Abstract
Synthesis of the hydrogen uptake (Hup) system in Rhizobium leguminosarum bv. viciae requires the function of an 18-gene cluster (hupSLCDEFGHIJK-hypABFCDEX). Among them, the hupE gene encodes a protein showing six transmembrane domains for which a potential role as a nickel permease has been proposed. In this paper, we further characterize the nickel transport capacity of HupE and that of the translated product of hupE2, a hydrogenase-unlinked gene identified in the R. leguminosarum genome. HupE2 is a potential membrane protein that shows 48% amino acid sequence identity with HupE. Expression of both genes in the Escherichia coli nikABCDE mutant strain HYD723 restored hydrogenase activity and nickel transport. However, nickel transport assays revealed that HupE and HupE2 displayed different levels of nickel uptake. Site-directed mutagenesis of histidine residues in HupE revealed two motifs (HX5DH and FHGX[AV]HGXE) that are required for HupE functionality. An R. leguminosarum double mutant, SPF22A (hupE hupE2), exhibited reduced levels of hydrogenase activity in free-living cells, and this phenotype was complemented by nickel supplementation. Low levels of symbiotic hydrogenase activity were also observed in SPF22A bacteroid cells from lentil (Lens culinaris L.) root nodules but not in pea (Pisum sativum L.) bacteroids. Moreover, heterologous expression of the R. leguminosarum hup system in bacteroid cells of Rhizobium tropici and Mesorhizobium loti displayed reduced levels of hydrogen uptake in the absence of hupE. These data support the role of R. leguminosarum HupE as a nickel permease required for hydrogen uptake under both free-living and symbiotic conditions.
Nickel is an essential microelement required for the synthesis of at least nine metalloenzymes, including hydrogenases and ureases (34). Most natural habitats contain very small amounts of available nickel, and bacteria require transport systems designed to take up and concentrate this element. However, an excess of free nickel can be toxic to cells by displacing essential metals from their native binding sites or by generating reactive oxygen species that cause oxidative DNA damage (14). For this reason, nickel uptake mechanisms are strictly regulated and balanced with efflux systems to avoid nickel overload in the cell.
Proteins involved in nickel transport have been described for different bacteria, in many cases in studies performed in connection to the enzymatic system requiring this element (hydrogenase, urease, or others). Nickel uptake systems are classified in two broad groups, single-component Ni2+ permeases and ATP-dependent binding cassette (ABC) transporters (39). The single-component permeases include three different groups, namely, the NiCoT, HupE/UreJ, and UreH families, whereas the multicomponent transporters include the NikMQO/CbiMQO and NikABCDE families.
Two nickel-dependent enzyme systems, hydrogenase and urease, have been described for Rhizobium leguminosarum (38, 48). However, no nickel transporter has been identified in this bacterial species, although one of the components of the hydrogenase gene cluster, hupE, encodes a candidate protein showing six putative transmembrane domains and a predicted signal peptide. Besides these features as a membrane protein, HupE does not show significant sequence similarities with NiCoT permeases such as HoxN from Ralstonia eutropha, HupN from Bradyrhizobium japonicum, or NixA from Helicobacter pylori (22, 23, 50). A homologous protein, UreJ, is encoded in several urease gene clusters, such as that from Bordetella bronchiseptica (33). The association of HupE/UreJ proteins with Ni-dependent enzymes has led to the assignment of a potential role as a nickel permease for these proteins (33). This possibility was further substantiated by experiments with heterologous expression of HupE from Rhodopseudomonas palustris and UreJ from Cupriavidus necator H16 in Escherichia coli (18). However, there is no previous evidence for a role for any of these proteins in the provision of nickel for nickel-dependent systems.
[NiFe] hydrogenases from diazotrophic bacteria catalyze the oxidation of molecular hydrogen evolved from nitrogenase, thus improving the energy efficiency of the nitrogen fixation process (43). In the case of R. leguminosarum, synthesis of the hydrogen-uptake (Hup) [NiFe] hydrogenase requires 18 genes (hupSLCDEFGHIJK-hypABFCDEX) (38). As in other hydrogenase systems, R. leguminosarum [NiFe] hydrogenase is a heterodimeric enzyme with two structural subunits encoded by the hupSL genes. HupL, the hydrogenase large subunit, contains a NiFe heterobimetallic center with a unique structure (21). Assembly of this NiFe center requires the concerted action of many of the hup and hyp gene products in a process that has been elucidated for E. coli (6). Incorporation of nickel into the active center requires the interaction of HypA and HypB, two nickel-binding proteins whose function can be substituted for by large amounts of nickel in the medium (5). An additional protein, SlyD, interacts with HypB to facilitate Ni release for hydrogenase synthesis in Escherichia coli (29). The final step in HupL maturation is the proteolytic processing of the C terminus of the large subunit precursor, carried out by the nickel-dependent protease HupD (40).
A particular feature of Rhizobium leguminosarum bv. viciae hydrogenase activity is its dependence on the plant host. In this bacterium, hydrogenase expression is induced during the nitrogen fixation process in bacteroid cells differentiating in pea root nodules. This symbiotic expression is driven by a NifA-dependent promoter controlling expression of structural and downstream genes (7). The obligate symbiotic expression of hydrogenase has hampered analysis of the hydrogenase system in R. leguminosarum. In order to allow hydrogenase expression in cultured vegetative cells, the NifA-dependent promoter of the R. leguminosarum hup operon was replaced with the FnrN-dependent promoter of the fixN gene, thus generating strain SPF25, where hydrogenase synthesis is induced when cells are exposed to microaerobic conditions (10).
Hydrogen uptake in pea bacteroids is limited by the availability of nickel to the plant, since addition of nickel chloride to the plant nutrient solution strongly increases levels of hydrogenase activity (9). Nickel limitation might be responsible for an additional level of plant-dependent control of hup expression. Legume hosts, such as lentil, which are nonpermissive for hydrogenase activity, have been described (32). This host effect, exerted at both transcriptional and posttranscriptional levels, is linked to a strong limitation of supply of nickel to the bacteroid in lentil nodules, suggesting the involvement of an unidentified metalloregulator (11).
