Abstract
The species Campylobacter jejuni is naturally competent for DNA uptake; nevertheless, nonnaturally transformable strains do exist. For a subset of strains we previously showed that a periplasmic DNase, encoded by dns, inhibits natural transformation in C. jejuni. In the present study, genetic factors coding for DNase activity in the absence of dns were identified. DNA arrays indicated that nonnaturally transformable dns-negative strains contain putative DNA/RNA nonspecific endonucleases encoded by CJE0566 and CJE1441 of strain RM1221. These genes are located on C. jejuni integrated elements 2 and 4. Expression of CJE0566 and CJE1441 from strain RM1221 and a homologous gene from strain 07479 in DNase-negative Escherichia coli and C. jejuni strains indicated that these genes code for DNases. Genetic transfer of the genes to a naturally transformable C. jejuni strain resulted in a decreased efficiency of natural transformation. Modeling suggests that the C. jejuni DNases belong to the Serratia nuclease family. Overall, the data indicate that the acquisition of prophage-encoded DNA/RNA nonspecific endonucleases inhibits the natural transformability of C. jejuni through hydrolysis of DNA.
Bacterial species display genetic diversity that can contribute to their capacity to adapt and survive in changing environments. One of the processes contributing to genetic diversity is horizontal gene transfer. This involves the acquisition of genetic material, ultimately resulting in insertion of the acquired DNA and/or deletion of existing genetic material (9). The ability to take up exogenous DNA is present in many bacteria (4, 30). In general, uptake of DNA during natural transformation involves binding of double-stranded DNA to bacterial surface components, followed by transport through the cytoplasmic membrane (5). Upon transport, one of the DNA strands is degraded into nucleotides, whereas the other strand enters the cytoplasm and may provide new characteristics to the host genome.
One of the bacterial species naturally competent for DNA uptake is the human pathogen Campylobacter jejuni (32). Worldwide, C. jejuni is one of the most frequent causes of human bacterial gastroenteritis (3). In C. jejuni horizontal gene transfer can occur within a host, as witnessed in chickens infected with two C. jejuni strains carrying distinct genetic markers (7). The ability to acquire exogenous DNA contributes to the generation of genetic diversity in C. jejuni, which is reflected by the genotypic variation seen among strains (8, 29, 33).
Thus far, several genes have been implicated in the process of natural transformation of C. jejuni (35). Yet not all C. jejuni strains are equally competent, since differences in natural transformation frequencies have been noted, and even nonnaturally transformable strains do exist (32, 34). Previously, we demonstrated that DNA-hydrolyzing activity inhibits natural transformation of C. jejuni and identified Dns as one of the responsible nucleases (13). Dns is encoded by a putative prophage present in C. jejuni strain RM1221, namely, C. jejuni integrated element 1 (CJIE1).
The presence of DNase activity in C. jejuni has long been known and was in fact one of the criteria for Lior's extended biotyping scheme for thermophilic campylobacters (21). Through identification of dns a genetic basis for DNase activity by a subset of C. jejuni strains has been provided, but the genetic factors responsible for DNase activity in dns-negative nonnaturally transformable C. jejuni strains are not yet known.
In this study, we attempted to identify and functionally characterize an additional DNase-encoding gene(s) present in a subset of nonnaturally transformable DNase+ C. jejuni strains. Comparative genomic hybridization for DNase+/dns+ and DNase+/ dns-negative C. jejuni strains revealed three highly homologous genes encoded by the putative phage-related integrated elements CJIE2 and CJIE4. Functional analysis showed that these genes encode DNases that reduce the natural transformability of C. jejuni through hydrolysis of exogenous DNA.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains used in this study are listed in Table 1. C. jejuni strains were grown on heart infusion (HI) agar plates (Difco) containing 5% sheep blood (HIS plates), at 37°C for 48 h under microaerobic conditions (6% O2, 7% CO2, 7% H2, and 80% N2). For electrotransformation and natural transformation, saponin plates were used, consisting of blood agar base no. 2 medium (Oxoid) containing 4% sheep blood lysed with 0.7% saponin. E. coli was cultured on Luria-Bertani agar or in Luria-Bertani broth at 37°C under aerobic conditions. When appropriate, media were supplemented with ampicillin (100 μg ml−1), chloramphenicol (12.5 μg ml−1), or kanamycin (30 μg ml−1).
TABLE 1.
