Abstract
To determine the relative importance of temperate bacteriophage in the horizontal gene transfer of fitness and virulence determinants of Enterococcus faecalis, a panel of 47 bacteremia isolates were treated with the inducing agents mitomycin C, norfloxacin, and UV radiation. Thirty-four phages were purified from culture supernatants and discriminated using pulsed-field gel electrophoresis (PFGE) and restriction mapping. From these analyses the genomes of eight representative phages were pyrosequenced, revealing four distinct groups of phages. Three groups of phages, ΦFL1 to 3, were found to be sequence related, with ΦFL1A to C and ΦFL2A and B sharing the greatest identity (87 to 88%), while ΦFL3A and B share 37 to 41% identity with ΦFL1 and 2. ΦFL4A shares 3 to 12% identity with the phages ΦFL1 to 3. The ΦFL3A and B phages possess a high DNA sequence identity with the morphogenesis and lysis modules of Lactococcus lactis subsp. cremoris prophages. Homologs of the Streptococcus mitis platelet binding phage tail proteins, PblA and PblB, are encoded on each sequenced E. faecalis phage. Few other phage genes encoding potential virulence functions were identified, and there was little evidence of carriage of lysogenic conversion genes distal to endolysin, as has been observed with genomes of many temperate phages from the opportunist pathogens Staphylococcus aureus and Streptococcus pyogenes. E. faecalis JH2-2 lysogens were generated using the eight phages, and these were examined for their relative fitness in Galleria mellonella. Several lysogens exhibited different effects upon survival of G. mellonella compared to their isogenic parent. The eight phages were tested for their ability to package host DNA, and three were shown to be very effective for generalized transduction of naive host cells of the laboratory strains OG1RF and JH2-2.
Enterococcus faecalis is a member of the natural flora of humans and colonizes the gastrointestinal and vaginal tracts and the oral cavity. In recent years it has emerged as an important opportunistic nosocomial pathogen and is a causative agent of bacteremia, infective endocarditis, and surgical wound and urinary tract infections. The accumulation of acquired antibiotic resistance determinants, in addition to its intrinsic resistance and tenacity, has given rise to the evolution of clinical isolates of E. faecalis that are therapeutically problematic (19). Greater notoriety was afforded to this species following the observed transfer of the conjugative transposon Tn1546 to Staphylococcus aureus, imparting vancomycin resistance (11). Subsequent analysis has revealed that multiple independent E. faecalis-dependent vanA transfers had occurred in the United States prior to 2007 (50). This places enterococci in an important and dynamic position within the health care system, warranting their increased study.
The specific determinants that are proposed to contribute to the virulence of E. faecalis are not universally present, and expression of the cognate genes is variable (21, 37). For example, in a recent study of 106 clonally diverse strains of E. faecalis the metallopeptidase gelatinase (GelE) was shown to be expressed in less than 60% of 106 genotypically positive isolates, whereas expression of cytolysin was less frequently observed (expression in ∼25% of isolates, with 30% being genotypically positive) (33). A proposed pathogenicity island identified with E. faecalis V583 (49) is composed of a variable gene set encoding the virulence determinants enterococcal surface protein, cytolysin, and aggregation substance. This highly variable 150-kb mobile element contains many components of unknown function that are hypothesized to facilitate survival and/or transmission in the health care setting (34, 40, 49).
Two sequenced and annotated genomes of E. faecalis have been completed and published to date. These are the blood isolate and first-observed vancomycin-resistant strain V583 (40) and the oral isolate OG1RF, used as a common laboratory strain (8). A major difference between these genomes is the presence in V583 of seven regions containing phage-associated sequences. In contrast, OG1RF contains only one phage remnant, which was proposed by McBride et al. (33) to form part of the core genome, a theory supported by the presence in OG1RF of this phage remnant region together with two CRISPR loci. CRISPR sequences provide sequence-specific resistance to bacteriophages via the assembly of phage DNA sequences interspersed as spacers between repeats, in concert with associated cas genes, which collectively operate as an RNA-based gene silencing mechanism (5, 6, 28, 30, 36, 42). This elegant heritable mechanism is proposed to limit horizontal gene transfer of bacteriophage, transposable elements, and conjugative plasmids (9, 10, 32).
Within the firmicute division of Gram-positive bacteria, temperate bacteriophages are key vectors for the horizontal transfer of virulence genes. In Staphylococcus aureus, bacteriophages encode and mobilize an impressive array of immune evasion genes (54, 55) and Panton-Valentine leukocidin (43). Several bacteriophage-encoded virulence determinants also contribute to pathogenesis in group A Streptococcus (2, 3, 4).
The role of bacteriophages in the virulence of E. faecalis is not clear. Encoded within seven phage-related sequences of strain V583, there are multiple reported homologs of the Streptococcus mitis platelet-binding proteins PblA and PblB (7) and a ferrochelatase (40). In contrast, the absence of mobile genetic elements (MGEs) in strain OG1RF led Bourgogne et al. (8) to speculate that they did not engender virulence in E. faecalis.
In this study we determined the morphology and complete genome sequences of eight induced bacteriophages purified from clinical isolates of E. faecalis. We sought to determine the potential carriage of genes that might contribute to the virulence or fitness of this organism and characterize the capacity of these phages to participate in transduction.
MATERIALS AND METHODS
Bacterial growth.
E. faecalis strains and plasmids used in this study are listed in Table 1. Bacterial strains were cultured in brain heart infusion (BHI) broth (Merck) or LB medium (Lab M). Chemicals were obtained from Sigma-Aldrich Co., United Kingdom, unless otherwise indicated. E. faecalis indicator strains were grown and maintained on LB medium supplemented with 1 mM CaCl2 at 37°C. Antibiotics were included in overnight cultures, when appropriate, at the following concentrations: 2 mg ml−1 kanamycin and 10 μg ml−1 erythromycin (Sigma).
TABLE 1.
Bacterial strains used in this study
| Strain or primer | Relevant characteristics or sequence | Reference and/or source |
|---|---|---|
| Strains | ||
| OG1RF | Lab strain, gelatinase producer (Rifr Fusr) | 15 |
| JH2-2 | Lab strain, gelatinase negative (Rifr Fusr) | 20 |
| V583 | Clinical isolate | 47 |
| NCTC775 | Type culture strain | NCTC, UK |
| TX5128 | OG1RF gelE sprE | 52 |
| TX5264 | OG1RF gelE | 51 |
| TX5243 | OG1RF sprE | 44 |
| LIV305 | OG1RF fsrB pTEX5249(pAT18-fsrABCD) | This study |
| LIV306 | nox::Tn917 | This study |
| LIV510 | JH2-2 pTEX5249(pAT18-fsrABCD) | This study |
| MMH594b | PAI::Cm | N. Shankar |
| LIV079-082 | Bacteremia isolates from UK | 60 |
| LIV162-167 | Bacteremia isolates from UK | 60 |
| LIV375-381 | Bacteremia isolates from Poland | M. Kawalec |
| LIV410-439 | MLST-typed bacteremia isolates from Spain | 45 |
| LIV1037 | JH2-2 ΦFL1A | This study |
| LIV1038 | JH2-2 ΦFL1B | This study |
| LIV1039 | JH2-2 ΦFL1C | This study |
| LIV1040 | JH2-2 ΦFL2A | This study |
| LIV1041 | JH2-2 ΦFL2B | This study |
| LIV1042 | JH2-2 ΦFL3A | This study |
| LIV1043 | JH2-2 ΦFL3B | This study |
| LIV1044 | JH2-2 ΦFL4A | This study |
| Primers | ||
| INT1-1 | TTGTGGTTTATCCAGATGG | This study |
| INT1-3 | AAACAGCTAACTGTTTCTG | This study |
| INT4-1 | GTTAAGACCCTAAACGAG | This study |
| INT4-3 | AGGACAGAGAGACAACTG | This study |
| EF0302 | GCTACGCAAGGGCTAGACTC | This study |
| EF0358 | GCGTCCTTTCGATACTTC | This study |
| EF1275 | CGGAAATCACATTGATGAGTTG | This study |
| EF1292 | CCACGTTCCTTCCATAACGG | This study |
| EF1416 | TAAACCAGGATTTGAAGAATTAGC | This study |
| EF1490 | GGAAGGAACTATTTGGAGTCCTA | This study |
| EF1987 | AATACCGCAAATCTGTGGTAA | This study |
| EF2044 | CGAAGGCTACATTACTAAAGAGCAG | This study |
| EF2083 | GGTAGAGCACCACCATGGTAAGG | This study |
| EF2146 | GATTCTGTAGGTAATTGTTTCATC | This study |
| EF2797 | CCTCATCTAGCGAGCTCAACTC | This study |
| EF2856 | CAAAAATATTGCCCACGTGAAAG | This study |
| EF2935 | GGTGAAACCAGCTAACATTTCTCG | This study |
| EF2956 | AGACACGCCAGATCGTATGTC | This study |
Induction, propagation, and purification of phages.