Mechanisms for metal provision in vegetative and symbiotic forms of bacterial cells are relevant for the synthesis of metalloenzymes. In this work, we describe the role of R. leguminosarum HupE and HupE2, two proteins involved in nickel transport for hydrogenase synthesis under both free-living and symbiotic conditions.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and growth conditions.
The bacterial strains and plasmids used in this work are listed in Table 1. For induction of hydrogenase activity in E. coli cells, strains MC4100 and HYD723 were grown at 28°C under anaerobic conditions in TGYEP medium (41) modified to contain glycerol (0.4%) and fumarate (20 mM). Antibiotics were added at the following concentrations (in μg·ml−1): for tetracycline, 10; for kanamycin, 50; and for ampicillin, 100. Rhizobium leguminosarum bv. viciae, Mesorhizobium loti, Rhizobium tropici, and Rhizobium etli strains were routinely grown at 28°C in tryptone-yeast extract (TY) (4) or yeast mannitol broth (YMB) medium (49). Strain SPF25 derives from R. leguminosarum UPM791 by replacement of the native NifA-dependent hupSL promoter with the fixN gene promoter controlled by FnrN (10). Cosmid pALPF1 is a pAL618 derivative carrying the same promoter modification as that in SPF25. Cosmid pALPFE is a pALPF1 derivative carrying a hupE gene deletion (see below). Plasmids pALPF1 and pALPFE were introduced into Rhizobium and Mesorhizobium strains by conjugation, and transconjugants were selected in Rhizobium minimal medium (Rm) (37) supplemented with tetracycline (5 μg·ml−1).
TABLE 1.
Bacterial strains and plasmidsa
| Strain or plasmid | Genotype or relevant characteristic(s) | Source or reference |
|---|---|---|
| E. coli | ||
| DH5α | recA1 endA1 gyrA96 thi hsdRl7 supE44 relA1 Δ(lacZYA-argF)U169 (φ80 lacZΔM15) deoR phoA | 27; Bethesda Research Laboratories |
| S17.1 | thi pro hsdR hsdM+ recA RP4::2-Tc::Mu-Kan::T7; Spr Smr | 46 |
| BW25113 | lacIqrrnBT14 ΔlacZWJ16 ΔhsdR514 ΔaraBADAH33 ΔrhaBADLD78 | 16 |
| MC4100 | F−araD139 Δ(argF-lac)U169 ptsF25 relA1 flb5301 rpsL150 | 12 |
| HYD723 | MC4100; nikA::MudI (Aprlac) | 51 |
| R. leguminosarum bv. viciae | ||
| UPM791 | 128C53 Strr; Nod+ Fix+ Hup+ | 30 |
| SPF25 | UPM791 with PfixN::hupSL | 10 |
| SPF2714 | SPF25 ΔhupE | This work |
| SPF228 | SPF25 ΔhupE2 | This work |
| SPF22A | SPF25 ΔhupE ΔhupE2 | This work |
| Rhizobium tropici CIAT899 | Wild type; Nod+ Fix+ Hup− | P. van Berkum (USDA, Beltsville, MD) |
| Rhizobium etli CE3 | CFN42; Nod+ Fix+ Hup−; Kanr | 36 |
| Mesorhizobium loti MAFF303099 | Wild type; Nod+ Fix+ Hup− | NIAS (Japan) |
| Plasmids | ||
| pCR2.1-TOPO | PCR product cloning vector; Apr Kanr | Invitrogenb |
| pRLH41 | pBluescriptSK containing a 5-kb DNA region of the UPM791 hup gene cluster | 31 |
| pAL618 | Cosmid pLAFR1 containing the UPM791 hup cluster; Tcr | 30 |
| pALPF1 | pAL618 with PfixN::hupSL | 10 |
| pALPFE | pALPF1 ΔhupE | This work |
| pK18mobsacB | pK18 derivative sacB; Kanr | 45 |
| pKPF1 | pK18mobsacB with PfixN::hupSL | 10 |
| pKE1 | pK18mobsacB with a deletion in hupE | This work |
| pKE2 | pK18mobsacB with a deletion in hupE2 | This work |
| pThupE2 | pCR2.1-TOPO with a 1.6-kb DNA of the hupE2 region | This work |
| pKD4 | Template plasmid harboring an FLP-mediated excision sequence flanking the Kanr gene | 16 |
| pCP20 | Plasmid for thermal induction of FLP synthesis; Cmr Apr | 16 |
| pBAD18-Kan | Expression vector carrying the arabinose PBAD promoter; Kanr | 26 |
| pBADE1 | pBAD18-Kan with the hupE gene | This work |
| pBADE2 | pBAD18-Kan with the hupE2 gene | This work |
| pBADE1.H series | pBADE1 with hupE mutated versions | This work |
Abbreviations: Ap, ampicillin; Kan, kanamycin; Str, streptomycin; Tc, tetracycline; Cm, chloramphenicol.
DNA manipulation techniques.
Plasmid DNA preparations, restriction enzyme digestions, agarose gel electrophoresis, DNA cloning, and transformation of DNA into Escherichia coli were carried out by standard protocols (44). R. leguminosarum genomic DNA was extracted as previously described (30). For Southern hybridizations, hupE and hupE2 DNA probes were generated by PCR with primers UE1/LE1 and hupE2.3/hupE2.4, respectively (Table 2), and labeled with digoxigenin (DIG) by using DIG-11-dUTP (Roche Molecular Biochemicals, Mannheim, Germany) at a 40 μM final concentration. Hybridizing bands were visualized using a chemiluminescence DIG detection kit as described by the manufacturer (Roche Molecular Biochemicals, Mannheim, Germany).
TABLE 2.