Bacterial strains used in the study
| Strain | Relevant genotype and/or phenotype | Source or reference |
|---|---|---|
| C. jejuni wild-type strains | ||
| C011300 | NT,a DNase+/dns+ | J. A. Frost (13) |
| C019165 | NT, DNase+/dns+ | J. A. Frost (13) |
| C012446 | NT, DNase+/dns+ | J. A. Frost (13) |
| C012599 | NT, DNase+/dns+ | J. A. Frost (13) |
| C356 (CNET076) | NT, DNase+/dns+ | W. F. Jacobs-Reitsma (13) |
| CCUG10950 | NT, DNase+/dns+ | Culture Collection, University of Gothenburg, Gothenburg, Sweden (13) |
| RM1221 | NT, DNase+/dns+ | 13, 22 |
| D3141 | NT, DNase+/dns negative | 13, 23 |
| D3226 | NT, DNase+/dns negative | 13, 23 |
| 40707L (CNET007) | NT, DNase+/dns negative | 13, 16 |
| 07479 | NT, DNase+/dns negative | 13, 16 |
| 41239B (CNET005) | NT, DNase+/dns negative | 13, 16 |
| C019168 | Naturally transformable, DNase− | J. A. Frost (13) |
| C. jejuni transformants | ||
| C019168(pWM1007Pr1492) | This study | |
| C019168(pWM1007Pr1492CJE0566RM1221) | This study | |
| C019168(pWM1007Pr1492CJE1441RM1221) | This study | |
| C019168(pWM1007Pr1492CJE056607479) | This study | |
| C019168hipO::cat | Contains chloramphenicol resistance cassette (cat) in hipO | This study |
| E. coli DH5α | F− φ80dlacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17(rK− mK+) phoA supE44 λ−thi-1 gyrA96 relA1 | Invitrogen, Breda, The Netherlands |
NT, nonnaturally transformable.
Microarray data analysis.
The comparative genomic hybridization data used were obtained in a previous study (13). Briefly, a whole C. jejuni genome open reading frame (ORF)-based DNA array, consisting of 1,530 ORFs (94%) from the naturally transformable strain NCTC 11168 and 227 unique ORFs from the nonnaturally transformable strain RM1221, was hybridized with chromosomal DNA isolated from six naturally transformable C. jejuni strains and 20 nonnaturally transformable strains. The microarrays were scanned and analyzed as described previously, and the status of a gene (present, divergent, or absent) was defined using comparative genomic indexing (13, 25).
In silico analysis.
The candidate genes obtained via comparative genomic indexing were used to search for homologues with Basic BLAST or BLAST Assembled Genomes (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The Blastn algorithm (nucleotide-nucleotide BLAST) was used to search for homologues based on nucleotide sequences, and the Blastp algorithm (protein-protein BLAST) was used to find homologues based on amino acid sequences. Alignments were assumed to be significant when the probability values obtained were <1. By performing pairwise alignments with EMBOSS-Align (http://www.ebi.ac.uk/Tools/emboss/align; covering whole length sequences) the overall similarities were determined (19, 24). The presence and location of a signal peptide cleavage site in the amino acid sequence were predicted by SignalP 3.0 (2).
Confirmation of the hybridization data by PCR.
The status (presence/absence) of CJE0566 and CJE1441 homologues was confirmed by PCR. To this end, chromosomal DNA was isolated from C. jejuni strains with the Gentra Puregene DNA isolation kit (Qiagen) according to the manufacturer's protocol for Gram-negative bacteria and used as a template in the PCR. The primer combinations used (CJE0566-Fint/CJE0566-Rint and CJE1441-Fint/CJE1441-Rint [Table 2]) were based on the coding sequences of CJE0566 and CJE1441 of strain RM1221 (NCBI reference sequence number NC_003912) (10).
TABLE 2.
