Abstract
Saccharophagus degradans strain 2-40 is a prominent member of newly discovered group of marine and estuarine bacteria that recycle complex polysaccharides. The S. degradans 2-40 genome codes for 15 extraordinary long polypeptides, ranging from 274 to 1,600 kDa. Five of these contain at least 52 cadherin (CA) and cadherin-like (CADG) domains, the types of which were reported to bind calcium ions and mediate protein/protein interactions in metazoan systems. In order to evaluate adhesive features of these domains, recombinant CA doublet domains (two neighboring domains) from CabC (Sde_3323) and recombinant CADG doublet domains from CabD (Sde_0798) were examined qualitatively and quantitatively for homophilic and heterophilic interactions. In addition, CA and CADG doublet domains were tested for adhesion to the surface of S. degradans 2-40. Results showed obvious homophilic and heterophilic, calcium ion-dependent interactions between CA and CADG doublet domains. Likewise, CA and CADG doublet domains adhered to the S. degradans 2-40 surface of cells that were grown on xylan from birch wood or pectin, respectively, as a sole carbon source. This research shows for the first time that bacterial cadherin homophilic and heterophilic interactions may be similar in their nature to cadherin domains from metazoan lineages. We hypothesize that S. degradans 2-40 cadherin and cadherin-like multiple domains contribute to protein-protein interactions that may mediate cell-cell contact in the marine environment.
Saccharophagus degradans strain 2-40 is the first free-living marine bacterium demonstrated to be capable of degrading cellulose of algal origin and higher plant material (29, 31). S. degradans 2-40 was isolated from decaying salt marsh cord grass, Spartina alterniflora, in the Chesapeake Bay watershed (3, 12). It is a pleomorphic, Gram-negative, aerobic, motile gammaproteobacterium, uniquely degrading at least 10 different complex polysaccharides, including agar, chitin, alginic acid, cellulose, β-glucan, laminarin, pectin, pullulan, starch, and xylan (9, 10, 15, 16, 30). These enzymatic capabilities suggest that the bacterium S. degradans may have a significant role in the marine carbon cycle, mediating the degradation of complex polysaccharides from plants, algae, and invertebrates (30, 31).
The S. degradans 2-40 genome has been sequenced (http://genome.jgi-psf.org/finished_microbes/micde/micde.home.html; GenBank accession numbers CP000282 and NC_007912). It has 4,008 genes in a single replicon consisting of 5.06 Mb. The genome codes for 15 polypeptides longer than 2,000 amino acids (aa), ranging from 274 to 1,600 kDa. Each contains multiple domains and motifs that are reported to bind calcium ions and mediate protein-protein interactions (30, 31). They are acidic, pI 3.5 to 4.9, and have a secretion signal; cadherin (CA) and cadherin-like (CADG) domains are prevalent.
Cadherins are a superfamily of transmembrane glycoproteins that mediate, Ca2+-dependent cell-cell adhesion in all solid tissues throughout the animal kingdom (25). At the molecular level, homotypic adhesion between cells arises from homophilic interactions between cadherin extracellular domains repeated in tandem (18, 19). Each of these domains, consisting of approximately 110 amino acids, forms a β-sandwich with Greek-key folding topology. The cadherin domains are mostly distributed in the metazoan lineage. According to the SMART database (accessed July 2009), there are 23,340 CA domains in 3,673 proteins, and of these 3,481 (94.7%) are metazoan proteins and only 186 (5.06%) are bacterial proteins (six other proteins with CA domains belong to Archaea).
Genomic analysis showed that CA domains and CADG domains are not uncommon in bacteria (7). Focusing on the proteins Sde_3323 and Sde_0798, we tested the notion that they would interact as in metazoans, binding via calcium-dependent homophilic and heterophilic interactions. We also examined the possibility that they would directly interact with the S. degradans 2-40 cell surface, playing a role in protein-protein interactions and cellular aggregation.
MATERIALS AND METHODS
Strains, media, and culture conditions.
S. degradans strain 2-40 or ATCC 43961 (Sde 2-40; formerly Microbulbifer degradans strain 2-40) was isolated from decaying salt marsh cord grass, S. alterniflora, in the Chesapeake Bay watershed (3). Escherichia coli strain XL-1 Blue was used for plasmid constructions and strain BL21(λDE3) pLysS was used for protein overexpression. S. degradans 2-40 was grown in minimal medium containing (per liter) 23 g of Instant Ocean salts (Aquarium Systems, Mentor, OH), 5 g of NH4Cl, 2 g of carbon source, and 50 mM Tris-HCl (pH 7.6); 20 g of agar per liter was used to prepare solid medium. The carbon sources used in this study included agar (Acumedia Manufacturers, Inc. Lansing, MI), agarose (Hispanagar, Burgos, Spain), cellulose (Sigma Aldrich, Munich, Germany), cellobiose (Sigma Aldrich, Munich, Germany), chitin from crab shells (practical grade; Sigma Aldrich, Munich, Germany), lichenan (Sigma Aldrich, Munich, Germany), pectin (Sigma Aldrich, Germany), starch (Sigma Aldrich, Munich, Germany), xylan from oat spelt (Sigma Aldrich, Munich, Germany), xylan from birch wood (Sigma Aldrich, Munich, Germany), marine broth 2216 (Difco Laboratories, MD), and glucose (Sigma Aldrich, Munich, Germany). All cultures were incubated at 30°C. E. coli XL1-Blue and BL21(λDE3) pLysS strains were grown in Luria-Bertani (LB) broth or agar supplemented with 50 μg/ml kanamycin and incubated at 37°C.
Cloning and molecular biology protocols.
All DNA manipulations and standard molecular biology protocols were performed as described by Sambrook et al. (27). Plasmid DNA was isolated using a plasmid purification kit (iNtRON Biotechnology, Inc., South Korea).
Plasmid constructions.
S. degradans strain 2-40 chromosomal DNA was isolated and used as a template for PCR amplifications of CA (amino acids 2288 to 2496) and CADG (amino acids 2261 to 2500) doublet domains (two neighboring domains) from CabC (Sde_3323) and CabD (Sde_0798), respectively. These domains were amplified using PCR and appropriately tailed primers (Table 1). The PCR-amplified products were digested by NcoI and NotI and cloned into pET28a (Novagen, Inc., Darmstadt, Germany) to obtain pET28a-CA and pET28a-CADG carrying DNA fragments encoding CA and CADG doublet domains, respectively. Two other PCR-amplified products were digested by BamHI and XhoI and cloned into pET28a-CBM3a (5) to obtain pET28a_CBM3a-CA and pET28a_CBM3a-CADG (CBM3a is a carbohydrate binding module of the Clostridium thermocellum scaffoldin protein CipA). The resulting recombinant plasmids were initially introduced into E. coli XL1-Blue cells and then retransformed into E. coli BL21(λDE3) pLysS cells.