Oligonucleotides used in this work
| Designation | Sequence (5′-3′) |
|---|---|
| hupE1.1 | CATCCATGAAGTATAGTAAGATTACGACGACGCTCGTGTAGGCTGGAGCTGCTTCG |
| hupE1.2 | TGCCAGGAAGCCCGCCACGTAGCCCGTCGCGTTTGACATATGAATATCCTCCTTAG |
| UE1 | CATATGAAGTATAGTAAGATTACG |
| LE1 | TCAGCCGACCAGCAGTGCCGCGCC |
| hupE2.1 | GGCGGCGACAACGAAGACGAA |
| hupE2.2 | GACGTGGCTGGATTTGACTTG |
| hupE2.3 | CGCCCGTTTGGTATTCGTCAT |
| hupE2.4 | CGAAAACACCGACCATCACCA |
| hupE2.5 | GGTGGTAAGCTTATGGTGATGGTCGGTGTTTT |
| hupE2.6 | GGTGGTAAGCTTGCCGTCAACGAAAACTCCAT |
| E2BAD1 | GCTAGCGAGGGAATCGAGCAATGAAA |
| E2BAD2 | CTTGACGTGGCTGGATTTGACT |
| EChupE1 | GCTAGCAGGAGGAATTCACCATGAAGTATAGTAAGAT |
| E1BAD | CGGGAGCCACGACGGAGCAG |
| H36I.1 | TGCGGGTCTCAACATTCCGTTCAGCG |
| H36I.2 | CGCTGAACGGAATGTTGAGACCCGCA |
| H43I.1 | GCGGCCTCGATATTATCCTGGCGATG |
| H43I.2 | CATCGCCAGGATAATATCGAGGCCGCT |
| H126I.1 | TGCGCGCTCTTCATCGGCCACGTCCAT |
| H126I.2 | GGACGTGGCCGATGAAGAGCGCGCAT |
| H128I.1 | CTCTTCCACGGCATCGTCCATGGCATC |
| H128I.2 | ATGCCATGGACGATGCCGTGGAAGAG |
| H22I.1 | CCCACGCCATTGTCGGGCTGCACGC |
| H22I.2 | GCGTGCAGCCCGACAATGGCGTGGG |
| H130I.1 | CACGGCCACGTCATCGGCATCGAGTTG |
| H130I.2 | CAACTCGATGCCGATGACGTTGGCCGTG |
| H155I.1 | ACGGTCATCCTGATCGTCCTCGGCATC |
| H155I.2 | GATGCCGAGGACGATCAGGATGACCGT |
| H26I.1 | GGGCTGATCGCCGATGGCACGCTTGC |
| H26I.2 | AGCGTGCCATCGGCGATCAGCCCGAC |
| H22I/H26I.1 | CCCACGCCATTGTCGGGCTGCACGCGGGCTGATCGCCGATGGCACGCTTGC |
| H22I/H26I.2 | GCGTGCAGCCCGACAATGGCGTGGGAGCGTGCCATCGGCGATCAGCCCGAC |
Cloning and sequencing the hupE2 gene of R. leguminosarum UPM791.
The DNA region containing the R. leguminosarum UPM791 hupE2 gene was amplified by PCR from genomic DNA with oligonucleotides hupE2.1 and hupE2.2. These primers were designed from the R. leguminosarum bv. viciae 3841 genomic sequence (Table 2). The 1.6-kb DNA fragment amplified was cloned in plasmid pCR2.1TOPO (Invitrogen BV, Groningen, The Netherlands) and sequenced with the following synthetic oligonucleotides: T7, Reverse, hupE2.3, hupE2.4, hupE2.5, and hupE2.6. DNA sequencing was carried out using a BigDye Terminator cycle sequencing ready reaction kit and an ABI377 automatic sequencer (PE Biosystems, Foster City, CA). DNA sequence was analyzed with the Sequencher 4.1 and DNA Strider 1.2 programs. Searches for homologous proteins were carried out with BLAST software (1). The 1.6-kb nucleotide sequence containing the R. leguminosarum UPM791 hupE2 gene has been deposited in GenBank (accession no. GQ420671). Transmembrane domains were predicted with the TMHMM2 (CBS, DTU, Denmark) and DAS (15) programs.
Mutant and plasmid constructions.
To generate a hupE mutant in strain UPM791, an in-frame deletion of 144 bp was created by taking advantage of a unique NdeI restriction site in plasmid pRLH41, which contains a 5-kb EcoRI fragment of the R. leguminosarum hup gene cluster. The EcoRI fragment carrying the hupE deletion was cloned in suicide vector pK18mobsac, and the resulting plasmid, pKE1, was introduced into R. leguminosarum UPM791 by conjugation. Homologous recombination was selected using the sacB system (45). The hupE mutant generated was named UPM791.27. The PNIFA promoter of UPM791.27 hydrogenase structural genes was replaced with the PFIXN promoter cloned in plasmid pKPF1, giving strain SPF2714.
An in-frame deletion of hupE was also generated in plasmid pALPF1 by following the one-step method of Datsenko and Wanner (16) based on the phage λ Red recombinase. Briefly, a 1.6-kb DNA fragment containing a kanamycin resistance gene flanked by recombinase target sites was PCR amplified from template plasmid pKD4 and primers hupE1.1 and hupE1.2. The 5′ oligonucleotide sequences were homologous to DNA regions adjacent to the hupE gene and the 3′ sequences to plasmid pKD4. The 1.6-kb fragment was introduced by electroporation in strain BW25113(pALPF1) to replace the hupE gene with the kanamycin resistance cassette. The resistance gene was removed by introduction of plasmid pCP20 expressing the FLP recombinase. The hupE in-frame deletion was confirmed by PCR amplification of the corresponding region in plasmid pALPF1 and sequencing.
To generate a deletion in the hupE2 gene, DNA regions corresponding to the 5′ and 3′ ends were amplified by PCR using plasmid pThupE2 as a template and oligonucleotides Reverse/hupE2.5 and T7/hupE2.6, respectively (Table 2). The resulting DNA fragments of ca. 900 and 600 bp were digested with BamHI/HindIII and EcoRI/HindIII, respectively, and cloned into pK18mobsac. The resulting plasmid (pKE2) was introduced into strains SPF25 and SPF2714ΔhupE by conjugation, and recombination of homologous DNA regions was selected by the sac system. The transconjugant strains carrying mutations in hupE2 and hupE hupE2 were named SPF228 and SPF22A, respectively. Deletions in the hupE and hupE2 genes were confirmed by Southern blot hybridization using DNA probes of the corresponding genes and genomic DNA from single and double mutants.
For construction of plasmids pBADE1 and pBADE2, the hupE and hupE2 genes were amplified from their ATG codons with primers EChupE1/E1BAD and E2BAD1/E2BAD2, using DNA from plasmid pRLH41 and from UPM791 genomic DNA, respectively, as templates. The amplified DNA fragments were cloned in plasmid pCR2.1TOPO, excised with NheI (a restriction site included in both upper primers) and XbaI, and cloned in vector pBAD18-Kan (26).