Primers and plasmids used in the study
| Primer or plasmid | Sequence (5′-3′) or relevant characteristic(s)a,b | Source or reference |
|---|---|---|
| Primers | ||
| CJE0566-Fint | CAAAGTTGTTCGCAAGTTTTAG | |
| CJE0566-Rint | CCTTTTAAGATTTTAATATAAGAG | |
| CJE1441-Fint | CAAAACTGCTCACAAGTTTTGG | |
| CJE1441-Rint | CCTTTTAAAATCTTAGTGTAAGAG | |
| CJE0566-F | GAAATGATTAAAGCAACAAGTTAAAC | |
| CJE0566-R | GTAAGCAAAAAGCACCCCATTC | |
| CJE1441-F | CAAATTGAAAGGGTATGAAAC | |
| CJE1441-R | TGCAATTTCCATTTTTACTCC | |
| CJE0566-Fexp | GATCTTCTAGAAAGGAGAATAAATGAAAAAACTCATAC | |
| CJE0566-Rexp | CCATGGCAATTGTTATATTTTATCACAATC | |
| CJE0566-1441-Fexp | AGATCTTCTAGAAAGGAGAATAAATGAAAAAACTTATAATC | |
| CJE1441-Rexp | CCATGGCAATTGTTAAAAATTGTCACATTTTAC | |
| Pr1492XbaI | AGATCTTCTAGACTCATTTAACGGTTGTCTCC | |
| Pr1492MfeI | ACTAGTCAATTGGCGATGGCCCTG | |
| pWM1007Pr1492-F | GCATATGAAGAGATTTTG | |
| pWM1007-R | CCAACTGGTAATGGTAG | |
| Plasmids | ||
| pCR2.1 | TA cloning vector, Ampr | Invitrogen |
| pCRCJE0566RM1221 | pCR2.1 containing CJE0566 of RM1221 | This study |
| pCRCJE1441RM1221 | pCR2.1 containing CJE1441 of RM1221 | This study |
| pCRCJE056607479 | pCR2.1 containing a CJE0566/CJE1441 homologue of 07479 | This study |
| pWM1007Pr1492 | C. jejuni expression plasmid containing the CJ1492 promoter, Kmr | 36 |
| pWM1007Pr1492CJE0566RM1221 | C. jejuni expression plasmid with CJE0566 of RM1221 | This study |
| pWM1007Pr1492CJE1441RM1221 | C. jejuni expression plasmid with CJE1441 of RM1221 | This study |
| pWM1007Pr1492CJE056607479 | C. jejuni expression plasmid with CJE0566/CJE1441 homologue of 07479 | This study |
| pHipO::cat | C. jejuni suicide vector with cat inserted in hipO, Cmr | 7 |
Endonuclease restriction sites introduced into the sequences are underlined.
Ribosomal binding sites are shown in bold.
Electrotransformation of C. jejuni.
C. jejuni mutant strains were constructed via electrotransformation as described previously (13). After recovery on nonselective saponin plates (5 h, 37°C, microaerobic conditions) the bacteria were harvested and plated onto selective saponin plates (48 h, 37°C, microaerobic conditions) to select for transformants.
Construction of DNase-encoding expression plasmids.
For gene expression in C. jejuni, the genes CJE0566 and CJE1441 from strain RM1221 and a CJE0566/CJE1441 homologue from strain 07479 were amplified by PCR using Taq polymerase with proofreading and primer sets CJE0566-F/CJE0566-R and CJE1441-F/CJE1441-R, respectively (Table 2). Obtained PCR fragments (706 bp with CJE0566 primers and 704 bp with CJE1441 primers) were cloned into pCR2.1 (Invitrogen, Breda, The Netherlands), resulting in pCRCJE0566RM1221, pCRCJE1441RM1221, and pCRCJE056607479 (4.6 kb). Verification of the inserts was done by sequence analysis (BaseClear, Leiden, The Netherlands). Subsequently, these three constructs were used as templates in PCRs to introduce a ribosomal binding site (AGGAG) and XbaI and MfeI restriction sites, using a different primer combination for each plasmid (CJE0566-Fexp/CJE0566-Rexp, CJE0566-1441-Fexp/CJE1441-Rexp, and CJE0566-1441-Fexp/CJE0566-Rexp [Table 2]). To introduce the same restriction sites into the kanamycin-resistant C. jejuni expression plasmid pWM1007Pr1492 (9.5 kb) (36), a PCR product was generated using the primers Pr1492XbaI and Pr1492MfeI (Table 2). After digestion with XbaI and MfeI, the resulting 671-bp fragments with CJE0566RM1221, CJE1441RM1221, and CJE056607479 were ligated into pWM1007Pr1492 to form pWM1007Pr1492CJE0566RM1221, pWM1007Pr1492CJE1441RM1221, and pWM1007Pr1492CJE056607479, respectively (10.2 kb). Verification of the size of the insert was done by PCR, using primers pWM1007Pr1492-F and pWM1007-R (Table 2) located on pWM1007Pr1492. The resulting C. jejuni expression plasmids were transferred into C. jejuni strain C019168 via electrotransformation (Table 1).
DNA hydrolysis assay.
DNase activity was detected using DNase agar (Oxoid) supplemented with 0.005% toluidine blue (15, 31). Cultures of E. coli grown in Luria-Bertani broth and of C. jejuni grown in HI broth, for 16 h at 37°C, were spun down (1.5 and 3 ml, respectively). The pelleted bacteria were suspended in 50 μl of 500 mM Tris-HCl (pH 7.5), incubated with 5 μl of polymyxin B (50 μg μl−1; 4°C, 15 min), followed by inoculation of the suspension into a well (5 mm in diameter) cut in the DNase agar plate after gelling. DNase I (2 μg; Roche Diagnostics, Indianapolis, IN) was included as a positive control, and 50 μl of 500 mM Tris-HCl (pH 7.5) was used as a negative control. The plate was incubated aerobically at 37°C for 24 h. Positive results for DNase activity were visible as colorless zones around the wells.
