Abstract
The filamentous cyanobacterium Anabaena sp. strain PCC 7120 forms nitrogen-fixing heterocysts in a periodic pattern in response to combined-nitrogen limitation in the environment. The master regulator of heterocyst differentiation, HetR, is necessary for both pattern formation and commitment of approximately every 10th cell of a filament to differentiation into a heterocyst. In this study, the individual contributions of four transcriptional start points (tsps) in regulation of transcription of hetR were assessed, and the effects of the regulatory genes patS, hetN, and patA on transcription from the tsps were determined. The tsp located at nucleotide −271 relative to the translational start site (−271 tsp) was the most tightly regulated tsp, and fluorescence from a −271 tsp-green fluorescent protein (GFP) reporter fusion was observed initially in groups of two cells and later in single cells arranged in a spatial pattern that mimicked the pattern of heterocysts that emerged. Conversely, the fluorescence from the −184 and −728/−696 tsp-GFP reporter fusions was uniform throughout filaments. Transcription from the −271 tsp was severely downregulated in a strain in which the patA gene, which encodes a positive regulator of differentiation, was deleted, and it was not detectable in strains overexpressing the genes encoding the negative regulators of differentiation, patS and hetN. In strains lacking the −271 tsp of hetR, pattern formation, the timing of commitment to differentiation, and the number of cells that differentiated into heterocysts were affected. Taken together, these results demonstrate the role of regulation of the −271 tsp of hetR in the genetic network that governs heterocyst pattern formation and differentiation.
The filamentous cyanobacterium Anabaena sp. PCC 7120 fixes dinitrogen in specialized cells called heterocysts. When the levels of fixed nitrogen are sufficient to support robust growth, the filaments are comprised entirely of undifferentiated vegetative cells. However, when the levels of fixed nitrogen become limiting (nitrogen starvation), certain cells of the filament develop into terminally differentiated heterocysts, where nitrogen fixation takes place. Heterocysts provide a microoxic environment for the oxygen-labile enzyme nitrogenase, which catalyzes the formation of ammonia from dinitrogen. The product of nitrogen fixation is transported from a heterocyst to the adjacent vegetative cells in the form of glutamine (18), and the heterocyst in return receives fixed carbon (28). Thus, the two cell types are metabolically interdependent and have different roles in the filament.
Approximately every 10th cell in a filament differentiates into a heterocyst to create a periodic pattern consisting of two cell types. Heterocysts do not divide, and the distance between two heterocysts increases as the intervening vegetative cells multiply. As the distance becomes greater, a cell approximately midway between two heterocysts differentiates, and thus the heterocyst pattern is maintained (12). This one-dimensional pattern of two cell types is one of the simplest and oldest patterns elaborated by a multicellular organism (27), making it an ideal system for investigation of developmental regulation in prokaryotes and definition of the minimal requirements for patterning and morphogenesis (22).
In Anabaena sp. PCC 7120 nitrogen starvation results in accumulation of 2-oxoglutarate because this strain lacks 2-oxoglutarate dehydrogenase (25). 2-Oxoglutarate is thought to bind to the global nitrogen regulator NtcA and enhances its capacity as a transcriptional regulator (15). NtcA in turn activates a range of genes involved in heterocyst formation and function (11). One of these genes is hetR. HetR is considered the master regulator of heterocyst differentiation. The level of expression of hetR in a cell correlates with the cell's developmental fate. Induction of expression of hetR is sufficient to trigger the differentiation process, even in the presence fixed nitrogen (5), and heterocyst formation does not take place in its absence (4). HetR positively autoregulates itself at the level of transcription (1). The HetR protein binds to its own promoter and to the promoters of other heterocyst-specific genes, suggesting that it functions as a regulator of transcription (13).
Transcription of hetR is both temporally and spatially regulated. Northern blot experiments have shown that hetR transcription levels begin to increase by 0.5 h after nitrogen starvation (5). Between 6 and 18 h during induction of heterocyst differentiation, there is up to a 5-fold increase in transcript levels (4). HetR also shows spatial patterning of transcription. In a filament of undifferentiated vegetative cells grown in the presence of combined nitrogen, a low level of transcription is observed for all of the cells of the filament (1). In contrast, by 3.5 h after removal of combined nitrogen there is patterned upregulation of transcription in groups of cells in the filament (1), and by 6 to 8 h transcription is localized to cells that are arranged in a periodic pattern and will differentiate into heterocysts (1, 6). Subsequently, upregulation of hetR transcription is observed only in cells that are midway between two heterocysts and will presumably also differentiate to preserve the pattern of heterocysts in a growing filament (1).
PatS is a negative regulator of heterocyst differentiation that is thought to mediate pattern formation by suppression of differentiation of neighboring cells by a source cell. Its C-terminal pentapeptide, RGSGR, has been shown to inhibit heterocyst differentiation (23, 30) and also prevent HetR from binding to DNA in the promoter region of hetR (13). HetN is another regulator of heterocyst differentiation that is thought to mediate pattern maintenance in a similar fashion. Heterocyst differentiation and the patterned expression of hetR are disrupted by overexpression of hetN (6). HetN and PatS function independently, although they converge functionally at or before hetR in the regulatory network controlling differentiation (2, 14). PatA is a positive regulator of differentiation that downregulates the inhibitory effects of PatS and HetN and promotes differentiation independent of its effects on PatS and HetN (2, 17). PatS, HetN, and PatA have each been shown to impact the spatial and temporal expression of hetR (5, 6, 13, 16).
The promoter region of hetR is complex. Four transcriptional start points (tsps) are located at nucleotides −728 (−728 tsp), −696, −271, and −184 relative to the translational start site, and each tsp gives rise to a distinct mRNA molecule (5). The transcript originating at the −184 tsp is present regardless of the nitrogen status (5, 19). Transcription from tsps −728 and −696 is induced upon nitrogen starvation (5). NtcA is required for transcription from tsp −728 (19), and its activity is mediated through NrrA, a response regulator of the OmpR family (7, 8). Transcription from tsp −271 is also induced upon nitrogen starvation. A functional copy of HetR is essential for transcription from tsp −271 (5). The HetR protein binds to a region of DNA around the −271 tsp (13), and this transcription start point is thought to be involved in positive autoregulation (5).