TABLE 1.
PCR oligonucleotide primers used in this study
| Primer namea | Sequence (5′ to 3′)b | Construct namec |
|---|---|---|
| CA_F | AACCATGGCCGCAGTAGAAGTAAATGTATTG | pET28a_CA |
| CA_R | TAGCGGCCGCGCCATCGATATCGTCATCATTAAC | |
| CADG_F | AACCATGGCTGTAAATGAAGATTCGGTG | pET28a_CADG |
| CADG_R | TTGCGGCCGCTATACTTTCCGATCCGTCGTGC | |
| CA_CBM3a_F | AAGGATCCGCAGTAGAAGTAAATGTA | pET28a_CBM3a-CA |
| CA_CBM3a_R | TTCTCGAGGCCATCGATATCGTCATCA | |
| CADG_CBM3a_F | AAGGATCCGCAGTAGAAGTAAATGTA | pET28a_CBM3a-CADG |
| CADG_CBM3a_R | TTCTCGAGACTTTCCGATCCGTCGT |
Forward (F) and reverse (R) primers are indicated.
Restriction sites are underlined.
See Materials and Methods.
Protein expression and purification.
An overnight culture of E. coli BL21(λDE3) pLysS cells was diluted to a cell density of 0.1 at 600 nm in LB medium containing kanamycin (50 μg/ml) and shaken vigorously at 37°C. When the culture reached a cell density of 0.6 at 600 nm, isopropyl-β-d-thiogalactopyranoside (IPTG) (Sigma Aldrich, St. Louis, MO) was added to a final concentration of 0.5 mM. The cells were incubated for an additional 3 h at 37°C and harvested by centrifugation at 4,000 × g for 20 min at 4°C. Then the pellet was suspended in sonication buffer consisting of 50 mM Na2HPO4 (pH 8.0), 0.3 M NaCl, and 10 mM imidazole. To the cell suspension, 1 mM phenylmethylsulfonyl fluoride (PMSF) was added, and the mixture was sonicated in an ultrasonic possessor (Misonics) until clear. The sonicate was centrifuged at 15,000 × g for 15 min at 4°C. The supernatant was mixed with His tag capture resin (Zephyr ProteomiX, Israel) equilibrated with sonication buffer. The resin was subsequently washed with wash buffer consisting of 50 mM Na2HPO4 (pH 8.0), 0.3 M NaCl, and 20 mM imidazole. The recombinant protein was absorbed onto the resin and was eluted with elution buffer containing 50 mM Na2HPO4 (pH 8.0), 0.3 M NaCl, and 250 mM imidazole. Further purification was performed as follows: 2 ml of the His-tagged proteins was injected into a gel filtration column (80-ml column volume; 1.6-cm diameter) and purified using gel filtration buffer consisting of 50 mM Tris-HCl (pH 7.5), 0.15 M NaCl, and 7.7 mM NaN3. The purified fractions were collected, and then the proteins were concentrated using a Centriprep centrifugal filter device YM 3 (Amicon Bioseparations Millipore, Billerica, MA) with a molecular-mass cutoff of 3 kDa. The concentrated samples were then stored at −20°C. The purified proteins were visualized by SDS-12.5% PAGE and stained by Coomassie brilliant blue.
Western blot analysis.
The recombinant proteins (His6-tagged CA [CAHis6] and CAGDHis6) were separated by SDS-PAGE (12.5%) and transferred onto a nitrocellulose membrane. The membrane was then blocked for 1 h with TBS-Ca-T buffer (25 mM Tris-HCl [pH 7.0], 150 mM NaCl, 7 mM CaCl2, and 0.04 mM Tween-20) and 0.04 mM bovine serum albumin ([BSA] Sigma Aldrich, St. Louis, MO). Recombinant CAHis6 and CADGHis6 fused to CBM3a (CBM3aCAHis6 and CBM3aCADGHis6, respectively) proteins (2 μg/ml) were added to blocking buffer to serve as protein probes. After an additional 1 h of incubation, the membrane was washed three times with TBS-Ca-T buffer and treated with rabbit anti-CBM3a serum (kindly provided by Ely Morag, Weizmann Institute, Israel), diluted 1:25,000 in blocking buffer for 1 h. The membrane was washed three times again and incubated with horseradish peroxidase (HRP)-conjugated AffiniPure goat anti-rabbit (Jackson ImmunoResearch Laboratories, Inc., Suffolk, United Kingdom) for 30 min, and washed three times more. The homophilic and heterophilic interactions of the CA and CADG domains were revealed by enhanced chemiluminescence substrate (SuperSignal West Pico; Pierce Biotechnology, Rockford, IL) according to the manufacturer's instructions.
To ensure the specificity of interactions, separate blots were run with identical samples and probed with normal rabbit IgG in place of the primary antibodies.
ELISA for quantitative analysis of protein-protein interactions.
The CA and CADG doublet domain interactions were assessed in microtiter plates by affinity analysis using matching fusion proteins (5). Enzyme-linked immunosorbent assay (ELISA) Nunc-Immuno plates (Nunc, Roskilde, Denmark) were coated with increasing amounts of CAHis6 or CADGHis6 (0 to 12.5 μg/well) in 0.1 M Na2CO3 (pH 9.0), for a final volume of 50 μl/well. The samples were incubated overnight at 4°C. After 24 h, the plates were blocked with 200 μl/well of blocking buffer containing TBS buffer, 0.04 mM Tween-20, and either 7 mM CaCl2·2H2O or 10 mM EDTA for 2 h at room temperature. This was followed by the addition of 100 μl/well of CBM3aCAHis6 or CBM3aCADGHis6, dissolved in blocking buffer, for 2 h at room temperature. The plates were washed three times with washing buffer containing TBS buffer, 0.04 mM Tween-20, and either 7 mM CaCl2·2H2O or 10 mM EDTA. Next, 50 μl/well of rabbit anti-CBM3a serum, diluted 1:25,000 in blocking buffer, was added and incubated for 1 h at room temperature. The plates were washed three times with washing buffer and incubated with 50 μl/well of HRP-conjugated AffiniPure goat anti-rabbit, diluted 1:10,000, for 30 min. After a washing step, the interactions were visualized by adding 100 μl/well of 3,3′,5,5′-tetramethylbenzidine (TMB) color reagent solution, prepared according to the manufacturer's instructions (Sigma Aldrich, St. Louis, MO). The reaction was stopped after 3 min with 50 μl/well of 1 M H2SO4. The optical density was directly measured at 450 nm by using an EL 800 Universal Microplate Reader (Bio-Tek Instruments, Inc., Belgium).