Natural transformation of C. jejuni.
For determination of the natural transformation frequencies of C. jejuni C019168 derivatives the biphasic method was used (32). Homologous chromosomal DNA (2 μg) isolated from C. jejuni strain C019168hipO::cat (Table 1) (7) served as donor DNA in the transformation (13). To determine the number of transformants per milliliter, 200-μl aliquots of the appropriate serial dilutions (100 to 10−5) were plated onto selective saponin plates. The CFU per milliliter was quantified using track dilution (18), whereby 10-μl aliquots of serial dilutions (10−4 to 10−8) were spotted onto nonselective saponin plates. For calculation of the natural transformation frequencies, the number of transformants per milliliter (selective plates) was divided by the total number of bacteria per milliliter (nonselective plates). All tests were performed in triplicate. Student's t test was used for statistical analysis.
Microarray series accession number.
The microarray data have been deposited in the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo) with the series accession number GSE18399.
RESULTS
Comparative genomic hybridization.
In our search for differences in genome content between dns+ and dns-negative C. jejuni strains that are nonnaturally transformable and display DNase activity, previously obtained comparative genomic hybridization data were used (13). To identify genes responsible for the DNase+ phenotype in dns-negative strains, the hybridization data were screened for genes absent in the majority of six nonnaturally transformable DNase+/dns+ C. jejuni strains but present in the majority of five nonnaturally transformable DNase+/dns-negative strains (see Table S1 in the supplemental material). A total of 34 genes were absent in almost all of the DNase+/dns+ strains (83% to 100%) and present in the majority of DNase+/dns-negative strains (60% to 100%), suggesting that one of these genes may be responsible for DNase activity in the DNase+/dns-negative strains. One DNase+/dns+ strain (C01965), however, exhibited hybridization patterns similar to the DNase+/dns-negative strains, i.e., it contained the majority of the 34 genes in addition to dns.
The majority of the identified genes (24 genes) hybridized with genes of CJIE2. Nine genes hybridized with genes of CJIE4; seven of these genes are located on a module also present in CJIE2 and are indistinguishable by comparative genomic hybridization (25).
Identification of nuclease-encoding genes.
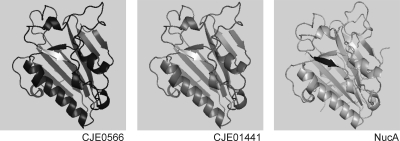
In silico analysis of the 34 identified genes present in the majority of DNase+/dns-negative strains using Blastn analysis confirmed the annotation of two genes (CJE0566 and CJE1441) from strain RM1221 as DNA/RNA nonspecific endonucleases. A homologue with the same annotation was found in Campylobacter lari strain RM2100 (Cla_0934). The analysis did not indicate a putative function of the remaining 32 genes in relation to natural transformability. Using Blastp and SignalP 3.0 (2), four putative nucleases were identified in C. jejuni strain RM1221 (CJE0556, CJE0566, CJE0575, and CJE1441), of which only CJE0566 and CJE1441 contained a functional signal peptide cleavage site, indicative of possible periplasmic localization. Both the CJE0566 and CJE1441 protein sequences contained a conserved domain of the NUC superfamily (SMART accession no. SM00477) characteristic for DNA/RNA nonspecific endonucleases (11, 12). Moreover, the translated sequences contained a DRGH motif (see Fig. S1 in the supplemental material) characteristic for the active site of members of the Serratia nuclease subfamily (26). Predicted structural models of CJE0566 and CJE1441 showed similarities with the crystal structure of NucA from Anabaena sp. (Fig. 1), which is also an extracellular nuclease of the Serratia nuclease family.
FIG. 1.
Comparison of predicted structural models of proteins encoded by CJE0566 and CJE1441 from C. jejuni strain RM1221 with the structural model of NucA from Anabaena sp. The structures were generated using Swiss-Model (1). The residues DRGH are shown in white (CJE0566 and CJE1441) or black (NucA).
Taken together, the analysis suggests that CJE0566 and CJE1441 may code for secreted nucleases that belong to the Serratia nuclease family.
DNA/RNA nonspecific endonucleases in C. jejuni.
Genome database analysis indicated CJE0566 and CJE1441 homologues in 11 out of the 12 available C. jejuni genome sequences, which included the DNase− strains NCTC 11168 (Cj0594c) and 260.94 (CJJ26094_0651) (13). In addition to CJE0566 and CJE1441, a third nuclease was found in strain RM1221 (CJE0697). Closer inspection of the putative nucleases indicated that they all code for proteins of 216 amino acids with a decreased probability of signal peptide cleavage (SignalP 3.0 [2]) and a mutation in the DRGH motif (D→T; see Fig. S1 in the supplemental material). These differences may explain the absence of DNase activity in the strains NCTC 11168 and 260.94. Overall, the results indicate that homologues of the putative nucleases encoded by CJE0566 and CJE1441 are widely spread throughout the C. jejuni species but that perhaps only a subset of these homologues is functional.
