Abstract
Legionella longbeachae causes most cases of legionellosis in Australia and may be underreported worldwide due to the lack of L. longbeachae-specific diagnostic tests. L. longbeachae displays distinctive differences in intracellular trafficking, caspase 1 activation, and infection in mouse models compared to Legionella pneumophila, yet these two species have indistinguishable clinical presentations in humans. Unlike other legionellae, which inhabit freshwater systems, L. longbeachae is found predominantly in moist soil. In this study, we sequenced and annotated the genome of an L. longbeachae clinical isolate from Oregon, isolate D-4968, and compared it to the previously published genomes of L. pneumophila. The results revealed that the D-4968 genome is larger than the L. pneumophila genome and has a gene order that is different from that of the L. pneumophila genome. Genes encoding structural components of type II, type IV Lvh, and type IV Icm/Dot secretion systems are conserved. In contrast, only 42/140 homologs of genes encoding L. pneumophila Icm/Dot substrates have been found in the D-4968 genome. L. longbeachae encodes numerous proteins with eukaryotic motifs and eukaryote-like proteins unique to this species, including 16 ankyrin repeat-containing proteins and a novel U-box protein. We predict that these proteins are secreted by the L. longbeachae Icm/Dot secretion system. In contrast to the L. pneumophila genome, the L. longbeachae D-4968 genome does not contain flagellar biosynthesis genes, yet it contains a chemotaxis operon. The lack of a flagellum explains the failure of L. longbeachae to activate caspase 1 and trigger pyroptosis in murine macrophages. These unique features of L. longbeachae may reflect adaptation of this species to life in soil.
Isolation of Legionella longbeachae was first reported in 1981 after isolation from patients with pneumonia in the United States (11, 59). Although L. longbeachae is not a common respiratory pathogen in either North America or Europe, where Legionella pneumophila infections are predominant, it accounts for more than 50% of legionellosis cases in Australia and is also prevalent in New Zealand and Thailand (10, 12, 60, 66, 68, 77, 93, 94). Legionnaires' disease induced by L. longbeachae infection is clinically indistinguishable from the disease caused by L. pneumophila (65). However, L. longbeachae infections have been associated with gardening and the use of potting soil, whereas the disease caused by other species is linked to freshwater sources (4, 65). L. longbeachae can survive for up to 9 months in moist potting soil at room temperature, in contrast to other Legionella species, which inhabit natural and manmade freshwater systems worldwide (34, 83, 84).
In addition to the differences in habitat, L. longbeachae differs from L. pneumophila in its virulence in murine models of infection. L. longbeachae replicates in the lungs of A/J, C57BL/6, and BALB/c mice (6), whereas most inbred mice, including C57BL/6 and BALB strains, are resistant to L. pneumophila (61). These differences in murine host susceptibility are likely due to different abilities to activate caspase 1-mediated pyroptosis in macrophages. While L. pneumophila rapidly triggers pyroptosis in C57BL/6 mouse macrophages, L. longbeachae does not do this (6).
Intracellular trafficking of L. longbeachae in mammalian macrophages also follows a route distinct from that of L. pneumophila. After phagocytosis, the L. pneumophila-containing vacuole (LCV) excludes early and late endosomal markers, such as early endosomal antigen 1 (EEA1), Rab5, LAMP-1, LAMP-2, and the mannose 6-phosphate receptor (M6PR) (5, 89). In L. pneumophila the Dot/Icm type IV secretion system is required for prevention of phagosome-lysosome fusion and for intracellular replication (47). Conversely, the L. longbeachae-containing vacuole acquires the early endosomal marker EEA1 and the late endosomal markers LAMP-2 and M6PR (5). It has been suggested that L. longbeachae intracellular trafficking resembles that of the facultative intracellular pathogen Brucella abortus, since a Brucella-containing vacuole also acquires early and late endosomal markers soon after infection (5). Despite the difference in intracellular trafficking between L. longbeachae and L. pneumophila, L. longbeachae rescues Dot/Icm-deficient L. pneumophila when these two organisms coinhabit LCV (5).
Results of the studies cited above indicate that L. longbeachae differs from other legionellae in terms of habitat, host specificity, and intracellular trafficking. In this paper, we describe an analysis of the sequenced and annotated genome of L. longbeachae clinical isolate D-4968 compared with published genomes of L. pneumophila strains Corby, Lens, Paris, and Philadelphia-1 (16, 17, 38). Specifically, we compared genes involved in gene regulation, protein secretion systems, and motility in order to identify genes responsible for making L. longbeachae unique among the legionellae.
MATERIALS AND METHODS
L. longbeachae isolate.
L. longbeachae serogroup 1 clinical isolate D-4968 was obtained from a 77-year-old woman diagnosed with legionellosis in Portland, OR, in May 2000 (4). It was selected from the CDC Legionella reference diagnostic library (Atlanta, GA) because it has been characterized with respect to intracellular trafficking (5), the ability to cause murine infections, and the ability to activate caspase 1 and caspase 3 in human and mouse macrophages (6, 39).
Bacterial growth and DNA isolation.
D-4968 was grown on buffered charcoal yeast extract (BCYE) agar plates for 72 h before DNA extraction. Genomic DNA was extracted using a QIAamp DNA mini kit (Qiagen Inc., Valencia, CA) according to the manufacturer's guidelines.
Whole-genome sequencing.
Legionella genome sequencing was conducted using Roche 454 GS-FLX pyrosequencing at the CDC Biotechnology Core Facility. A shotgun library was produced by the method of Margulies et al. (57) and was sequenced with an LR70 sequencing kit. After trimming and refiltering to recover short high-quality reads, the sequencing run provided 251,332 reads, a draft de novo assembly was produced using the Newbler assembler (v 2.0). This assembly resulted in a draft genome consisting of 4.05 Mb in 89 contigs.
Optical mapping.
A methanol-treated culture of D-4968 was submitted to OpGen Technologies, Madison, WI, for construction of optical maps. The immobilized genomic DNA of D-4968 was digested with either the XbaI or PvuII restriction enzyme, and the sizes and order of digested fragments were determined. The sequence contigs were converted to in silico restriction maps using the MapIt software (OpGen) and were aligned with the optical maps based on their restriction fragment patterns.
Gap closure.
Contig arrangements predicted by the optical map were verified by PCR amplification of DNA fragments connecting adjusted contigs using Platinum Taq DNA polymerase (Invitrogen, Carlsbad, CA). Primers that annealed to the 5′ and 3′ regions of each contig were designed using the PrimerSelect program of DNASTAR Lasergene 7 (DNASTAR, Inc., Madison, WI). PCR products were purified using ExoSAP-IT (USB Corporation, Cleveland, OH). DNA sequencing was performed with an ABI BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems, Foster City, CA), and the products were analyzed with a model 3130xl ABI PRISM genetic analyzer (Applied Biosystems). DNASTAR SeqMan was used to assemble amplified DNA fragments with the contigs. Where gaps between adjusted contigs were predicted to be larger than 3 kb, either the Elongase enzyme mix (Invitrogen) or a Qiagen LongRange PCR kit (Qiagen) was used for PCR amplification. Sequencing directly from genomic DNA was also conducted for large gaps by modifying the reaction conditions using the method of Heiner et al. (41) and increasing the number of cycles to 100.
Plasmid purification.
Plasmid DNA was purified from L. longbeachae D-4968, L. pneumophila strain Lens, and L. pneumophila strain Paris grown on BCYE agar plates for 96 h using a HiSpeed plasmid midi kit (Qiagen) according to the manufacturer's guidelines.
Screening L. longbeachae isolates for the presence of flagellar biosynthesis and chemotaxis genes.
Primers were designed to amplify internal DNA fragments of the flaA, fliI, flgH, and flgG flagellar biosynthesis genes of L. pneumophila strain Philadelphia-1 and of genes annotated as cheR, cheA, and cheB in the genome of L. longbeachae D-4968 (see Table S1 in the supplemental material). DNA from L. longbeachae serogroup 1 and serogroup 2 isolates from the CDC Legionella reference diagnostic library (Atlanta, GA) was extracted using an InviMag bacterial DNA kit (Invitek, Berlin, Germany) and a KingFisher workstation. The extracted DNA was used for PCR amplification of flagellar and chemotaxis gene fragments, followed by sequencing as described above.
Annotation and analysis.
The D-4968 DNA sequence was submitted to the J. Craig Venter Institute (JCVI) Annotation Service, where it was subjected to JCVI's prokaryotic annotation pipeline, including gene finding with Glimmer, BLAST-extend-repraze (BER) searches, HMM searches, TMHMM searches, SignalP predictions, and automatic annotations from AutoAnnotate. The annotation tool Manatee was used to manually review the output (downloaded from SourceForge [manatee.sourceforge.net] in November 2007).
The Rapid Annotation using Subsystem Technology (RAST) service was used for fully automated annotation of the D-4968 genome and comparison with genomes of L. pneumophila strains (7). The integrated microbial genomes (IMG) system was used for searches of currently available microbial genomes for the presence of protein domains and motifs (58). Alignment of L. longbeachae and L. pneumophila genomes was conducted using the Mauve genome alignment software (23). The sequences used for comparative genomic analysis were the sequences of L. pneumophila strains Corby, Lens, Paris, and Philadelphia-1 (GenBank accession numbers NC_009494.1, NC_006369.1, NC_006368.1, and NC_002942.5, respectively).
Nucleotide sequence accession number.
The Whole-Genome Shotgun Project of D-4968 has been deposited in the DDBJ/EMBL/GenBank database under accession ACZG00000000. The version described in this paper is the first version (accession no. ACZG01000000).
RESULTS AND DISCUSSION
General features of the genome.
Pyrosequencing of the L. longbeachae D-4968 genome revealed that it consists of 4.05 × 106 bp, contains 3,821 protein-encoding genes, and has an average G+C content of 37% (Table 1) based on the data for all contigs. A comparison of the L. longbeachae genome with four previously described L. pneumophila genomes indicated that L. longbeachae is substantially larger and has a higher coding capacity (Table 1). Eighty-nine contigs produced by Newbler assembly were arranged into two large scaffolds consisting of 2.5 × 106 bp and 1.4 × 106 bp using two optical maps and primer walking (see Fig S1A in the supplemental material; data not shown). Both optical maps suggested that the D-4968 chromosome has a circular topology. Thus, the two scaffolds are connected.
TABLE 1.
Comparison of the sizes and compositions of the L. longbeachae and L. pneumophila genomes
| Parameter | L. longbeachae D-4968b | L. pneumophila strain Corby | L. pneumophila strain Lens | L. pneumophila strain Paris | L. pneumophila strain Philadelphia-1 |
|---|---|---|---|---|---|
| Genome length (bp)a | 4,050,119 | 3,576,470 | 3,405,519 | 3,635,495 | 3,397,754 |
| G+C content (%) | 37 | 38 | 38 | 38 | 38 |
| No. of protein-encoding genes | 3,821 | 3,206 | 2,934 | 3,166 | 2,942 |
| No of tRNAs | 43 | 44 | 44 | 44 | 43 |
| Mobile plasmid-like element(s) (integration site) | Contains lvh (tRNAPro) | Trb-1 and Trb-2 (tRNAPro and tmRNA, respectively) | Contains lvh (tmRNA) | pP36 containing lvh (tmRNA) | pLP45 containing lvh (tRNAArg) |
| Plasmid (size [kb]) | Yes (>49)c | Yes (60) | Yes (132) | ||
| Topology | Circular | Circular | Circular | Circular | Circular |
Includes the plasmid in L. pneumophila strains Lens and Paris.
Based on data for all 13 contigs.
The size assumes that contig 3 is part of the plasmid.