Site-directed mutagenesis of the hupE gene was carried out in plasmid pCR2.1TOPO containing the hupE gene. To do this, two 26- to 28-mer complementary oligonucleotides with centered nucleotide substitutions were designed to generate a His-to-Ile substitution for each His residue in HupE. PCR amplification with complementary primers and DNA from pCR2.1TOPO-hupE as a template generated complete plasmids that potentially harbored the corresponding mutation. The resulting plasmids were treated with DpnI and transformed into DH5α cells. Substitution of target nucleotides as well as absence of additional mutations was verified by sequencing. Mutated hupE versions were excised with the NheI/XbaI restriction enzymes and cloned into vector pBAD18-Kan for further analysis.
Plant tests.
Pea (Pisum sativum L. cv. Frisson), lentil (Lens culinaris cv. Magda), lotus (Lotus corniculatus), and bean (Phaseolus vulgaris cv. Torcaza INIA) seeds were surface sterilized, germinated on 1% water agar plates, and planted in Leonard jar-type assemblies under bacteriologically controlled conditions (42). Plants were grown on a nitrogen-free plant nutrient solution that was supplemented, when indicated, with 85 μM NiCl2 on day 10 after seedling inoculation (9).
Hydrogenase activity assays.
Hydrogenase activity in R. leguminosarum, R. tropici, R. etli, and M. loti bacteroid suspensions from 21-day-old plants was measured using an amperometric method with oxygen as an electron acceptor (42). Hydrogenase activity in R. leguminosarum vegetative cells was induced under stoppered-flask conditions with 50-ml cultures. Strains were aerobically grown in YMB medium to an optical density at 600 nm of 0.4. From these cultures, a 1:10 dilution was done in fresh YMB medium. When indicated, NiCl2 was added to give a 1 μM final concentration. The 200-ml flasks were tightly capped, flushed five times with a 0.8% O2 atmosphere, and incubated at 28°C for 16 h. Cell cultures were centrifuged and suspended in 5 ml Dixon buffer (32 mM K2HPO4, 24 mM KH2PO4, and 0.24 mM MgCl2) before amperometric determination. For induction of hydrogenase activity in E. coli cells, strains MC4100 and HYD723 were grown in LB medium at 28°C for 16 h. A 1:20 dilution in a modified TGYEP medium was carried out, and arabinose was added to give a 200 μM final concentration. The 25-ml cultures were then capped, flushed five times with a N2 atmosphere, and incubated at 28°C for 5 h. After incubation, cultures were centrifuged, resuspended in 5 ml Dixon buffer, and assayed for hydrogenase activity as described above. For quantification of hydrogenase activity, the protein content of bacteroids and vegetative cells was determined by the bicinchoninic acid method (47), with the modifications described by Brito et al. (9).
Nickel transport assays.
E. coli strain HYD723 carrying plasmid pBAD18-Kan and derivatives were grown to mid-exponential growth phase under aerobic conditions in LB medium. Then, l-arabinose was added up to 200 μM and the culture incubated at 37°C for 1 h. Cells were harvested by centrifugation, washed, and resuspended in potassium phosphate buffer (65 mM KH2PO4-K2HPO4, pH 6.8) supplemented with 11 mM glucose and 10 mM MgCl2 to give a final concentration of 0.2 to 0.4 mg of bacteria·ml−1 (dry weight). Nickel uptake was initiated by adding 150 nM 63NiCl2 (7.52 mCi·ml−1 and 1.2 mg Ni·ml−1; Amersham). Samples (100 μl) were taken at regular time intervals, vacuum filtered through cellulose nitrate membrane filters (0.45-μm pore size and 25-mm diameter; Millipore), and washed twice with 2.5 ml potassium phosphate buffer containing 10 mM nickel chelator EDTA. Filters were dried and placed in scintillation vials with 5 ml of scintillation fluid (OCS; Amersham) for counting with a Packard liquid scintillation counter. Similar growth and arabinose induction conditions were used for the analysis of membrane protein fractions. Cell cultures (250 ml) were centrifuged after induction, resuspended in 3 ml of buffer W (100 mM Tris-HCl, pH 8, 150 mM NaCl), mechanically disrupted by 3 passages through a French press cell (SLM Aminco, Silver Spring, MD), centrifuged at 6,000 rpm to remove cell debris, and ultracentrifuged at 135,000 × g for 1 h. Membrane pellets were resuspended in 600 μl of buffer W and ultracentrifuged again. Finally, membrane pellets were resuspended in 200 μl of buffer W and quantified for protein content (47). SDS-PAGE analysis was performed with 60 μg per sample in 15% acrylamide gels run at 95 V for ca. 1 h and Coomassie blue stained after the run.
RESULTS
Two hupE genes are involved in hydrogen oxidation in R. leguminosarum bv. viciae.