Cloning and sequencing of three C. jejuni DNA/RNA nonspecific endonucleases.
The presence of CJE0566 and CJE1441 homologues in the nonnaturally transformable DNase+/dns-negative C. jejuni strains was confirmed by PCR using primer sets based on the coding sequences of both genes from strain RM1221 (data not shown). Detailed genomic indexing of all CJIE2 and CJIE4 genes of strain RM1221 in all nonnaturally transformable DNase+/dns-negative C. jejuni strains revealed that the genes flanking CJE0566 and CJE1441 in RM1221 were not present in these strains, with the exception of CJE0565 and CJE0567 in strain 07479 (see Table S2 in the supplemental material). From this strain, gene CJE0566 was successfully amplified by PCR. This gene codes for a protein of 217 amino acids and contains a DRGH motif and a signal peptide with a high cleavage probability (see Fig. S1 in the supplemental material).
To functionally characterize CJE0566 from strain 07479 and the homologues from strain RM1221, the three genes were cloned and introduced into E. coli strain DH5α. DNA hydrolysis assays using DNase agar with toluidine blue demonstrated that each of the plasmids converted the E. coli phenotype from DNase− into DNase+ (data not shown), indicating that CJE0566 and CJE1441 from strain RM1221 and CJE0566 from strain 07479 indeed code for functional DNases.
Inhibition of natural transformation by DNA/RNA nonspecific endonucleases.
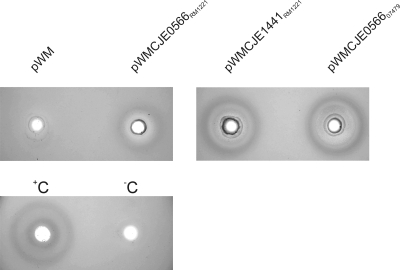
To investigate the effect of different DNases on the process of natural transformation in C. jejuni, the three genes encoding the proteins were introduced via electrotransformation into the naturally transformable DNase− C. jejuni strain C019168 using the expression plasmid pWM1007Pr1492 (36). As a control, the same vector without an insert was introduced. DNA hydrolysis assays demonstrated the presence of DNA-hydrolyzing activities for the transformants carrying the CJE0566RM1221, CJE1441RM1221, and CJE056607479 plasmids, but not for the control strain carrying the empty vector (Fig. 2).
FIG. 2.
Effects of CJE0566RM1221, CJE1441RM1221, and CJE056607479 on the DNase activity of C. jejuni. The three genes were placed on expression plasmid pWM1007Pr1492 (pWM) and introduced into C. jejuni strain C019168. DNase activity of polymyxin B-treated C. jejuni transformants was detected using DNase agar supplemented with toluidine blue. +C, with DNase I; −C, with 500 mM Tris-HCl (pH 7.5).
To determine the effect of the presence of DNase activity on natural transformation, the derivatives of C. jejuni strain C019168 were transformed with homologous chromosomal DNA with a chloramphenicol resistance cassette in hipO (C019168hipO::cat). This construct demonstrated a 35- to 40-fold decrease in the natural transformation frequencies for the DNase-positive strains compared to the control strain carrying the empty plasmid (P < 0.01) (Table 3). These results indicate that the nucleases encoded by CJE0566 and CJE1441 from C. jejuni strain RM1221 and their homologue from strain 07479 impair natural transformation in DNase+/dns-negative C. jejuni strains.
TABLE 3.
Effects of CJE0566RM1221, CJE1441RM1221, and CJE056607479 on the natural transformation frequency of C. jejuni
| Strain | Natural transformation frequencya |
|---|---|
| C019168(pWM1007Pr1492) | (1.1 ± 0.1) × 10−4 |
| C019168(pWM1007Pr1492CJE0566RM1221) | (3.1 ± 1.0) × 10−6b |
| C019168(pWM1007Pr1492CJE1441RM1221) | (2.8 ± 2.0) × 10−6b |
| C019168(pWM1007Pr1492CJE056607479) | (2.6 ± 3.3) × 10−6b |
The natural transformation frequency was determined by dividing the number of transformants per milliliter by the total number of CFU per milliliter. The values are the averages of three experiments.
P < 0.01 by Student's t test for the comparison with the control strain carrying the empty expression plasmid (pWM1007Pr1492).