Cell-based ELISA for quantitative analysis of protein-bacterial cell surface interactions.
The cell-based enzyme-linked immunosorbent assay was employed, essentially as described by Rosok et al. (26). ELISA Nunc-Immuno plates were coated with 50 μl/well of 10 μg/ml poly-l-lysine for 30 min at 25°C. Next, the plates were coated with 50 μl/well of S. degradans 2-40 cells from cultures grown in different sole carbon sources, diluted to an optical density of 0.2 at 600 nm, and incubated for 1 h at 37°C. The plates were then blocked for 1 h at 25°C with Instant Ocean salts buffer (0.04 mM Tween-20 and 50 mM Tris-HCl, pH 7.6) containing, per liter, 23 g of Instant Ocean salts, 5 g of NH4Cl, and 30 g of BSA. Recombinant CAHis6 and CADGHis6 proteins (100 μg/ml) were added to the blocking buffer and served as probes for 1 h at 30°C. The plates were washed three times with Instant Ocean salts buffer. Next, 50 μl/well purified mouse anti-His (horseradish peroxidase-labeled monoclonal mouse antibody; Sigma), diluted 1:1,000 in Instant Ocean salts buffer, was added and incubated for 1 h at 25°C. Finally, after three washes with Instant Ocean salts buffer, detection was performed using 100 μl/well TMB color reagent solution. The reaction was stopped after 3 min with 50 μl/well 1 M H2SO4, and the optical density was measured at 450 nm using an EL 800 Universal Microplate Reader.
TEM.
Cells were harvested during the logarithmic growth stage by centrifugation, washed twice in Instant Ocean salts buffer, and resuspended in 1/2 volume of Instant Ocean salts buffer. For transmission electron microscopy (TEM), a 10-μl drop of cell suspension was adsorbed onto 200 carbon-coated cuprum grids for 30 s and stained for 25 s with 1% (wt/vol) uranyl acetate. Specimens were viewed on a transmission electron microscope (JEM-1200EX; Japan) and images were taken by camera (Mega View ΙΙΙ, SIS, Germany).
Bioinformatics analysis of protein sequences.
Protein domains were identified using the Simple Modular Architecture Tool (SMART) (20, 28) (http://www.smart.embl-heidelberg.de), the Pfam protein families database (11) (http://pham.sanger.ac.uk), integrated resource of Protein Domains (InterPro [http://www.ebi.ac.uk/interpro/]), and the database of protein families and domains PROSITE (http://www.expasy.ch/prosite/). Similarity searches were performed using the BLAST algorithm at the NCBI server (http://www.ncbi.nih.nlm.gov) (2). Type II secretion signals were identified using the iPSORT program (http://www.hypothesiscreator.net/iPSORT) (4) and the SignalP, version 1.1, program (http://www.cbs.dtu.dk/services/SignalP) (22). Pairwise and multiple sequence alignments were performed with the ClustalW program, version 1.84 (14), with the EMBL ClustalW2 server (http://www.ebi.ac.uk/Tools/clustalw2/index.html) and the Network Protein Sequence Analysis server (http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsaclustalw.html). Estimated protein molecular masses were calculated using the Peptide Mass Tool at the ExPASy server of the Swiss Institute of Bioinformatics (http://www.us.expasy.org).
RESULTS
Identification and modular structure of cadherin and cadherin-like encoding representative proteins in the S. degradans 2-40 genome.
Upon the release of the complete S. degradans 2-40 genome sequence (GenBank NC_007912), our systematic analysis revealed that the S. degradans 2-40 genome contains at least 52 cadherin (CA) and cadherin-like (CADG) domains in five large, secreted proteins (Sde_0798, Sde_1294, Sde_2834, Sde_3233, and Sde_3323) (see Table S1 in the supplemental material). This research focused on the CA doublet domains from CabC (Sde_3323) and on CADG doublet domains from CabD (Sde_0798). The sequences were analyzed using the protein database as described above in Materials and Methods section.
CabC and CabD were found to be putative paralogous secreted proteins with an extraordinarily complex modular architecture (Fig. 1 and Table 2; see also Table S3 in the supplemental material). CabD (Sde_0798) is a 3,474-aa protein with an estimated pI of 3.57 and molecular mass of 359.6 kDa. It contains two CADHERIN_2 (PS50268) domains and eight CADG (SM00736) domains. In addition, CabD contains prominent adhesive domains: five He_PIG, five VCBS repeats, and FG-GAP (Fig. 1). CabC (Sde_3323) is a 3,477-aa protein with an estimated pI of 3.51 and molecular mass of 348.5 kDa. It contains three CADHERIN_2 (PS50268) domains, two CADG (SM00736) domains, and six additional modules resembling cadherins (only one was identified by InterPro as cadherin, and five others were predicted in this study) (Fig. 1). Independent protein databases define cadherins by different names as cadherin, CADHERIN_2, or CA, while cadherin-like domains are defined as cadherin-like or CADG. In addition, CabC contains other domains that are reported to have adhesive function including one Calx-beta, one PKD, four CheC, and one He_PIG (Table 2).
FIG. 1.
Schematic diagram of the S. degradans CabC (Sde_3323) and CabD (Sde_0798) main domains. The CabC and CabD proteins were analyzed using the Pfam, SMART, InterPro, and PROSITE databases. The domains are color coded according to the legend on the figure. The cadherin terminology is still evolving and is defined as follows: CA (SMART accession number SM00112), CADHERIN_2 (InterPro PS50268), cadherin (InterPro, SSF49313), and CADG (SMART SM00736). Other domains include He_PIG (Pfam PF05345), OMP b-brl (InterPro G3DSA:2.40.160.20) and VCBS repeats (InterPro TIGR01965). All of them have been suggested to play a role in adhesion. Numbers shown below selected domains indicate numbering of amino acid residues. Protein domains analyzed in this study are framed with a dashed line.
TABLE 2.