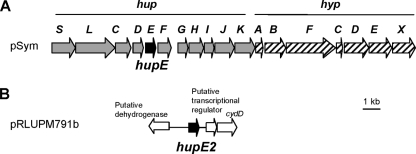
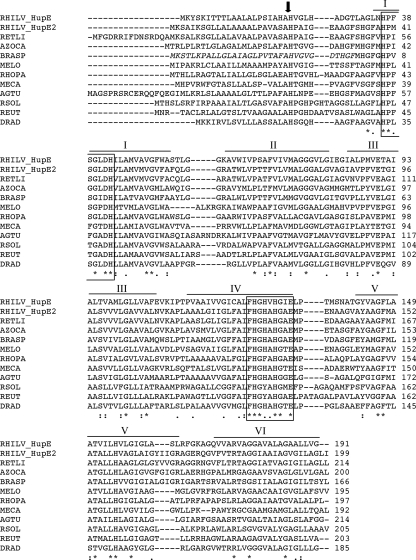
The role of hupE, a gene located into the R. leguminosarum hydrogenase gene cluster, was investigated for SPF25, a strain that expresses hydrogenase activity in microaerobic (1% O2) free-living cells (10). For this purpose, SPF2714, a nonpolar hupE mutant with a 144-bp deletion, was generated (see Materials and Methods). Analysis of hydrogen oxidation in microaerobic free-living cells showed that SPF2714 expressed hydrogenase activity to a level similar to that observed in the wild-type strain (see Table 4), suggesting that either HupE was not necessary for hydrogen uptake or another protein was replacing its role. Inspection of the genome sequence of R. leguminosarum bv. viciae strain 3841 revealed the presence of an open reading frame, pRL90274, whose predicted product is described as a putative transmembrane hydrogenase-related protein. This protein showed 48% sequence identity with R. leguminosarum UPM791 HupE (Table 3). The pRL90274 gene appears as a monocistronic operon flanked by a potential transcriptional regulator adjacent to the cydDCAB operon and a gene encoding a putative dehydrogenase (Fig. 1) (52). We investigated the presence of a pRL90274 orthologous gene in UPM791. As described in Materials and Methods, a 1.6-kb DNA fragment was amplified by PCR using genomic DNA of R. leguminosarum UPM791 as a template. Sequence analysis of this region revealed the presence of an open reading frame, named hupE2, which appeared as a monocistronic operon (Fig. 1). Since previous work in our laboratory had led to the identification of the cydDCAB operon in plasmid pRLUPM791b of R. leguminosarum UPM791 (unpublished data), we assume this location for the newly identified hupE2 gene. hupE2 encodes a 199-amino-acid-residue protein that showed 92% identity with the predicted product of pRL90274 and 48% with the R. leguminosarum UPM791 HupE protein. Comparison of the HupE and HupE2 amino acid sequences with the HupE/UreJ proteins from other organisms (Table 3) showed higher percentages of identity for HupE2 (up to 92%) than for HupE (up to 48%). Protein sequence analysis of HupE2 suggested a cleavage site for the signal peptidase and six potential transmembrane domains in the mature protein, with both N- and C-terminal ends exposed to the periplasm (Fig. 2).
TABLE 4.
Hydrogenase activity in microaerobic free-living cells from R. leguminosarum hupE- and hupE2-deficient mutants
| Strain | Hydrogenase activitya obtained with: |
||
|---|---|---|---|
| No metal added | NTAb | NiCl2b | |
| SPF25 | 820 ± 160 | 920 ± 70 | 1,000 ± 260 |
| SPF2714 ΔhupE | 890 ± 170 | 520 ± 50 | 1,140 ± 100 |
| SPF228 ΔhupE2 | 930 ± 240 | 1,120 ± 20 | 1,110 ± 270 |
| SPF22A ΔhupE ΔhupE2 | 140 ± 70 | 40 ± 20 | 1,300 ± 100 |
Values are expressed in nmol H2 h−1 mg prot−1. Hydrogenase activities were obtained using oxygen as the electron acceptor. Values are averages of results from four experiments ± SE.
Nickel chloride and nitrilotriacetate were added at 1 μM and 50 μM, respectively.
TABLE 3.
Sequence conservation between HupE-like proteins from different bacterial groups and R. leguminosarum bv. viciae HupE and HupE2
| Microorganism | Gene index designation | Valuea for R. leguminosarum UPM791 |
|
|---|---|---|---|
| HupE | HupE2 | ||
| R. leguminosarum bv. viciae 3841 | 116249722 | 48.0 (61.0) | 92.0 (96.0) |
| Rhizobium etli CFN42 | 86360024 | 42.3 (56.7) | 79.9 (86.4) |
| A. caulinodans ORS571 | 46392545 | 43.3 (61.6) | 54.4 (73.5) |
| Mesorhizobium loti MAFF303099 | 13474132 | 41.1 (54.3) | 47.0 (60.4) |
| Rhodopseudomonas palustris CGA009 | 39934041 | 48.3 (62.7) | 47.3 (62.7) |
| Deinococcus radiodurans R1 | 15807977 | 50.3 (62.3) | 46.7 (59.8) |
| Methylococcus capsulatus strain Bath | 53802532 | 42.8 (57.7) | 40.9 (58.1) |
| Agrobacterium tumefaciens C58 | 159185133 | 42.7 (56.0) | 40.1 (55.0) |
| Ralstonia solanacearum GMI1000 | 17546753 | 43.8 (55.8) | 39.3 (54.5) |
| Ralstonia eutropha H16 | 113867102 | 41.5 (58.0) | 38.6 (55.3) |
Values represent the percentages of identity (similarity) between amino acid sequences of HupE-like proteins from each bacterium and HupE/HupE2 from R. leguminosarum UPM791.
FIG. 1.
Genomic context of hupE and hupE2 genes in R. leguminosarum bv. viciae UPM791. (A) The hupE gene is part of the hupSLCDEF operon located in the symbiotic plasmid. Expression of hupE is controlled by the hydrogenase structural gene promoter. (B) The hupE2 gene is found in plasmid pRLUPM791b and bears an independent promoter. Black arrows highlight the positions of the hupE and hupE2 genes. Gray and hatched arrows show the hup and hyp genes, respectively. White arrows indicate genes adjacent to hupE2 unlinked to hydrogenase biosynthesis.
FIG. 2.
Sequence alignment of HupE/UreJ proteins. Asterisks indicate identical amino acids. Horizontal lines correspond to transmembrane domains, designated by roman numerals, as predicted using TMHMM2 software. The vertical arrow shows the predicted cleavage site for a signal peptidase. Boxed residues correspond to conserved regions in HupE/UreJ proteins. RHILV, R. leguminosarum bv. viciae 3841 (GI|116249722); RETLI, R. etli CFN42 (GI|86360024); AZOCA, Azorhizobium caulinodans ORS571 (GI|46392545); BRASP, Bradyrhizobium sp. BTAi (GI|148252085); MELO, Mesorhizobium loti MAFF303099 (GI|13474132); RHOPA, Rhodopseudomonas palustris CGA009 (GI|39934041); DRAD, Deinococcus radiodurans R1 (GI|15807977); MECA, Methylococcus capsulatus strain Bath (GI|53802532); AGTU, Agrobacterium tumefaciens C58 (GI|159185133); RSOL, Ralstonia solanacearum GMI1000 (GI|17546753); REUT, Ralstonia eutropha H16 (GI|113867102). Inspection of the sequence corresponding to BRASP HupE revealed an annotation mistake in the database. The corrected sequence includes 33 additional residues (shown in italics) at the N terminus.