Eleven small contigs (size range, 0.8 to 49 kb) either fill two gaps between the scaffolds (see Fig S1A in the supplemental material) or constitute a plasmid (see Fig S1B in the supplemental material). Contig 3, which was not placed in the scaffolds, contains a number of genes that are highly homologous to genes located on the plasmid of L. pneumophila strain Paris (see Table S2 in the supplemental material). This indicates that contig 3 most likely constitutes a plasmid, which may be exchanged between different Legionella species.
Figure 1 shows multiple alignments of four L. pneumophila genomes with the D-4968 scaffolds. The results indicate that L. longbeachae has a very limited synteny with L. pneumophila, suggesting that there is a substantial evolutionary distance between these species. On the other hand, the L. longbeachae and L. pneumophila genomes share a number of small-scale clusters of homology where the gene order is highly conserved. Most of these clusters contain genes involved in essential metabolic processes and protein secretion apparatus biosynthesis.
FIG. 1.
Comparison of the genomes of L. longbeachae and L. pneumophila: Mauve alignment of two scaffolds of the L. longbeachae D-4968 genome with the genomes of L. pneumophila strains Corby, Lens, Paris, and Philadelphia-1 presented horizontally. The colored boxes represent homologous segments completely free of genomic rearrangements. These homologous regions are connected by lines between genomes. Blocks below the center line indicate regions with inverse orientation. Regions outside blocks lack homology between genomes. Within each block there is a similarity profile of the DNA sequences, and white areas indicate the sequences specific to a genome. The Corby strain has more large-scale genomic rearrangements than the Lens, Paris, and Philadelphia-1 strains, which are almost colinear. The D-4968 genome composition is substantially different from that of all L. pneumophila genomes.
Gene regulation.
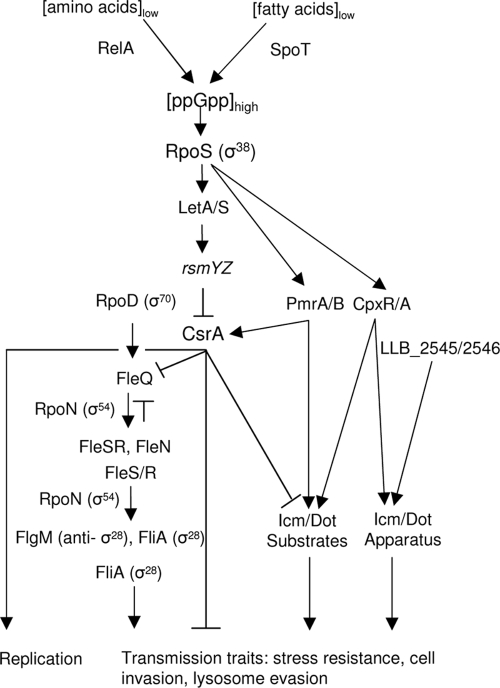
A search of the D-4968 genome for regulatory genes revealed that despite the differences in habitat, host range, and genome size and order, the regulatory networks of L. longbeachae and L. pneumophila appear to be very similar. This could be due to the fact that both species are intracellular parasites and have rather small regulatory gene composition (16). Both L. pneumophila and D-4968 encode six sigma factors, RpoD, RpoH, RpoS, RpoN, RpoE, and FliA (Fig. 2; see Table S3 in the supplemental material) (16). There are also comparable numbers of two-component systems, including 11 histidine kinases, 16 response regulators, and three putative hybrid proteins with both sensory box histidine kinase and response regulator domains (see Table S3 in the supplemental material). Similar to L. pneumophila, the most abundant type of regulator encoded by D-4968 is the family of proteins with GGDEF and EAL domains (16) (see Table S3 in the supplemental material). These domains are involved in synthesis (GGDEF) and hydrolysis (EAL) of bis-(3′-5′)-cyclic dimeric GMP, a novel global second messenger used for regulation of motility, production of extracellular matrix components, and cell-to-cell communication in other bacteria (72). Interestingly, there are only 13 genes encoding GGDEF-EAL family proteins in D-4968, whereas the L. pneumophila genomes contain from 21 (Lens) to 23 (Paris and Corby) such genes. In contrast, the obligatory intracellular pathogen Coxiella burnetii has only two GGDEF domain proteins. If the number of these regulatory proteins correlates with the diversity of environmental stimuli encountered by bacteria (35), the smaller number of GGDEF and EAL proteins in L. longbeachae than in L. pneumophila may suggest that the former species inhabits a less diverse environment.
FIG. 2.
Model for the regulatory network of L. longbeachae. This model is based on proposed L. pneumophila regulatory cascades (1, 44, 49, 64, 69, 78, 85) and the availability of homologs of L. pneumophila regulators in the D-4968 genome. When nutrients are abundant inside the cell, CsrA represses transmission traits and promotes replication. Nutrient starvation stimulates the RelA and SpoT enzymes to produce the alarmone ppGpp. A high alarmone concentration activates the major regulator RpoS, which in turn activates the two-component LetA/S system. When stimulated, LetA/S activates transcription of the small noncoding RNAs RsmY and RsmZ, which sequester CsrA and stop its repressor activity, resulting in expression of transmission traits. Specifically, elimination of CsrA repression activates the master regulator FleQ, which, together with the alternative sigma factor RpoN, activates expression of fleN and fleSR. FleN may be an anti-activator of FleQ. FleS/R is a two-component regulatory system in which the sensor kinase FleS activates the response regulator FleR by phosphorylation. In turn, FleR∼P, together with RpoN, induces expression of the fliA and flgM genes. The alternative sigma factor FliA may positively regulate expression of genes involved in cell invasion (49, 63). High levels of ppGpp may also lead to activation of the Dot/Icm type IV secretion system regulators PmrA and CpxR.
The biphasic L. pneumophila life cycle was first described by Rowbotham (76). Bacteria cycle between an intracellular, acid-resistant, nonmotile, replicative form and a free-living, highly motile, transmissive form, during which they express numerous virulence factors, such as the Icm/Dot secretion system (for a review see reference 64). The L. pneumophila life cycle can be modeled in broth cultures, where bacteria in the exponential growth phase have many attributes of the replicative form, while stationary-phase cells have traits of the transmissive form. Cellular differentiation is coordinated by a regulatory circuit that includes components of the stringent response mechanism, two-component regulatory systems, and alternative sigma factors (64). The L. longbeachae genome encodes all the major parts of this proposed regulatory circuit, including the recently described small noncoding RNAs RsmY and RsmZ (44, 69, 78) (Fig. 2; see Table S3 in the supplemental material). However, there are differences between phase regulation in L. longbeachae and phase regulation in L. pneumophila. In contrast to intracellular replication of L. pneumophila, the ability of L. longbeachae to replicate intracellularly is independent of the growth phase of the infecting bacteria (5). L. longbeachae may have additional components that alter the differentiation-controlling network, or the interactions between conserved elements of the network may differ in L. longbeachae and L. pneumophila. Several putative regulatory genes specific to L. longbeachae are described below.
Protein secretion systems.
L. pneumophila genomes encode multiple protein secretion systems, including the putative type I Lss secretion machinery, type II PilD-dependent Lsp, type IVA Lvh, and type IVB Icm/Dot, as well as a putative type V secretion pathway (25). Gram-negative bacterial pathogens and endosymbionts use these specialized secretion systems to transport toxins and effectors to the extracellular environment or directly into the host cell to modify host physiology and promote interactions (91). It has been established that type II and type IVB secretion systems are essential for L. pneumophila virulence (25). Genes encoding type I, II, IVA, and IVB secretion system homologs were also found in the D-4968 genome, but there were several noteworthy differences.
Type I secretion system.
Type I secretion systems allow secretion of substrates into the extracellular space in a one-step process, without a stable periplasmic intermediate (37). The L. longbeachae genome includes lssX, lssY, lssZ, and lssA genes that share high levels of identity with genes of the L. pneumophila lssXYZABD locus encoding a putative type I secretion system (48). The loci in L. longbeachae and L. pneumophila have similar gene orders and compositions upstream of the lssXYZA operon, but they differ downstream of lssA (see Fig S2 in the supplemental material). L. longbeachae does not contain lssB and lssD, which appear to be essential for the secretion process as they encode a protein of the ATP-binding cassette transporter and the membrane fusion protein, respectively (1). Detection of lssA and lssY but not lssD in non-L. pneumophila Legionella species has been reported previously (48). D-4968 contains 19 genes encoding ATP-binding cassette transporters (LssB orthologs) and 13 genes encoding membrane fusion proteins with a “HlyD” Pfam domain (LssD orthologs) dispersed throughout the genome (see Table S4 in the supplemental material). However, most of these genes have homologs in the L. pneumophila genome and/or are associated with putative multidrug efflux systems. Thus, L. longbeachae may not encode a functional type I secretion system. The presence of the lssXYZA operon in L. longbeachae in the absence of contiguous type I machinery suggests that lssXYZA-encoded proteins may function independently from lssBD and are either not secreted or utilize different secretion machinery.
Type II secretion system.
In many Gram-negative bacteria type II secretion is used to translocate proteins across the outer membrane via multisubunit complexes inserted into both the inner and outer membranes (37). To date, L. pneumophila is the only intracellular pathogen known to possess a functional type II secretion system (T2SS) (19). The L. pneumophila T2SS has been extensively studied and has been shown to be required for optimal growth on bacteriologic media at 12 to 25°C, survival in tap water at 4 to 17°C, establishment of biofilms, sliding motility, intracellular infection of protozoans and mammalian macrophages, and persistence in the lungs of A/J mice (24, 54, 55, 73, 75, 82, 87). Rossier et al. identified T2SS genes in several non-L. pneumophila Legionella species and hypothesized that the T2SS occurs throughout the genus Legionella (75). Indeed, the L. longbeachae genome includes all 12 lsp (Legionella secretion pathway) genes, which are organized in five loci that encode the core components of the T2SS apparatus (Table 2) (19). The L. pneumophila T2SS translocates multiple enzymes, including the major zinc metalloprotease ProA, the type II RNase SrnA, the lipases LipA and LipB, the aminopeptidases LapA and LapB, the lysophospholypase A PlaA, and the chitinase ChiA (24, 25, 73, 74). To date, a total of 25 T2SS substrates have been identified in L. pneumophila, but it has been predicted that the L. pneumophila T2SS may be involved in the secretion of up to 60 proteins (18, 24). The L. longbeachae genome encodes 13 proteins homologous to L. pneumophila enzymes demonstrated to be secreted in a T2SS manner (Table 2) (18). However, a number of T2SS substrates identified in L. pneumophila are not encoded by the D-4968 genome, such as a chiA-encoded chitinase, which was shown to promote L. pneumophila persistence in the lungs of A/J mice (24). Based on these findings and those of Rossier et al. (75), we hypothesize that a conserved T2SS apparatus is encoded by all legionellae, but the number and type of T2SS substrates vary depending on the ecological niche occupied by the species.
TABLE 2.
