions about the very high level of identity observed between PrhG and HrpG, especially in the helix-turn-helix domain which is predicted to bind DNA operator sequences. This suggests that the homologous PrhG and HrpG proteins are able to discriminate between specific target promoter sequences and that the hrpB promoter is unique in being a confirmed target of both regulators.
How is hrpB control jointly achieved by PrhG and HrpG? The fact that several OmpR/PhoB family regulators bind to DNA as dimers (13, 19) raised the possibility that an active PrhG/HrpG heterodimer controls hrpB transcription in minimal medium. Our complementation data (Fig. 5) do not fit this hypothesis: both regulators provided in trans on the same backbone vector are individually able to activate hrpB expression at similar levels. The use of the pLP5 construct containing prhG and hrpG on the same vector also revealed that there may be no cooperative binding on DNA since in the ΔhrpG ΔprhG mutant background, complementation experiments with pLP5 did not lead to a significantly higher level of hrpB expression. The slight increase observed compared to complementation with only one of the regulators probably corresponds to an additive effect due to the presence of multiple copies of both regulatory genes. Although the mechanism of action and the DNA-binding specificities of PrhG and HrpG are still unknown, our data suggest that these two related regulators are able to activate hrpB independently. Simultaneous control of key T3SS promoters by distinct regulatory proteins has also been reported in other bacterial pathogens, such as Salmonella (11).
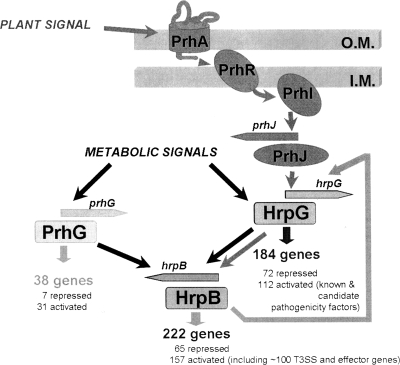
Because HrpG is the regulator acting just upstream of HrpB in the Hrp regulatory cascade, we determined whether PrhG could influence hrpG transcription (and could therefore indirectly control hrpB) or if, on the other hand, it was itself under the transcriptional control of HrpG. Our data show that the expression of each regulator is independent of that of the other. Interestingly, in the course of this investigation, we confirmed that hrpG expression is partially dependent on hrpB under minimal medium conditions. This had been previously observed through the transcriptomic profiling analysis of an hrpB deletion mutant, which showed decreased hrpG transcription (26). This retropositive regulatory loop illustrates the complex interplay between T3SS gene regulators in R. solanacearum (Fig. 7).
FIG. 7.
Model describing the regulation network involved in the control of R. solanacearum pathogenicity. HrpB is the regulator mainly devoted to the control of the T3SS and effector gene transcription. PrhG and HrpG both regulate the expression of hrpB but belong to different signaling pathways and integrate at least two distinct inducing signals. The nature of the activating plant signal(s) is unknown, but it requires physical contact between bacteria and plant cells (1). The metabolic signals indicated here are those perceived by bacteria grown under minimal medium conditions. HrpG regulates, independently of HrpB, the expression of a subset of genes that includes several virulence and pathogenicity factors (33). The specific subset of genes regulated by PrhG is smaller and contains only one other target regulated by HrpG, RSp0201. The feedback loop by which HrpB induces hrpG (see text) is also shown. O.M., outer membrane; I.M., inner membrane.
Since prhG is not crucial during plant pathogenesis, the biological relevance of this novel hrp regulatory component remains elusive. Surprisingly, transcriptomic profiling analyses revealed that, besides the hrpB regulon, prhG controls very few specific targets compared to hrpG (39 versus 184). Most of these prhG-specific genes were under positive control (32 out of 39), which makes unlikely a role for PrhG as a direct transcriptional repressor. The examination of the specifically activated genes does not provide any clue about the role of prhG during the R. solanacearum life cycle. In particular, none of these genes are clustered or organized in defined operons. Although eight of them (RSc0695, RSc1853, RSc2942, RSc3114, RSp0142, RSp0312, RSp0934, and RSp1577) are predicted to be involved in type I or II secretion processes, the significance of this is unknown.