Domain organization of CabC (Sde_3323) and CabD (Sde_0798) proteins
| Protein and domain prediction site (accession no.)a | Amino acid position | Predicted domain |
|---|---|---|
| CabC | ||
| PS50268 | 346-482 | CADHERIN_2 |
| 2077-2179 | ||
| 2289-2373 | ||
| PF00801 | 2946-3004 | PKD |
| SM00089 | 2920-3007 | |
| PF03160 | 2839-2870 | Calx-beta |
| SM00736 | 2565-2658 | CADG |
| 2911-3011 | ||
| PF04509 | 1463-1486 | CheC |
| 1567-1582 | ||
| 1663-1678 | ||
| 1855-1870 | ||
| PF05345 | 2597-2640 | He_PIG |
| G3DSA:2.40. 160.20 | 3308-3467 | OMP-b-brl |
| SSF49313 | 2563-2657 | Cadherin |
| CabD | ||
| PS50268 | 1208-1305 | CADHERIN_2 |
| 1977-2061 | ||
| SM00112 | 936-1015 | CA |
| 1509-1583 | ||
| 1695-1769 | ||
| SM00736 | 1305-1396 | CADG |
| 1397-1489 | ||
| 1490-1582 | ||
| 1583-1675 | ||
| 1676-1768 | ||
| 2251-2343 | ||
| 2345-2436 | ||
| 2534-2620 | ||
| PF05345 | 1320-1379 | He_PIG |
| 1521-1564 | ||
| 1707-1750 | ||
| 2282-2313 | ||
| 2375-2418 | ||
| TIGR01965 | 693-794 | VCBS repeat |
| 1320-1411 | ||
| 1786-1879 | ||
| 2077-2169 | ||
| 2171-2266 | ||
| G3DSA:2.40.160.20 | 3279-3469 | OMP-b-brl |
| PF01839 | 3016-3054 | FG-GAP |
| SSF49313 | 1301-1393 | Cadherin |
| 1394-1486 | ||
| 1487-1579 | ||
| 1580-1672 | ||
| 1673-1767 | ||
| 2249-2340 | ||
| 2341-2435 | ||
| G3DSA:2.130.10.130 | 2965-3097 |
The domains of CabC and CabD were predicted by several protein databases including SMART, Pham, InterPro, and PROSITE.
Systematic analysis of bacterial genomes revealed that Shewanella species, which are taxonomically close to S. degradans, contain a putative CabC/CabD orthologous protein (see Fig. S1 in the supplemental material). Intriguingly, only a genome of the dissimilatory metal ion-reducing Shewanella oneidensis MR-1 has a very similar organization of a locus encoding either CabC- or CabD-like protein (see Fig. S1). In both S. degradans and S. oneidensis, cabC and cabD genes form operons consisting of two additional genes, one of which encodes a putative serine protease (see Fig. S1). A CabC/CabD orthologous protein in S. oneidensis (SO_0189) was annotated as a fibronectin type III domain-containing protein. SO_0189 is a 2,522-aa protein (256.8 kDa; pI 4.06) containing four CA domains that overlap with four CADG domains, four FN3 domains, He_PIG, and OMP_b-brl. Moreover, SO_0189 is also a putative secreted protein because it contains a signal peptide motif.
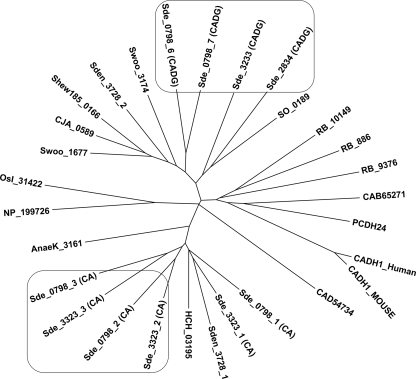
Sequences of cadherin and cadherin-like domains from representative bacterial and eukaryotic species (see Table S2 in the supplemental material) were subjected to multiple sequence analysis, and their putative evolutionary relationship was viewed in a phylogenetic tree (Fig. 2). This analysis revealed a high sequence divergence of CA and CADG modules between eukaryotic and prokaryotic species, as well as among bacteria themselves (Fig. 2).
FIG. 2.
Phylogeny of cadherins inferred by the minimum evolution. CA and CADG domains derived from deduced amino acid sequences of prokaryotic and eukaryotic species were aligned using the ClustalW program, which served to create an unrooted tree using the MEGA, version 4.1, program. The terminology for cadherin and cadherin-like domains and the accession numbers for their parent proteins are given in Fig. 1 and Table S2 in the supplemental material, respectively.
The S. degradans 2-40 CA and CADG modules are located on separate branches. The sequence alignment of the cadherin doublet domain, from amino acid 2288 to 2496 of Sde_3323, and the cadherin-like doublet domain, from amino acid 2261 to 2500 of Sde_0798, had 23% identity (see Fig. S2 in the supplemental material). In addition these domains contain a conserved calcium binding motif, designated DXDXDG, for its core of conserved aspartic acid and glycine residues (see Fig. S2). The two CA modules (CA doublet domain) from CabC (Sde_3323) and two CADG modules (CADG doublet domain) from CabD (Sde_0798) were chosen for further study of their adhesive features. We considered a doublet domain in order to ensure a functional unit with a calcium binding site between two cadherin repeats. Attempts to express and investigate interactions between more than two cadherin domains were unsuccessful because such constructs formed insoluble inclusion bodies.
Examination of homophilic and heterophilic interactions between cadherin and cadherin-like doublet domains.
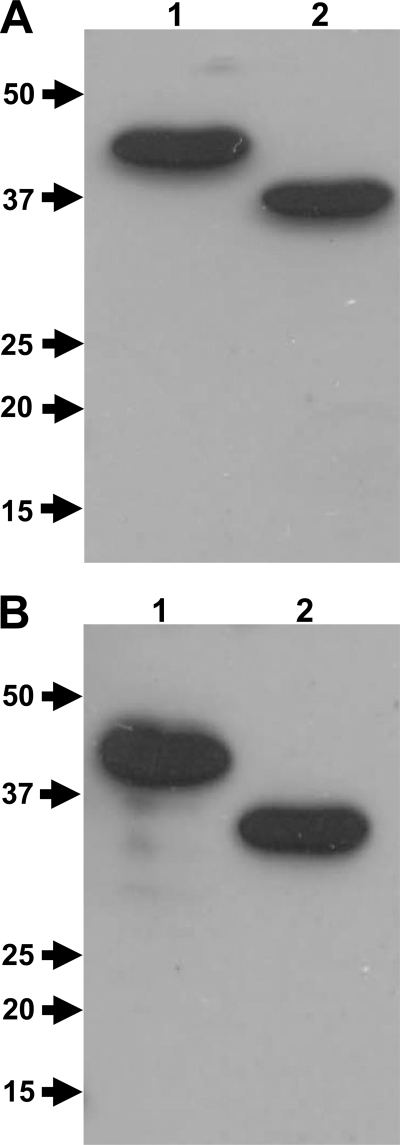
The recombinant CA doublet domains from CabC and the recombinant CADG doublet domains from CabD were tested by Western blot analysis for homophilic interactions between CA-CA doublet domains and CADG-CADG doublet domains, as well as for heterophilic interaction between CA and CADG doublet domains.