Two E. faecalis strains were used as hosts for propagation: JH2-2 and NCTC775 (Table 1). For the induction of phages, three inducing agents were used (mitomycin C and norfloxacin at 1 to 3 μg ml−1 or UV), with bacterial culture on LB supplemented with either 1 mM MgSO4 or 1 mM CaCl2. Overnight cultures were diluted 10-fold in 10 ml of fresh broth, grown to an optical density at 600 nm (OD600) of 0.5 to 0.6 prior to the addition of inducing agent, and incubated for 2 to 3 h. Supernatants were filtered (0.2-μm membrane), and 10 to 50 μl of the filtrate was mixed with 100 μl of host cells and added to 5 ml of top agar (0.4% [wt/vol] agar) at 40 to 45°C and poured on agar plates.
To induce phages using UV radiation, cultures were harvested when the OD600 was 0.4 to 0.5. Cells were washed, resuspended in 1 mM CaCl2, poured into petri dishes, and exposed to UV (366 nm) for 40 to 60 s. One ml of the treated cells was added into 10 ml of fresh LB supplemented with CaCl2 and grown for 2 h at 37°C prior to plaque assay. Induced phages were further propagated and purified using the soft agar overlay method (48).
Host range determination.
The host range of different phages was determined using 49 different isolates of E. faecalis, including strains OG1RF and JH2-2. Bacteria were grown to exponential phase, and 100 μl of the bacterial culture was mixed in 5 ml of top agar and poured onto the LB agar plates. Spot assays were performed using 5 to 10 μl of the purified phage (12). Plates were incubated at 37°C for 24 h, and clear zones were recorded as a positive test.
Extraction of phage DNA.
Purified phages were precipitated with PEG-8000, centrifuged, and resuspended in SM buffer (48). Concentrated phages were repeatedly treated with DNase/RNase (Ambion, United Kingdom) to remove host nucleic acids prior to phage genomic DNA isolation by organic extraction. Purified DNA was precipitated with isopropanol and sodium acetate before it was washed in 70% [vol/vol] ethanol and suspended in deionized water.
PFGE.
Purified phage DNA was mixed with DNA sample buffer and separated using a CHEF-DR III system (Bio-Rad) at 6 V cm−1 and pulse ramps from 0.5 to 10 s for 12 h at 14°C in 0.5× Tris-borate-EDTA (TBE) buffer. Following pulsed-field gel electrophoresis (PFGE), the DNA was visualized by staining with ethidium bromide.
Electron microscopy.
Purified bacteriophages were used to produce fresh particles using the soft-agar overlay method, and single plaques were picked and mixed in 20 μl of sterilized distilled water, and then 10 μl of the suspension was absorbed onto Formvar-coated copper grids for 10 to 15 min and negatively stained with 3% (wt/vol) of uranyl acetate (pH 4). Morphology of the phages was observed, and images were obtained using a Tecnai G2 twin transmission electron microscope (TEM).
DNA sequencing and bioinformatic analysis.
Pyrosequencing of bacteriophage genomes was performed by generating standard fragment template DNA libraries, which were multiplex identifier (MID) tagged to allow multiple samples to be run in a single plate region, using the GS-FLX DNA library preparation kit (Roche Applied Sciences, United States); amplifying by emPCR; and sequencing on a GS-FLX (454 Life Sciences, United States). The 454 reads were assembled with Newbler (version 2.0) (Roche, United States) using default assembly parameters. Protein-coding sequences (CDS) were identified with Glimmer (13) and GeneMark (29). Putative functions were inferred using BLAST against the National Center for Biotechnology Information databases (1) and InterProScan (18). Artemis version 11 was used to organize data and facilitate annotation (46). Orthologs were defined using OrthoMCL (27). Comparative genome analysis was performed using MUMmer version 3 (24) and visualized using CIRCOs (23).
Lysogen generation.
Lysogens of JH2-2 were generated after phage infection of the strain by use of the eight sequenced phages by harvesting colonies from semiconfluent lysis plates. The clones were propagated for several rounds of growth on agar plates, and their ability to generate phages after UV induction, as described above, was tested. Released phages were tested for their host range, which was determined using the hosts tested in the original phage isolations and by PCR amplification of the endolysin gene. Confirmed lysogens were cultured and observed to grow with growth rates and viable counts identical to those of the isogenic parent strain JH2-2 (data not shown). Chromosomal integration sites of phages were determined via two PCR screens. First, JH2-2 lysogen DNA was amplified using primers for integrase (primers int1-1 or int1-3 for phages FL1A to C, ΦFL2A and B, and ΦFL3A and B, or primers int4-1 or int4-3 for phage ΦFL4A) (Table 1) with primers for genes adjacent to phage insertion sites that were identified by comparative genome hybridization studies (26, 34). Second, amplification was performed across the phage junction sites using primers for EF0302/EF0358, EF1275/EF1292, EF1416/EF1490, EF1987/EF2044, EF2083/EF2146, EF2797/EF2856, and EF2935/EF2956 (Table 1) for the JH2-2 lysogens and JH2-2.
Galleria mellonella infection.
Caterpillars of Galleria mellonella were infected as described previously (25, 38). Briefly, caterpillars (2.5 to 3 cm, 300 mg) were inoculated subcutaneously using a sterile syringe needle and washed cells from exponential-phase cultures (OD600 = 0.5) of JH2-2 grown in BHI. Cells were resuspended in sterile water containing 25 μg ml−1 rifampin (Sigma) before injecting a 10-μl volume, equivalent to 8 × 106 CFU, through the last proleg. Bacillus subtilis 168 was used as an avirulent bacterium control, and buffer containing rifampin was used as an inoculation control. No loss of viability in the control groups was observed with each assay repeat. Triplicate experiments were conducted, and each comprised for each strain being tested 20 caterpillars, which were monitored for viability at four hourly time intervals postinoculation during incubation at 37°C. Statistical analysis was performed using Student's t test.
Transduction using identified phages.