DISCUSSION
The species C. jejuni is naturally transformable, but the efficiency of transformation is variable, and even nontransformable strains exist (32, 34). Many nonnaturally transformable C. jejuni strains express DNase activity. For a subset of strains this is caused by a periplasmic DNase encoded by dns (13). A second subset of nonnaturally transformable C. jejuni strains, however, contains DNase activity in the absence of dns (13). The present study provides evidence that these DNase+/dns-negative strains contain genes coding for DNA/RNA nonspecific endonucleases that are homologous to CJE0566 and CJE1441 from C. jejuni strain RM1221. These genes confer DNase activity to E. coli and inhibit natural transformation of a naturally transformable C. jejuni strain. These results demonstrate that these genes code for DNases and that besides Dns, which is a member of the endonuclease I family, also DNases of the DNA/RNA nonspecific endonuclease family influence the natural transformability of C. jejuni.
The novel C. jejuni DNases were identified by analysis of comparative genomic hybridization of dns-positive and dns-negative nonnaturally transformable DNase+ strains. This showed that the integrated elements CJIE2 and CJIE4 contained genes that are present in the dns-negative strains but absent in the dns+ strains. CJIE2 and CJIE4 are two of the three phage-related integrated elements present in C. jejuni strain RM1221 but absent in strain NCTC 11168 (10). The presence of these integrated elements in other C. jejuni strains and their diversity between strains (see Table S2 in the supplemental material) are in agreement with earlier reports (6, 25). The majority of CJIE2 and CJIE4 genes present in DNase+/dns-negative C. jejuni strains encode hypothetical proteins, but the homologues of CJE0566 (CJIE2) and CJE1441 (CJIE4) are annotated as DNA/RNA nonspecific endonucleases, a family of endonucleases that is widely distributed among prokaryotic and eukaryotic species (26). Indeed, the predicted protein structures of CJE0566 and CJE1441 resembled the three-dimensional (3D) structure of NucA from Anabaena sp. (Fig. 1), a member of the Serratia nuclease family which belongs to the group of DNA/RNA nonspecific endonucleases (14, 28). CJE0566 and CJE1441 are very similar at the amino acid level and display considerable similarity to Cla_0934 from C. lari RM2100. In contrast to CJE0566 and CJE1441, Cla_0934 is not located on integrated element CLIE1 in C. lari, although CLIE1 is very similar to CJIE4 of C. jejuni (10).
Members of the Serratia nuclease family contain a so-called DRGH motif {DRGH[QLIM]-X(3)-[AG]; accession no. PDOC00821} in which histidine is the active site residue (11, 12). Although the amino acid sequences and the predicted 3D structures of CJE0566 and CJE1441 share similarity with members of this family, the sequences deviate from the DRGH motif in their fifth residue (Q→T). Evidently, the presence of a threonine at this position in the gene products of CJE0566 and CJE1441 does not affect activity, as expression of these genes in DNase− E. coli renders the cells DNase positive. The observed inhibition of natural transformation after introduction of the DNase-encoding genes into the DNase-negative C. jejuni provides functional evidence that, besides dns (13), additional DNases may impair genetic transfer via natural transformation (Table 3). It should be noted that although functionally similar, Dns does not belong to the Serratia nuclease protein family, as evidenced by the absence of the Serratia nuclease-specific DRGH motif.
In silico analysis of C. jejuni genomes revealed that 11 out of 12 C. jejuni genomes available in the public databases contain homologues of CJE0566/CJE1441, including the DNase− strains NCTC 11168 and 260.94 (13). The DNase− phenotype of these strains may be caused by differences in protein expression levels but more likely can be attributed to amino acid changes. We expect the nucleases from DNase− strains to be inactive, as the first amino acid of the conserved DRGH motif is missing. Serratia nuclease mutagenesis of this aspartic acid results in reduced enzyme activity (11, 12). In fact, when searching the SMART nonredundant database (http://smart.embl-heidelberg.de) (20, 27) for proteins with NUC domains (accession no. SM00477), it appeared that among 183 entries of bacterial origin approximately 20% are predicted to be inactive due to amino acid substitutions in the catalytic site. Besides the incomplete DRGH motif, the inactive C. jejuni proteins also appear to contain a degenerated signal peptide sequence which may hamper transport to the periplasm. Changes in protein localization (from cytoplasmic to periplasmic) can be beneficial to a host for some properties (17). However, we consider it unlikely that nucleases from DNase− strains are functionally expressed in the cytoplasm, as induced high-level expression in E. coli resulted in the cessation of growth (unpublished observation). The apparent degeneration of DNase-encoding genes in some C. jejuni strains may indicate that the presence of this trait brings no selective advantage to these strains.
Finally, our findings indicate that C. jejuni strains have acquired phage-like integrated elements that code for different types of periplasmic DNases. These elements are genetically variable and widely spread through the C. jejuni population, thereby contributing to genetic diversity of the species. On the other hand, the encoded DNases limit horizontal gene transfer in C. jejuni through inhibition of natural transformation. This suggests that bacteriophages may both create and restrain genetic diversity of the species C. jejuni.