To investigate the role of HupE2 in hydrogenase activity, single hupE2 (SPF228) and double hupE hupE2 (SPF22A) mutant strains were generated as described in Materials and Methods. Levels of hydrogenase activity in microaerobic cells of the mutant SPF228 were similar to those in the wild-type strain SPF25 (Table 4). In contrast, the double mutant SPF22A showed a drastic reduction of hydrogenase activity. Supplementation of medium with 1 μM NiCl2 resulted in complete reversion of the hydrogenase-defective phenotype of SPF22A (Table 4). We also analyzed the effect of nitrilotriacetate (NTA) on hydrogenase activity in the hupE and hupE2 mutants. NTA is a metal chelator that shows a higher binding affinity for Ni2+ than for other metal ions, such as Co2+, Mn2+, and Zn2+ (25). The presence of 50 μM NTA did not have an effect on hydrogen oxidation in the wild-type or hupE2 mutant strains. However, cells from the hupE mutant SPF2714 reached only 50% of the hydrogenase activity (Table 4). These results indicate that HupE and HupE2 have similar functions, likely related to nickel transport, that are required for hydrogenase activity in microaerobic free-living cells. However, HupE2 is apparently less efficient in providing nickel to hydrogenase when this metal is present at low concentrations, as is the case when the NTA chelator is present.
Since nickel is also a constituent of urease, a metalloenzyme present in R. leguminosarum UPM791 (48), we also checked the effect of the inactivation of both hupE genes on the ability of the cells to grow on minimal medium modified to contain urea as the only nitrogen source. In these experiments, we found no significant differences in growth between the hupE hupE2 double mutant and the wild type, whereas a urease-deficient mutant (URE4) (48) showed a strong reduction in growth (data not shown). These data suggest that the residual levels of nickel uptake responsible for the very low hydrogenase activity observed in the double mutant are likely sufficient to sustain urease-based growth.
HupE and HupE2 restore hydrogenase activity and nickel transport in an E. coli nik mutant.
To investigate the role of R. leguminosarum HupE and HupE2 proteins in hydrogenase biosynthesis, transport of 63Ni2+ in strains SPF25 and SPF22A was assayed under aerobic conditions as well as under the microaerobic conditions that induce hydrogen oxidation. For these assays, cells were incubated with 100 nM or 1 μM 63Ni2+ in a magnesium-containing buffer. However, no conclusive results could be obtained, since controls with disrupted cells of wild-type and mutant strains showed levels of nickel accumulation similar to those in the corresponding cell cultures (data not shown), suggesting a high level of nonspecific Ni2+ binding to the bacterial surface.
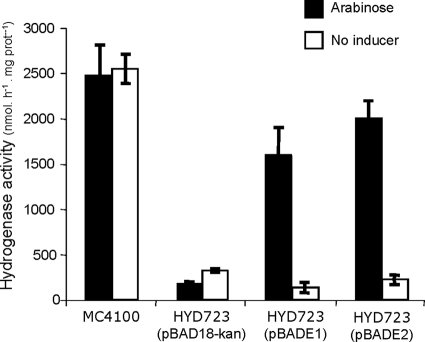
For E. coli, the ABC-type nickel transport system nikABCDE and the phenotype of the HYD723 Nik− mutant have been well characterized (35). In the mutant, nickel transport is impaired, and hence, no hydrogenase activity is detected in a nickel-depleted culture medium. We first investigated complementation of hydrogen oxidation in HYD723 through expression of the hupE and hupE2 genes. Plasmids pBADE1 and pBADE2 containing the hupE and hupE2 gene coding sequences, respectively, under the arabinose PBAD promoter system were introduced into HYD723. To induce hydrogen oxidation, strains were grown under anaerobic conditions in a TGYEP modified medium containing glycerol and fumarate. Under these conditions, high levels of hydrogenase activity were observed in wild-type strain MC4100, whereas nik mutant HYD723 carrying control plasmid pABD18-Kan showed very low levels of activity (Fig. 3). This defective phenotype was reverted by supplementation of Ni into the medium (data not shown). High levels of hydrogen oxidation were also detected in cultures of strains HYD723(pBADE1) and HYD723(pBADE2) under low-Ni conditions, but only when arabinose was added as an inducer to the culture medium (Fig. 3). These results indicate that HupE and HupE2 can individually complement the function of the NikABCDE nickel transport system, which is lacking in strain HYD723, thus restoring hydrogenase activity in E. coli.
FIG. 3.
Effect of HupE and HupE2 on the hydrogenase activity of the E. coli HYD723 nik mutant. Wild-type strain MC4100 and nik mutant HYD723 expressing pBAD-derivative plasmid pBADE1 (hupE) or pBADE2 (hupE2) or control plasmid pBAD18-Kan were assayed for hydrogenase activity in anaerobic cultures grown in a glycerol-fumarate medium for 5 h with 200 μM arabinose or without inducer. Values for hydrogenase activities are averages of results from four independent assays, with error bars corresponding to standard errors (SE). prot, protein.
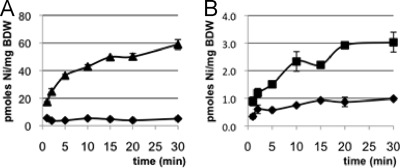
We then investigated the nickel transport capability of R. leguminosarum HupE and HupE2 in E. coli cells by measuring 63Ni2+ uptake in the complemented strains. Strain HYD723 derivatives carrying plasmids pBADE1, pBADE2, and pBAD18-Kan were incubated aerobically with 150 nM 63Ni2+ and arabinose as an inducer of PBAD promoter expression. In these experiments, expression of HupE led to much higher levels of nickel accumulation than those observed with the E. coli mutant strain carrying control plasmid pBAD18-Kan (Fig. 4). Surprisingly, expression of HupE2 (HYD723 carrying plasmid pBADE2), which was shown to fully complement the nik mutation for hydrogenase activity, resulted in Ni accumulation levels that were ca. 20-fold lower than those mediated by HupE although still significantly higher than those detected in cells carrying control plasmid pBAD18-Kan. These results indicate that both HupE and HupE2 are nickel permeases. Both proteins, however, appear to differ in their affinities or efficiencies in transporting this metal. Analysis of the expression levels of both HupE and HupE2 in arabinose-induced E. coli cells was attempted by SDS-PAGE of membrane fractions followed by Coomassie blue staining. In these experiments, we were not able to detect any protein band associated with arabinose-inducible HupE expression (data not shown), suggesting that very low levels of HupE protein are required for full hydrogenase activity in E. coli. Since we could not detect HupE, we cannot exclude the possibility that HupE2 is produced at lower levels.