The overexpressed recombinant CA and CADG doublet domains, fused to a His6 tag, were equivalently subjected to SDS-PAGE (12%), transferred to nitrocellulose membranes, and probed with CA doublet domains fused to CBM3a tag and with CADG doublet domains fused to CBM3a tag. The CBM3a protein alone was checked for any interactions with CA and CADG; none was observed (data not shown). The Western blotting analysis revealed obvious homophilic interactions between CAHis6 and CBM3aCAHis6 and also revealed heterophilic interactions between CADGHis6 and CBM3aCAHis6 (when the membrane was probed with CBM3aCAHis6) (Fig. 3A). Additionally, there were homophilic interactions between CADGHis6 and CBM3aCADGHis6 and heterophilic interactions between CAHis6 and CBM3aCADGHis6 (when the membrane was probed with CBM3aCADGHis6) (Fig. 3B).
FIG. 3.
Homophilic and heterophilic interactions between cadherin and cadherin-like doublet domains shown by Western blotting analysis. The recombinant cadherin (CAHis6) doublet domain (lane 1) and cadherin-like (CADGHis6) doublet domain (lane 2) were subjected to SDS-PAGE (12%) and blotted to a nitrocellulose membrane. The membrane was probed with the recombinant CBM3aCAHis6 doublet domain construct from CabC (Sde_3323) (A) and with recombinant CBM3aCADGHis6 doublet domain construct from CabD (Sde_0798) (B). The interactions were revealed by using sequential incubation with primary rabbit anti-CBM3a antibody and HRP-labeled goat-anti-rabbit secondary antibody. The protein molecular mass markers (in kDa) are on the left side of each figure.
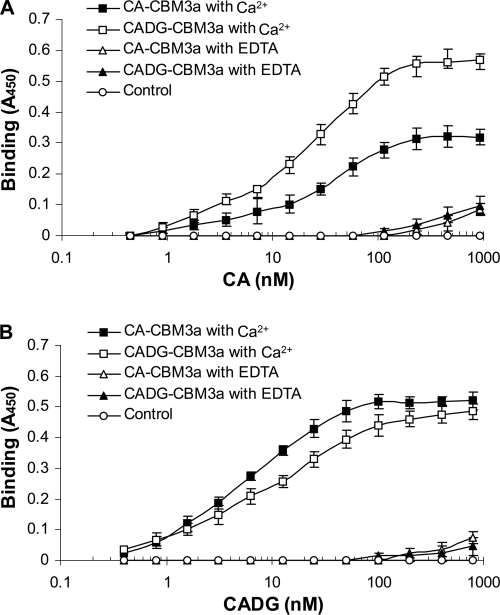
These observations were the first experimental evidence that bacterial cadherin and cadherin-like domains may also have homophilic and heterophilic interactions that are similar to those of cadherin domains from metazoan lineages. Thus, in order to further investigate CA and CADG doublet domain homophilic and heterophilic interactions, ELISAs were performed. ELISA plates were coated with incremental concentrations of CAHis6 (Fig. 4A) and CADGHis6 doublet domains (Fig. 4B) and probed with CBM3aCAHis6 and CBM3aCADGHis6 constructs. The interactions were detected using CBM3a antibody in the presence of calcium or EDTA.
FIG. 4.
Homophilic and heterophilic interactions between cadherin and cadherin-like doublet domains demonstrated by ELISA. Interactions between the CA doublet domain from CabC (Sde_3323) and CADG doublet domain from CabD (Sde_0798) were carried out by ELISA. The microtiter plates were coated with CAHis6 (A) and CADGHis6 (B) at incremental concentrations. The proteins were probed with recombinant cadherin and cadherin-like doublet domains (CBM3aCAHis6 and CBM3aCADGHis6, respectively) in the presence of Ca2+ and EDTA, as indicated. The interactions were revealed by using sequential incubation with primary rabbit anti-CBM3a antibody and HRP-labeled goat-anti-rabbit secondary antibody. Data are means ± standard deviations for at least three independent determinations.
The results of the ELISA supported those of the Western blot analysis clearly showing homophilic and heterophilic interactions between CA and CADG doublet domains in a Ca2+-dependent manner (Fig. 4). In addition, the negative logarithms of the concentrations that precede the half-maximal chromogenic responses (pEC50 values) were determined from the respective binding curves (21). The pEC50 for CAHis6 interactions with CBM3aCAHis6 was 7.34 ± 0.15. The pEC50 for CAHis6 interactions with CBM3aCADGHis6 was 7.5 ± 0.25, the pEC50 for CADGHis6 interactions with CBM3aCAHis6 was 7.75 ± 0.2, and the pEC50 for CADGHis6 interactions with CBM3aCADGHis6 7.64 ± 0.1. All interactions were observed only in the presence of calcium. When these were assayed with EDTA, CA and CADG doublet domains did not interact.
Examination of cadherin and cadherin-like domain adhesive features to the cell surfaces of S. degradans strain 2-40.