The fact that prhG is an ancestral gene broadly conserved among the taxonomically diverse R. solanacearum phylotypes is an argument supporting the view that this gene is not an evolutionary remnant of the hrp regulatory system and may have conserved a specific function. The prhG gene was indeed detected through comparative genomic hybridization in a selection of 18 strains representative of the biodiversity of the species (17), also suggesting that prhG is not the result of a recent duplication of hrpG. It is interesting that, unlike hrpG, which is universally conserved among Xanthomonas spp. and R. solanacearum as a T3SS regulator component, prhG appears to be specific to R. solanacearum, as no direct counterpart could be detected in any of the Xanthomonas sp. genomes sequenced to date. The analysis of phylogenetic relationships suggests that the orthologous hrpG genes from R. solanacearum and Xanthomonas spp. have diverged before the prhG/hrpG separation in R. solanacearum (Fig. 1B), thus raising the possibility that Xanthomonas HrpG proteins could combine the specificities of both the HrpG and PrhG proteins of R. solanacearum.
The findings that prhG expression is specifically induced under minimal medium conditions and that the prhG-dependent regulation of hrpB follows this prhG expression profile strongly suggest that prhG has an important role in activating the T3SS before contact with host cells, which is the signal triggering the activation of hrpG (1). In the case of hrp gene expression, minimal medium conditions have been proposed to mimic the plant apoplast environment (3, 30, 32), and this probably corresponds to an early phase of the plant-bacterium interaction before the translocation of type III effectors inside plant cells. It is tempting to speculate that minimal medium conditions reproduce the conditions under which the bacteria first perceive some host physicochemical signals necessary to induce the setting up of a functional T3SS before a second, stronger, plant cell wall-specific signal specifically transduced through hrpG would act as a type III effector translocation-triggering signal. Since a prhG mutant is not affected or very slightly affected in pathogenicity on tomato and Arabidopsis plants, this scenario implies that the first step could also be mediated through hrpG. It is possible that prhG requirement may be more important under other conditions such as on other host plants (considering the very wide host range of the bacterium) or respond specifically to some unknown signals in the environment that were absent under the experimental conditions of the present study. Further work is necessary to define the mechanistic basis of hrpB regulation by PrhG and obtain a better definition of the environmental signals involved.
Supplementary Material
Acknowledgments
We are very grateful to Christian Boucher for critical reading of the manuscript and fruitful discussions, Alice Guidot and Nemo Peeters for help with protein sequence analysis, and Patrick Barberis and Laure Bapaume for technical assistance.
This work was funded by grants from the French Ministry of Science to L.P. and by an ANR-05-242-798 grant to S.G., as well a European reintegration grant (MERG030948) and grant BIO06-0919 from the Spanish Ministry of Science and Technology to M.V.
Footnotes
Published ahead of print on 11 December 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Aldon, D., B. Brito, C. Boucher, and S. Genin. 2000. A bacterial sensor of plant cell contact controls the transcriptional induction of Ralstonia solanacearum pathogenicity genes. EMBO J. 19:2304-2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alfano, J. R., and A. Collmer. 1997. The type III (Hrp) secretion pathway of plant pathogenic bacteria: trafficking harpins, Avr proteins, and death. J. Bacteriol. 179:5655-5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arlat, M., C. L. Gough, C. Zischek, P. A. Barberis, A. Trigalet, and C. Boucher. 1992. Transcriptional organization and expression of the large hrp gene cluster of Pseudomonas solanacearum. Mol. Plant-Microbe Interact. 5:187-193. [DOI] [PubMed] [Google Scholar]
- 4.Arlat, M., F. Van Gijsegem, J. C. Huet, J. C. Pernollet, and C. A. Boucher. 1994. PopA1, a protein which induces a hypersensitivity-like response on specific Petunia genotypes, is secreted via the Hrp pathway of Pseudomonas solanacearum. EMBO J. 13:543-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1989. Current protocols in molecular biology. Greene Publishing Associates & Wiley Interscience, New York, NY.