Type II secretion system
| Locus | Gene | Gene product | Homolog in L. pneumophila strain Philadelphia-1 | Comments |
|---|---|---|---|---|
| Type II secretion apparatus | ||||
| LLB_0056 | pilD | Type IV prepilin peptidase PilD | lpg1524 | Prepilin peptidase required for both the T2SS and type IV pilus biogenesis |
| LLB_0202 | lspF | Type II secretory pathway protein LspF | lpg1363 | Inner membrane protein |
| LLB_0203 | lspG | Type II secretory pathway protein LspG | lpg1362 | Major pseudopilin |
| LLB_0204 | lspH | Type II secretory pathway protein LspH | lpg1361 | Minor pseudopilin |
| LLB_0205 | lspI | Type II secretory pathway protein LspI | lpg1360 | Minor pseudopilin |
| LLB_0206 | lspJ | Type II secretory pathway protein LspJ | lpg1359 | Minor pseudopilin |
| LLB_0207 | lspK | Type II secretory pathway protein LspK | lpg1358 | Minor pseudopilin |
| LLB_0407 | lspL | Type II secretory pathway protein LspL | lpg1874 | Inner membrane platform |
| LLB_0408 | lspM | Type II secretory pathway protein LspM | lpg1875 | Inner membrane platform |
| LLB_0510 | lspD | Type II secretory pathway protein LspD | lpg1320 | Outer membrane secretin |
| LLB_0511 | lspE | Type II secretory pathway protein LspE | lpg1319 | ATPase |
| LLB_0880 | lspC | Type II secretory pathway protein LspC | lpg0895 | Substrate recognition and/or secretin interactions |
| Type II effectorsa | ||||
| LLB_0177 | Putative lipoprotein | lpg1385 | ||
| LLB_0340 | plcA | Phospholipase C PlcA | lpg1455 | |
| LLB_0700 | map | Major acid phosphatase Map | lpg1119 | |
| LLB_1497 | icmX | IcmX | lpg2689 | |
| LLB_1611 | lapA | Leucine aminopeptidase LapA | lpg2814 | |
| LLB_1661 | plaC | Acyltransferase PlaC | lpg2837 | |
| LLB_1671 | srnA | Secreted RNase A | lpg2848 | |
| LLB_2032 | Cellulase family protein | lpg1918 | ||
| LLB_2271 | Putative N-acetylmuramoyl-l-alanine amidase | lpg0264 | ||
| LLB_2504 | plaA | Lysophospholipase A PlaA | lpg2343 | |
| LLB_2607 | proA | Zinc metalloproteinase ProA/Msp | lpg0467 | |
| LLB_2608 | lipA | Lipase A | lpg0468 | |
| LLB_2928 | lipB | Lipase B | lpg1157 | |
| LLB_3547 | lapB | Leucine aminopeptidase LapB | lpg0032 | Authentic frameshift |
Based on homology to 25 L. pneumophila type II secretion system substrates listed in the review of Cianciotto (18).
Type IVA and type IVB secretion systems.
Type IV secretion systems (T4SSs) are membrane-associated transporter complexes believed to have evolved from bacterial conjugation machinery and to be capable of transporting both proteins and nucleic acids into plant, animal, and bacterial cells (37, 91). T4SSs are used by many Gram-negative bacteria for exchange of genetic material, spread of conjugative plasmids, and injection of virulence factors into eukaryotic host cells (8). T4SSs have been grouped into subclasses, including type IVA, which is similar to the Agrobacterium tumefaciens Vir system, and type IVB, which is similar to the Tra/Trb bacterial conjugation systems (25).
L. pneumophila strains Paris, Lens, and Philadelphia-1 express a type IVA secretion system (T4ASS), Lvh (Legionella vir homologues) (16, 17, 80). L. pneumophila strain Corby lacks Lvh, but it contains two novel T4ASSs, Trb-1 and Trb-2 (38), whose roles are not known. The Lvh system of L. pneumophila is required for plasmid conjugation, as well as for low-temperature host cell entry and replication (9, 71, 80). L. longbeachae D-4968 contains all 11 lvh genes (lvhB2 to lvhB11 and lvhD4) arranged in the same order (see Table S5 in the supplemental material), implying that the function is similar. Homologs of the Corby Trb-1 or Trb-2 T4ASSs were not found in L. longbeachae, which is consistent with the observation that the lvh and trb-tra gene clusters do not coexist in the same genome (38).
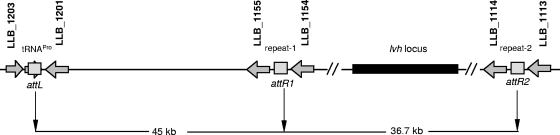
In L. pneumophila, the T4ASS-encoding gene clusters are located on plasmid-like elements which also contain genes encoding mobility factors and enzymes, such as phage integrases and transposases (27, 38). These plasmid-like elements are either integrated into the chromosome at sites in the tmRNA, tRNAPro, and tRNAArg genes or excised as multicopy plasmids (Table 1). The D-4968 genome contains two 45-bp sequences, which are identical to the 3′ end of the tRNAPro gene, in the intergenic regions flanking the lvh gene cluster (Fig. 3; see Fig S3 in the supplemental material). There are also several genes that are phage related, encode mobility factors, or are components of a restriction system located in the region between the tRNAPro gene and the second sequence repeat (see Table S5 in the supplemental material). The 3′ end of the tRNAPro gene and the two direct sequence repeats flanking the lvh locus may represent the chromosome-plasmid junction sites attL, attR1, and attR2 (Fig. 3).We hypothesize that this region could be a plasmid-like element that exists in either integrated or excised form and that mobility of the lvh region may be widespread in the genus Legionella and contribute to interspecies exchange of genetic information.
FIG. 3.
Type IVA Lvh secretion system: schematic diagram of the integrated form of the putative plasmid-like element. The gray boxes indicate the predicted att sites, and the arrows indicate the open reading frames flanking these sites. The black rectangle indicates the position of the lvh locus.
In addition to type IVA secretion systems, L. pneumophila contains several type IVB secretion systems (T4BSSs), including the Icm (intracellular multiplication)/Dot (defective organelle trafficking) system present in all strains and Tra-like systems identified in selected strains of this species (25). In L. pneumophila, the Icm/Dot system is encoded in two genomic regions; region I contains seven genes (icmV, icmW, icmX, dotA, dotB, dotC, and dotD), and region II contains 18 genes (icmT, icmS, icmR, icmQ, icmP/dotM, icmO/dotL, icmN/dotK, icmM/dotJ, icmL/dotI, icmK/dotH, icmE/dotG, icmG/dotF, icmC/dotE, icmD/dotP, icmJ/dotN, icmB/dotO, icmF, and icmH/dotU) (for a review, see reference 1). The gene organization of both regions is highly conserved in all four L. pneumophila strains. It has been proposed that the Icm/Dot proteins form a multicomponent translocation apparatus that spans both bacterial membranes (92). The inner membrane proteins IcmG/DotF and IcmE/DotG and the outer membrane proteins DotC, DotD, and IcmK/DotH interact with each other, forming the core transmembrane complex of the Icm/Dot system (92). Icm/Dot components are required for intracellular replication in protozoans or macrophages and to cause disease in animals (29, 81). The Icm/Dot system contributes to avoidance of association with markers of endosome-lysosome fusion, remodeling of LCV by endoplasmic reticulum (ER)-derived vesicles, and modulation of host signal transduction processes (30).
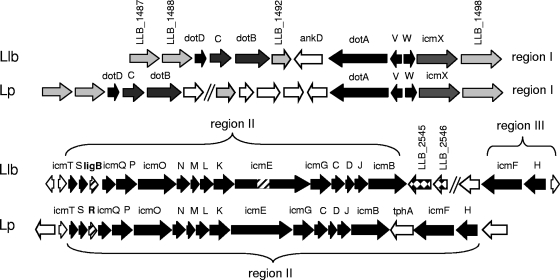
The L. longbeachae D-4968 genome contains 24 out of 25 icm/dot genes identified in L. pneumophila (see Table S6 in the supplemental material). These genes are located in three genomic regions and are generally homologous, and the orders of the genes are similar, with three exceptions (Fig. 4). In D-4968, the icmF and icmH/dotU genes of L. pneumophila region II are separated from the rest of the icmT-icmB/dotO gene cluster. The icmE/dotG gene of L. longbeachae is 1,431 bp longer than that of L. pneumophila, mostly due to insertion of bases encoding 390 amino acids that form several pentapeptide repeats (Pfam accession number PF00805) (data not shown). Since IcmE/DotG is an inner membrane protein comprising the core of the Icm/Dot transmembrane complex (92), the considerably larger L. longbeachae IcmE/DotG protein suggests that the size and/or structure of the L. longbeachae Icm/Dot apparatus may be different from the size and/or structure of the L. pneumophila Icm/Dot apparatus. L. longbeachae contains ligB, a functional homologue of icmR (fir gene family) that corresponds in position but does not share sequence homology with L. pneumophila icmR and functions in a similar manner with the corresponding icmQ gene during infection (32, 33). The L. longbeachae icmS, ligB, and icmQ genes are required for intracellular growth in human macrophages (32). The regulatory region of ligB in D-4968 contains the CpxR consensus binding site GTAAANNNNNNGAAAG but no PmrA-binding site. This finding corroborates a previous hypothesis that proposed that L. longbeachae lost the ancestral PmrA regulatory element in the fir promoter as a response to specific environmental conditions (31).
FIG. 4.
Organization and open reading frames of the Icm/Dot genomic regions in L. longbeachae (Llb) (regions I, II, and III) and L. pneumophila (Lp) (regions I and II). The icm/dot genes shared by the two species are indicated by black arrows. Functional homologs ligB and icmR are indicated by striped arrows. Sequences adjacent to the Icm/Dot genes that are homologous in the two species are indicated by gray arrows, whereas genes that do not have a homolog in this vicinity are indicated by open arrows. The sequence encoding the 370-amino-acid region specific to L. longbeachae IcmE (see text) is indicated by a striped box. The LLB_2545 and LLB_2546 genes encoding a putative sensor kinase and a response regulator, respectively, of a two-component signal transduction system are proposed to be involved in icm/dot regulation and are indicated by checkered arrows.
Interestingly, the icmB gene of dot/icm genomic region II in L. longbeachae is adjacent to a pair of genes encoding a two-component sensor kinase and response regulator (Fig. 4). The putative sensor kinase, encoded by LLB_2545, shares 33% sequence identity with the C-terminal region of the L. longbeachae two-component sensor kinase CpxA, while the putative response regulator, encoded by LLB_2546, shares 42% identity with the Escherichia coli response regulator OmpR and 35% identity with the L. longbeachae response regulator CpxR. The LLB_2545-LLB_2546-encoded two-component regulatory system appears to be unique to L. longbeachae. Sequence similarity to CpxAR and the close proximity of the LLB_2545 and LLB_2546 genes to the icm/dot genomic region suggest involvement in regulation of icm/dot genes. However, the heterogeneity of the N-terminal signal input domain sequences of the LLB_2545 and CpxA proteins suggests that they may respond to different environmental signals. Further studies of the icm/dot region in other Legionella species are needed to determine if there is a correlation between loss of the PmrA regulatory element in fir genes and acquisition of additional two-component regulators, such as the LLB_2545 and LLB_2546 regulators. The high level of conservation of Icm/Dot system components in the D-4968 genome suggests that this T4BSS plays an essential role in the life cycle of the bacterium, but its regulation might differ from that of its counterpart in L. pneumophila.
The L. longbeachae and L. pneumophila Icm/Dot secretion systems also differ in the set of effectors secreted via these T4SSs. To date, 140 L. pneumophila proteins have been determined to be Icm/Dot effectors, and it is predicted that the total number of Icm/Dot substrates approaches 300 (14, 47). The function of the majority of these effectors remains unknown since most of them are individually dispensable during intracellular growth or for replication vacuole morphology (30). We used a combination of the lists of L. pneumophila Icm/Dot effectors from the Icm/Dot effector database (http://microbiology.columbia.edu/shuman/effectors.html) and two recent publications (14, 47) to search for homologs in L. longbeachae using a local BLAST search. L. pneumophila and L. longbeachae share such well-characterized effectors as RalF, LepA, LepB, and SdhA (for a review, see reference 47). Yet only 43 of 140 homologs were identified in D-4968, although multiple paralogs were found for SidE and LidA (see Table S7 in the supplemental material). DrrA/SidM, which exhibits both guanine nucleotide exchange factor (GEF) and GDP dissociation inhibitor displacement factor (GDF) activities with the small GTPase Rab1 promoting its recruitment to the LCV (46, 56), was conspicuously not present in L. longbeachae. However, DrrA/SidM also is not present in L. pneumophila strain Paris (1), supporting the general trend of individual dispensability of Icm/Dot effectors. Several of the L. pneumophila Icm/Dot effectors that are not present in L. longbeachae are those with ankyrin repeats and Legionella effectors identified by machine learning (Lem) (14), and only 4/13 and 6/23 of these effectors have been found in D-4968, respectively (see Table S7 in the supplemental material).