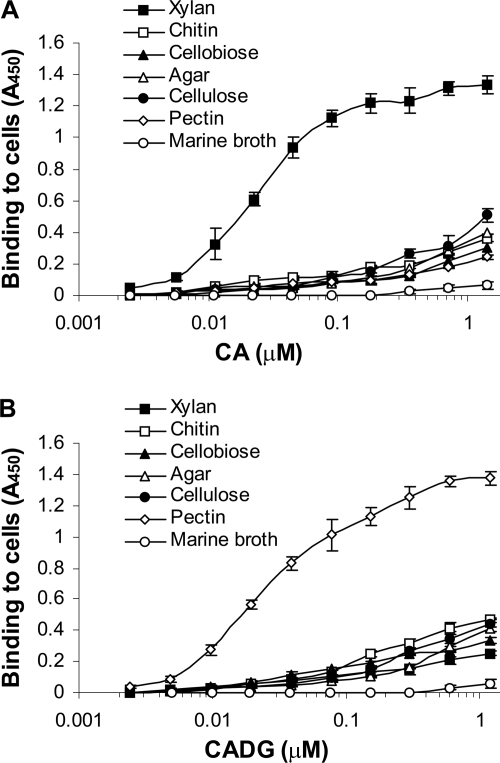
To investigate the possibility that CA and CADG may function in cell-cell contact, these bacterial cadherins were tested for their adhesion to the S. degradans 2-40 cell surface by cell-based ELISA. S. degradans 2-40 was cultivated in minimal medium with sole carbon sources including cellobiose, cellulose, agar, pectin, chitin, and xylan from birch wood. The control culture was grown on complete medium (marine broth). The ELISA plates were coated with poly-l-lysine and then with live S. degradans 2-40. The cells were probed with the CAHis6 doublet domain (Fig. 5A) and with the CADGHis6 doublet domain (Fig. 5B).
FIG. 5.
Interaction of cadherin and cadherin-like doublet domains with the S. degradans 2-40 cell surface. The microtiter plates containing attached S. degradans 2-40 cells from minimal medium cultures with different carbon sources (xylan from birch wood, chitin, cellobiose, agar, cellulose, and pectin), were probed with the cadherin (CAHis6) doublet domain from CabC (Sde_3323) and cadherin-like (CADGHis6) doublet domain from CabD (Sde_0798) at incremental concentrations and labeled with mouse anti-His6-horseradish peroxidase antibodies. S. degradans 2-40 cells grown on complete medium containing marine broth, attached to microtiter plates, and probed with the same probes were used as a control. Data are means ± standard deviations for at least three independent determinations.
The CA and CADG domains bound directly to the bacterial cell surface. Interestingly, the binding affinities of cadherins to S. degradans 2-40 were dependent on the sole carbon source growth medium (Fig. 5). The CA binding affinity was clearly highest on S. degradans 2-40 that was grown on xylan from birch wood. The CADG was highest on S. degradans 2-40 grown on pectin. There were reduced interactions in S. degradans 2-40 grown on other sole carbon sources, and no interactions were detected in S. degradans 2-40 grown on complete medium (marine broth).
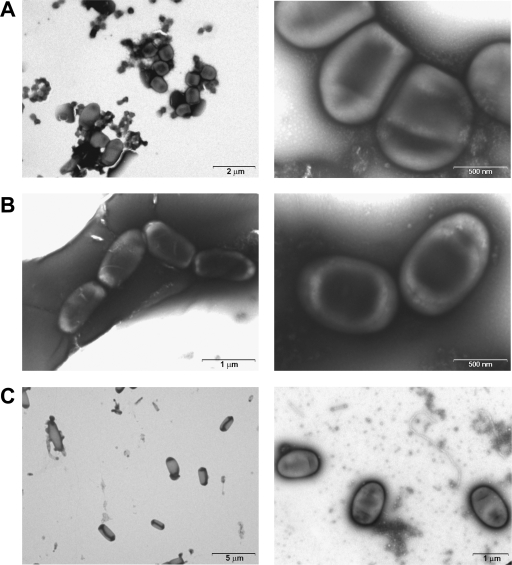
Electron and phase-contrast microscopic observations and micrographs revealed significant, S. degradans 2-40 aggregation and/or chain formation among cultures grown in single, xylan, and pectin complex carbon source media but not among cells actively growing in medium with glucose as the sole carbon source (Fig. 6) or in complete medium (marine broth) (data not shown). These observations were consistent with the results of cell-based ELISAs.
FIG. 6.
TEM of cells grown with xylan from birch wood (A), pectin (B), and glucose (C). The micrographs of the experimental cultures grown to logarithmic phase are shown at higher magnification to reveal aggregation architecture and cell-to-cell bonding. The glucose controls (C) are representative cultures viewed at lower magnification to reveal their typical nonaggregative state. Cellular contact appears to involve densely staining surface features in cells grown with xylan from birch wood and with pectin. The glucose-grown cultures displayed minimal or no cell-to-cell binding. S. degradans frequently manifests bipolar staining under both light and electron microscopy.
DISCUSSION
Cadherins are a family of adhesive molecules that mediate Ca2+-dependent cell-cell adhesion in solid tissues of higher organisms and modulate a wide variety of cellular processes including polarization and migration (18). Cadherins were reported to be most widely distributed in the metazoan lineage (18, 25). The genome of the unicellular choanoflagellate Monosiga brevicollis, a planktonic marine organism, was very recently reported to encode the premetazoan ancestor of the cadherins (1). Cadherins and cadherin-like domains are also present in bacteria (7). According to analysis via the SMART database, the number of known cadherin domain-containing proteins increased from 73 to only 185 during the last few years. Thus, cadherin domains were thought to be relatively rare among bacteria.
However, this research showed that CA and CADG domains are common in certain bacteria, especially among species of the order Alteromonadales, including S. degradans 2-40 and Shewanella spp. (13, 17, 24). Moreover, the genomes of at least four species of a novel bacterial phylum Planctomycetes (Blastopirellula marina, Gemmata obscuriglobus, Planctomyces maris, and Rhodopirellula baltica) encode hypothetical proteins that have putative cadherins and cadherin-like domains (Fig. 2). In most cases, annotations of bacterial proteins containing cadherin and cadherin-like domains are not particularly informative, with most descriptions stating “hypothetical proteins” or “proteins with unknown function.” Thus, a role of cadherins in bacteria remains obscure.
Our analyses show that the S. degradans 2-40 genome contains ≥52 CA and CADG domains in five bacterial proteins, ranging in size from 348 to 1,331 kDa. The calculated relative abundance of cadherin and cadherin-like domains in S. degradans 2-40 genome is 0.12%. We were surprised to discover that such abundance is comparable to cadherin abundance in some, but not all, metazoan genomes: arthropod Drosophila melanogaster (0.13%), cnidarian Nematostella vectensis (0.12%), Mus musculus (0.39%), and choanoflagellate M. brevicollis (0.26%) (1). Interestingly, the major group of cadherin-containing prokaryotes are aquatic, as are some of the metazoans or metazoans at some stages of development, for example, M. brevicollis, a proposed cadherin-containing ancestor. It is possible that these adhesive domains may bind molecules and small organisms in aquatic environments where dilution would be problematic.