- 6.Brito, B., D. Aldon, P. Barberis, C. Boucher, and S. Genin. 2002. A signal transfer system through three compartments transduces the plant cell contact-dependent signal controlling Ralstonia solanacearum hrp genes. Mol. Plant-Microbe Interact. 15:109-119. [DOI] [PubMed] [Google Scholar]
- 7.Brito, B., M. Marenda, P. Barberis, C. Boucher, and S. Genin. 1999. prhJ and hrpG, two new components of the plant signal-dependent regulatory cascade controlled by PrhA in Ralstonia solanacearum. Mol. Microbiol. 31:237-251. [DOI] [PubMed] [Google Scholar]
- 8.Cunnac, S., C. Boucher, and S. Genin. 2004. Characterization of the cis-acting regulatory element controlling HrpB-mediated activation of the type III secretion system and effector genes in Ralstonia solanacearum. J. Bacteriol. 186:2309-2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cunnac, S., A. Occhialini, P. Barberis, C. Boucher, and S. Genin. 2004. Inventory and functional analysis of the large Hrp regulon in Ralstonia solanacearum: identification of novel effector proteins translocated to plant host cells through the type III secretion system. Mol. Microbiol. 53:115-128. [DOI] [PubMed] [Google Scholar]
- 10.Denny, T. P. 2006. Plant Pathogenic Ralstonia species, p. 573-644. In S. S. Gnanamanickam (ed.), Plant-associated bacteria. Springer Publishing, Dordrecht, The Netherlands.
- 11.Feng, X., D. Walthers, R. Oropeza, and L. J. Kenney. 2004. The response regulator SsrB activates transcription and binds to a region overlapping OmpR binding sites at Salmonella pathogenicity island 2. Mol. Microbiol. 54:823-835. [DOI] [PubMed] [Google Scholar]
- 12.Galán, J. E., and H. Wolf-Watz. 2006. Protein delivery into eukaryotic cells by type III secretion machines. Nature 444:567-573. [DOI] [PubMed] [Google Scholar]
- 13.Gao, R., and A. M. Stock. 2009. Biological insights from structures of two-components proteins. Annu. Rev. Microbiol. 63:133-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Genin, S., and C. Boucher. 2002. Ralstonia solanacearum: secrets of a major pathogen unveiled by analysis of its genome. Mol. Plant Pathol. 3:111-118. [DOI] [PubMed] [Google Scholar]
- 15.Genin, S., B. Brito, T. P. Denny, and C. Boucher. 2005. Control of the Ralstonia solanacearum type III secretion system (Hrp) genes by the global virulence regulator PhcA. FEBS Lett. 579:2077-2081. [DOI] [PubMed] [Google Scholar]
- 16.Genin, S., C. L. Gough, C. Zischek, and C. A. Boucher. 1992. Evidence that the hrpB gene encodes a positive regulator of pathogenicity genes from Pseudomonas solanacearum. Mol. Microbiol. 6:3065-3076. [DOI] [PubMed] [Google Scholar]
- 17.Guidot, A., P. Prior, J. Schoenfeld, S. Carrere, S. Genin, and C. Boucher. 2007. Genomic structure and phylogeny of the plant pathogen Ralstonia solanacearum inferred from gene distribution analysis. J. Bacteriol. 189:377-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huynh, T. V., D. Dahlbeck, and B. J. Staskawicz. 1989. Bacterial blight of soybean: regulation of a pathogen gene determining host cultivar specificity. Science 245:1374-1377. [DOI] [PubMed] [Google Scholar]
- 19.Kenney, L. J. 2002. Structure/function relationships in OmpR and other winged-helix transcription factors. Curr. Opin. Microbiol. 5:135-141. [DOI] [PubMed] [Google Scholar]
- 20.Lewis, J. D., D. S. Guttman, and D. Desveaux. 2009. The targeting of plant cellular systems by injected type III effector proteins. Semin. Cell Dev. Biol. 20:1055-1063. [DOI] [PubMed] [Google Scholar]
- 21.Lindgren, P. B., R. C. Peet, and N. J. Panopoulos. 1986. Identification and cloning of genes involved in phaseolotoxin production by Pseudomonas syringae pv. “phaseolicola.” J. Bacteriol. 166:1096-1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marenda, M., B. Brito, D. Callard, S. Genin, P. Barberis, C. Boucher, and M. Arlat. 1998. PrhA controls a novel regulatory pathway required for the specific induction of Ralstonia solanacearum hrp genes in the presence of plant cells. Mol. Microbiol. 27:437-453. [DOI] [PubMed] [Google Scholar]