FIG. 4.
Nickel transport of the E. coli HYD723 nik mutant as a function of HupE and HupE2 expression. Strain HYD723 derivatives harboring plasmid pBADE1 (hupE; ▴ in panel A) or pBADE2 (hupE2; ▪ in panel B) or control plasmid pBAD18-Kan (⧫ in both panels) were grown in Luria-Bertani medium under aerobic conditions at mid-log phase (OD600, ≈0.5). Then, arabinose (final concentration, 200 μM) was added to induce PBAD expression. The uptake assay was performed in the presence of 150 nM 63Ni2+ by using a cell density equal to that in the original culture for pBADE1 (A) or 10-fold higher for pBADE2 (B). Aliquots (100 μl) were filtered at the indicated times. The intracellular concentration of 63Ni per milligram of bacteria (dry weight) (BDW) was determined. The values shown represent a typical experiment and are averages of results from two replicates, with error bars indicating standard deviations. Error bars smaller than the symbols are not shown.
Identification of amino acid residues required for HupE activity.
Expression of HupE in the E. coli heterologous system was used to investigate HupE amino acid residues required for nickel transport. Analysis of amino acid sequences of R. leguminosarum HupE and HupE-like proteins from other microorganisms revealed the presence of a histidine domain (HX5DH [motif I]) located in transmembrane domain I (Fig. 2). This domain is highly similar to a domain (HX4DH) that has been reported as essential for nickel transport in permeases of the NiCoT family, such as HoxN of Cupriavidus necator (formerly Ralstonia eutropha) and NixA of Helicobacter pylori (17, 23, 24). An additional histidine-rich sequence (FHGX[AV]HGXE [motif II]), located in transmembrane domain IV, was highly conserved in HupE/UreJ proteins from different microorganisms (Fig. 2). To analyze the importance of these sequences in HupE functionality, conserved histidine residues H36 and H43, located in motif I; H126 and H130, present in motif II; and H22 and H155, located at the N-terminal and C-terminal parts of the protein, respectively, were replaced by isoleucine through site-directed mutagenesis. In addition, single mutants were generated in nonconserved histidine residues H26 and H128 (see Materials and Methods for details). Expression of mutated variants of HupE under the PBAD promoter system was induced in the E. coli nik mutant HYD723, and hydrogenase activity was measured under the anaerobic conditions previously established. Expression of the different mutant forms of HupE protein led to three different phenotypes (Table 5). Variants of HupE mutated in residues H22, H26, H128, and H155 showed wild-type levels of hydrogen oxidation. In contrast, hydrogenase activity was drastically reduced in strain HYD723 expressing versions H36I, H43I, and H130I of HupE, whereas an intermediate phenotype was associated with the H126I HupE variant, which retained 35% of the hydrogenase activity. We also generated a double mutant involving residues H22 and H26, located in the N-terminal part of HupE and predicted to be exposed to the periplasm. Strain HYD723 expressing this variant protein showed reduced (42%) hydrogenase activity in comparison to the strain complemented with wild-type plasmid pBADE1. These data indicate that conserved histidine residues located in motifs I and II are required for the function or stability of the HupE protein.
TABLE 5.
Hydrogenase activity in E. coli HYD723 expressing pBADE1 or derivative plasmids carrying mutations in histidine residues of HupE
| Plasmid | Hydrogenase activitya obtained with: |
|
|---|---|---|
| 200 μM arabinose | No inducer | |
| pBADE1 | 1,700 ± 420 | 20 ± 20 |
| pBAD18-Kan | 100 ± 80 | 40 ± 30 |
| pBADE1.H22I | 2,450 ± 670 | 70 ± 60 |
| pBADE1.H26I | 1,390 ± 200 | 100 ± 30 |
| pBADE1.H22I/H26I | 720 ± 240 | 40 ± 10 |
| pBADE1.H36I | 70 ± 20 | 210 ± 10 |
| pBADE1.H43I | 260 ± 130 | 210 ± 100 |
| pBADE1.H126I | 610 ± 190 | 30 ± 30 |
| pBADE1.H128I | 1,960 ± 510 | 60 ± 40 |
| pBADE1.H130I | 190 ± 100 | 140 ± 40 |
| pBADE1.H155I | 1,190 ± 60 | 40 ± 10 |
Hydrogenase activities (expressed in nmol H2 h−1 mg prot−1) in anaerobic cells were measured, using oxygen as an electron acceptor. Values are means of results from three independent assays ± SE.
HupE is required for symbiotic hydrogen oxidation.
Symbiotic hydrogen uptake is a particular trait present only in certain strains of the Rhizobium group (2). We investigated the role of HupE and HupE2 in hydrogen oxidation in bacteroid cells of strains SPF25, SPF2714 (hupE), SPF228 (hupE2), and SPF22A (hupE hupE2) obtained from 21-day-old pea nodules. In contrast to what was observed for the vegetative cells, no differences in hydrogenase activity were observed among bacteroids of wild-type and mutant strains from plants grown without nickel supplementation (Table 6). In order to increase the nickel demand inside the bacteroid cell, extra copies of the hydrogenase gene cluster were introduced. To this effect, plasmids pALPF1, containing the R. leguminosarum hup cluster, and pALPFE, a pALPF1 derivative containing a nonpolar hupE mutation (see Materials and Methods), were introduced into strains SPF25 and SPF22A, respectively. Pea bacteroids from SPF25(pALPF1) and SPF22A(pALPFE) were assayed for hydrogenase activity (Table 6). Despite the extra copies of the hup gene cluster, no reduction in hydrogen oxidation was observed under any plant growth condition tested (i.e., nutrient solution supplemented or not supplemented with nickel). We then analyzed symbiotic hydrogenase activity in the R. leguminosarum SPF25-Lens culinaris association. This symbiosis requires larger amounts of nickel in the plant nutrient solution for hydrogenase activity (11). As shown in Table 6, bacteroids of strain SPF22A(pALPFE) obtained from lentil plants grown on a standard nutrient solution were strongly impaired in hydrogen oxidation in comparison to those of strain SPF25(pALPF1). Hydrogenase activity in SPF22A(pALPFE) bacteroids was restored by cultivation of lentil plants on a nutrient solution containing 85 μM NiCl2.