We focused on CA doublet domains from CabC (Sde_3323) and CADG doublet domains from CabD (Sde_0798). Although, CA and CADG doublet domains show significant sequence similarity, they are phylogenetically distant, but both belong to the broad cadherin family that is divided into subfamilies based on molecular characteristics. These include type 1 cadherins, type 2 cadherins, desmosomal cadherins, protocadherins, and flamingo cadherins. Metazoan E-cadherins and flamingo cadherins are bound by pathogenic bacteria which exploit them as extracellular tethers during host cell invasion (1). The choanoflagellate cadherins were proposed to fill an equivalent role in binding bacterial prey for recognition or capture (1). We propose that S. degradans 2-40 cadherins and cadherin-like domains are involved in protein-protein interactions and probably mediate cell-cell contact.
Bacterial cadherins from S. degradans 2-40 interacted homophilically and heterophilically. The Ca2+ had significant influence on these interactions, similar to the situation found in eukaryotic cadherins (19, 25). The estimated pEC50 values of these interactions are in good correlation with apparent dissociation constants (Kds) of ∼10−8 M that were calculated by double-reciprocal plots of binding curves. Thus, prokaryotic interactions may be 3 to 5 orders of magnitude stronger, while mammalian cadherin interactions had Kds ranging from 10−3 to 10−5 M (8, 18), requiring numerous interactions for strong adhesion (18). There are still disagreements about how classical cadherins dimerize as well as about the structural basis of cadherin recognition (6, 23). Moreover, there are few data about bacterial cadherins since they were not previously explored. The involvement of at least two types of mammalian cadherin-cadherin contacts in adhesion has been deduced from electron microscopy experiments (6). These involve contacts between extracellular cadherin domains belonging to cadherins of the same cell (cis interaction) and contacts between cadherins of interacting cells (trans interaction). Due to a very high degree of sequence homology between the various classical cadherins, it was initially thought that homophilic interactions were strongly favored. More recent data show that heterophilic interactions can be formed and suggest that selectivity results from the collective effects of many individual dimer interactions (6). We hypothesize that S. degradans 2-40 cadherins and cadherin-like domains also may produce cis and trans interactions due to their adhesive features.
Evidence from ELISA studies revealed that CA and CADG doublet domains directly bind to bacterial cell surfaces and that such adhesion is mediated by Ca2+. The same results were obtained in our laboratory in the process of studying R. baltica cadherin domains (data not shown). The CA and CADG domains were best expressed in xylan and pectin as sole carbon sources and not in logarithmically growing S. degradans in glucose. Thus, the expression of these proteins appears to be regulated.
This biochemical evidence (Fig. 5) coincides with observations of S. degradans cellular interactions (Fig. 6). Parallel in vivo observations revealed that cells growing with pectin or xylan as the sole carbon source formed long chains and/or aggregates. The logarithmically growing cells in glucose did not exhibit such cell-to-cell interaction. It is hypothesized that one or more of these cadherin-containing mega-proteins may be exported and anchored to the cell surface, where it may mediate cellular aggregation or attachment to other S. degradans 2-40 cells or other prokaryotes, algae, and/or invertebrates in the marine environment. Such cellular interactions would enhance the complex carbohydrate deconstruction abilities of S. degradans 2-40 either by adhering it to its substrate, in the case of eukaryotic attachments, or by achieving a quorum/critical mass of carbohydrases, necessary for an efficient attack on complex carbohydrates, in the case of cellular aggregation/chain formation.
Supplementary Material
Acknowledgments
This research was supported by grant 2003041 from the United States-Israel Binational Science Foundation.
We are grateful to Larry E. Taylor II, Erez Matalon, and Hamutal Kahel-Raifer for many helpful discussions as well as to Daphna Shimon for technical assistance.
Footnotes
Published ahead of print on 18 December 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Abedin, M., and N. King. 2008. The premetazoan ancestry of cadherins. Science 319:946-948. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andrykovitch, G., and I. Marx. 1988. Isolation of a new polysaccharide-digesting bacterium from a salt marsh. Appl. Environ. Microbiol. 54:1061-1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bannai, H., Y. Tamada, O. Maruyama, K. Nakai, and S. Miyano. 2002. Extensive feature detection of N-terminal protein sorting signal. Bioinformatics 18:298-305. [DOI] [PubMed] [Google Scholar]
- 5.Barak, Y., T. Handelsman, D. Nakar, R. Mechaly, R. Lamed, Y. Shoham, and E. A. Bayer. 2005. Matching fusion-protein systems for affinity analysis of two interacting families of proteins: the cohesin-dockerin interaction. J. Mol. Recognit. 18:491-501. [DOI] [PubMed] [Google Scholar]
- 6.Cailliez, F., and R. Lavery. 2006. Dynamics and stability of E-cadherin dimmers. Biophys. J. 91:3964-3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cao, L., X. Yan, C. W. Borysenko, H. C. Blair, C. Wu, and L. Yu. 2005. CHDL: a cadherin-like domain in Proteobacteria and Cyanobacteria. FEMS Microbiol. Lett. 251:203-209. [DOI] [PubMed] [Google Scholar]
- 8.Chen, P. C., S. Posy, A. Ben-Shaul, L. Shapiro, and B. H. Honig. 2005. Specificity of cell-cell adhesion by classical cadherins: critical role for low-affinity dimerization through β-strand swapping. Proc. Natl. Acad. Sci. U. S. A. 102:8531-8536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ekborg, N. A., L. E. Taylor, A. G. Longmire, B. Henrissat, R. M. Weiner, and S. W. Hutchenson. 2006. Genomic and proteomic analysis of the agarolytic system expressed by Saccharophagus degradans 2-40. Appl. Environ. Microbiol. 72:3396-3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ensor, L. A., S. K. Stosz, and R. M. Weiner. 1999. Expression of multiple complex polysaccharide-degrading enzyme systems by marine bacterium strain 2-40. J. Ind. Microbiol. Biotechnol. 23:123-126. [DOI] [PubMed] [Google Scholar]