Transduction experiments were performed using methods modified from those described previously (17). Lysates of donor strains containing antibiotic resistance markers were generated by mixing 5 ml of cells (OD600 = 0.05) cultured in LB with 5 ml of phage buffer (50 mM Tris-HCl, pH 7.8; 4 mM CaCl2; 1 mM MgSO4;10 mM NaCl; 1% [wt/vol] gelatin) and 50 μl of phage stock (∼1010 PFU ml−1) and incubating them at 30°C until complete lysis was observed. A total of 0.5 ml of recipient cells (JH2-2 or OG1RF), consisting of 10-fold concentrated overnight culture in LB, was mixed with 0.5 ml of sterile, filtered donor lysate, and 1 ml of LB containing 10 mM CaCl2. After 20 min incubation at 37°C, the cultures were shaken at 250 rpm for 15 min. Cells were then pelleted, washed, and resuspended in 1 ml of 10 mM sodium citrate and incubated on ice for 20 min. Aliquots were spread onto LB agar containing 20 mM sodium citrate, incubated at 37°C for 60 to 90 min, and overlaid with 5 ml of molten LB with 0.7% [wt/vol] agar containing selective concentrations of antibiotic. Plates were incubated at 37°C for 24 to 48 h. PCR amplification of kanamycin or erythromycin genes was performed to confirm transduction using DNA primers described previously (22).
Nucleotide sequence accession number.
DNA sequence data of the phages have been deposited in GenBank under accession numbers no. GQ478081 (ΦFL1A), GQ478082 (ΦFL1B), GQ478083 (ΦFL1C), GQ478084 (ΦFL2A), GQ478085 (ΦFL2B), GQ478086 (ΦFL3A), GQ478087 (ΦFL3B), and GQ478088 (ΦFL4A).
RESULTS AND DISCUSSION
Prophage induction and distribution.
The presence of inducible temperate bacteriophages in the genomes of 47 E. faecalis isolates was investigated by exposing them to chemical and physical inducing agents. Plaque assays were performed using the induced culture supernatants and determined that 17 (36%) of the tested isolates were lysogenic, and based upon differences in plaque size and appearance, 12 (26%) of these isolates were polylysogenic. Each distinguishable plaque type was picked and propagated independently, using strain JH2-2 as host, for three successive rounds of infection to ensure phage homogeneity. Ultimately, 34 phages were identified from the 17 lysogens. DNA was then purified from high-titer stocks of each phage to discriminate them further by using PFGE to size the genomes and restriction fragment length polymorphism (RFLP) with HaeIII and RsaI endonucleases. RFLP indicated that there were four major groups of phages present among the 34 tested, based upon clear similarities that were observed in the fragmentation patterns (data not shown).
The abilities of the 34 phages to lyse different E. faecalis hosts were assayed in spot plaque tests using, as indicator strains, the original 47 bacterial isolates that were screened for lysogeny, plus the laboratory strains OG1RF and JH2-2. This revealed that the laboratory strain OG1RF was not a productive host, since none of the phages were capable of consistent replication to produce plaques, in support of the identification of CRISPR loci (8). A small amount of lysis could be observed with many phages at high multiplicities of infection (MOIs), indicating that OG1RF was susceptible to adhesion and infection but that phage replication was limited. Twelve strains (26%) were identified that did not contain inducible prophage and did not plaque any of the 34 purified phages. To identify reasons for this apparent resistance to infection, the isolates were tested, using PCR, for the presence of the CRISPR 1 to 7 and CRISPR 8 to 14 sequences identified with OG1RF by Bourgogne et al. (8), using their published primer sets. None of the 12 bacterial isolates were positive for either CRISPR amplicon, while OG1RF was confirmed to contain these regions (data not shown). This suggests that these specific CRISPR loci do not contribute to the observed phage resistance in these 12 strains. In contrast to OG1RF, the laboratory strain JH2-2 was determined to be a universal indicator strain under the conditions studied here (Table 2). None of the clinical isolates tested was universally sensitive to all 34 phages; however, LIV166 and LIV427 were sensitive to all but two of the 34 phages tested (Table 2; data not shown).
TABLE 2.
Bacteriophage sensitivity of E. faecalis indicator strainsa
| Indicator strain | Observed lysis with purified bacteriophage |
|||||||
|---|---|---|---|---|---|---|---|---|
| ΦFL1A | ΦFL1B | ΦFL1C | ΦFL2A | ΦFL2B | ΦFL3A | ΦFL3B | ΦFL4A | |
| LIV413 (ΦFL1A) | − | − | − | − | − | − | − | + |
| LIV412 (ΦFL1B) | − | − | − | − | + | − | + | + |
| LIV378 (ΦFL1C)* | − | − | − | − | − | − | − | + |
| LIV167 (ΦFL2A)* | − | − | − | − | − | − | − | − |
| LIV165 (ΦFL2B)* | − | − | − | − | − | − | − | − |
| LIV379 (ΦFL3A)* | + | + | − | − | − | − | − | + |
| LIV165 (ΦFL3B)* | − | − | − | − | − | − | − | − |
| LIV080 (ΦFL4A)* | − | − | − | − | − | − | − | − |
| OG1RF | − | − | − | − | − | − | − | − |
| JH2-2 | + | + | + | + | + | + | + | + |
| LIV166 | + | + | + | + | + | + | + | + |
| LIV427 | + | + | + | + | + | + | + | − |
+ indicates a clear zone of lysis. * indicates a polylysogen host. PCR of CRISPR loci was negative for all strains except OG1RF.
Phage morphology.
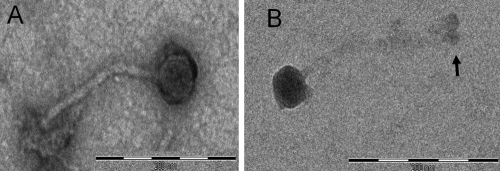
Based upon differences in their host range (Table 2), genome size, and RFLP profiles, eight phages were selected for further study. Their morphology was determined using transmission electron microscopy (Fig. 1), which revealed that they belonged to the Siphoviridae, having isometric capsids approximately 50 nm in breadth and with long noncontractile tails that were 200 to 240 nm in length (Table 3). At the distal end of the tail tip of each phage was a putative cell adhesion, base plate protein structure.
FIG. 1.
Scanning electron micrographs of purified E. faecalis phages induced from their original host strains. (A) ΦFL1A; (B) ΦFL4A. Bar, 200 nm. Arrow indicates the likely tail base plate protein structure.
TABLE 3.
Characteristics of the eight genome-sequenced E. faecalis bacteriophagesa
| Property | Bacteriophages |
|||||||
|---|---|---|---|---|---|---|---|---|
| ΦFL1A | ΦFL1B | ΦFL1C | ΦFL2A | ΦFL2B | ΦFL3A | ΦFL3B | ΦFL4A | |
| Ref no. | 1 | 3 | 20 | 12 | 30 | 4 | 29 | 10 |
| Strain ID | LIV413 | LIV412 | LIV378 | LIV167 | LIV165 | LIV379 | LIV165 | LIV80 |
| Inducing agent | Nor | UV | Nor | Nor | UV | Nor | UV | Mc |
| Genome size (bp) | 38,764 | 38,989 | 38,721 | 36,270 | 36,826 | 39,576 | 40,275 | 37,856 |
| No. of predicted ORFs | 61 | 63 | 74 | 63 | 59 | 64 | 66 | 55 |
| G+C content (mol%) | 36.7 | 34 | 34 | 34.6 | 34.6 | 34.5 | 34.5 | 37.8 |
| Capsid diam (nm) | 48 | 53 | 48 | 48 | 50 | 49 | 51 | 48 |
| Tail length (nm) | 225 | 205 | 243 | 229 | 221 | 220 | 225 | 204 |
Nor, norfloxacin; Mc, mitomycin C; UV, 366 nm; ID, identifier; ORFs, open reading frames.
Genome sequencing of E. faecalis phages.