Differential gene expression has long been recognized as a factor that has an important role in regulating heterocyst differentiation (29). We investigated the temporal and spatial transcription patterns of the four transcription start sites of hetR and found that only the −271 tsp is regulated spatially. We also analyzed the role played by the heterocyst regulators PatA, PatS, and HetN in controlling transcription and found that each regulator affects transcription from a subset of tsps.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Anabaena sp. PCC 7120 (wild type) and its derivatives are described in Table 1. The growth conditions used and regulation of expression of the petE promoter (PpetE) have been described previously (22). For analysis of the temporal transcription profile of hetR, Anabaena strains were grown in BG-11 medium buffered with 30 mM N-tris(hydroxymethyl)methyl-2-aminoethanesulfonic acid (TES) (pH 8.0) and supplemented with 10 mM NH4Cl and 5 mM NaHCO3. Cells were washed twice in medium lacking a fixed-nitrogen source (BG-110) and transferred to BG-110. In all other experiments, Anabaena strains were grown in BG-11 medium buffered with 10 mM HEPES (pH 8.0) and supplemented with 2 mM (NH4)2SO4 and 5 mM NaHCO3 prior to transfer to BG-110, when appropriate. Escherichia coli strains bearing plasmids were grown in medium supplemented with 50 μg/ml kanamycin sulfate or 100 μg/ml spectinomycin sulfate. Strains of Anabaena bearing plasmids were grown in medium supplemented with 45 μg/ml neomycin sulfate, and strains of Anabaena bearing suicide plasmids recombined into the chromosome were grown in medium supplemented with 2.5 μg/ml spectinomycin sulfate and streptomycin sulfate. Bright-field microscopy and fluorescence microscopy were performed as previously described (2). All fluorescence images were captured using the same exposure and sensitivity camera settings. Plasmids were introduced into different strains of Anabaena by conjugation as previously described (9).
TABLE 1.
Strains, plasmids, and PCR primers used in this study
| Strain, plasmid, or primer | Relevant characteristics | Source or reference |
|---|---|---|
| Anabaena sp. strains | ||
| PCC 7120 | Wild type | Pasteur Culture Collection |
| UHM101 | ΔpatA | 20 |
| 120PN | PpetE-hetN | 6 |
| UHM103 | ΔhetR | 2 |
| HM156 | Nucleotides −295 to −268 of promoter of hetR deleted | This study |
| UHM157 | Nucleotides −317 to −268 of promoter of hetR deleted | This study |
| Plasmids | ||
| pNC101 | Shuttle vector derived from pAM504 for replication in E. coli and Anabaena with promoterless lacZ gene; Kmr Nmr | 22 |
| pAM504 | Shuttle vector for replication in E. coli and Anabaena; Kmr Nmr | 26 |
| pAM1956 | Shuttle vector for replication in E. coli and Anabaena with promoterless gfp gene; Kmr Nmr | 30 |
| pAM1951 | Shuttle vector with PpatS-gfp; Kmr Nmr | 30 |
| pSMC127 | pAM504 carrying PhetR-gfp | 6 |
| pDR211 | Shuttle vector derived from pAM504 with PpetE-patS | This study |
| pRR139 | pAM1956 with −728/−696 tsp region fused to gfp | This study |
| pRR149 | pAM1956 with −728/−696(+132) tsp region fused to gfp | This study |
| pRR140 | pAM1956 with −271 tsp region fused to gfp | This study |
| pRR150 | pAM1956 with −271/−184 tsp region fused to gfp | This study |
| pRR151 | pAM1956 with −184 tsp region fused to gfp | This study |
| pRR142 | pNC101 with −728/−696 tsp region fused to lacZ | This study |
| pRR143 | pNC101 with −271 tsp region fused to lacZ | This study |
| pRR152 | pNC101 with −728/−696(+132) tsp region fused to lacZ | This study |
| pDR325 | Suicide vector derived from pRL277 containing a 3,605-bp fragment starting 1,691 bp upstream and ending 1,014 bp downstream of the hetR coding region | This study |
| pRR164 | Suicide vector derived from pDR325 lacking nucleotides −295 to −268 in the hetR promoter region | This study |
| pRR165 | Suicide vector derived from pDR325 lacking nucleotides −317 to −268 in the hetR promoter region | This study |
| Primers (5′-3′) | ||
| hetR5′for | TTTAGATCTGCTGTCGTTCTCAGCCACAGAGATTTGTCC | |
| hetR-Chr-SacI-R | ATATAGAGCTCATGTCTTGGCTCAGTCGCGGATGATGG | |
| Sac GFP L-F3 | CACACGAGCTCGACAAAGGACTTATAATCAAAACAAATG | |
| Kpn GFP-L-R2 | ATATGGTACCCTTAGCAATACCTTTTATTTCC | |
| Sac GFP-S-F3 | ATTATGGAGCTCGGAAATAAAAGGTATTGCTAAGTTGG | |
| Kpn GFP-S-R2 | ATATGGTACCTTACTGGCGAACTTTATGGTTC | |
| Sac GFP-C-F | TTATGAGCTCCATAAAGTTCGCCAGTAATTGACAAATG | |
| Kpn GFP M R3 | AAATGGTACCATTACAAATAGTTGAATAGCACGC | |
| Kpn GFP L R3 | ATATGGTACCCTCACTAACGCCCATTGAATC | |
| lacZ Bam 696 Fwd | CACACGGATCCGACAAAGGACTTATAATCAAAACAAATG | |
| lacZ Sma 696 Rev2 | ATATCCCGGGCTTAGCAATACCTTTTATTTCC | |
| lacZ Bam 271 Fwd | ATTATGGGATCCGGAAATAAAAGGTATTGCTAAGTTGG | |
| lacZ Sma 271 R2 | ATATCCCGGGTTACTGGCGAACTTTATGGTTC | |
| lacZ Sma 696 R3 | ATATCCCGGGCTCACTAACGCCCATTGAATC | |
| 696new2 | AATCGCACAATCTCTCCTTGAGGTG | |
| 696new1 | TTTGCTCTTATGGCAGTGTAGGTCG | |
| new184nesrev | CGCTTGATCAGATCGATGTCGTTACTC | |
| 184new1 | CATTGCACTGGGGCCAAGACGCTT | |
| hetRcf-NdeI | CATATGAGTAACGACATCGATCTGATC | |
| hetRcr-SacI | GAGCTCTTAATCTTCTTTTCTACCAAACACC | |
| Xho-Bam-PpetE-F | TATATCTCGAGGGATCCGCTGAGGTACTGAGTACACAGC | |
| PpetE-Nde-Pst-R | ATATACTGCAGCATATGGTTCTCCTAACCTGTAGTTTTATTTTTC | |
| patS-NdeI-F | CATATGAAGGCAATTATGTTAGTGAATTTCTGTGATGAG | |
| patS-SalI-SpeI-R | ACTAGTGTCGACCTATCTACCACTACCGCGCTCATCACAG | |
| 271 tsp Fwd | CAGATAAGTTCCGGATAATAGGGCCATAAAGTTCGCCAGTA | |
| 271 tsp Rev | TACTGGCGAACTTTATGGCCCTATTATCCGGAACTTATCTG | |
| Fwd-N | CAAATTTATTGACGCTATTAAAGAGCCATAAAGTTCGCCAGTA | |
| Rev-N | TACTGGCGAACTTTATGGCTCTTTAATAGCGTCAATAAATTTG | |
| PhetR-BamHI-F | ATATAGGATCCAACCCTTATGACAAAGGAC | |
| hetR-SacI-SpeI-R | TATATAGAGCTCACTAGTACTTTTATTCACTCTGGGTGC |
Construction of strains and plasmids.