Many effectors identified in L. pneumophila contain domains that are present mostly in eukaryotes (47). Therefore, we searched the D-4968 genome for genes predicted to encode proteins with eukaryote-like motifs, including ankyrin repeats, leucine-rich repeats, F-boxes, U-boxes, Sel-1 domains, and serine-threonine protein kinase domains (1, 16). As a result, we identified 29 genes that encode proteins with domains preferentially found in eukaryotic proteins (Table 3). Ankyrin repeats mediate protein-protein interactions in eukaryotes and are involved in many cellular processes, including cell motility, cell signaling, and transcriptional regulation (36). L. longbeachae contains a large family of ankyrin repeat-containing proteins, like such intracellular pathogens as L. pneumophila, C. burnetii, Wolbachia pipientis, and Rickettsia felis. In addition to four homologs of Icm/Dot effectors, AnkH, AnkK, AnkC, and AnkD (see Table S7 in the supplement material), we identified 18 other ankyrin repeat-containing proteins (Table 3), 2 of which are homologous to the L. pneumophila Corby Lpc_1606 protein and the rest of which are unique to L. longbeachae. Ankryin repeat-containing proteins in L. longbeachae are likely to be Icm/Dot effectors, as they are in L. pneumophila.
TABLE 3.
Genes encoding proteins with eukaryote-like domainsa
| Locus | Gene | Gene product | Homolog in L. pneumophilab | L. pneumophila locus |
|---|---|---|---|---|
| Ankyrin repeat | ||||
| LLB_0015 | Ankyrin repeat-containing protein | N | ||
| LLB_0262 | Ankyrin repeat-containing protein | N | ||
| LLB_0287 | Ankyrin repeat-containing protein | N | ||
| LLB_0528 | Ankyrin repeat-containing protein | N | ||
| LLB_0557 | Ankyrin repeat-containing protein | N | ||
| LLB_0670 | Ankyrin repeat-containing protein | N | ||
| LLB_0847 | Ankyrin repeat-containing protein | N | ||
| LLB_1621 | Ankyrin repeat-containing protein | N | ||
| LLB_1762 | Ankyrin repeat-containing protein | N | ||
| LLB_1797 | Ankyrin repeat-containing protein | Y | LPC_1606 | |
| LLB_1853 | Ankyrin repeat-containing protein | N | ||
| LLB_1992 | Ankyrin repeat-containing protein | Y (partial) | lpp0750 | |
| LLB_2238 | Ankyrin repeat-containing protein | N | ||
| LLB_2283 | Ankyrin repeat-containing protein | N | ||
| LLB_2882 | Ankyrin repeat-containing protein | N | ||
| LLB_3543 | Ankyrin repeat-containing protein | Y | LPC_1606 | |
| LLB_3657 | Ankyrin repeat-containing protein | N | ||
| LLB_3762 | Ankyrin repeat-containing protein | N | ||
| F-box domain | ||||
| LLB_0157 | Putative choline kinase | Y | lpp1363 | |
| LLB_3234 | F-box domain-containing protein | Y | lpp1330 | |
| LLB_3296 | F-box domain-containing protein | Y | lpp1330 | |
| U-box domain | ||||
| LLB_1403 | U-box domain-containing protein | N | ||
| Sel-1 domain | ||||
| LLB_0208 | TPR repeat family protein | Y | lpp1310 | |
| LLB_0879 | Sel-1 domain repeat-containing protein | Y | lpp0957 | |
| LLB_2685 | enhC | Enhanced entry protein EnhC | Y | lpp2629 |
| Leucine-rich repeat | ||||
| LLB_1764 | Leucine-rich repeat-containing protein | N | ||
| Serine threonine protein kinases | ||||
| LLB_0562 | Putative serine/threonine protein kinase | N | ||
| LLB_3272 | Conserved hypothetical protein | Y (weak) | lpp1439 | |
| LLB_3444 | Serine/threonine protein kinase domain protein | Y (weak) | lpp2626 | |
| LLB_3727 | Serine/threonine-protein kinase | Y | lpp1439 |
Homologs of Icm/Dot effectors listed in Table S7 in the supplemental material are not included.
N, no; Y, yes.
F-box- and U-box-domain containing proteins of L. pneumophila were predicted to interact with the ubiquitin machinery of eukaryotic cells (1). We identified three L. longbeachae genes predicted to encode F-box-containing proteins, all of which have homologs in L. pneumophila (Table 3). For a long time the L. pneumophila LegU2/LubX Icm/Dot effector has been described as the only protein known in prokaryotes to contain U-box domains (16). However, a second gene with a putative U-box domain was recently found in “Candidatus Protochlamydia amoebophila” (3). A U-box domain is present in eukaryotic E3 ubiquitin ligases, and recently it has been demonstrated that LegU2/LubX has ubiquitin ligase activity since it can catalyze polyubiquitination of eukaryotic Cdc2-like kinase 1 (52). L. longbeachae does not have a LegU2/LubX homolog, but it encodes a novel U-box domain protein, the LLB_1403 protein, which does not share identity with any known protein in the database. The U-box of the LLB_1403 protein appears to contain all conserved hydrophobic residues critical for ubiquitin conjugating enzyme binding (52) (data not shown). This suggests that the LLB_1403 protein may be an Icm/Dot effector with ubiquitin ligase activity. Additional studies are needed to determine which of the 29 proteins listed in Table 3 are actual substrates of the Icm/Dot system.
Type V secretion system.
It is hypothesized that L. pneumophila strain Paris possesses a type V secretion system because it encodes a putative autotransporter protein, Lpp0779 (16). L. longbeachae does not encode a homolog of Lpp0779 or any other putative autotransporter. Therefore, it is unlikely that L. longbeachae employs this type of protein secretion.
Eukaryote-like proteins.
The L. pneumophila genome encodes a large number and wide variety of eukaryote-like proteins (1, 26, 40). The acquisition of eukaryote-like genes can be explained by eukaryote-to-prokaryote horizontal gene transfer occurring during intracellular growth of naturally competent L. pneumophila in aquatic protozoans (26, 40). The eukaryote-like proteins likely interfere with host cellular processes by mimicking functions of eukaryotic enzymes (1). A list of 30 L. pneumophila proteins with the highest similarity scores with eukaryotic proteins (1) was used to search the genome of L. longbeachae D-4968, which resulted in identification of 14 homologs (Table 4). For example, LLB_0444 encodes ectonucleoside triphosphate diphosphohydrolase (apyrase), which shares 58% identity with Lpg1905, a secreted apyrase recently shown to be important for L. pneumophila replication in eukaryotic cells and for infection in A/J mice (79). It is conceivable that LLB_0444 may also be important for pathogenesis of L. longbeachae. On the other hand, a gene encoding a second L. pneumophila apyrase homolog, Lpg0971, was not found in the L. longbeachae genome. Nor does D-4968 encode a homolog of L. pneumophila sphingosine-1-phosphate lyase, which is predicted to influence host cell survival and apoptosis (40).
TABLE 4.
Genes encoding homologs of L. pneumophila proteins with the highest similarity scores with eukaryotic proteins
| Locus | Gene | Gene product | Homolog in L. pneumophilaa | L. pneumophila locus |
|---|---|---|---|---|
| LLB_0243 | mucD | Serine protease MucD | Y | lpp0965 |
| LLB_0255 | SPFH domain/band 7 family protein | Y | plpp0050 | |
| LLB_0444 | Ectonucleoside triphosphate diphosphohydrolase I (apyrase) | Y | lpp1880 | |
| LLB_0796 | Thiamine biosynthesis protein NMT-1 homolog | Y | lpp1522 | |
| LLB_1582 | nuoE | NADH-quinone oxidoreductase, E subunit | Y | lpp2832 |
| LLB_1699 | Conserved hypothetical protein | Y | lpp2923 | |
| LLB_1873 | udk | Uridine kinase (uridine monophosphokinase) (cytidine monophosphokinase) | Y (weak) | lpp1167 |
| LLB_2065 | purC | Phosphoribosylaminoimidazole-succinocarboxamide synthase | Y | lpp1647 |
| LLB_2129 | Phytanoyl-coenzyme A dioxygenase family protein | Y | lpp0578 | |
| LLB_2523 | Glucoamylase homolog | Y | lpp0489 | |
| LLB_2782 | xth | Exodeoxyribonuclease III | Y | lpp0702 |
| LLB_2976 | Putative calcium-transporting ATPase | Y | lpp1127 | |
| LLB_3015 | Methyltransferase domain family protein | Y | lpp0358 | |
| LLB_3698 | Putative uracil DNA glycosylase | Y | lpp1665 |
Y, yes; N, no.
While L. longbeachae does not share all eukaryote-like homologs with L. pneumophila, it may encode additional eukaryote-like proteins unique to it. Therefore, we adapted the criteria used by Albert-Weissenberger et al. in the search for eukaryote-like proteins in L. pneumophila genomes (1) to search the D-4968 genome for genes that have no homologs in L. pneumophila. Over 70 such genes were identified in L. longbeachae. Table 5 lists proteins with 35% or greater sequence identity to eukaryotic proteins and no counterparts in L. pneumophila. Some of the proteins listed have homologs in other bacteria, including many intracellular pathogens such as Coxiella and Brucella, while others have homologs that are found exclusively in eukaryotic organisms. Some examples of the genes include LLB_2177, which encodes a protein phosphatase 2C (PP2C) domain protein that shares sequence identity only with eukaryotic protein phosphatases. The PP2C family proteins are Mg2+/Mn2+-dependent serine/threonine phosphatases (50, 86). In eukaryotes, PP2Cs are involved in several cell regulation and cell signaling processes, including p53 activation (86), implying that L. longbeachae may use the serine/threonine phosphatase activity of the LLB_2177 protein to modulate similar processes in its host cells. Three L. longbeachae genes, LLB_2053, LLB_3045, and LLB_3686 (Table 5 and data not shown), encode proteins containing the Ras family motif (Pfam00071), a small GTP-binding domain rarely found in bacteria. A search of 1,284 bacterial genomes using the IMG website (58) yielded only 22 other bacterial species encoding proteins with a Ras family motif. L. pneumophila employs a number of Icm/Dot effectors to subvert the functions of the host small GTPases Arf1 and Rab1 in order to facilitate remodeling of the LCV by ER-derived vesicles (30). The unique L. longbeachae small GTPase homologs may mimic host small GTPase activity and participate directly in the recruitment of ER-derived vesicles to the LCV, competing with Arf1 and Rab1.
TABLE 5.