Pyrosequencing was performed to obtain the genome sequence of the eight selected phages. The sequence was obtained by tagging separately the DNA fragment libraries with unique ligation MIDs, and these MID-tagged samples were pooled and sequenced simultaneously. The correct assembly of the contigs constituting the genomes of each phage was confirmed by PCR amplification across adjoining contigs and by comparing restriction maps predicted in silico with those generated from restriction enzyme analysis (data not shown).
General features of E. faecalis phage genomes.
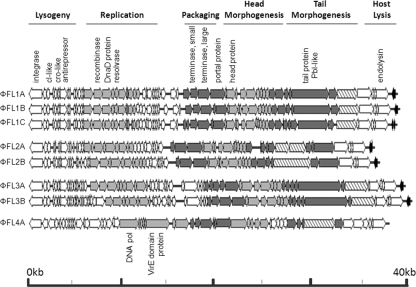
A comparison of the eight phage genome sequences reveals four distinct groups of phages (ΦFL1, ΦFL2, ΦFL3, and ΦFL4) (Table 3), with multiple nonidentical genome sequences exhibiting >97% sequence identity within each of three phage groups (ΦFL1A to C, ΦFL2A and B, and ΦFL3A and B) (Table 4). The eight genomes comprise 36 to 40 kb double-stranded DNA (dsDNA), with an average G+C content of 34 to 38 mol% and between 55 and 74 coding sequences. Their organization is very similar to that of protein coding sequences, forming functional clusters typical of temperate bacteriophages (14, 16) (Fig. 2). The region encoding lysogeny control proteins is transcribed mostly in the direction opposite to those sequences encoding proteins for replication and packaging and that are structural components of the virion and host lysis.
TABLE 4.
Comparative analysis of E. faecalis bacteriophage genomesa
| Phage | % DNA identity |
|||||||
|---|---|---|---|---|---|---|---|---|
| ΦFL1A | ΦFL1B | ΦFL1C | ΦFL2A | ΦFL2B | ΦFL3A | ΦFL3B | ΦFL4A | |
| ΦFL1A | 100 | 99 | 99 | 88 | 87 | 37 | 38 | 5 |
| ΦFL1B | 100 | 99 | 88 | 87 | 37 | 38 | 4 | |
| ΦFL1C | 100 | 88 | 87 | 37 | 38 | 4 | ||
| ΦFL2A | 100 | 99 | 40 | 40 | 12 | |||
| ΦFL2B | 100 | 41 | 41 | 12 | ||||
| ΦFL3A | 100 | 99 | 3 | |||||
| ΦFL3B | 100 | 3 | ||||||
| ΦFL4A | 100 | |||||||
Percentage of identity was determined using Fasta3.
FIG. 2.
Comparative genome alignments of the eight E. faecalis phages. Modular organization is highlighted with gray shading to reveal grouped functions associated with the phage life cycle. Shading indicates functional modules and the tail protein with similarity to the Streptococcus mitis phage M1 platelet-binding protein PblA (stippled). EF1419-like protein(s) is rightward of endolysin in the ΦFL1A to C, ΦFL2A and B, and ΦFL3A and B genomes (shaded black). Genomes are drawn to scale.
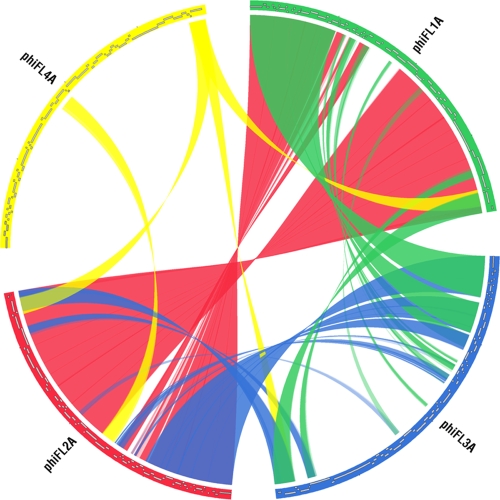
Between the four identified phage groups there is a variable degree of DNA identity (Table 4). ΦFL1 and ΦFL2 share a high degree of DNA identity (87 to 88%), which is spread nonuniformly across the genome. The major variation between the ΦFL1 and ΦFL2 phages resides in the region between the transcribed clusters of genes with putative functions in DNA replication and packaging (Fig. 2 and 3). This variable region consists of different combinations of <10 genes encoding proteins with high levels of sequence identity to those encoded by the EF_1417-EF1489 (phage03) and EF_2084-EF_2145 (phage05) regions of the E. faecalis V583 genome sequence (26, 33, 40). The V583 phage03 and phage05 regions appear to be intact prophages, thereby suggesting that extensive recombination between temperate phages in E. faecalis has generated hybrid phage genomes. Phages ΦFL3A and B and ΦFL4A exhibit a lesser degree of sequence identity with ΦFL1A to C and ΦFL2A and B (37 to 41% and 5 to 12% identity, respectively), and ΦFL3A and B and ΦFL4A share only 3% sequence identity with each other (Table 4; Fig. 3). Although there are lower levels of DNA sequence identity between the ΦFL3A and B and the ΦFL1 and ΦFL2 phages, there was more extensive identity between the encoded proteins of these phage groups (Fig. 3; see Tables SA1 to SA8 in the supplemental material), indicating conserved function. Moreover, the modular structure of the various ΦFL1, ΦFL 2,and ΦFL3 genomes (Fig. 2) was clearly evident in their similar organizations and the high sequence identity of the lysogeny, replication, and lysis modules (Fig. 3) (41).
FIG. 3.
Circos comparison plot of the E. faecalis bacteriophage genome groups conducted using the NUCmer/MUMmer software. Genomes are displayed clockwise and the transparent color ribbons between genomes indicate regions of shared homology between phages ΦFL1A (green), ΦFL2A (red), ΦFL3A (blue), and ΦFL4A (yellow). Coordinates were generated using NUCmer with the parameters breaklen (200), maxgap (90), mincluster (65), and minmatch (20).
ΦFL4A differs markedly from the ΦFL1, ΦFL2, and ΦFL3 phages, in terms of its very low level of DNA sequence identity and the number of unique genes. The E. faecalis V583 strain is proposed to be a lysogen, and its genome contains seven prophage/prophage-like sequences (phage01 to phage07) (26, 33, 40). Of these, ΦFL4A corresponds to the phage01 region, with which it shares 87% DNA sequence identity. The other phages identified in this study have much lower levels of sequence identity (20 to 25%) with the other prophage regions of E. faecalis V583. Induction of strain V583 using the conditions tested in this study did not result in released bacteriophage, as judged by plaque assay with the indicator strains JH2-2 and NCTC775 (data not shown).
Comparative genome analysis of the E. faecalis phages using BLASTn against the GenBank database identified that the ΦFL3A and ΦFL3B phages shared 34% and 31% sequence coverage identity with prophages identified in the Lactobacillus lactis subsp. cremoris SK11 (31) and MG1363 (59) strains, respectively. This sequence identity is confined mostly to the morphogenesis and lysis modules (see Tables SA1 to SA8 in the supplemental material). Recent analyses revealed that the virulent lactococcal phage P087 encodes a morphogenesis module that is similar to the E. faecalis V583 prophage phage05 (57), which led the authors to speculate that recombination could occur between phages infecting these low G+C bacteria. The observation here of identities between prophages of lactococci and enterococci lends support to this hypothesis and indicates that this might have occurred between multiple different phage types. Overall, within the DNA sequences of the eight E. faecalis phages reported here, regions of identity were observed (4 to 8% coverage of genome with >70% identity) with phages from several different low-G+C genera, including Streptococcus pyogenes, Streptococcus agalactiae, and Staphylococcus aureus. The presence of putative DNA cytosine and adenine methyltransferases in both ΦFL3 genomes could help to reduce host barriers to successful infection and potentially facilitate intergenus infection.