- 23.Marx, C. J., and M. E. Lidstrom. 2002. Broad host range cre-lox system for antibiotic marker recycling in Gram-negative bacteria. Biotechniques 33:1062-1067. [DOI] [PubMed] [Google Scholar]
- 24.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 25.Mole, B. M., D. A. Baltrus, J. L. Dangl, and S. R. Grant. 2007. Global virulence regulation networks in phytopathogenic bacteria. Trends Microbiol. 15:363-371. [DOI] [PubMed] [Google Scholar]
- 26.Occhialini, A., S. Cunnac, N. Reymond, S. Genin, and C. Boucher. 2005. Genome-wide analysis of gene expression in Ralstonia solanacearum reveals that the hrpB gene acts as a regulatory switch controlling multiple virulence pathways. Mol. Plant-Microbe Interact. 18:938-949. [DOI] [PubMed] [Google Scholar]
- 27.Poueymiro, M., S. Cunnac, P. Barberis, L. Deslandes, N. Peeters, A. C. Cazale-Noel, C. Boucher, and S. Genin. 2009. Two type III secretion system effectors from Ralstonia solanacearum GMI1000 determine host range specificity on tobacco. Mol. Plant-Microbe Interact. 22:538-550. [DOI] [PubMed] [Google Scholar]
- 28.Poueymiro, M., and S. Genin. 2009. Secreted proteins from Ralstonia solanacearum: a hundred tricks to kill a plant. Curr. Opin. Microbiol. 12:44-52. [DOI] [PubMed] [Google Scholar]
- 29.Qian, W., Z.-J. Han, and C. He. 2008. Two-component signal transduction systems of Xanthomonas spp.: a lesson from genomics. Mol. Plant-Microbe Interact. 21:151-161. [DOI] [PubMed] [Google Scholar]
- 30.Rahme, L. G., M. N. Mindrinos, and N. J. Panopoulos. 1992. Plant and environmental sensory signals control the expression of hrp genes in Pseudomonas syringae pv. phaseolicola. J. Bacteriol. 174:3499-3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Salanoubat, M., S. Genin, F. Artiguenave, J. Gouzy, S. Mangenot, M. Arlat, A. Billault, P. Brottier, J. C. Camus, L. Cattolico, M. Chandler, N. Choisne, C. Claudel-Renard, S. Cunnac, N. Demange, C. Gaspin, M. Lavie, A. Moisan, C. Robert, W. Saurin, T. Schiex, P. Siguier, P. Thebault, M. Whalen, P. Wincker, M. Levy, J. Weissenbach, and C. A. Boucher. 2002. Genome sequence of the plant pathogen Ralstonia solanacearum. Nature 415:497-502. [DOI] [PubMed] [Google Scholar]
- 32.Tang, X., Y. Xiao, and J.-M. Zhou. 2006. Regulation of the type III secretion system in phytopathogenic bacteria. Mol. Plant-Microbe Interact. 19:1159-1166. [DOI] [PubMed] [Google Scholar]
- 33.Valls, M., S. Genin, and C. Boucher. 2006. Integrated regulation of the type III secretion system and other virulence determinants in Ralstonia solanacearum. PLoS Pathog. 2:e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Van Gijsegem, F., J. Vasse, J. C. Camus, M. Marenda, and C. Boucher. 2000. Ralstonia solanacearum produces hrp-dependent pili that are required for PopA secretion but not for attachment of bacteria to plant cells. Mol. Microbiol. 36:249-260. [DOI] [PubMed] [Google Scholar]
- 35.Vasse, J., S. Genin, P. Frey, C. Boucher, and B. Brito. 2000. The hrpB and hrpG regulatory genes of Ralstonia solanacearum are required for different stages of the tomato root infection process. Mol. Plant-Microbe Interact. 13:259-267. [DOI] [PubMed] [Google Scholar]
- 36.Wengelnik, K., and U. Bonas. 1996. HrpXv, an AraC-type regulator, activates expression of five of the six loci in the hrp cluster of Xanthomonas campestris pv. vesicatoria. J. Bacteriol. 178:3462-3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wengelnik, K., G. Van den Ackerveken, and U. Bonas. 1996. HrpG, a key hrp regulatory protein of Xanthomonas campestris pv. vesicatoria is homologous to two-component response regulators. Mol. Plant-Microbe Interact. 9:704-712. [DOI] [PubMed] [Google Scholar]
- 38.Yoshimochi, T., Y. Hikichi, A. Kiba, and K. Ohnishi. 2009. The global virulence regulator PhcA negatively controls the Ralstonia solanacearum hrp regulatory cascade by repressing expression of the PrhIR signalling proteins. J. Bacteriol. 191:3424-3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou, J. M., and J. Chai. 2008. Plant pathogenic bacterial type III effectors subdue host responses. Curr. Opin. Microbiol. 11:179-185. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.