Supplementary Material
Acknowledgments
This work was supported by the Product Boards of Livestock, Meat, and Eggs, Zoetermeer, The Netherlands. E.J.G. received a travel grant from The Netherlands Organization for Scientific Research. C.T.P. and M.R.G. were supported by the United States Department of Agriculture, Agricultural Research Service CRIS project 5325-42000-045.
We thank J. A. Frost, W. F. Jacobs-Reitsma, and M. M. S. M. Wösten for kindly supplying C. jejuni strains and vectors, L. van der Graaf-van Bloois and A. G. de Boer for technical support, and M. R. de Zoete for helpful advice.
Footnotes
Published ahead of print on 18 December 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Arnold, K., L. Bordoli, J. Kopp, and T. Schwede. 2006. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22:195-201. [DOI] [PubMed] [Google Scholar]
- 2.Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783-795. [DOI] [PubMed] [Google Scholar]
- 3.Butzler, J. P. 2004. Campylobacter, from obscurity to celebrity. Clin. Microbiol. Infect. 10:868-876. [DOI] [PubMed] [Google Scholar]
- 4.Chen, I., P. J. Christie, and D. Dubnau. 2005. The ins and outs of DNA transfer in bacteria. Science 310:1456-1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, I., and D. Dubnau. 2004. DNA uptake during bacterial transformation. Nat. Rev. Microbiol. 2:241-249. [DOI] [PubMed] [Google Scholar]
- 6.Clark, C. G., and L. K. Ng. 2008. Sequence variability of Campylobacter temperate bacteriophages. BMC Microbiol. 8:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Boer, P., J. A. Wagenaar, R. P. Achterberg, J. P. M. van Putten, L. M. Schouls, and B. Duim. 2002. Generation of Campylobacter jejuni genetic diversity in vivo. Mol. Microbiol. 44:351-359. [DOI] [PubMed] [Google Scholar]
- 8.Dingle, K. E., F. M. Colles, D. R. A. Wareing, R. Ure, A. J. Fox, F. E. Bolton, H. J. Bootsma, R. J. L. Willems, R. Urwin, and M. C. J. Maiden. 2001. Multilocus sequence typing system for Campylobacter jejuni. J. Clin. Microbiol. 39:14-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dutta, C., and A. Pan. 2002. Horizontal gene transfer and bacterial diversity. J. Biosci. 27:27-33. [DOI] [PubMed] [Google Scholar]
- 10.Fouts, D. E., E. F. Mongodin, R. E. Mandrell, W. G. Miller, D. A. Rasko, J. Ravel, L. M. Brinkac, R. T. DeBoy, C. T. Parker, S. C. Daugherty, R. J. Dodson, A. S. Durkin, R. Madupu, S. A. Sullivan, J. U. Shetty, M. A. Ayodeji, A. Shvartsbeyn, M. C. Schatz, J. H. Badger, C. M. Fraser, and K. E. Nelson. 2005. Major structural differences and novel potential virulence mechanisms from the genomes of multiple Campylobacter species. PLoS Biol. 3:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Friedhoff, P., O. Gimadutdinow, and A. Pingoud. 1994. Identification of catalytically relevant amino acids of the extracellular Serratia marcescens endonuclease by alignment-guided mutagenesis. Nucleic Acids Res. 22:3280-3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Friedhoff, P., B. Kolmes, O. Gimadutdinow, W. Wende, K. L. Krause, and A. Pingoud. 1996. Analysis of the mechanism of the Serratia nuclease using site-directed mutagenesis. Nucleic Acids Res. 24:2632-2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaasbeek, E. J., J. A. Wagenaar, M. R. Guilhabert, M. M. S. M. Wösten, J. P. M. van Putten, L. van der Graaf-van Bloois, C. T. Parker, and F. J. van der Wal. 2009. A DNase encoded by integrated element CJIE1 inhibits natural transformation of Campylobacter jejuni. J. Bacteriol. 191:2296-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghosh, M., G. Meiss, A. M. Pingoud, R. E. London, and L. C. Pedersen. 2007. The nuclease A-inhibitor complex is characterized by a novel metal ion bridge. J. Biol. Chem. 282:5682-5690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hänninen, M. L. 1989. Rapid method for the detection of DNase of campylobacters. J. Clin. Microbiol. 27:2118-2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harrington, C. S., F. M. Thomson-Carter, and P. E. Carter. 1999. Molecular epidemiological investigation of an outbreak of Campylobacter jejuni identifies a dominant clonal line within Scottish serotype HS55 populations. Epidemiol. Infect. 122:367-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hofreuter, D., V. Novik, and J. E. Galán. 2008. Metabolic diversity in Campylobacter jejuni enhances specific tissue colonization. Cell Host Microbe 4:425-433. [DOI] [PubMed] [Google Scholar]
- 18.Jett, B. D., K. L. Hatter, M. M. Huycke, and M. S. Gilmore. 1997. Simplified agar plate method for quantifying viable bacteria. Biotechniques 23:648-650. [DOI] [PubMed] [Google Scholar]
- 19.Kruskal, J. B. 1983. An overview of sequence comparison, p. 1-44. In D. Sankoff and J. B. Kruskal (ed.), Time warps, string edits, and macromolecules: the theory and practise of sequence comparison. Addison Wesley, Reading, MA.