Lysogeny maintenance module.
The integration of phage DNA into the host bacterium's chromosome at the attB site is mediated by the phage-encoded integrase. The identified putative integrases of the seven phages in the ΦFL1, ΦFL2, and ΦFL3 groups all belong to the serine recombinase family and possess identical amino acid sequences. In contrast, ΦFL4A encodes an integrase, which belongs to the Tn1545-related family of conjugative transposon integrases. The ΦFL4A integrase displays 99% amino acid identity to the EF0303 integrase protein of the prophage phage01 identified by Paulsen et al. (40). The contributions of prophages in a polylysogen with respect to E. faecalis pathogenesis and fitness will be important to determine. The observation that one quarter of strains tested in this study contained multiple inducible phages reveals that polylysogeny is common, and several strains were found to contain up to 5 distinct inducible phages, as determined by host range.
Lysogenic conversion genes in E. faecalis phages.
Typically genes that are known or proposed to endow enhanced fitness or virulence upon their host and that are carried by many temperate phage of low-G+C bacteria, are located between the gene encoding endolysin and the right phage attachment site, e.g., S. aureus immune escape and Panton-Valentine leukocidin (PVL) genes (14, 43, 54, 55). Examination of the eight E. faecalis phage genomes reveals that ΦFL1 to 3, but not ΦFL4A, possess either one or two coding sequences located in this region. These coding sequences appeared as either a full-length protein or two rearranged segments (C-terminal followed by N-terminal portions) with very high overall sequence identity to each other and the E. faecalis V583 gene EF1419. Sequence analysis of the N-terminal portion reveals no clear functional information other than a potential membrane-spanning domain. The C-terminal portion contains a putative nucleic acid binding domain, with matches to Zn-ribbon structure transcription factors with conservation of the four Zn binding cysteines. Moreover, significant sequence identity is observed between this potential lysogenic conversion protein domain and proteins in the lysogeny control locus (between the integrase and cI) of Streptococcus pneumoniae phage MM1 (39) and Streptococcus thermophilus phage ΦO1205 (53). The proposed DNA binding function of this domain is consistent with a role in lysogeny maintenance and suggests that these coding sequences are potentially transcribed as part of the lysogeny control cluster. This would indicate that within the eight E. faecalis phages identified, constituting four phage groups, there are no lysogenic conversion genes located beyond the endolysin gene that influence host fitness or virulence. Within the E. faecalis V583 genome, several of the prophage regions (phages 03, 04, and 05) (26, 40) contain potential lysogenic conversion genes, notably cold shock protein (cspC) and ferrochelatase (hemH) genes. Thus, bacteriophages are likely to participate in the mobilization of converting activities located in this terminal phage genome region, but these were not evident in the phages studied here.
Potential phage-encoded virulence factors.
The genome sequence of E. faecalis V583 contains multiple homologs of the Streptococcus mitis phage M1 platelet-binding tail protein PblA; each of these is located within putative prophage structural gene clusters in strain V583, namely, phage01 (EF0348), phage04 (EF2003), and phage06 (EF2813) (33, 40). Each of the phages identified in this study encoded a tail protein with significant sequence identity to either the Streptococcus mitis PblA protein (ΦFL1A to C, ΦFL2A and B, and ΦFL4A) or PblB protein (ΦFL3A and B) (Fig. 2; see also Tables SA1 to SA8 in the supplemental material). Since the sequence-specific determinants contributing to platelet adhesion are yet to be defined for PblA and PblB, it remains to be determined whether the E. faecalis homologs similarly mediate attachment to α2-8-linked sialic acid residues on platelet membrane gangliosides (35).
Examination of the E. faecalis phage genomes reveals few encoded proteins that might contribute to virulence. The presence of a lipoprotein in the lysogeny maintenance region of ΦFL4A, discussed above, could potentially modify the E. faecalis cell surface to increase adhesion. In addition, FL4A also encodes a VirE domain protein; VirE1 and VirE2 of Agrobacterium tumefaciens are virulence-associated proteins associated with the type IV secretion pathway (61). The ΦFL1A to C, ΦFL2A and B, and ΦFL3A and B phages each encode a YopX domain protein; Yop proteins are of plasmid origin, and in Yersinia pestis several of these are effectors delivered via the type III secretion system into the cytosol of the host cell (56). Equally, these identified proteins might be concerned with the bacteriophage life cycle.
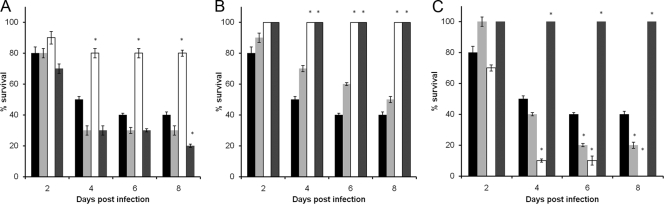
Impact of lysogeny upon Galleria mellonella survival.
Phage lysogens of E. faecalis strain JH2-2 were generated as a means of evaluating the potential contribution of the identified phages to host fitness and virulence. Lysogens of all eight phages were isolated by selecting colonies from semiconfluent lysis plates (Table 1). The lysogens were confirmed by UV induction, which produced progeny phages with a host range identical to those identified from the originating host strain (data not shown). Caterpillars of the greater wax moth, Galleria mellonella, were inoculated subcutaneously with 8 × 106 CFU of bacteria to compare the JH2-2 lysogens with their isogenic parent strain, JH2-2. Very reproducible results were obtained from multiple infection experiments, which revealed that lysogeny resulted in altered survival in most, but not all, of the lysogens in this model (Fig. 4). Lysogens of JH2-2 containing ΦFL1B, ΦFL2B, or ΦFL4A showed no killing of the caterpillars compared to JH2-2 (P < 0.01 after 4, 6, and 8 days). In contrast, lysogens of ΦFL3A and ΦFL3B were observed to contribute to an increase in caterpillar death by their JH2-2 host (P < 0.05 after 6 and 8 days). These experiments reveal that lysogeny with the identified phages engenders a capacity to alter the in vivo fitness and behavior of E. faecalis, at least in the model used here. Their actual contribution to infection awaits more defined experiments with animals and a systematic analysis of the impact that lysogeny has upon gene expression during growth. The integration sites of the phages were performed by PCR using primers for the integrase of each phage and genes adjacent to phage integration sites identified by comparative genome hybridization studies (26, 34). In addition PCR amplifications across these integration sites were performed using the primer pairs listed in Table 1. This confirmed that JH2-2 has one integrated partial phage inserted between EF1275 and EF1293, labeled phage02 in the comparative E. faecalis genome studies (26, 34). Phages ΦFL1A to C, ΦFL2A and B, and ΦFL3A and B possess an integrase with identical amino acid sequences, and in the JH2-2 lysogens LIV1037 to 1043 these are integrated between EF2798 and EF2856, as judged by PCR amplification using primers INT1-1/EF2856 and INT1-3/EF2856 DNA. Phage ΦFL4A was integrated between EF0302 and EF0355 in JH2-2 lysogen LIV1044 using PCR amplification with primers INT4-1/EF0302 and INT4-3/EF0302 (data not shown). These sites correspond to those of phage06 and phage01, respectively, in the previous comparative genome studies (26, 34).
FIG. 4.