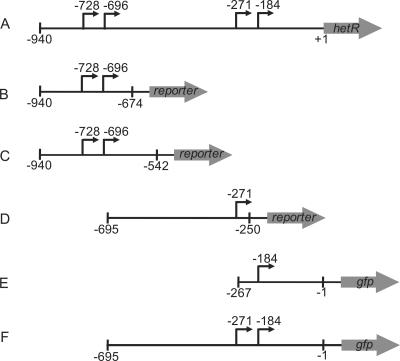
Strains, plasmids, and primers are listed in Table 1. PCR-derived constructs were sequenced to verify their integrity. For creation of tsp-reporter fusions, the promoter region of hetR was separated into five regions bearing one or more transcriptional start points of hetR. The different tsp regions were cloned in front of the coding regions of either gfp (encoding green fluorescent protein [GFP]) or lacZ. The nucleotide boundaries of the tsp regions for each fusion are shown in Fig. 1.
FIG. 1.
Schematic diagrams of reporter fusions with tsp regions of the hetR promoter. (A) hetR promoter and coding region with its four transcriptional start points. (B) −728/−696 tsp-reporter fusion. (C) −728/−696(+132) tsp-reporter fusion. (D) −271 tsp-reporter fusion. (E) −184 tsp-reporter fusion. (F) −271/−184 tsp-reporter fusion. The arrows indicate tsps, and the vertical lines indicate the boundaries of sequences from the promoter region of hetR that were included in each of the fusions. The numbers are positions relative to the translational start site of hetR.
The tsp-gfp fusions were generated by inserting the appropriate tsp regions into plasmid pAM1956 (30) as SacI-KpnI fragments. The plasmids that contained the gfp fusions and the primers (in parentheses) used to generate the corresponding regions of the hetR promoter are as follows: pRR139 for the −728/−696 tsp-gfp fusion (Sac GFP L-F3 and Kpn GFP-L-R2); pRR149 for the −728/−696(+132) tsp-gfp fusion [the −728/−696(+132) tsp fusion includes an additional 132 bases of the hetR promoter downstream of those in the initial construct] (Sac GFPL-F3 and Kpn GFP L R3); pRR140 for the −271 tsp-gfp fusion (Sac GFP-S-F and Kpn GFP-S-R2); pRR151 for the −184 tsp-gfp fusion (Sac GFP C-F and Kpn GFP-M-R3); and pRR150 for the −271/−184 tsp-gfp fusion (Sac GFP-S-F3 and Kpn GFP-M-R3). The tsp-lacZ fusions were generated by inserting the appropriate tsp region into plasmid pNC101 (22) as a BamHI-SmaI fragment. The plasmids that contained the lacZ fusions and the primers (in parentheses) used to generate the corresponding regions of the hetR promoter are as follows: pRR142 for the −728/−696 tsp-lacZ fusion (lacZ Bam 696 Fwd and lacZ Sma 696 Rev2); pRR152 for the −728/−696(+132) tsp-lacZ fusion (lacZ Bam 696 Fwd and lacZ Sma 696 R3); and pRR143 for the −271 tsp-lacZ fusion (lacZ Bam 271 Fwd and lacZ Sma 271 R2).
Plasmid pDR211 is a mobilizable shuttle vector derived from pAM504 (26) and contains the patS gene under the control of a copper-inducible promoter (PpetE). Primers Xho-Bam-PpetE-F and PpetE-Nde-Pst-R were used to amplify PpetE, and the resultant DNA fragment was cloned into pGEM-T (Promega). PpetE was subsequently moved into pBluescript SK(+) (Stratagene) as an XhoI-PstI fragment. The patS gene was then inserted downstream of PpetE to generate PpetE-patS. The patS gene was amplified using primers patS-NdeI-F and patS-SalI-SpeI-R, and the resultant DNA fragment was cloned into pGEM-T before it was moved as an NdeI-SpeI fragment behind PpetE in pBluescript. PpetE-patS was then moved into pAM504 as a BamHI-SacI fragment to obtain plasmid pDR211.
Plasmid pDR325 is a suicide vector derived from pRL277 (1) and contains a 3,605-bp fragment starting 1,691 bp upstream of the hetR coding region and ending 1,014 bp downstream of the hetR coding region. This 3,605-bp fragment was amplified by PCR using PCC 7120 chromosomal DNA as the template with primers hetR5′for and hetR-Chr-SacI-R and was cloned into pRL277 as a BglII-SacI fragment using restriction sites introduced into the primers to create pDR325. The hetR promoter and coding region in pDR325 was replaced with one of two versions lacking either nucleotides −295 to −268 or nucleotides −317 to −268 using NcoI and SpeI to obtain plasmids pRR164 and pRR165, respectively. The segment of DNA bearing the deletions was generated by overlap extension PCR using primers PhetR-BamHI-F, 271 tsp Rev, 271 tsp Fwd, and hetR-SacI-SpeI-R to create pRR164 and using primers PhetR-BamHI-F, Rev-N, Fwd-N, and hetR-SacI-SpeI-R to create pRR165. Strains UHM156 and UHM157 lack nucleotides −295 to −268 and nucleotides −317 to −268 of the hetR promoter, respectively, and were created by allelic replacement as previously described (20) using plasmids pRR164 and pRR165, respectively.
RNA isolation and analysis.
Cells grown for RNA isolation were flash-frozen in liquid nitrogen at specified time intervals and stored at −80°C until they were processed. TRIzol (Invitrogen) was used for RNA extraction as previously described (22). Three micrograms of RNA was used for rapid amplification of cDNA ends (RACE) as previously described (22). Primers designed to bind downstream of the −184 tsp were used to amplify the −271 and −184 transcripts (primers 184new1 and new184nesrev for cDNA synthesis and RACE, respectively), and primers designed to bind downstream of nucleotide −696 were used to amplify the −728 and −696 transcripts (primers 696new1 and 696new2 for cDNA synthesis and RACE, respectively). RACE products were sequenced to verify their integrity. Ten micrograms of RNA was used for Northern blotting as previously described (5). Primers hetRcf-NdeI and hetRcr-SacI were used to make the hetR-specific probe. Northern blot and RACE experiments were done two or more times.
β-Galactosidase assays.
Cells were prepared and β-galactosidase activity was determined as previously described (22). Enzyme activity is expressed as nanomoles of o-nitrophenol per minute per milligram of protein.
RESULTS
Expression from the −271 tsp of hetR is tightly regulated.