- 20.Letunic, I., T. Doerks, and P. Bork. 2009. SMART 6: recent updates and new developments. Nucleic Acids Res. 37:D229-D232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lior, H. 1984. New, extended biotyping scheme for Campylobacter jejuni, Campylobacter coli, and “Campylobacter laridis.” J. Clin. Microbiol. 20:636-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller, W. G., A. H. Bates, S. T. Horn, M. T. Brandl, M. R. Wachtel, and R. E. Mandrell. 2000. Detection on surfaces and in Caco-2 cells of Campylobacter jejuni cells transformed with new gfp, yfp, and cfp marker plasmids. Appl. Environ. Microbiol. 66:5426-5436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Misawa, N., B. M. Allos, and M. J. Blaser. 1998. Differentiation of Campylobacter jejuni serotype O19 strains from non-O19 strains by PCR. J. Clin. Microbiol. 36:3567-3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Needleman, S. B., and C. D. Wunsch. 1970. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol. 48:443-453. [DOI] [PubMed] [Google Scholar]
- 25.Parker, C. T., B. Quinones, W. G. Miller, S. T. Horn, and R. E. Mandrell. 2006. Comparative genomic analysis of Campylobacter jejuni strains reveals diversity due to genomic elements similar to those present in C. jejuni strain RM1221. J. Clin. Microbiol. 44:4125-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rangarajan, E. S., and V. Shankar. 2001. Sugar non-specific endonucleases. FEMS Microbiol. Rev. 25:583-613. [DOI] [PubMed] [Google Scholar]
- 27.Schultz, J., F. Milpetz, P. Bork, and C. P. Ponting. 1998. SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. U. S. A. 95:5857-5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shlyapnikov, S. V., V. V. Lunin, M. Perbandt, K. M. Polyakov, V. Y. Lunin, V. M. Levdikov, C. Betzel, and A. M. Mikhailov. 2000. Atomic structure of the Serratia marcescens endonuclease at 1.1 Å resolution and the enzyme reaction mechanism. Acta Crystallogr. D Biol. Crystallogr. 56:567-572. [DOI] [PubMed] [Google Scholar]
- 29.Suerbaum, S., M. Lohrengel, A. Sonnevend, F. Ruberg, and M. Kist. 2001. Allelic diversity and recombination in Campylobacter jejuni. J. Bacteriol. 183:2553-2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thomas, C. M., and K. M. Nielsen. 2005. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat. Rev. Microbiol. 3:711-721. [DOI] [PubMed] [Google Scholar]
- 31.Waller, J. R., S. L. Hodel, and R. N. Nuti. 1985. Improvement of two toluidine blue O-mediated techniques for DNase detection. J. Clin. Microbiol. 21:195-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang, Y., and D. E. Taylor. 1990. Natural transformation in Campylobacter species. J. Bacteriol. 172:949-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wassenaar, T. M., B. N. Fry, A. J. Lastovica, J. A. Wagenaar, P. J. Coloe, and B. Duim. 2000. Genetic characterization of Campylobacter jejuni O:41 isolates in relation with Guillain-Barre syndrome. J. Clin. Microbiol. 38:874-876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wassenaar, T. M., B. N. Fry, and B. A. M. van der Zeijst. 1993. Genetic manipulation of Campylobacter: evaluation of natural transformation and electro-transformation. Gene 132:131-135. [DOI] [PubMed] [Google Scholar]
- 35.Wiesner, R. S., and V. J. DiRita. 2008. Natural competence and transformation in Campylobacter, p. 559-570. In I. Nachamkin, C. M. Szymanski, and M. J. Blaser (ed.), Campylobacter, 3rd ed. ASM Press, Washington, DC.
- 36.Wösten, M. M. S. M., C. T. Parker, A. van Mourik, M. R. Guilhabert, L. van Dijk, and J. P. M. van Putten. 2006. The Campylobacter jejuni PhosS/PhosR operon represents a non-classical phosphate-sensitive two-component system. Mol. Microbiol. 62:278-291. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.