Relative survival of E. faecalis JH2-2 phage lysogens compared to their isogenic parent strain. Caterpillars were inoculated with ∼8 × 106 CFU JH2-2 (black bars); JH2-2 ΦFL1A (light gray bars), JH2-2 ΦFL1B (white bars), or JH2-2 ΦFL1C (dark gray bars) (A); JH2-2 ΦFL2A (light gray bars), JH2-2 ΦFL2B (white bars), B. subtilis control (dark gray bars) (B); or JH2-2 ΦFL3A (light gray bars), JH2-2 ΦFL3B (white bars), JH2-2 ΦFL4A (dark gray bars) (C). B. subtilis control and solution controls did not result in any deaths in any of the experiments. Viability was monitored every 4 h for 8 days. * indicates significant difference (P < 0.05) relative to JH2-2.
Transduction using identified phages.
The eight sequenced phages were tested for their ability to package and transduce chromosomal and extrachromosomal DNA. E. faecalis donor strains of OG1RF and JH2-2, which contained chromosome or plasmid-located markers, were infected at a high MOI with phages to generate lysates. The following antibiotic resistance-marked loci were tested for transduction into recipient cells of OG1RF and JH2-2: chromosomal insertional inactivations mediated by Tn917 or the suicide plasmid pTEX4577; a pathogenicity-associated island (PAI) marked with chloramphenicol resistance in strain MMH594b; and the extrachromosomal replicative plasmid pAT18 (Table 1). Transduction was successfully achieved for all of the markers attempted, with the exception of the PAI, most probably due to the 120-kb size of the PAI element being larger than the ∼40-kb packaging size of the eight E. faecalis phages (Table 5). Each of the eight phages were found to be capable of generalized transduction, with ΦFL1C, ΦFL2A, and ΦFL3B being identified as the phages producing consistently high numbers of transductants, with either OG1RF or JH2-2 as recipient cells. The successful transduction of the gelE and nox markers to recipient OG1RF was confirmed by selecting for the resistance marker and confirming that levels of protease activity matched the altered levels present in the donor strain (Table 5) (58; J. Shankar and M. J. Horsburgh, unpublished data). Phage-mediated transfer of the fsrABCD operon-containing plasmid into the fsrABCD-negative recipient strain JH2-2 was confirmed by demonstrating gelatinase activity in recipient cells. The successful transduction of the antibiotic markers was confirmed further via PCR amplification of the corresponding resistance gene in each experiment; no transductants were observed with control transduction plates (data not shown). Each of the eight phages contain putative terminase and portal protein functions, indicative of capsid packaging of DNA being achieved using the head-full mechanism.
TABLE 5.
Measurement of transduction capabilities of E. faecalis bacteriophagesa
| Donor | Transductants per ml for indicated phage and recipient |
|||||
|---|---|---|---|---|---|---|
| ΦFL1C |
ΦFL2A |
ΦFL3B |
||||
| JH2-2 | OG1RF | JH2-2 | OG1RF | JH2-2 | OG1RF | |
| OG1RF gelE::pTEX4577 | 2,025 | 610 | 1,700 | 2,580 | 1,660 | 180 |
| OG1RF nox::Tn917 | 310 | ND | 280 | ND | 910 | ND |
| JH2-2 pTEX5249 (pAT18::fsrABCD) | 1,150 | 1,125 | 1,580 | 650 | 1,145 | 1,105 |
| MMH594b PAI::cat | 0 | 0 | 0 | 0 | 0 | 0 |
| OG1RF (control) | 0 | 0 | 0 | 0 | 0 | 0 |
Data show typical results from one of three experiments producing very similar data. Resistance markers were transduced from the indicated donor strains to the recipient strains using the phage described. ND, not determined.
The identification of bacteriophage capable of generalized transduction in E. faecalis will be of benefit to the molecular genetics of this organism. In particular, genetic linkage of phenotypes to transposon-mediated insertions will be particularly welcome, since presently this requires the production of defined allelic insertion or replacement mutants, typically using suicide plasmids. A more rapid determination of linkage will be possible using the identified phages. The transduction of OG1RF with marked loci indicates that the phages recognize receptors on the surface of the enterococcal strain, allowing the introduction of nucleic acid into the recipient cell by the phage. The CRISPR loci are known to impart phage DNA resistance to the recipient cell at the post-DNA injection stages of phage infection, which is sequence specific and thus does not occur during the generalized transduction described here.
This study predicts a more limited involvement of phages in the carriage of phage-encoded virulence factors among enterococcal strains than has previously been reported for other low-GC Gram-positive pathogenic bacteria. However, the transducing abilities of these phages, together with their shared sequence homology with those infecting low-GC Gram-positive bacteria, hints at a potential role in the transfer of genetic information between the different genera. Thus, phages potentially play an important role in the acknowledged spread of antibiotic resistance genes from enterococcal strains to pathogenic bacteria. Future studies will investigate the effects of single and multiple lysogeny upon host cell gene expression and fitness.
Supplementary Material
Acknowledgments
Azra Yasmin was supported by a Higher Education Commission of Pakistan Postdoctoral Fellowship. J.K. was supported by the BBSRC grant BB/D003563/1 awarded to M.J.H.
We are grateful to Daniel Rigden for bioinformatics discussions. We thank Barbara Murray, Michael Gilmore, Neil Woodford, Magda Kawalec, Patricia Ruiz-Garabajosa, Nathan Shankar, and Willem van Wamel for kindly providing many of the strains utilized in this study.
Footnotes
Published ahead of print on 11 December 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Aziz, R. K., S. A. Ismail, H. W. Park, and M. Kotb. 2004. Post-proteomic identification of a novel phage-encoded streptodornase, Sda1, in invasive M1T1 Streptococcus pyogenes. Mol. Microbiol. 54:184-197. [DOI] [PubMed] [Google Scholar]
- 3.Aziz, R. K., R. A. Edwards, W. W. Taylor, D. E. Low, A. McGeer, and M. Kotb. 2005. Mosaic prophages with horizontally acquired genes account for the emergence and diversification of the globally disseminated M1T1 clone of Streptococcus pyogenes. J. Bacteriol. 187:3311-3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Banks, D. J., B. Lei, and J. M. Musser. 2003. Prophage induction and expression of prophage-encoded virulence factors in group A Streptococcus serotype M3 strain MGAS315. Infect. Immun. 71:7079-7086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barrangou, R., C. Fremaux, H. Deveau, M. Richards, P. Boyaval, S. Moineau, D. A. Romero, and P. Horvath. 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315:1709-1712. [DOI] [PubMed] [Google Scholar]