Eukaryote-like genes not present in L. pneumophila
| Locus | Gene | Gene-product | Best hit in Blast-extend-repraze searchesa | Homolog in Coxiella burnetiib | Homolog in Brucella abortisb |
|---|---|---|---|---|---|
| LLB_0304 | Conserved hypothetical protein | XP_820|438.1, Trypanosoma cruzi strain CL Brener | N | N | |
| LLB_0308 | katA | Catalase KatA | Q9PWF7, Rana rugosa | N | N |
| LLB_0349 | Putative 7-dehydrocholesterol reductase | ABA01480.1, Gossypium hirsutum | Y | N | |
| LLB_0720 | Putative esterase | 1XKL_D, Nicotiana tabacum | N | N | |
| LLB_1007 | Phosphoserine phosphatase | CAA71318.1, Homo sapiens | Y | N | |
| LLB_1398 | Putative aconitate hydratase | P39533, Saccharomyces cerevisiae | N | N | |
| LLB_1476 | NAD-dependent epimerase/dehydratase family protein | XP_001111155.1, Macaca mulatta | N | N | |
| LLB_1736 | GNAT family acetyltransferase | Q6P8J2, Mus musculus | N | N | |
| LLB_1848 | Putative serine carboxypeptidase | XP_001431633.1, Paramecium tetraurelia | N | N | |
| LLB_2053 | Ras family small GTP-binding protein domain-containing protein | CAA66447.1, Lotus japonicus | N | N | |
| LLB_2155 | Methyltransferase domain protein | XP_001584020.1, Trichomonas vaginalis G3 | N | N | |
| LLB_2177 | Protein phosphatase 2C domain protein | XP_001500411.1, Equus caballus | N | N | |
| LLB_2182 | Putative 8-amino-7-oxononanoate synthase | Q9NUV7, Homo sapiens | N | N | |
| LLB_2246 | Putative quinone oxidoreductase | XP_643354.1, Dictyostelium discoideum | N | N | |
| LLB_2275 | add | Adenosine deaminase | Q6FP78, Candida glabrata | N | N |
| LLB_2365 | Short-chain dehydrogenase/reductase family oxidoreductase | XP_001637773.1, Nematostella vectensis | N | N | |
| LLB_2379 | gad | Glutamate decarboxylase | NP_178310.1, Arabidopsis thaliana | N | N |
| LLB_2449 | Catalytic LigB subunit of aromatic ring-opening dioxygenase family protein | CAE47100.1, Beta vulgaris | N | N | |
| LLB_2496 | l-Lactate dehydrogenase | NP_956777.1, Danio rerio | N | N | |
| LLB_2553 | ahpC | Alkylhydroperoxide reductase | Q26695, Trypanosoma brucei | N | Y |
| LLB_2797 | Putative Hsp20 family heat shock protein | Q943E7, Oryza sativa subsp. japonica | Y | N | |
| LLB_2807 | Cyclic nucleotide-binding domain protein | XP_667983.1, Cryptosporidium hominis | N | N | |
| LLB_2814 | Putative asparaginase | XP_641813.1, Dictyostelium discoideum | N | N | |
| LLB_2830 | Cytosine deaminase | XP_001265161.1, Neosartorya fischeri | N | N | |
| LLB_2867 | Aldehyde dehydrogenase (NAD) family protein | P51648, Homo sapiens | N | N | |
| LLB_2876 | S-(Hydroxymethyl)glutathione dehydrogenase/ class III alcohol dehydrogenase | NP_571924.2, Danio rerio | N | N | |
| LLB_2877 | fghA | S-Formylglutathione hydrolase | XP_855986.1, Canis lupus familiaris | N | N |
| LLB_2924 | Acyl-coenzyme A dehydrogenase domain protein | NP_001086464.1, Xenopus laevis | N | N | |
| LLB_3045 | Ras family small GTP-binding protein domain-containing protein | P11620, Schizosaccharomyces pombe | N | N | |
| LLB_3084 | Fatty acid desaturase domain protein | Q9LVZ3, Arabidopsis thaliana | N | N | |
| LLB_3125 | Conserved hypothetical protein | XP_001117848.1, Macaca mulatta | N | N | |
| LLB_3192 | Putative tubulin-tyrosine ligase family protein | XP_001601458.1, Nasonia vitripennis | N | N | |
| LLB_3222 | Putative alanine-glyoxylate aminotransferase | Q9SR86, Arabidopsis thaliana | N | N | |
| LLB_3287 | Fatty acid desaturase family protein | Q949X0, Arabidopsis thaliana | N | N | |
| LLB_3303 | Putative membrane protein | NP_001054330.1, Oryza sativa subsp. japonica | N | N | |
| LLB_3421 | Putative NAD-dependent epimerase/dehydratase | NP_564095.1, Arabidopsis thaliana | N | Y | |
| LLB_3455 | Aromatic ring-opening dioxygenase family protein | Q949R4, Arabidopsis thaliana | N | N | |
| LLB_3743 | Formate dehydrogenase | Q07511, Solanum tuberosum | N | N | |
| LLB_3816 | Endoglucanase-like protein | XP_001030472.1, Tetrahymena thermophila | N | N | |
| LLB_3818 | Short-chain dehydrogenase/reductase family oxidoreductase | Q3T046, Bos taurus | N | Y |
Blast-extend-repraze searches were conducted by the JCVI Annotation Service.
N, no; Y, yes.
In summary, L. longbeachae contains a large number of eukaryote-like proteins, some of which have homologs in L. pneumophila, but most of these proteins are unique to L. longbeachae. These proteins are likely involved in modulation of host cellular functions during the intracellular phase of L. longbeachae growth. The difference between eukaryote-like proteins in L. pneumophila and eukaryote-like proteins in L. longbeachae may be due to the differences in the protozoan hosts that are specific for each species, since these hosts are the most likely the sources for acquisition of these proteins. It would be interesting to see how many eukaryote-like proteins are shared by different Legionella species and how many of them are species specific.
Flagella, motility, and chemotaxis.
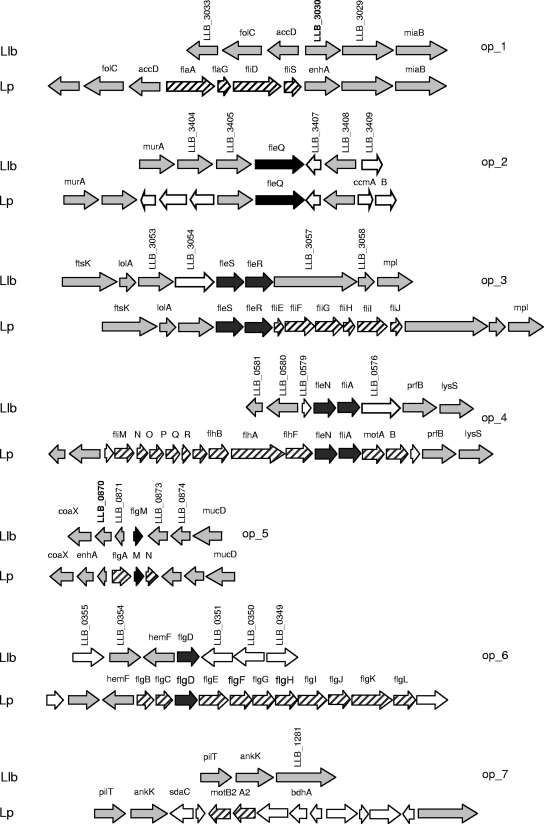
In L. pneumophila, flagellar structural and regulatory genes are organized in seven operons: (i) flaAG fliDS, (ii) fleQ, (iii) fleSR fliEFGHLI, (iv) fliMNOPOR flhBAF fleN fliA motAB, (v) flgAMN, (vi) flgBCDEFGHIJKL, and (vii) motB2A2 (43). Even though these operons are very similar to those of Pseudomonas and Salmonella, the L. pneumophila motility system lacks an important component of most bacterial flagellar systems: a chemotaxis system. The regulatory network that governs L. pneumophila flagellum-mediated motility includes RpoN (σ54) and its regulators FleQ, FleN, FleS, and FleR, as wells as FliA (σ28) and its anti-sigma factor FlgM (Fig. 2) (1). In contrast, the L. longbeachae genome contains a complete set of chemotaxis genes but only a few flagellar genes, most of which are predicted to perform regulatory functions (Fig. 5; see Table S8 in the supplement material). Comparison of the L. pneumophila regions containing flagellar operons with the L. longbeachae regions containing flagellar operons showed that the content and the order of genes adjacent to the flagellar loci are well conserved between the two species (Fig. 5). However, L. longbeachae lacks the majority of flagellar structural genes. The absence of transposon elements and pseudogenes in these chromosomal regions of L. longbeachae suggests that the loss of flagellar genes was not a recent event.
FIG. 5.
Comparison of the genomic regions containing flagellar operons in L. pneumophila (Lp) with the corresponding regions in L. longbeachae (Llb). Flagellar genes shared by the two species are indicated by black arrows. L. pneumophila flagellar genes that do not have counterparts in L. longbeachae are indicated by striped arrows. Sequences adjacent to flagellar operon genes that are homologous in the two species are indicated by gray arrows, whereas genes that do not have homologs in this vicinity are indicated by open arrows. The enhA homologs LLB_0870 and LLB_3030, which are proposed to constitute the FliA regulon in D-4968 (see text), are indicated by bold type.
Previous papers contain conflicting reports regarding expression of flagella by L. longbeachae. Heuner et al. did not detect the flagellin-encoding flaA sequence in L. longbeachae serogroup 1 using Southern hybridization, nor did they visualize flagella on the surface by electron microscopy (42). On the other hand, Asare et al. reported that D-4968 and several other L. longbeachae serogroup 1 strains express flagella in the postexponential phase (6). However, extensive microscopic examination did not confirm motility for D-4968 or six other L. longbeachae isolates (both serogroup 1 and serogroup 2) from the CDC culture collection when they were tested in our laboratory (data not shown). Nor did PCR-based screening of 50 L. longbeachae isolates belonging to both serogroups detect flaA in any isolate (data not shown). PCR amplification of fragments of the less polymorphic fliI, flgG, and flgH genes from eight L. longbeachae isolates (six serogroup 1 isolates and two serogroup 2 isolates) was also unsuccessful. Amplification of the mip gene as a positive control for PCR was possible with all isolates (data not shown). The failure to detect flagellar genes in other L. longbeachae isolates could be due to decreased specificity of PCR primers designed based on L. pneumophila sequences. However, the complete absence of flagellar biogenesis genes in D-4968, in the recently sequenced L. longbeachae D-5243 isolate from the CDC culture collection (N. A. Kozak et al., unpublished data), and in four other L. longbeachae isolates sequenced in France (15) supports the hypothesis that the majority of L. longbeachae isolates do not possess flagellar genes.
It has been shown previously that L. pneumophila flagellin is required for caspase 1 activation in murine macrophages (53, 62, 70).The lack of a flagellum in D-4968 and other L. longbeachae isolates provides an explanation for the failure of this species to activate caspase 1 and trigger rapid pyroptosis in C57BL/6 and BALB/c mouse macrophages, allowing establishment of infection in mice resistant to L. pneumophila (6). The ability to replicate in various mouse strains and the fact that D-4968 was isolated from a Legionnaires' disease patient (4) indicate that the flagellum is not a factor that is required for L. longbeachae virulence.
The detection of flagellar regulatory genes without structural genes in the D-4968 genome raises several questions. The first question is whether ancestors of this L. longbeachae isolate carried the full set of flagellar structural genes and, if so, what selective pressure caused their loss and by what mechanism were they lost. Second, the presence of an intact fliA gene implies functionality, but the nature of the genes that FliA regulates has yet to be determined. Studies on L. pneumophila demonstrated that FliA, in addition to flagellar genes, regulates factors that contribute to virulence-associated traits, including the inhibition of phagosome maturation (63). A recent transcriptome study of L. pneumophila strain Paris identified several genes of the FliA regulon that do not play a role in flagellum biogenesis or regulation, including lpp0972 and lpp1290 that encode enhanced entry proteins (EnhA) (13). In D-4968, LLB_0870 and LLB_3030 are highly homologous to lpp0972 and lpp1290, respectively. Both LLB_0870 and LLB_3030 are located in chromosomal regions that correspond to loci adjacent to the flagellar operons in L. pneumophila (Fig. 5). This suggests that LLB_0870 and LLB_3030 may constitute the FliA regulon of D-4968. Further studies should help identify other FliA-regulated genes in L. longbeachae, which may be more numerous than FliA-regulated genes in L. pneumophila.