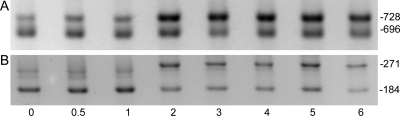
Northern blotting and primer extension have been used to show that the levels of transcription from hetR tsp −184 are similar under nitrogen-replete and nitrogen starvation conditions, while expression from the hetR −728, −696, and −271 tsps is induced after deprivation of combined nitrogen (5, 19). Because transcription from tsp −271 has been implicated in direct positive autoregulation of hetR, which has the potential to rapidly amplify transcription of hetR, we were interested in examining transcription from each of the tsps using a more sensitive approach. To examine the tightness of regulation of transcription from the individual tsps, RACE, a very sensitive procedure, was used with RNA extracted from the wild type before induction of heterocyst differentiation and at 0.5, 1, 2, 3, 4, 5, and 6 h after induction of differentiation in BG-110, which lacks combined nitrogen. Under the conditions tested, transcription from tsps −728, −696, and −184 was detected at all eight time points (Fig. 2). However, the transcript originating from the −271 tsp was not detected in nitrogen-replete conditions or at 0.5 and 1 h. Transcription was detected for the −271 tsp 2 h after removal of combined nitrogen or later. A band for a third RACE product slightly smaller than the −271 tsp RACE product was observed at 0, 0.5, and 1 h (Fig. 2B). This band has been observed previously and was shown to correspond to the −184 tsp, suggesting that nonspecific priming, not another tsp, is responsible for this product (22). Isolation and sequencing in this case indicated that the corresponding bands in Fig. 2B also corresponded to transcription from the −184 tsp. These results indicate that transcription originates from the −728 and −696 tsps even under nitrogen-replete conditions. Conversely, transcripts originating from the −271 tsp are not detectable prior to 2 h after nitrogen step-down, even with the ultrasensitive RACE assay, which includes PCR amplification of products.
FIG. 2.
Temporal patterns of transcription from the tsps of hetR. (A) RACE products corresponding to the −728 and −696 tsps. (B) RACE products corresponding to the −271 and −184 tsps. The times (in hours) after removal of combined nitrogen are indicated below the gels.
Transcription from the −271 tsp is spatially regulated.
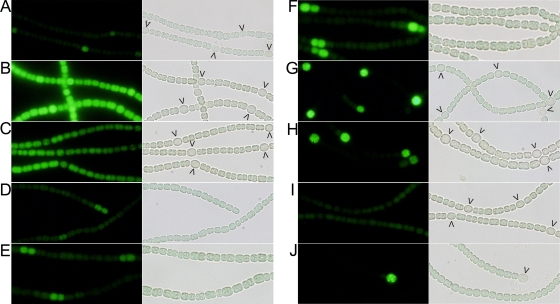
To determine which of the transcriptional start points of hetR contribute to patterned expression of hetR and which transcriptional start points contribute to the uniform background expression in filaments, the promoter region of hetR was separated into five regions with one or more of the hetR transcriptional start points, each region was fused individually to gfp, and the resulting fusions were introduced into the wild-type strain using a replicating plasmid. The individual tsp regions and resulting fusions are shown in Fig. 1. Consistent with previous reports on the spatial regulation of expression of hetR, expression, as indicated with the full-length promoter of hetR fused to gfp, was observed in all cells, was induced in certain cells prior to differentiation, and continued in mature heterocysts (1, 6) (Fig. 3A). In contrast, the wild-type strain bearing the −728/−696 tsp-gfp fusion exhibited considerable fluorescence throughout the filament under both nitrogen-replete and nitrogen starvation conditions (Fig. 3B). There was no appreciable difference in fluorescence between vegetative cells, proheterocysts, and heterocysts. The GFP fluorescence from this fusion was considerably brighter than that from the full-length promoter-transcriptional fusion of hetR to gfp. Similar levels of fluorescence were also observed in vegetative cells, proheterocysts, and heterocysts of the wild-type strain with the −184 tsp fusion (Fig. 3C). There was not a notable difference in the levels of GFP fluorescence between nitrogen-replete and nitrogen starvation conditions.
FIG. 3.
Fluorescence micrographs (left images) and light transmission micrographs (right images) of Anabaena sp. PCC 7120 bearing (A) PhetR-gfp in pSMC127, (B) the −728/−696 tsp-gfp fusion in pRR139, (C) the −184 tsp-gfp fusion in pRR151, (D to H) the −271 tsp-gfp fusion in pRR140, and (I) the −728/-696(+132) tsp-gfp fusion in pRR149 and (J) the patA mutant UHM101 bearing the −271 tsp-gfp fusion in pRR140. The images in panels A, B, C, G, I, and J were obtained 18 h after combined nitrogen was removed from the culture. The images in panels D, E, F, and H were obtained 4, 8, 12, and 24 h, respectively, after combined nitrogen was removed from the culture. The arrowheads indicate proheterocysts and heterocysts.
The wild-type strain bearing the −271 tsp-gfp fusion showed spatially patterned transcription (Fig. 3D to H). Temporal and spatial aspects of transcription from this fusion were examined under nitrogen-replete conditions and at specific time points after induction of heterocyst differentiation. GFP fluorescence was barely detectable under nitrogen-replete conditions. Upon induction, fluorescence increased by 4 h in groups of cells, most of which were in pairs, and the intensity of fluorescence increased with time. Localization of fluorescence to primarily single cells was seen at 18 h after nitrogen step-down and continued in proheterocysts and heterocysts. Vegetative cells between proheterocysts or heterocysts were dark except for the occasional cells midway between two heterocysts, which would presumably also differentiate in a subsequent round of heterocyst formation. The same progression of fluorescence from the −271 tsp-gfp fusion was observed even when ammonia was added to a concentration of 2 mM 14 h after nitrogen starvation began, just after the time of commitment to heterocyst differentiation (data not shown).
The gfp fusion with both the −271 and −184 tsps showed fluorescence in vegetative cells, proheterocysts, and heterocysts, and the fluorescence was brighter in the proheterocysts and heterocysts (data not shown). These results indicate that transcription from tsp −271 alone is regulated in a cell-type-specific manner and suggest that patterned induction of hetR is primarily, if not solely, attributable to this autoregulated transcriptional start site. A control strain bearing a promoterless version of the fusion plasmid was dark under conditions similar to those used for the GFP fusions (data not shown).
A region in the hetR promoter downstream of tsps −728 and −696 may have a negative regulatory role in transcription.