- 6.Beloglazova, N., G. Brown, M. D. Zimmerman, M. Proudfoot, K. S. Makarova, M. Kudritska, S. Kochinyan, S. Wang, M. Chruszcz, W. Minor, E. V. Koonin, A. M. Edwards, A. Savchenko, and A. F. Yakunin. 2008. A novel family of sequence-specific endoribonucleases associated with the clustered regularly interspaced short palindromic repeats. J. Biol. Chem. 283:20361-20371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bensing. B. A., I. R. Siboo, and P. M. Sullam. 2001. Proteins PblA and PblB of Streptococcus mitis, which promote binding to human platelets, are encoded within a lysogenic bacteriophage. Infect. Immun. 69:6186-6192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bourgogne, A., D. A. Garsin, X. Qin, K. V. Singh, J. Sillanpaa, S. Yerrapragada, Y. Ding, S. Dugan-Rocha, C. Buhay, H. Shen, G. Chen, G. Williams, D. Muzny, A. Maadani, K. A. Fox, J. Gioia, L. Chen, Y. Shang, C. A. Arias, S. R. Nallapareddy, M. Zhao, V. P. Prakash, S. Chowdhury, H. Jiang, R. A. Gibbs, B. E. Murray, S. K. Highlander, and G. M. Weinstock. 2008. Large scale variation in Enterococcus faecalis illustrated by the genome analysis of strain OG1RF. Genome Biol. 9:R110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brouns, S. J., M. M. Jore, M. Lundgren, E. R. Westra, R. J. Slijkhuis, A. P. Snijders, M. J. Dickman, K. S. Makarova, E. V. Koonin, and J. van der Oost. 2008. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321:960-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carte, J., R. Wang, H. Li, R. M. Terns, and M. P. Terns. 2008. Cas6 is an endoribonuclease that generates guide RNAs for invader defense in prokaryotes. Genes Dev. 22:3489-3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang, S., D. M. Sievert, J. C. Hageman, M. L. Boulton, F. C. Tenover, F. P. Downes, S. Shah, J. T. Rudrik, G. R. Pupp, W. J. Brown, D. Cardo, and S. K. Fridkin. 2003. Infection with vancomycin-resistant Staphylococcus aureus containing the vanA resistance gene N. Engl. J. Med. 348:1342-1347. [DOI] [PubMed] [Google Scholar]
- 12.Chopin, M. C., A. Chopin, and C. Roux. 1976. Definition of bacteriophage groups according to their lytic action on mesophilic lactic streptococci. Appl. Environ. Microbiol. 32:741-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delcher, A., D. Harmon, S. Kasif, O. White, and S. Salzberg. 1999. Improved microbial gene identification with glimmer. Nucleic Acids Res. 27:4636-4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Desiere, F., S. Lucchini, C. Canchaya, M. Ventura, and H. Brüssow. 2002. Comparative genomics of phages and prophages in lactic acid bacteria. Antonie Van Leeuwenhoek 82:73-79. [PubMed] [Google Scholar]
- 15.Dunny, G. M., B. L. Brown, and D. B. Clewell. 1978. Induced cell aggregation and mating in Streptococcus faecalis: evidence for a bacterial sex pheromone. Proc. Natl. Acad. Sci. U. S. A. 75:3479-3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hatfull, G. F., S. G. Cresawn, and R. W. Hendrix. 2008. Comparative genomics of the mycobacteriophages: insights into bacteriophage evolution. Res. Microbiol. 159:332-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Horsburgh, M. J., J. L. Aish, I. J. White, L. Shaw, J. K. Lithgow, and S. J. Foster. 2002. σB modulates virulence determinant expression and stress resistance: characterization of a functional rsbU strain derived from Staphylococcus aureus 8325-4. J. Bacteriol. 184:5457-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hunter, S., R. Apweiler, T. K. Attwood, A. Bairoch, A. Bateman, D. Binns, P. Bork, U. Das, L. Daugherty, L. Duquenne, R. D. Finn, J. Gough, D. Haft, N. Hulo, D. Kahn, E. Kelly, A. Laugraud, I. Letunic, D. Lonsdale, R. Lopez, M. Madera, J. Maslen, C. McAnulla, J. McDowall, J. Mistry, A. Mitchell, N. Mulder, D. Natale, C. Orengo, A. F. Quinn, J. D. Selengut, C. J. A. Sigrist, M. Thimma, P. D. Thomas, F. Valentin, D. Wilson, C. H. Wu, and C. Yeats. 2009. Interpro: the integrative protein signature database. Nucleic Acids Res. 37:D211-D215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huycke, M. M., D. F. Sahm, and M. S. Gilmore. 1998. Multiple-drug resistant enterococci: the nature of the problem and an agenda for the future. Emerg. Infect. Dis. 4:239-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jacob, A. E., and S. J. Hobbs. 1974. Conjugal transfer of plasmid-borne multiple antibiotic resistance in Streptococcus faecalis var. zymogenes. J. Bacteriol. 117:360-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jett, B. D., M. M. Huycke, and M. S. Gilmore. 1994. Virulence of enterococci. Clin. Microbiol. Rev. 7:462-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kenny, J. G., D. Ward, E. Josefsson, I. M. Jonsson, J. Hinds, H. H. Rees, J. A. Lindsay, A. Tarkowski, and M. J. Horsburgh. 2009. The Staphylococcus aureus response to unsaturated long chain free fatty acids: survival mechanisms and virulence implications. PLoS One 4:e4344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krzywinski, M., J. Schein, I. Birol, J. Connors, R. Gascoyne, D. Horsman, S. J. Jones, and M. A. Marra. 2009. Circos: an information aesthetic for comparative genomics. Genome Res. 19:1639-1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kurtz, S., A. Phillippy, A. L. Delcher, M. Smoot, M. Shumway, C. Antonescu, and S. L. Salzberg. 2004. Versatile and open software for comparing large genomes. Genome Biol. 5:R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lebreton, F., E. Riboulet-Bisson, P. Serror, M. Sanguinetti, B. Posteraro, R. Torelli, A. Hartke, Y. Auffray, and J. C. Giard. 2009. ace, which encodes an adhesin in Enterococcus faecalis, is regulated by Ers and is involved in virulence. Infect. Immun. 77:2832-2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lepage, E., S. Brinster, C. Caron, C. Ducroix-Crepy, L. Rigottier-Gois, G. Dunny, C. Hennequet-Antier, and P. Serror. 2006. Comparative genomic hybridization analysis of Enterococcus faecalis: identification of genes absent from food strains. J. Bacteriol. 188:6858-6868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li, L., C. J. Stoeckert, and D. S. Roos. 2003. Orthomcl: identification of ortholog groups for eukaryotic genomes. Genome Res. 13:2178-2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lillestøl, R. K., P. Redder, R. A. Garrett, and K. Brugger. 2006. A putative viral defence mechanism in archaeal cells. Archaea 2:59-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lukashin, A., and M. Borodovsky. 1998. GeneMark.hmm: new solutions for gene finding. Nucleic Acids Res. 26:1107-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Makarova, K. S., N. V. Grishin, S. A. Shabalina, Y. I. Wolf, and E. V. Koonin. 2006. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol. Direct 1:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Makarova, K., A. Slesarev, Y. Wolf, A. Sorokin, B. Mirkin, E. Koonin, A. Pavlov, N. Pavlova, V. Karamychev, N. Polouchine, V. Shakhova, I. Grigoriev, Y. Lou, D. Rohksar, S. Lucas, K. Huang, D. M. Goodstein, T. Hawkins, V. Plengvidhya, D. Welker, J. Hughes, Y. Goh, A. Benson, K. Baldwin, J. H. Lee, I. Díaz-Muñiz, B. Dosti, V. Smeianov, W. Wechter, R. Barabote, G. Lorca, E. Altermann, R. Barrangou, B. Ganesan, Y. Xie, H. Rawsthorne, D. Tamir, C. Parker, F. Breidt, J. Broadbent, R. Hutkins, D. O'Sullivan, J. Steele, G. Unlu, M. Saier, T. Klaenhammer, P. Richardson, S. Kozyavkin, B. Weimer, and D. Mills. 2006. Comparative genomics of the lactic acid bacteria. Proc. Natl. Acad. Sci. U. S. A. 103:15611-15616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marraffini, L. A., and E. J. Sontheimer. 2008. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science 322:1843-1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McBride, S. M., V. A. Fischetti, D. J. LeBlanc, R. C. Moellering, Jr., and M. S. Gilmore. 2007. Genetic diversity among Enterococcus faecalis. PLoS One 2:e582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McBride, S. M., P. S. Coburn, A. S. Baghdayan, R. J. Willems, M. J. Grande, N. Shankar, and M. S. Gilmore. 2009. Genetic variation and evolution of the pathogenicity island of Enterococcus faecalis. J. Bacteriol. 191:3392-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mitchell, J., and P. M. Sullam. 2009. Streptococcus mitis phage-encoded adhesins mediate attachment to α2-8-linked sialic acid residues on platelet membrane gangliosides. Infect. Immun. 77:3485-3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mojica, F. J., C. Diez-Villasenor, J. Garcia-Martinez, and E. Soria. 2005. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J. Mol. Evol. 60:174-182. [DOI] [PubMed] [Google Scholar]