- 11.Finn, R. D., J. Tate, J. Mistry, P. C. Coggill, S. J. Sammut, H.-R. Hotz, G. Ceric, K. Forslund, S. R. Eddy, E. L. L Sonnhammer, and A. Bateman. 2008. The Pfam protein families database. Nucleic Acids Res. 36:D281-D288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gonzales, J. M., and R. M. Weiner. 2000. Phylogenetic characterization of marine bacterium strain 2-40, a degrader of complex polysaccharides. Int. J. Syst. Evol. Microbiol. 8:831-834. [DOI] [PubMed] [Google Scholar]
- 13.Heidelberg, J. F., I. T. Paulsen, K. E. Nelson, E. J. Gaidos, W. C. Nelson, T. D. Read, J. A. Eisen, R. Seshadri, N. Ward, B. Methe, R. A. Clayton, T. Meyer, A. Tsapin, J. Scott, M. Beanan, L. Brinkac, S. Daugherty, R. T. DeBoy, R. J. Dodson, A. S. Durkin, D. H. Haft, J. F. Kolonay, R. Madupu, J. D. Peterson, L. A. Umayam, O. White, A. M. Wolf, J. Vamathevan, J. Weidman, M. Impraim, K. Lee, K. Berry, C. Lee, J. Mueller, H. Khouri, J. Gill, T. R. Utterback, L. A. McDonald, T. V. Feldblyum, H. O. Smith, J. C. Venter, K. H. Nealson, and C. M. Fraser. 2002. Genome sequence of the dissimilatory metal ion-reducing bacterium Shewanella oneidensis. Nat. Biotechnol. 20:1118-1123. [DOI] [PubMed] [Google Scholar]
- 14.Higgins, D. G., J. D. Thompson, and T. J. Gibson. 1996. Using Clustal for multiple sequence alignment. Methods Enzymol. 266:383-402. [DOI] [PubMed] [Google Scholar]
- 15.Howard, M. B., N. A. Ekborg, L. E. Taylor, R. M. Weiner, and S. W. Hutcheson. 2003. Genomic analysis and initial characterization of the chitinolytic system of Microbulbifer degradans strain 2-40. J. Bacteriol. 185:3352-3360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Howard, M. B., N. A. Ekborg, L. E. Taylor, R. M. Weiner, and S. W. Hutcheson. 2004. Chitinase B of “Microbulbifer degradans” 2-40 contains two catalytic domains with two different chitinolytic activities. J. Bacteriol. 186:1297-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ivanova, E. P., S. Flavier, and R. Christen. 2004. Phylogenetic relationships among marine Alteromonas-like proteobacteria: emended description of the family Alteromonadaceae and proposal of Pseudoalteromonadaceae fam. nov., Colwelliaceae fam. nov., Shewanellaceae fam. nov., Moritellaceae fam. nov., Ferrimonadaceae fam. nov., Idiomarinaceae fam. nov. and Psychromonadaceae fam. nov. Int. J. Syst. Evol. Microbiol. 54:1773-1788. [DOI] [PubMed] [Google Scholar]
- 18.Koch, A. W., K. L. Manzur, and W. Shan. 2004. Structures based models of cadherin mediated cell adhesion: the evolution continues. Cell Mol. Life Sci. 61:1884-1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leckband, D., and A. Prakasam. 2006. Mechanism and dynamics of cadherin adhesion. Annu. Rev. Biomed. Eng. 8:259-287. [DOI] [PubMed] [Google Scholar]
- 20.Letunic, I., R. R. Copley, S. Schmidt, F. D. Ciccarelli, T. Doerks, J. Schultz, C. P. Ponting, and P. Bork. 2004. SMART 4.0: towards genomic data integration. Nucleic Acids Res. 32:142-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Motulsky, H. J., and A. Cristopoulos. 2003. Fitting models to biological data using linear and nonlinear regression: a practical guide to curve fitting. GraphPad Software, Inc., San Diego, CA.
- 22.Nielson, H., J. Englebrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 23.Patel, S. D., C. Ciatto, C. P. Chen, F. Bahna, M. Rajebhosale, N. Arkus, I. Schieren, T. M. Jessell, B. Honig, S. R. Price, and L. Shapiro. 2006. Type II cadherin ectodomain structures: implications for classical cadherin specificity. Cell 124:1255-1268. [DOI] [PubMed] [Google Scholar]
- 24.Pinchuk, G. E., C. Ammons, D. E. Culley, S. M. W. Li, J. S. McLean, M. F. Romine, K. H. Nelson, J. K. Fredrickson, and A. S. Beliaev. 2008. Utilization of DNA as a sole source of phosphorus, carbon and energy by Shewanella spp.: ecological and physiological implications for dissimilatory metal reduction. Appl. Environ. Microbiol. 74:1198-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rigden, D. J., and M. Y. Galperin. 2004. The DxDxDG motif for calcium binding: multiple structural contexts and implications for evolution. J. Mol. Biol. 343:971-984. [DOI] [PubMed] [Google Scholar]
- 26.Rosok, M. J., M. R. Stebbins, K. Connelly, M. E. Lostrom, and A. W. Siadak. 1990. Generation and characterization of murine antiflagellum monoclonal antibodies that are protective against lethal challenge with Pseudomonas aeruginosa. Infect. Immun. 58:3819-3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sambrook, J., and D. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 28.Schultz, J., R. R. Copley, T. Doerks, C. P. Ponting, and P. Bork. 2000. SMART: a web-based tool for the study of genetically mobile domains. Nucleic Acids Res. 28:231-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taylor, L. E., B. Henrissat, P. M. Coutinho, N. A. Ekborg, S. W. Hutcheson, and R. M. Weiner. 2006. Complete cellulase system in the marine bacterium Saccharophagus degradans strain 2-40. J. Bacteriol. 188:3849-3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weiner, R. M., L. E. Taylor, N. A. Ekborg, and L. Whitehead. 2001. Degradosomes: potential importance in the ocean's carbon cycle and in algal culture and aquaculture, p. 259-264. In N. K. Saxena (ed.), Recent advances in marine science and technology, 2000. PACON International, Honolulu, HI.
- 31.Weiner, R. M., L. E. Taylor, B. Henrissat, L. Hauser, M. Land, P. M. Coutinho, C. Rancurel, E. H. Saunders, A. G. Longmire, H. Zhang, E. A. Bayer, H. J. Gilbert, F. Larimer, I. B. Zhulin, N. A. Ekborg, R. Lamed, P. M. Richardson, I. Borovok, and S. W. Hutcheson. 2008. Complete genome sequence of the complex carbohydrate-degrading marine bacterium, Saccharophagus degradans strain 2-40. PLoS Genet. 4:e1000087. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.