As mentioned above, the level of GFP fluorescence observed with the −728/−696 tsp fusion was greater than the level of GFP fluorescence observed with the full-length promoter fusion of hetR to gfp. We speculated that the region of DNA downstream of the two tsps may have a role in downregulating transcription. To verify this, the −728/−696(+132) tsp fusion, which included an additional 132 bases of the hetR promoter downstream of the initial construct (Fig. 1), was introduced into the wild-type strain on a replicating plasmid, and GFP fluorescence was examined. As predicted, the GFP fluorescence from this fusion was greatly reduced under both nitrogen-replete and nitrogen starvation conditions compared to the GFP fluorescence from the original −728/−696 tsp fusion (Fig. 3I). A low level of fluorescence was observed in both vegetative cells and heterocysts. In an attempt to quantify the difference in transcription in the two tsp fusions, corresponding fusions to lacZ were constructed (Fig. 1) and introduced on a plasmid into the wild-type strain. However, there were fewer transconjugants containing the −728/−696 tsp-lacZ fusion, and they grew more slowly than other strains, including the wild-type strain containing the −728/−696(+132) tsp-lacZ fusion. In addition, transconjugants exhibited dissimilar levels of β-galactosidase activity, which varied by more than 2 orders of magnitude or was absent in some cases. In contrast, transconjugants containing the lacZ fusion containing the additional downstream 132-bp region grew normally and exhibited similar levels of β-galactosidase activity that were roughly 20- to 60-fold lower than the levels exhibited by some of the strains carrying the −728/−696 tsp-lacZ fusion. These results suggest that transcription from the −728/−696 tsp-lacZ fusion negatively affects growth and that the presence of the extra 132-bp region limits transcriptional reporter activity.
PatA is required for transcription from tsp −271 in intercalary cells.
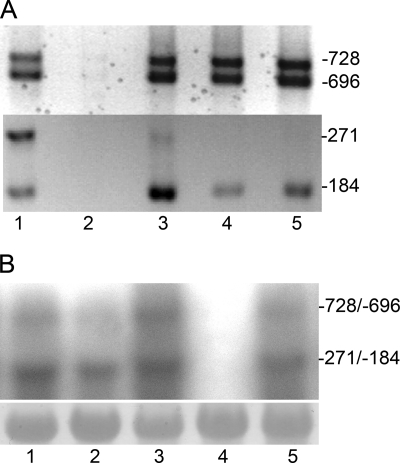
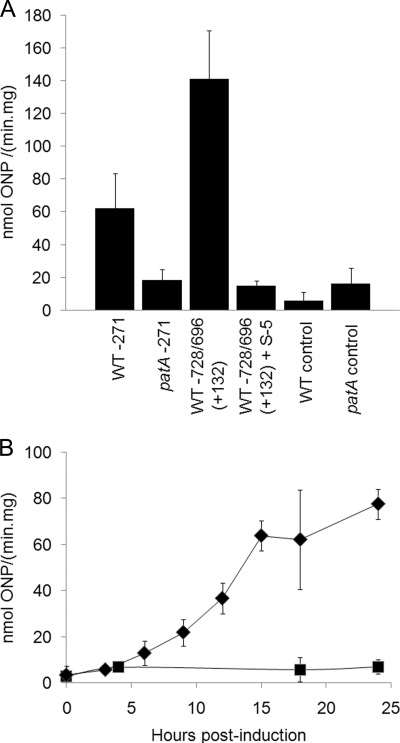
In order to determine if the heterocyst regulator PatA plays a role in the regulation of transcription from one or more of the tsps of hetR, transcription from the hetR tsps was analyzed in a patA mutant. Transcription from all four tsps was detected by RACE with RNA from both the wild type and the patA mutant, but not in the negative control, a strain lacking hetR. However, the intensity of the band corresponding to the −271 transcript was lighter in the patA mutant (Fig. 4A). Because RACE is an amplification procedure and therefore could not be relied upon to report quantitative differences in transcription, other more quantitative approaches were used. Transcripts originating from the −271 and −184 tsps typically cannot be resolved on a Northern blot (4) (Fig. 4B). Therefore, the −271 tsp-lacZ fusion was introduced into the wild type and into the patA mutant on a replicating plasmid. Differences in β-galactosidase activity between the two strains clearly showed that the level of transcription from tsp −271 was significantly lower in a patA mutant (Fig. 5A). There was substantial upregulation of transcription from tsp −271 in the wild type after nitrogen step-down up to about 15 h after induction of differentiation, after which the level of transcription started to level off (Fig. 5B). The patA mutant, however, showed only a modest increase in transcription from this tsp. A patA mutant produces primarily terminal heterocysts. The reduced level of transcription from the −271 tsp in the patA mutant correlates with the lower percentage of heterocysts in this strain. The lower levels of transcription could have been caused by insufficient induction of transcription from tsp −271 in the intercalary cells of the filaments. To verify our hypothesis, the −271 tsp-gfp fusion was introduced on a replicating plasmid into the patA mutant, and the spatial transcription pattern was examined. GFP fluorescence was not detected under nitrogen-replete conditions. After induction of heterocyst differentiation, GFP expression was seen in the terminal cells of the filaments, which is where heterocyst differentiation primarily occurs in this strain (Fig. 3J). Other than occasional intercalary cells, which presumably would also differentiate into heterocysts, GFP fluorescence was not discernible in intercalary cells of the filaments. However, the gfp expression from the −728/−696 tsp fusion or the −184 tsp fusion was similar to that observed in the wild type (data not shown). This demonstrates that PatA is required for normal patterned transcription from tsp −271 and underscores the tight nature of the control of transcription from this tsp.
FIG. 4.
Effects of PatA, PatS, and HetN on hetR transcription. (A) RACE results. Lane 1, wild-type strain; lane 2, hetR deletion strain UHM103; lane 3, patA deletion strain UHM101; lane 4, strain 7120PN, which overexpresses hetN from the petE promoter; lane 5, wild-type strain bearing pDR211, which overexpresses patS from the petE promoter. The upper panel shows products from the −728 and −696 tsps, and the lower panel shows products from the −271 and −184 tsps. (B) Northern blot results obtained using RNA isolated after 18 h of growth in BG110. Lane 1, strain 7120PN, which overexpresses hetN from the petE promoter; lane 2, wild type bearing plasmid pDR211, which overexpresses patS from the petE promoter; lane 3, wild type; lane 4, hetR deletion strain UHM103; lane 5, patA deletion stain UHM101. The lower panel shows the results for methylene blue-stained RNA as loading control.
FIG. 5.
(A) β-Galactosidase activity 18 h after induction of heterocyst differentiation for the wild type bearing the −271 tsp-lacZ fusion in plasmid pRR143, the patA mutant UHM101 bearing the −271 tsp-lacZ fusion in plasmid pRR143, the wild type bearing the −728/−696(+132) tsp-lacZ fusion in plasmid pRR152, the wild type bearing the −728/−696(+132) tsp-lacZ fusion in plasmid pRR152 with 6 μM PatS-5 included in the medium, the wild type bearing the control plasmid pNC101, and the patA mutant UHM101 bearing control plasmid pNC101. WT, wild type; S-5, PatS-5; ONP, o-nitrophenol. (B) Transcription from the −271 tsp at different times after induction of heterocyst differentiation. Diamonds, β-galactosidase activity of the wild type bearing the −271 tsp-lacZ fusion; squares, β-galactosidase activity of the wild type bearing control plasmid pNC101.
PatS and HetN affect transcription from multiple tsps of hetR.