Identification of a homolog of the anti-sigma factor FlgM gene in the genome of D-4968 presents an interesting paradox. Typically, FlgM inhibits activity of FliA by directly binding to this sigma factor, and FliA is released only after complete assembly of a flagellar intermediate that functions as a secretion channel for FlgM (45). However, D-4968 does not have the genes required for complete synthesis of the flagellar intermediate, so FlgM cannot be secreted in this way. If the LLB_0872 protein, which shares 46% identity with L. pneumophila FlgM, does function as an FliA-specific anti-sigma factor, L. longbeachae must have a novel mechanism for release of inhibition.
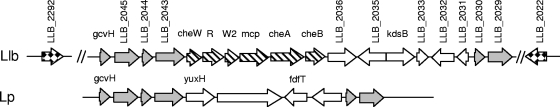
L. longbeachae D-4968 contains a chemotaxis operon with six genes encoding homologs of a methyl-accepting chemotaxis protein, the histidine kinase CheA, the methylesterase CheB, the methyltransferase CheR, and two CheW scaffold proteins (Fig. 6). A gene encoding the response regulator CheY is usually included in a chemotaxis operon (2, 90), but a cheY homolog was not found in the operon of D-4968. Two genes, LLB_2022 and LLB_2292 located outside the chemotaxis operon, encode CheY-like proteins, which may function together with the gene products of the chemotaxis operon. There are no homologs of either LLB_2022 or LLB_2292 in L. pneumophila. PCR-based screening of eight L. longbeachae isolates from the CDC culture collection (six serogroup 1 isolates and two serogroup 2 isolates) indicated that all of them contained cheR, cheA, and cheB genes (data not shown). Therefore, it is likely that many L. longbeachae isolates contain chemotaxis genes.
FIG. 6.
Organization and open reading frames of the L. longbeachae chemotaxis operon and comparison of the genomic region containing this operon with the homologous genomic region in L. pneumophila. Chemotaxis genes are indicated by striped arrows. Sequences adjacent to chemotaxis operon genes that are homologous in the two species are indicated by gray arrows, whereas genes that do not have a homolog in this vicinity are indicated by open arrows. MCP, methyl accepting chemotaxis protein. LLB_2002 and LLB_2292 that encode CheY-like proteins and may function together with the gene products of the chemotaxis operon are indicated by checkered arrows.
It is paradoxical that the genome of L. pneumophila has a complete set of flagellum genes and no chemotaxis genes, whereas the genome of L. longbeachae lacks flagellar genes but contains a chemotaxis operon. L. pneumophila flagellum-mediated motility is not regulated by a typical chemotaxis pathway, and the L. longbeachae chemotaxis genes are likely involved in regulation of a different process. Recent studies of the chemotaxis-like systems in different bacteria showed that many systems differ from the E. coli chemotaxis system and are not involved in the control of flagellum-mediated motility. These so-called chemosensory systems were shown to regulate type IV pilus-based motility, flagellar and type IV pilus expression, cell differentiation, and biofilm formation (51). The chemotaxis-like operon in D-4968 may be involved in the regulation of type IV pilus-based motility (see below), yet L. pneumophila also possesses type IV pili and exhibits twitching motility (22). It is more likely that twitching motility is regulated in similar ways in the two species and thus is che independent. According to RAST analysis, the composition of the chemotaxis-like operon in D-4968 is similar to the composition of one of the che operons in Geobacter uraniireducens and Magnetococcus. In G. uraniireducens the che cluster that is similar to the D-4968 che operon is predicted to regulate processes involving cell-cell interactions or social motility (90). Future studies of G. uraniireducens, Magnetococcus, and L. longbeachae should indicate whether the che operons in these organisms control similar functions.
Type IV pili.
L. pneumophila type IV pili (Tfp) promote attachment to mammalian and protozoan cells, are important for biofilm formation, and are required for natural competence (55, 79, 88). A recent study reported that Tfp are also required for twitching motility at 37°C but not at 27°C (22). Similar to other bacteria producing type IVa pili, L. pneumophila contains multiple Tfp biogenesis genes that are scattered throughout the genome (67). The core components necessary for Tfp biogenesis are considered to be the major pilin PilE, the prepilin peptidase PilD, the assembly ATPase PilB, the retraction ATPase PilT, the inner membrane protein PilC, and the secretin PilQ (see Table S8 in the supplemental material) (67). L. longbeachae D-4968 contains all of the Tfp biogenesis genes present in L. pneumophila, except for one: the gene encoding the prepilin, PilX, which has a stop codon in the middle of the reading frame (see Table S8 in the supplemental material). Regardless of PilX reading frame disruption, this protein is not a core component of Tfp, and its absence may not affect Tfp expression in L. longbeachae. L. longbeachae most likely produces Tfp, which, in the absence of flagella, may be the sole motility organ. Additional studies are needed to determine whether L. longbeachae exhibits Tfp-mediated twitching motility and whether the chemotaxis-like operon described above is involved in its regulation. Perhaps the loss of flagellum and retention of Tfp by L. longbeachae were adaptations for survival in soil, in contrast to other Legionella species that inhabit freshwater environments.
Other virulence factors.
L. longbeachae D-4968 contains a number of genes which encode homologs of L. pneumophila virulence factors or putative virulence factors that are not described above (see Table S9 in the supplemental material). For example, LLB_3347 encodes a macrophage infectivity potentiator (Mip). L. longbeachae mip mutants have a reduced capacity to infect and multiply within amoeba and do not cause death in guinea pigs, unlike the wild-type bacteria (28). D-4968 also encodes a weak homolog of major outer membrane protein MOMP, which has been shown to bind to the complement component CR3 of human monocytes in L. pneumophila and thus is important for phagocytosis (for a review, see reference 1). D-4968 contains a number of enh homologs, including genes encoding seven paralogs of EnhA, two paralogs of EnhB, and one paralog of EnhC, involved in L. pneumophila entry into host cells (21) (see Table S9 in the supplemental material). However, D-4968 lacks an RtxA-encoding gene that was identified together with the enh genes in L. pneumophila in a screen for the enhanced-entry phenotype and has been shown to be involved in cell entry, adherence, cytotoxicity, and pore formation (20, 21). The D-4968 genome encodes a homolog of the integral membrane protein MviN, which is important for Salmonella enterica serovar Typhimurium virulence and is predicted to be a virulence determinant in L. pneumophila (1). Finally, D-4968 homologs of the regulators BipA and Fis have been predicted to be involved in virulence gene regulation in L. pneumophila (1).
In conclusion, a comparison of the L. longbeachae D-4968 genome with four sequenced and annotated genomes of L. pneumophila strains showed that the majority of virulence factors are shared by these species. The major difference between the species is the absence of flagellar biosynthesis genes in L. longbeachae. Even though the type II secretion apparatus and the type IV secretion apparatus are conserved in L. longbeachae and L. pneumophila, we predict that the sets of effectors secreted via these systems differ and reflect the adaptation of these species to different host environments and extracellular ecological niches. Further studies should help determine whether all L. longbeachae strains have L. longbeachae D-4968-specific properties.
Supplementary Material
Acknowledgments
We thank JCVI for providing the JCVI Annotation Service, including the automatic annotation data and the manual annotation tool Manatee. We also thank Awdhesh Kalia and Yousef Abu Kwaik for helpful suggestions at the project-planning stage.
Footnotes
Published ahead of print on 11 December 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Albert-Weissenberger, C., C. Cazalet, and C. Buchrieser. 2007. Legionella pneumophila—a human pathogen that co-evolved with fresh water protozoa. Cell. Mol. Life Sci. 64:432-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexandre, G., and I. B. Zhulin. 2003. Different evolutionary constraints on chemotaxis proteins CheW and CheY revealed by heterologous expression studies and protein sequence analysis. J. Bacteriol. 185:544-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Angot, A., A. Vergunst, S. Genin, and N. Peeters. 2007. Exploitation of eukaryotic ubiquitin signaling pathways by effectors translocated by bacterial type III and type IV secretion systems. PLoS Pathog. 3:e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anonymous. 2000. Legionnaires' disease associated with potting soil—California, Oregon, and Washington, May-June 2000. MMWR Morb. Mortal. Wkly. Rep. 49:777-778. [PubMed] [Google Scholar]
- 5.Asare, R., and Y. Abu Kwaik. 2007. Early trafficking and intracellular replication of Legionella longbeachae within an ER-derived late endosome-like phagosome. Cell. Microbiol. 9:1571-1587. [DOI] [PubMed] [Google Scholar]
- 6.Asare, R., M. Santic, I. Gobin, M. Doric, J. Suttles, J. E. Graham, C. D. Price, and Y. Abu Kwaik. 2007. Genetic susceptibility and caspase activation in mouse and human macrophages are distinct for Legionella longbeachae and L. pneumophila. Infect. Immun. 75:1933-1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aziz, R. K., D. Bartels, A. A. Best, M. DeJongh, T. Disz, R. A. Edwards, K. Formsma, S. Gerdes, E. M. Glass, M. Kubal, F. Meyer, G. J. Olsen, R. Olson, A. L. Osterman, R. A. Overbeek, L. K. McNeil, D. Paarmann, T. Paczian, B. Parrello, G. D. Pusch, C. Reich, R. Stevens, O. Vassieva, V. Vonstein, A. Wilke, and O. Zagnitko. 2008. The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Backert, S., and T. F. Meyer. 2006. Type IV secretion systems and their effectors in bacterial pathogenesis. Curr. Opin. Microbiol. 9:207-217. [DOI] [PubMed] [Google Scholar]
- 9.Bandyopadhyay, P., S. Liu, C. B. Gabbai, Z. Venitelli, and H. M. Steinman. 2007. Environmental mimics and the Lvh type IVA secretion system contribute to virulence-related phenotypes of Legionella pneumophila. Infect. Immun. 75:723-735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Begg, K., P. Roche, R. Owen, C. Liu, M. Kaczmarek, A. Hii, S. Stirzaker, A. McDonald, G. Fitzsimmons, P. McIntyre, R. Menzies, I. East, D. Coleman, and K. O'Neil. 2008. Australia's notifiable diseases status, 2006: annual report of the National Notifiable Diseases Surveillance System. Commun. Dis. Intell. 32:139-207. [DOI] [PubMed] [Google Scholar]
- 11.Bibb, W. F., R. J. Sorg, B. M. Thomason, M. D. Hicklin, A. G. Steigerwalt, D. J. Brenner, and M. R. Wulf. 1981. Recognition of a second serogroup of Legionella longbeachae. J. Clin. Microbiol. 14:674-677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blumer, C., P. Roche, J. Spencer, M. Lin, A. Milton, C. Bunn, H. Gidding, J. Kaldor, M. Kirk, R. Hall, T. Della-Porta, R. Leader, and P. Wright. 2003. Australia's notifiable diseases status, 2001: annual report of the National Notifiable Diseases Surveillance System. Commun. Dis. Intell. 27:1-78. [DOI] [PubMed] [Google Scholar]
- 13.Bruggemann, H., A. Hagman, M. Jules, O. Sismeiro, M. A. Dillies, C. Gouyette, F. Kunst, M. Steinert, K. Heuner, J. Y. Coppee, and C. Buchrieser. 2006. Virulence strategies for infecting phagocytes deduced from the in vivo transcriptional program of Legionella pneumophila. Cell. Microbiol. 8:1228-1240. [DOI] [PubMed] [Google Scholar]
- 14.Burstein, D., T. Zusman, E. Degtyar, R. Viner, G. Segal, and T. Pupko. 2009. Genome-scale identification of Legionella pneumophila effectors using a machine learning approach. PLoS Pathog. 5:e1000508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cazalet, C., L. Gomez-Valero, M. Lomma, C. Rusniok, N. Zidane, C. Bouchier, S. Jarraud, J. Etienne, and C. Buchrieser. 2009. Legionella pneumophila and Legionella longbeachae pathogenesis: new insights gained from comparative and functional genomics, p. 61. Legionella 2009, Paris, France.