- 37.Murray, B. E. 1990. The life and times of the Enterococcus. Clin. Microbiol. Rev. 3:46-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mylonakis, E., R. Moreno, J. B. El Khoury, A. Idnurm, J. Heitman, S. B. Calderwood, F. M. Ausubel, and A. Diener. 2005. Galleria mellonella as a model system to study Cryptococcus neoformans pathogenesis. Infect. Immun. 73:3842-3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Obregón, V., J. L. García, E. García, R. López, and P. García. 2003. Genome organization and molecular analysis of the temperate bacteriophage MM1 of Streptococcus pneumoniae. J. Bacteriol. 185:2362-2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paulsen, I. T., L. Banerjei, G. S. Myers, K. E. Nelson, R. Seshadri, et al. 2003. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science 299:2071-2074. [DOI] [PubMed] [Google Scholar]
- 41.Pedulla, M. L., M. E. Ford, J. M. Houtz, T. Karthikeyan, C. Wadsworth, J. A. Lewis, D. Jacobs-Sera, J. Falbo, J. Gross, N. R. Pannunzio, W. Brucker, V. Kumar, J. Kandasamy, L. Keenan, S. Bardarov, J. Kriakov, J. G. Lawrence, W. R. Jacobs, Jr., R. W. Hendrix, and G. F. Hatfull. 2003. Origins of highly mosaic mycobacteriophage genomes. Cell 113:171-182. [DOI] [PubMed] [Google Scholar]
- 42.Pourcel, C., G. Salvignol, and G. Vergnaud. 2005. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology 151:653-663. [DOI] [PubMed] [Google Scholar]
- 43.Prévost, G., B. Cribier, P. Couppie, P. Petiau, G. Supersac, V. Finck-Barbancon, H. Monteil, and Y. Piemont. 1995. Panton-Valentine leucocidin and gamma-hemolysin from Staphylococcus aureus ATCC 49775 are encoded by distinct genetic loci and have different biological activities. Infect. Immun. 63:4121-4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qin, X., K. V. Singh, G. M. Weinstock, and B. E. Murray. 2000. Effects of Enterococcus faecalis fsr genes on production of gelatinase and a serine protease and virulence. Infect. Immun. 68:2579-2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ruiz-Garbajosa, P., M. J. Bonten, D. A. Robinson, J. Top, S. R. Nallapareddy, C. Torres, T. M. Coque, R. Cantón, F. Baquero, B. E. Murray, R. del Campo, and R. J. Willems. 2006. Multilocus sequence typing scheme for Enterococcus faecalis reveals hospital-adapted genetic complexes in a background of high rates of recombination. J. Clin. Microbiol. 44:2220-2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rutherford, K., J. Parkhill, J. Crook, T. Horsnell, P. Rice, M. A. Rajandream, and B. Barrell. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16:944-945. [DOI] [PubMed] [Google Scholar]
- 47.Sahm, D. F., J. Kissinger, M. S. Gilmore, P. R. Murray, R. Mulder, J. Solliday, and B. Clarke. 1989. In vitro susceptibility studies of vancomycin-resistant Enterococcus faecalis. Antimicrob. Agents Chemother. 33:1588-1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sambrook, J., E. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 49.Shankar, N., A. S. Baghdayan, and M. S. Gilmore. 2002. Modulation of virulence within a pathogenicity island in vancomycin-resistant Enterococcus faecalis. Nature 417:746-750. [DOI] [PubMed] [Google Scholar]
- 50.Sievert, D. M., J. T. Rudrik, J. B. Patel, L. C. McDonald, M. J. Wilkins, and J. C. Hageman. 2008. Vancomycin-resistant Staphylococcus aureus in the United States, 2002-2006. Clin. Infect. Dis. 46:668-674. [DOI] [PubMed] [Google Scholar]
- 51.Sifri, C. D., E. Mylonakis, K. V. Singh, X. Qin, D. A. Garsin, B. E. Murray, F. M. Ausubel, and S. B. Calderwood. 2002. Virulence effect of Enterococcus faecalis protease genes and the quorum-sensing locus fsr in Caenorhabditis elegans and mice. Infect. Immun. 70:5647-5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Singh, K. V., X. Qin, G. M. Weinstock, and B. E. Murray. 1998. Generation and testing of mutants of Enterococcus faecalis in a mouse peritonitis model. J. Infect. Dis. 178:1416-1420. [DOI] [PubMed] [Google Scholar]
- 53.Stanley, E., G. F. Fitzgerald, C. Le Marrec, B. Fayard, and D. van Sinderen. 1997. Sequence analysis and characterization of phi O1205, a temperate bacteriophage infecting Streptococcus thermophilus CNRZ1205. Microbiology 143:3417-3429. [DOI] [PubMed] [Google Scholar]
- 54.Tormo, M. A., M. D. Ferrer, E. Maiques, C. Ubeda, L. Selva, I. Lasa, J. J. Calvete, R. P. Novick, and J. R. Penadés. 2008. Staphylococcus aureus pathogenicity island DNA is packaged in particles composed of phage proteins. J. Bacteriol. 190:2434-2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van Wamel, W. J., S. H. Rooijakkers, M. Ruyken, K. P. van Kessel, and J. A. van Strijp. 2006. The innate immune modulators staphylococcal complement inhibitor and chemotaxis inhibitory protein of Staphylococcus aureus are located on beta-hemolysin-converting bacteriophages. J. Bacteriol. 188:1310-1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Viboud, G. I., and J. B. Bliska. 2005. Yersinia outer proteins: role in modulation of host cell signaling responses and pathogenesis. Annu. Rev. Microbiol. 59:69-89. [DOI] [PubMed] [Google Scholar]
- 57.Villion, M., M. C. Chopin, H. Deveau, S. D. Ehrlich, S. Moineau, and A. Chopin. 2009. P087, a lactococcal phage with a morphogenesis module similar to an Enterococcus faecalis prophage. Virology 388:49-56. [DOI] [PubMed] [Google Scholar]
- 58.Waters, C. M., M. H. Antiporta, B. E. Murray, and G. M. Dunny. 2003. Role of the Enterococcus faecalis GelE protease in determination of cellular chain length, supernatant pheromone levels, and degradation of fibrin and misfolded surface proteins. J. Bacteriol. 185:3613-3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wegmann, U., M. O'Connell-Motherway, A. Zomer, G. Buist, C. Shearman, C. Canchaya, M. Ventura, A. Goesmann, M. J. Gasson, O. P. Kuipers, D. van Sinderen, and J. Kok. 2007. Complete genome sequence of the prototype lactic acid bacterium Lactococcus lactis subsp. cremoris MG1363. J. Bacteriol. 189:3256-3270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Woodford, N., R. Reynolds, J. Turton, F. Scott, A. Sinclair, A. Williams, and D. Livermore. 2003. Two widely disseminated strains of Enterococcus faecalis highly resistant to gentamicin and ciprofloxacin from bacteraemias in the UK and Ireland. J. Antimicrob. Chemother. 52:711-714. [DOI] [PubMed] [Google Scholar]
- 61.Zhao, Z., E. Sagulenko, Z. Ding, and P. J. Christie. 2001. Activities of virE1 and the VirE1 secretion chaperone in export of the multifunctional VirE2 effector via an Agrobacterium type IV secretion pathway. J. Bacteriol. 183:3855-3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.