The regulatory proteins PatS and HetN impact the spatiotemporal expression of HetR (6, 13, 16). To determine how each protein specifically impacts transcription from the four tsps of hetR, RACE was performed using RNA isolated 18 h after induction from a strain overexpressing hetN from the chromosome (strain 7120PN) or from the wild-type strain bearing plasmid pDR211, which was used to overexpress patS from a copper-inducible promoter. The transcripts from tsps −184, −696, and −728 were detected in both strains (Fig. 4A). The band corresponding to transcripts from tsp −271 was absent in both 7120PN and the strain overexpressing patS, showing that PatS and HetN prevent detectable levels of transcription from tsp −271.
In a Northern blot containing the RNA obtained from these strains, the lower band, which corresponded to transcripts from tsps −271 and −184, was detected in both strains (Fig. 4B). Since the RACE results show that tsp −271 is turned off in these two strains, this band likely corresponded to transcripts originating from tsp −184. The upper band, which corresponded to transcripts from tsps −728 and −696, was similar to the band from the wild type for the strain overexpressing hetN. However, this band was lighter in the strain overexpressing patS.
In order to corroborate the Northern blot results, the wild-type strain bearing the −728/−696(+132) tsp-lacZ fusion on a replicating plasmid was grown in medium supplemented with 6 μM PatS pentapeptide (PatS-5) (31), and β-galactosidase assays were performed. The β-galactosidase activity was significantly lower in the wild-type strain grown with PatS-5 added exogenously than in the same strain grown without PatS-5 (Fig. 5A). These results suggest that in addition to its negative impact on transcription from tsp −271, PatS downregulates transcription from one or both of the two distal tsps as well.
The −271 tsp is required for normal numbers of heterocysts, the timing of patterning, and commitment to differentiation.
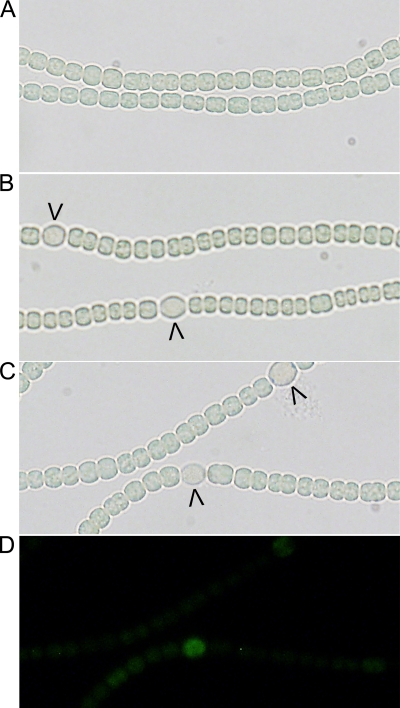
To investigate whether transcription from the −271 tsp is involved in creation of the periodic pattern of heterocysts or instead is a consequence of an existing pattern, two strains that lacked regions of DNA around the −271 tsp were created. Plasmids containing the hetR coding and promoter regions lacking either nucleotides −295 to −268 or nucleotides −317 to −268 were constructed and recombined into the chromosomal hetR locus of a hetR deletion strain. The results obtained with the two single-recombinant strains were indistinguishable, so these strains are referred to below collectively as the −271 tsp deletion strains. RACE using RNA extracted 48 h after transfer to fixed-nitrogen-free media indicated that transcription from the −271 tsp did not occur in the −271 tsp deletion strains, whereas the transcription from the −184, −696, and −728 tsps, as indicated by RACE, was similar to that of the wild type (data not shown). In both strains morphologically distinct heterocysts did not appear until 48 h after nitrogen starvation began (Fig. 6A and B), compared to 24 h for the wild type, and addition of ammonia to a final concentration of 2 mM 14 h after nitrogen starvation began prevented differentiation of heterocysts (data not shown). In contrast, the wild type forms cells committed to differentiation in a pattern prior to 14 h, after which addition of a source of fixed nitrogen does not prevent differentiation (30). The percentage of heterocysts formed by the −271 tsp deletion strains was about 2.5% at 48 h, compared to 9% for the wild type. In addition, induction of transcription from the −271 tsp in isolated cells was not observed from the −271 tsp deletion strains with the PhetR-gfp fusion on a plasmid 24 h after removal of combined nitrogen (data not shown). An increase in GFP fluorescence was observed in some cells 36 h after combined nitrogen was removed (Fig. 6C and D). Patterned fluorescence in the wild type with the same construct was visible 8 h after combined nitrogen was removed. Subsequently, the corresponding double-recombinant strains UHM156 and UHM157, which differ from the wild type by having nucleotides −295 to −268 and nucleotides −317 to −268, respectively, of the hetR promoter deleted cleanly from the chromosome, were isolated and shown to have phenotypes indistinguishable from those of the single-recombinant strains used in the experiment described above. These results indicate that the −271 tsp is required for pattern formation, commitment to differentiation, and differentiation of a number of heterocysts similar to that seen in the wild type.
FIG. 6.
Deletion of the −271 tsp region from the hetR promoter. (A and B) Photomicrographs of the −271 tsp deletion strain lacking nucleotides −295 to −268 of the hetR promoter region 24 h (A) and 48 h (B) after induction of heterocyst differentiation. (C and D) Light transmission micrograph (C) and fluorescence micrograph (D) obtained 48 h after combined nitrogen was removed from the −271 tsp deletion strain lacking nucleotides −295 to −268 of the hetR promoter region bearing PhetR-gfp in a plasmid (pSMC127). The arrowheads indicate proheterocysts and heterocysts.
DISCUSSION
The spatiotemporal regulation of transcription from the four transcriptional start sites of hetR was investigated to determine how each tsp affects cellular differentiation and pattern formation in Anabaena sp. PCC 7120. Induction of hetR expression through use of inducible promoters, such as PpetE (5) and Pnir (23), on a multicopy plasmid triggers heterocyst differentiation even under conditions under which differentiation does not normally take place. Under such conditions, however, the frequency and pattern of heterocysts are atypical. Differentiation of heterocysts in a wild-type pattern requires first that expression of HetR is localized to select cells and then that the levels of expression are sufficient to irreversibly commit cells to differentiate into heterocysts. Differential expression of hetR in response to the timing of nitrogen starvation and the relative position of a cell in a filament is facilitated by multiple transcription start points.
When fixed nitrogen in a form that can be readily incorporated into carbon skeletons, such as ammonia, is present at a concentration sufficient for robust growth, expression of hetR is primarily from the −184 tsp (5, 19). Presumably, a low, basal level of hetR expression maintains the cells of the filament in a state primed to respond quickly to nitrogen starvation. It is not likely that transcription from the other tsps of hetR has a significant role in maintaining basal hetR expression levels. Transcription is not detected from the −271 tsp under nitrogen-replete conditions. The RACE experiments demonstrated that transcription occurs from the −728 and −696 tsps even under nitrogen-replete conditions. However, the level of transcription is quite low, as demonstrated by other workers using Northern blots and primer extension (5, 19) and as demonstrated in this study with reporter fusions. The dramatic differences in transcription observed in nitrogen-replete conditions with the −728/−696 and −728/−696(+132) tsp-reporter fusions indicate that a region of DNA downstream of these tsps is necessary for limiting transcription from the tsps under nitrogen-replete conditions.