- 16.Cazalet, C., C. Rusniok, H. Bruggemann, N. Zidane, A. Magnier, L. Ma, M. Tichit, S. Jarraud, C. Bouchier, F. Vandenesch, F. Kunst, J. Etienne, P. Glaser, and C. Buchrieser. 2004. Evidence in the Legionella pneumophila genome for exploitation of host cell functions and high genome plasticity. Nat. Genet. 36:1165-1173. [DOI] [PubMed] [Google Scholar]
- 17.Chien, M., I. Morozova, S. Shi, H. Sheng, J. Chen, S. M. Gomez, G. Asamani, K. Hill, J. Nuara, M. Feder, J. Rineer, J. J. Greenberg, V. Steshenko, S. H. Park, B. Zhao, E. Teplitskaya, J. R. Edwards, S. Pampou, A. Georghiou, I. C. Chou, W. Iannuccilli, M. E. Ulz, D. H. Kim, A. Geringer-Sameth, C. Goldsberry, P. Morozov, S. G. Fischer, G. Segal, X. Qu, A. Rzhetsky, P. Zhang, E. Cayanis, P. J. De Jong, J. Ju, S. Kalachikov, H. A. Shuman, and J. J. Russo. 2004. The genomic sequence of the accidental pathogen Legionella pneumophila. Science 305:1966-1968. [DOI] [PubMed] [Google Scholar]
- 18.Cianciotto, N. P. 2009. Many substrates and functions of type II secretion: lessons learned from Legionella pneumophila. Future Microbiol. 4:797-805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cianciotto, N. P. 2005. Type II secretion: a protein secretion system for all seasons. Trends Microbiol. 13:581-588. [DOI] [PubMed] [Google Scholar]
- 20.Cirillo, S. L., L. E. Bermudez, S. H. El-Etr, G. E. Duhamel, and J. D. Cirillo. 2001. Legionella pneumophila entry gene rtxA is involved in virulence. Infect. Immun. 69:508-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cirillo, S. L., J. Lum, and J. D. Cirillo. 2000. Identification of novel loci involved in entry by Legionella pneumophila. Microbiology 146:1345-1359. [DOI] [PubMed] [Google Scholar]
- 22.Coil, D. A., and J. Anne. 2009. Twitching motility in Legionella pneumophila. FEMS Microbiol. Lett. 293:271-277. [DOI] [PubMed] [Google Scholar]
- 23.Darling, A. C., B. Mau, F. R. Blattner, and N. T. Perna. 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14:1394-1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DebRoy, S., J. Dao, M. Soderberg, O. Rossier, and N. P. Cianciotto. 2006. Legionella pneumophila type II secretome reveals unique exoproteins and a chitinase that promotes bacterial persistence in the lung. Proc. Natl. Acad. Sci. U. S. A. 103:19146-19151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De Buck, E., J. Anne, and E. Lammertyn. 2007. The role of protein secretion systems in the virulence of the intracellular pathogen Legionella pneumophila. Microbiology 153:3948-3953. [DOI] [PubMed] [Google Scholar]
- 26.de Felipe, K. S., S. Pampou, O. S. Jovanovic, C. D. Pericone, S. F. Ye, S. Kalachikov, and H. A. Shuman. 2005. Evidence for acquisition of Legionella type IV secretion substrates via interdomain horizontal gene transfer. J. Bacteriol. 187:7716-7726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Doleans-Jordheim, A., M. Akermi, C. Ginevra, C. Cazalet, E. Kay, D. Schneider, C. Buchrieser, D. Atlan, F. Vandenesch, J. Etienne, and S. Jarraud. 2006. Growth-phase-dependent mobility of the lvh-encoding region in Legionella pneumophila strain Paris. Microbiology 152:3561-3568. [DOI] [PubMed] [Google Scholar]
- 28.Doyle, R. M., T. W. Steele, A. M. McLennan, I. H. Parkinson, P. A. Manning, and M. W. Heuzenroeder. 1998. Sequence analysis of the mip gene of the soilborne pathogen Legionella longbeachae. Infect. Immun. 66:1492-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Edelstein, P. H., M. A. Edelstein, F. Higa, and S. Falkow. 1999. Discovery of virulence genes of Legionella pneumophila by using signature tagged mutagenesis in a guinea pig pneumonia model. Proc. Natl. Acad. Sci. U. S. A. 96:8190-8195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ensminger, A. W., and R. R. Isberg. 2009. Legionella pneumophila Dot/Icm translocated substrates: a sum of parts. Curr. Opin. Microbiol. 12:67-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feldman, M., and G. Segal. 2007. A pair of highly conserved two-component systems participates in the regulation of the hypervariable FIR proteins in different Legionella species. J. Bacteriol. 189:3382-3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Feldman, M., and G. Segal. 2004. A specific genomic location within the icm/dot pathogenesis region of different Legionella species encodes functionally similar but nonhomologous virulence proteins. Infect. Immun. 72:4503-4511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feldman, M., T. Zusman, S. Hagag, and G. Segal. 2005. Coevolution between nonhomologous but functionally similar proteins and their conserved partners in the Legionella pathogenesis system. Proc. Natl. Acad. Sci. U. S. A. 102:12206-12211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fields, B. S., R. F. Benson, and R. E. Besser. 2002. Legionella and Legionnaires' disease: 25 years of investigation. Clin. Microbiol. Rev. 15:506-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Galperin, M. Y., A. N. Nikolskaya, and E. V. Koonin. 2001. Novel domains of the prokaryotic two-component signal transduction systems. FEMS Microbiol. Lett. 203:11-21. [DOI] [PubMed] [Google Scholar]
- 36.Gaudet, R. 2008. A primer on ankyrin repeat function in TRP channels and beyond. Mol. Biosyst. 4:372-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gerlach, R. G., and M. Hensel. 2007. Protein secretion systems and adhesins: the molecular armory of Gram-negative pathogens. Int. J. Med. Microbiol. 297:401-415. [DOI] [PubMed] [Google Scholar]
- 38.Glockner, G., C. Albert-Weissenberger, E. Weinmann, S. Jacobi, E. Schunder, M. Steinert, J. Hacker, and K. Heuner. 2008. Identification and characterization of a new conjugation/type IVA secretion system (trb/tra) of Legionella pneumophila Corby localized on two mobile genomic islands. Int. J. Med. Microbiol. 298:411-428. [DOI] [PubMed] [Google Scholar]
- 39.Gobin, I., M. Susa, G. Begic, E. L. Hartland, and M. Doric. 2009. Experimental Legionella longbeachae infection in intratracheally inoculated mice. J. Med. Microbiol. 58:723-730. [DOI] [PubMed] [Google Scholar]
- 40.Gomez-Valero, L., C. Rusniok, and C. Buchrieser. 2009. Legionella pneumophila: population genetics, phylogeny and genomics. Infect. Genet. Evol. 9:727-739. [DOI] [PubMed] [Google Scholar]
- 41.Heiner, C. R., K. L. Hunkapiller, S. M. Chen, J. I. Glass, and E. Y. Chen. 1998. Sequencing multimegabase-template DNA with BigDye terminator chemistry. Genome Res. 8:557-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Heuner, K., L. Bender-Beck, B. C. Brand, P. C. Luck, K. H. Mann, R. Marre, M. Ott, and J. Hacker. 1995. Cloning and genetic characterization of the flagellum subunit gene (flaA) of Legionella pneumophila serogroup 1. Infect. Immun. 63:2499-2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Heuner, K., and M. Steinert. 2003. The flagellum of Legionella pneumophila and its link to the expression of the virulent phenotype. Int. J. Med. Microbiol. 293:133-143. [DOI] [PubMed] [Google Scholar]
- 44.Hovel-Miner, G., S. Pampou, S. P. Faucher, M. Clarke, I. Morozova, P. Morozov, J. J. Russo, H. A. Shuman, and S. Kalachikov. 2009. σS controls multiple pathways associated with intracellular multiplication of Legionella pneumophila. J. Bacteriol. 191:2461-2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hughes, K. T., and K. Mathee. 1998. The anti-sigma factors. Annu. Rev. Microbiol. 52:231-286. [DOI] [PubMed] [Google Scholar]
- 46.Ingmundson, A., A. Delprato, D. G. Lambright, and C. R. Roy. 2007. Legionella pneumophila proteins that regulate Rab1 membrane cycling. Nature 450:365-369. [DOI] [PubMed] [Google Scholar]
- 47.Isberg, R. R., T. J. O'Connor, and M. Heidtman. 2009. The Legionella pneumophila replication vacuole: making a cosy niche inside host cells. Nat. Rev. Microbiol. 7:13-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jacobi, S., and K. Heuner. 2003. Description of a putative type I secretion system in Legionella pneumophila. Int. J. Med. Microbiol. 293:349-358. [DOI] [PubMed] [Google Scholar]
- 49.Jules, M., and C. Buchrieser. 2007. Legionella pneumophila adaptation to intracellular life and the host response: clues from genomics and transcriptomics. FEBS Lett. 581:2829-2838. [DOI] [PubMed] [Google Scholar]
- 50.Kennelly, P. J. 2002. Protein kinases and protein phosphatases in prokaryotes: a genomic perspective. FEMS Microbiol. Lett. 206:1-8. [DOI] [PubMed] [Google Scholar]
- 51.Kirby, J. R. 2009. Chemotaxis-like regulatory systems: unique roles in diverse bacteria. Annu. Rev. Microbiol. 63:45-59. [DOI] [PubMed] [Google Scholar]
- 52.Kubori, T., A. Hyakutake, and H. Nagai. 2008. Legionella translocates an E3 ubiquitin ligase that has multiple U-boxes with distinct functions. Mol. Microbiol. 67:1307-1319. [DOI] [PubMed] [Google Scholar]
- 53.Lightfield, K. L., J. Persson, S. W. Brubaker, C. E. Witte, J. von Moltke, E. A. Dunipace, T. Henry, Y. H. Sun, D. Cado, W. F. Dietrich, D. M. Monack, R. M. Tsolis, and R. E. Vance. 2008. Critical function for Naip5 in inflammasome activation by a conserved carboxy-terminal domain of flagellin. Nat. Immunol. 9:1171-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liles, M. R., V. K. Viswanathan, and N. P. Cianciotto. 1998. Identification and temperature regulation of Legionella pneumophila genes involved in type IV pilus biogenesis and type II protein secretion. Infect. Immun. 66:1776-1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lucas, C. E., E. Brown, and B. S. Fields. 2006. Type IV pili and type II secretion play a limited role in Legionella pneumophila biofilm colonization and retention. Microbiology 152:3569-3573. [DOI] [PubMed] [Google Scholar]
- 56.Machner, M. P., and R. R. Isberg. 2007. A bifunctional bacterial protein links GDI displacement to Rab1 activation. Science 318:974-977. [DOI] [PubMed] [Google Scholar]