The two distal tsps of hetR appear to form the nitrogen-status-sensing and response arm of the hetR transcription apparatus. The signal for nitrogen starvation is transmitted to hetR by NtcA via NrrA, resulting in induction of transcription from the tsps located at nucleotides −728 and −696 (7, 8). The data for tsp-gfp fusions suggest that transcription from these two tsps occurs at roughly the same level along the filament and is not spatially regulated, although when the data obtained for these fusions are interpreted, it should be borne in mind that each of these tsp fusions lacks parts of the promoter that may contribute to its regulation. On the other hand, our results demonstrated that the −271 tsp is differentially regulated based on a cell's position in a filament. Soon after induction, clusters of fluorescent cells are present in filaments carrying the −271 tsp-gfp fusion, demonstrating that transcription from the −271 tsp is upregulated in several groups of contiguous cells prior to resolution of these clusters to produce single cells. This pattern of transcription is similar to that exhibited by patS (30). Between 12 and 18 h after the onset of nitrogen starvation, transcription from this start site is localized to developing heterocysts. This time roughly coincides with the commitment of cells to differentiation. Subsequently, transcription from the −271 tsp is seen only in vegetative cells that are midway between heterocysts, and these cells presumably differentiate in a secondary round of differentiation. These results suggest that the patterned induction of hetR observed upon nitrogen deprivation is attributable primarily to this transcriptional start site. Intriguingly, when nitrate, which must be reduced before it can be incorporated into an organic molecule, is used as the fixed-nitrogen source rather than ammonia, filaments are maintained in a state in which clusters of cells express hetR from the −271 tsp (data not shown), demonstrating that the spatial induction of transcription from the −271 tsp can be maintained in an intermediate state. Our results are consistent with constitutive expression from the −184 tsp, which supplies a basal level of hetR mRNA under all growth conditions.
The network of genes involved in regulating differentiation and pattern formation has the potential to act as a genetic switch that converts a temporary input signal, such as nitrogen starvation, into a stable “on” or “off” gene expression response, leading to permanent alteration of cells (10). In genetic switches positive and/or negative feedback loops contribute to memory by the system. HetR forms positive and negative feedback loops with itself and with other regulatory proteins. In strains in which the −271 tsp was deleted from the chromosome, heterocyst differentiation was delayed along with patterned transcription from a PhetR-gfp reporter fusion. The percentage of heterocysts was also lower in these strains. Taken together, these results indicate that the −271 tsp is necessary for normal timing of pattern formation and morphological differentiation.
Heterocyst differentiation is an endpoint process leading to permanent genetic, morphological, and physiological changes in the differentiating cell. The heterocyst does not divide and cannot revert to being a vegetative cell. Between 9 and 14 h after induction of differentiation, cells become committed to differentiate, even if the initial starvation signal is removed (30). By using mathematical modeling, Brandman et al. (3) predicted that many biological systems use multiple interlinked loops because they generate reliable all-or-none switches that are both rapidly induced and more resistant to noise in the upstream signaling pathway than single positive feedback loops. When one of two loops regulating a pathway is slow and the other is fast, the fast loop is critical to the speed of transition from one state to the other, while the slow loop is essential for the stability of the “on” state (3). We suggest that the NtcA-mediated activation of transcription from the −728 and −696 tsps generates a quick response to nitrogen starvation, while induction of transcription from the −271 tsp takes place at a lower rate and is necessary for the normal timing of irreversible commitment to differentiation.
Our results show that PatA, HetN, and PatS all regulate transcription from the −271 tsp, which in turn should affect the direct positive feedback loop discussed above. HetN and PatS have a negative effect on transcription from the −271 tsp, and PatA has a positive effect. These findings are consistent with the suggestion that PatA attenuates the inhibitory activities of PatS and HetN. It has been proposed that PatA mediates the protein-protein interaction between HetR and HetF (22). It has also been proposed that HetF, a predicted protease, modifies HetR into a form that is less sensitive to inhibition by PatS and HetN. In the absence of hetF, transcription from the −271 tsp is not detected, and overexpression of hetF in a patA mutant results in bypass of the terminal heterocyst phenotype (22). Thus, it seems likely that PatA works through HetF to effect transcription from tsp −271. It has been proposed that PatS acts as a diffusible inhibitor of differentiation and interacts with HetR in pattern formation. Upregulation of patS transcription is dependent on hetR (13). It is likely that competition between HetR and PatS plays a significant role in localization of transcription from tsp −271. Although HetN does not play a major role in pattern formation, it appears to stabilize the pattern and regulate the sites of new heterocysts between existing heterocysts to maintain the pattern as a filament lengthens by preventing transcription from tsp −271. Transcription from the start sites of hetR is differentially regulated based on fixed-nitrogen levels in the medium and the relative position of a cell in a filament. Several genes involved in differentiation, such as ntcA (19, 21), hetR (5, 19), glnA (24), patS (30), and possibly the hetZ-patU cluster (31), are transcribed from multiple transcription start points. The use of multiple tsps rather than differential regulation of a single tsp appears to be a common theme for the regulation of genes involved in the differentiation and patterning of heterocysts.
Acknowledgments
We thank Guntram Christiansen, Hongwei Li, and members of the Li lab for help with RNA work, Douglas Risser for providing plasmids pDR211 and pDR325, and Kelly Higa for help in constructing strains.
This work was supported by grant MCB-0343998 from the National Science Foundation.
Footnotes
Published ahead of print on 11 December 2009.
REFERENCES
- 1.Black, T. A., Y. Cai, and C. P. Wolk. 1993. Spatial expression and autoregulation of hetR, a gene involved in the control of heterocyst development in Anabaena. Mol. Microbiol. 9:77-84. [DOI] [PubMed] [Google Scholar]
- 2.Borthakur, P. B., C. C. Orozco, S. S. Young-Robbins, R. Haselkorn, and S. M. Callahan. 2005. Inactivation of patS and hetN causes lethal levels of heterocyst differentiation in the filamentous cyanobacterium Anabaena sp. PCC 7120. Mol. Microbiol. 57:111-123. [DOI] [PubMed] [Google Scholar]
- 3.Brandman, O., J. E. J. Ferrell, R. Li, and T. Meyer. 2005. Interlinked fast and slow positive feedback loops drive reliable cell decisions. Science 310:496-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buikema, W. J., and R. Haselkorn. 1991. Characterization of a gene controlling heterocyst development in the cyanobacterium Anabaena 7120. Genes Dev. 5:321-330. [DOI] [PubMed] [Google Scholar]
- 5.Buikema, W. J., and R. Haselkorn. 2001. Expression of the Anabaena hetR gene from a copper-regulated promoter leads to heterocyst differentiation under repressing conditions. Proc. Natl. Acad. Sci. U. S. A. 98:2729-2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Callahan, S. M., and W. J. Buikema. 2001. The role of HetN in maintenance of the heterocyst pattern in Anabaena sp. PCC 7120. Mol. Microbiol. 40:941-950. [DOI] [PubMed] [Google Scholar]
- 7.Ehira, S., and M. Ohmori. 2006. NrrA directly regulates expression of hetR during heterocyst differentiation in the cyanobacterium Anabaena sp. strain PCC 7120. J. Bacteriol. 188:8520-8525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ehira, S., and M. Ohmori. 2006. NrrA, a nitrogen-responsive response regulator facilitates heterocyst development in the cyanobacterium Anabaena sp. strain PCC 7120. Mol. Microbiol. 59:1692-1703. [DOI] [PubMed] [Google Scholar]
- 9.Elhai, J., and C. P. Wolk. 1988. Conjugal transfer of DNA to cyanobacteria. Methods Enzymol. 167:747-754. [DOI] [PubMed] [Google Scholar]
- 10.Ferrell, J. E. J. 2002. Self-perpetuating states in signal transduction: positive feedback, double-negative feedback and bistability. Curr. Opin. Cell Biol. 14:140-148. [DOI] [PubMed] [Google Scholar]
- 11.Frías, J. E., E. Flores, and A. Herrero. 1994. Requirement of the regulatory protein NtcA for the expression of nitrogen assimilation and heterocyst development genes in the cyanobacterium Anabaena sp. PCC 7120. Mol. Microbiol. 14:823-832. [DOI] [PubMed] [Google Scholar]
- 12.Golden, J. W., and H.-S. Yoon. 2003. Heterocyst development in Anabaena. Curr. Opin. Microbiol. 6:557-563. [DOI] [PubMed] [Google Scholar]
- 13.Huang, X., Y. Dong, and J. Zhao. 2004. HetR homodimer is a DNA-binding protein required for heterocyst differentiation, and the DNA-binding activity is inhibited by PatS. Proc. Natl. Acad. Sci. U. S. A. 101:4848-4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khudyakov, I. Y., and J. W. Golden. 2004. Different functions of HetR, a master regulator of heterocyst differentiation in Anabaena sp. PCC 7120, can be separated by mutation. Proc. Natl. Acad. Sci. U. S. A. 101:16040-16045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laurent, S., H. Chen, S. Bedu, F. Ziarelli, L. Peng, and C.-C. Zhang. 2005. Nonmetabolizable analogue of 2-oxoglutarate elicits heterocyst differentiation under repressive conditions in Anabaena sp. PCC 7120. Proc. Natl. Acad. Sci. U. S. A. 102:9907-9912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li, B., X. Huang, and J. Zhao. 2002. Expression of hetN during heterocyst differentiation and its inhibition of hetR up-regulation in the cyanobacterium Anabaena sp. PCC 7120. FEBS Lett. 517:87-91. [DOI] [PubMed] [Google Scholar]
- 17.Liang, J., L. Scappino, and R. Haselkorn. 1992. The patA gene product, which contains a region similar to CheY of Escherichia coli, controls heterocyst pattern formation in the cyanobacterium Anabaena 7120. Proc. Natl. Acad. Sci. U. S. A. 89:5655-5659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meeks, J. C., C. P. Wolk, W. Lockau, N. Schilling, P. W. Shaffer, and W.-S. Chien. 1978. Pathways of assimilation of [13N]N2 and 13NH4+ by cyanobacteria with and without heterocysts. J. Bacteriol. 134:125-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muro-Pastor, A. M., A. Valladares, E. Flores, and A. Herrero. 2002. Mutual dependence of the expression of the cell differentiation regulatory protein HetR and the global nitrogen regulator NtcA during heterocyst development. Mol. Microbiol. 44:1377-1385. [DOI] [PubMed] [Google Scholar]
- 20.Orozco, C. C., D. D. Risser, and S. M. Callahan. 2006. Epistasis analysis of four genes from Anabaena sp. strain PCC 7120 suggests a connection between PatA and PatS in heterocyst pattern formation. J. Bacteriol. 188:1808-1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramasubramanian, T. S., T.-F. Wei, A. K. Oldham, and J. W. Golden. 1996. Transcription of the Anabaena sp. strain PCC 7120 ntcA gene: multiple transcripts and NtcA binding. J. Bacteriol. 178:922-926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Risser, D. D., and S. M. Callahan. 2008. HetF and PatA control levels of HetR in Anabaena sp. strain PCC 7120. J. Bacteriol. 190:7645-7654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Risser, D. D., and S. M. Callahan. 2007. Mutagenesis of hetR reveals amino acids necessary for HetR function in the heterocystous cyanobacterium Anabaena sp. strain PCC 7120. J. Bacteriol. 189:2460-2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Valladares, A., A. M. Muro-Pastor, A. Herrero, and E. Flores. 2004. The NtcA-dependent P1 promoter is utilized for glnA expression in N2-fixing heterocysts of Anabaena sp. strain PCC 7120. J. Bacteriol. 186:7337-7343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vázquez-Bermúdez, M., A. Herrero, and E. Flores. 2000. Uptake of 2-oxoglutarate in Synechococcus strains transformed with the Escherichia coli kgtP gene. J. Bacteriol. 182:211-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wei, T.-F., R. Ramasubramanian, and J. W. Golden. 1994. Anabaena sp. strain PCC 7120 ntcA gene required for growth on nitrate and heterocyst development. J. Bacteriol. 176:4473-4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wolk, C. P. 1996. Heterocyst formation. Annu. Rev. Genet. 30:59-78. [DOI] [PubMed] [Google Scholar]
- 28.Wolk, C. P. 1968. Movement of carbon from vegetative cells to heterocysts in Anabaena cylindrica. J. Bacteriol. 96:2138-2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wolk, C. P., A. Ernst, and J. Elhai. 1994. Heterocyst metabolism and development, p. 769-823. In D. A. Bryant (ed.), The molecular biology of cyanobacteria, vol. 1. Kluwer Academic Publishers, Dordrecht, The Netherlands. [Google Scholar]
- 30.Yoon, H.-S., and J. W. Golden. 2001. PatS and products of nitrogen fixation control heterocyst pattern. J. Bacteriol. 183:2605-2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang, W., Y. Du, I. Khudyakov, Q. Fan, H. Gao, D. Ning, C. P. Wolk, and X. Xu. 2007. A gene cluster that regulates both heterocyst differentiation and pattern formation in Anabaena sp. strain PCC 7120. Mol. Microbiol. 66:1429-1443. [DOI] [PubMed] [Google Scholar]