- 57.Margulies, M., M. Egholm, W. E. Altman, S. Attiya, J. S. Bader, L. A. Bemben, J. Berka, M. S. Braverman, Y. J. Chen, Z. Chen, S. B. Dewell, L. Du, J. M. Fierro, X. V. Gomes, B. C. Godwin, W. He, S. Helgesen, C. H. Ho, G. P. Irzyk, S. C. Jando, M. L. Alenquer, T. P. Jarvie, K. B. Jirage, J. B. Kim, J. R. Knight, J. R. Lanza, J. H. Leamon, S. M. Lefkowitz, M. Lei, J. Li, K. L. Lohman, H. Lu, V. B. Makhijani, K. E. McDade, M. P. McKenna, E. W. Myers, E. Nickerson, J. R. Nobile, R. Plant, B. P. Puc, M. T. Ronan, G. T. Roth, G. J. Sarkis, J. F. Simons, J. W. Simpson, M. Srinivasan, K. R. Tartaro, A. Tomasz, K. A. Vogt, G. A. Volkmer, S. H. Wang, Y. Wang, M. P. Weiner, P. Yu, R. F. Begley, and J. M. Rothberg. 2005. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437:376-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Markowitz, V. M., E. Szeto, K. Palaniappan, Y. Grechkin, K. Chu, I. M. Chen, I. Dubchak, I. Anderson, A. Lykidis, K. Mavromatis, N. N. Ivanova, and N. C. Kyrpides. 2008. The integrated microbial genomes (IMG) system in 2007: data content and analysis tool extensions. Nucleic Acids Res. 36:D528-D533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McKinney, R. M., R. K. Porschen, P. H. Edelstein, M. L. Bissett, P. P. Harris, S. P. Bondell, A. G. Steigerwalt, R. E. Weaver, M. E. Ein, D. S. Lindquist, R. S. Kops, and D. J. Brenner. 1981. Legionella longbeachae species nova, another etiologic agent of human pneumonia. Ann. Intern. Med. 94:739-743. [DOI] [PubMed] [Google Scholar]
- 60.Miller, M., P. Roche, K. Yohannes, J. Spencer, M. Bartlett, J. Brotherton, J. Hutchinson, M. Kirk, A. McDonald, and C. Vadjic. 2005. Australia's notifiable diseases status, 2003 annual report of the National Notifiable Diseases Surveillance System. Commun. Dis. Intell. 29:1-61. [DOI] [PubMed] [Google Scholar]
- 61.Miyamoto, H., K. Maruta, M. Ogawa, M. C. Beckers, P. Gros, and S. Yoshida. 1996. Spectrum of Legionella species whose intracellular multiplication in murine macrophages is genetically controlled by Lgn1. Infect. Immun. 64:1842-1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Molofsky, A. B., B. G. Byrne, N. N. Whitfield, C. A. Madigan, E. T. Fuse, K. Tateda, and M. S. Swanson. 2006. Cytosolic recognition of flagellin by mouse macrophages restricts Legionella pneumophila infection. J. Exp. Med. 203:1093-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Molofsky, A. B., L. M. Shetron-Rama, and M. S. Swanson. 2005. Components of the Legionella pneumophila flagellar regulon contribute to multiple virulence traits, including lysosome avoidance and macrophage death. Infect. Immun. 73:5720-5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Molofsky, A. B., and M. S. Swanson. 2004. Differentiate to thrive: lessons from the Legionella pneumophila life cycle. Mol. Microbiol. 53:29-40. [DOI] [PubMed] [Google Scholar]
- 65.O'Connor, B. A., J. Carman, K. Eckert, G. Tucker, R. Givney, and S. Cameron. 2007. Does using potting mix make you sick? Results from a Legionella longbeachae case-control study in South Australia. Epidemiol. Infect. 135:34-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Owen, R., P. W. Roche, K. Hope, K. Yohannes, A. Roberts, C. Liu, S. Stirzaker, F. Kong, M. Bartlett, B. Donovan, I. East, G. Fitzsimmons, A. McDonald, P. B. McIntyre, and R. I. Menzies. 2007. Australia's notifiable diseases status, 2005: annual report of the National Notifiable Diseases Surveillance System. Commun. Dis. Intell. 31:1-70. [DOI] [PubMed] [Google Scholar]
- 67.Pelicic, V. 2008. Type IV pili: e pluribus unum? Mol. Microbiol. 68:827-837. [DOI] [PubMed] [Google Scholar]
- 68.Phares, C. R., P. Wangroongsarb, S. Chantra, W. Paveenkitiporn, M. L. Tondella, R. F. Benson, W. L. Thacker, B. S. Fields, M. R. Moore, J. Fischer, S. F. Dowell, and S. J. Olsen. 2007. Epidemiology of severe pneumonia caused by Legionella longbeachae, Mycoplasma pneumoniae, and Chlamydia pneumoniae: 1-year, population-based surveillance for severe pneumonia in Thailand. Clin. Infect. Dis. 45:e147-e155. [DOI] [PubMed] [Google Scholar]
- 69.Rasis, M., and G. Segal. 2009. The LetA-RsmYZ-CsrA regulatory cascade, together with RpoS and PmrA, post-transcriptionally regulates stationary phase activation of Legionella pneumophila Icm/Dot effectors. Mol. Microbiol. 72:995-1010. [DOI] [PubMed] [Google Scholar]
- 70.Ren, T., D. S. Zamboni, C. R. Roy, W. F. Dietrich, and R. E. Vance. 2006. Flagellin-deficient Legionella mutants evade caspase-1- and Naip5-mediated macrophage immunity. PLoS Pathog. 2:e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ridenour, D. A., S. L. Cirillo, S. Feng, M. M. Samrakandi, and J. D. Cirillo. 2003. Identification of a gene that affects the efficiency of host cell infection by Legionella pneumophila in a temperature-dependent fashion. Infect. Immun. 71:6256-6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Romling, U., M. Gomelsky, and M. Y. Galperin. 2005. C-di-GMP: the dawning of a novel bacterial signalling system. Mol. Microbiol. 57:629-639. [DOI] [PubMed] [Google Scholar]
- 73.Rossier, O., J. Dao, and N. P. Cianciotto. 2009. A type II secreted RNase of Legionella pneumophila facilitates optimal intracellular infection of Hartmannella vermiformis. Microbiology 155:882-890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rossier, O., J. Dao, and N. P. Cianciotto. 2008. The type II secretion system of Legionella pneumophila elaborates two aminopeptidases, as well as a metalloprotease that contributes to differential infection among protozoan hosts. Appl. Environ. Microbiol. 74:753-761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rossier, O., S. R. Starkenburg, and N. P. Cianciotto. 2004. Legionella pneumophila type II protein secretion promotes virulence in the A/J mouse model of Legionnaires' disease pneumonia. Infect. Immun. 72:310-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rowbotham, T. J. 1986. Current views on the relationships between amoebae, legionellae and man. Isr J. Med. Sci. 22:678-689. [PubMed] [Google Scholar]
- 77.Ruehlemann, S. A., and G. R. Crawford. 1996. Panic in the potting shed. The association between Legionella longbeachae serogroup 1 and potting soils in Australia. Med. J. Aust. 164:36-38. [DOI] [PubMed] [Google Scholar]
- 78.Sahr, T., H. Bruggemann, M. Jules, M. Lomma, C. Albert-Weissenberger, C. Cazalet, and C. Buchrieser. 30 March 2009. Two small ncRNAs jointly govern virulence and transmission in Legionella pneumophila. Mol. Microbiol. doi: 10.1111/j.1365-2958.2009.06677.x. [DOI] [PMC free article] [PubMed]
- 79.Sansom, F. M., P. Riedmaier, H. J. Newton, M. A. Dunstone, C. E. Muller, H. Stephan, E. Byres, T. Beddoe, J. Rossjohn, P. J. Cowan, A. J. d'Apice, S. C. Robson, and E. L. Hartland. 2008. Enzymatic properties of an ecto-nucleoside triphosphate diphosphohydrolase from Legionella pneumophila: substrate specificity and requirement for virulence. J. Biol. Chem. 283:12909-12918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Segal, G., J. J. Russo, and H. A. Shuman. 1999. Relationships between a new type IV secretion system and the icm/dot virulence system of Legionella pneumophila. Mol. Microbiol. 34:799-809. [DOI] [PubMed] [Google Scholar]
- 81.Segal, G., and H. A. Shuman. 1999. Legionella pneumophila utilizes the same genes to multiply within Acanthamoeba castellanii and human macrophages. Infect. Immun. 67:2117-2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Soderberg, M. A., O. Rossier, and N. P. Cianciotto. 2004. The type II protein secretion system of Legionella pneumophila promotes growth at low temperatures. J. Bacteriol. 186:3712-3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Steele, T. W., J. Lanser, and N. Sangster. 1990. Isolation of Legionella longbeachae serogroup 1 from potting mixes. Appl. Environ. Microbiol. 56:49-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Steele, T. W., C. V. Moore, and N. Sangster. 1990. Distribution of Legionella longbeachae serogroup 1 and other legionellae in potting soils in Australia. Appl. Environ. Microbiol. 56:2984-2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Steinert, M., K. Heuner, C. Buchrieser, C. Albert-Weissenberger, and G. Glockner. 2007. Legionella pathogenicity: genome structure, regulatory networks and the host cell response. Int. J. Med. Microbiol. 297:577-587. [DOI] [PubMed] [Google Scholar]
- 86.Stern, A., E. Privman, M. Rasis, S. Lavi, and T. Pupko. 2007. Evolution of the metazoan protein phosphatase 2C superfamily. J. Mol. Evol. 64:61-70. [DOI] [PubMed] [Google Scholar]
- 87.Stewart, C. R., O. Rossier, and N. P. Cianciotto. 2009. Surface translocation by Legionella pneumophila: a form of sliding motility that is dependent upon type II protein secretion. J. Bacteriol. 191:1537-1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Stone, B. J., and Y. Abu Kwaik. 1998. Expression of multiple pili by Legionella pneumophila: identification and characterization of a type IV pilin gene and its role in adherence to mammalian and protozoan cells. Infect. Immun. 66:1768-1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Swanson, M. S., and B. K. Hammer. 2000. Legionella pneumophila pathogenesis: a fateful journey from amoebae to macrophages. Annu. Rev. Microbiol. 54:567-613. [DOI] [PubMed] [Google Scholar]
- 90.Tran, H. T., J. Krushkal, F. M. Antommattei, D. R. Lovley, and R. M. Weis. 2008. Comparative genomics of Geobacter chemotaxis genes reveals diverse signaling function. BMC Genomics. 9:471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tseng, T. T., B. M. Tyler, and J. C. Setubal. 2009. Protein secretion systems in bacterial-host associations, and their description in the Gene Ontology. BMC Microbiol. 9(Suppl. 1):S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Vincent, C. D., J. R. Friedman, K. C. Jeong, E. C. Buford, J. L. Miller, and J. P. Vogel. 2006. Identification of the core transmembrane complex of the Legionella Dot/Icm type IV secretion system. Mol. Microbiol. 62:1278-1291. [DOI] [PubMed] [Google Scholar]
- 93.Yohannes, K., P. Roche, C. Blumer, J. Spencer, A. Milton, C. Bunn, H. Gidding, M. Kirk, and T. Della-Porta. 2004. Australia's notifiable diseases status, 2002: annual report of the National Notifiable Diseases Surveillance System. Commun. Dis. Intell. 28:6-68. [DOI] [PubMed] [Google Scholar]
- 94.Yohannes, K., P. W. Roche, A. Roberts, C. Liu, S. M. Firestone, M. Bartlett, I. East, B. P. Hull, M. D. Kirk, G. L. Lawrence, A. McDonald, P. B. McIntyre, R. I. Menzies, H. E. Quinn, and C. Vadjic. 2006. Australia's notifiable diseases status, 2004, annual report of the National Notifiable Diseases Surveillance System. Commun. Dis. Intell. 30:1-79. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.