TABLE 6.
Hydrogenase activity in bacteroid cells as a function of R. leguminosarum hupE and hupE2 genes
| Host | Strain | Hydrogenase activitya obtained with: |
|
|---|---|---|---|
| No NiCl2 added | 85 μM NiCl2 | ||
| Pisum sativum | SPF25 | 2,010 ± 500 | 5,580 ± 650 |
| SPF2714 ΔhupE | 2,820 ± 160 | 5,740 ± 440 | |
| SPF228 ΔhupE2 | 3,460 ± 140 | 4,380 ± 680 | |
| SPF22A ΔhupE ΔhupE2 | 2,200 ± 350 | 5,430 ± 910 | |
| SPF25(pALPF1) | 2,480 ± 210 | 11,120 ± 1,150 | |
| SPF22A(pALPFE) | 2,710 ± 530 | 10,250 ± 700 | |
| Lens culinaris | SPF25(pALPF1) | 1,100 ± 350 | 2,690 ± 450 |
| SPF22A(pALPFE) | 360 ± 120 | 2,080 ± 650 | |
| Lotus corniculatus | MAFF303099(pALPF1) | 1,910 ± 320 | 3,930 ± 200 |
| MAFF303099(pALPFE) | 1,100 ± 60 | 3,570 ± 560 | |
| Phaseolus vulgaris | CIAT899(pALPF1) | 1,210 ± 290 | 1,350 ± 180 |
| CIAT899(pALPFE) | 360 ± 140 | 940 ± 240 | |
| CE3(pALPF1) | 6,730 ± 1,540 | 5,410 ± 320 | |
| CE3(pALPFE) | 5,600 ± 1,610 | 4,890 ± 920 | |
Values are expressed in nmol H2 h−1 mg protein−1. Hydrogenase activities were measured using oxygen as an electron acceptor. Values were determined for bacteroids from 21-day-old plants and represent averages of results from at least two experiments ± SE.
The relevance of HupE in symbiotic hydrogen oxidation was further investigated for other rhizobium-legume symbioses (Table 6). Plasmids pALPF1 and pALPFE were introduced into Mesorhizobium loti MAFF303099, an endosymbiont of different Lotus species, and Rhizobium tropici CIAT899 and Rhizobium etli CE3, which establish symbiosis with Phaseolus vulgaris. Bacteroids of MAFF303099(pALPFE) extracted from 21-day-old Lotus corniculatus plants grown without nickel addition showed a 42% reduction in hydrogenase activity in comparison to the level for the wild-type strain MAFF303099(pALPF1). Addition of nickel to the nutrient solution restored hydrogen oxidation to wild-type levels. In the case of Rhizobium-bean symbioses, hydrogen uptake of CIAT899(pALPFE) bacteroid cells decreased more than 70% compared to that of CIAT899(pALPF1), and again, this phenotype was reverted by incorporation of nickel in the plant nutrient solution. In contrast, no difference in hydrogenase activity was observed between CE3(pALPF1) and CE3(pALPFE) bacteroids obtained from bean plants grown in plant nutrient solutions supplemented or not supplemented with nickel. The latter result can be explained by the presence in R. etli CE3 of a hupE orthologous gene, showing 42% sequence identity with R. leguminosarum HupE, that might complement its function in bean bacteroids (Table 3). In conclusion, these results indicate that HupE and HupE2 are involved in symbiotic hydrogenase activity providing nickel to the bacteroid cell inside the nodule. The relevance of their role depends on the particular legume-rhizobium symbiosis, suggesting the occurrence of alternative mechanisms in the host plant that replace the function of HupE and HupE2.
DISCUSSION
In this paper, we characterize the hydrogenase-related protein HupE as a nickel transporter required for hydrogenase activity in microaerobic free-living cells and bacteroids of R. leguminosarum bv. viciae UPM791. We have also identified HupE2, a HupE-homolog that displays a similar role in this strain. HupE and HupE2 belong to the HupE/UreJ family of single component metal permeases. An increasing number of hupE-like genes have been identified as a result of bacterial genome sequencing efforts. Some of them are linked to hydrogenase or urease gene clusters, but many appear as isolated genes in the bacterial genome. A recent database search identified 103 transporters of this family in a wide diversity of bacteria, including members of several proteobacterial subdivisions and cyanobacteria (53). Members of the HupE/UreJ family of transporters have been postulated as nickel transporters on the basis of their features as membrane proteins, the presence of amino acid sequence motifs involved in nickel transport in other proteins, and their association with nickel-containing systems such as hydrogenase or urease (3, 33). Functional evidence for a direct involvement of HupE in nickel transport had been obtained by measuring nickel accumulation in E. coli cells that express the R. palustris HupE protein (18). However, no reports had demonstrated the physiological role of a HupE-based transport system in the activity of the associated metalloenzyme. In this work, we identified two hupE-like genes whose simultaneous disruption led to a drastic reduction of hydrogenase activity in R. leguminosarum vegetative cells. Hydrogenase activity was recovered upon the addition of nickel to the culture medium, indicating that the defect in the double mutant was at the level of provision of nickel to the enzyme. The presence of residual activity in the double mutant indicates that nickel can enter the cell via some other mechanisms, such as nonspecific magnesium transporters (50). Nickel-dependent complementation of metalloenzyme activity has previously been described. Nickel transport mediated by the Ralstonia eutropha HoxN nickel permease, a member of the NiCoT family, could be demonstrated in an E. coli heterologous system involving urease activity (50). In R. palustris, a HupE-deficient mutant was impaired in hydrogenase activity, but this phenotype was not recovered by an excess of nickel (18). In contrast, a HupE-deficient mutant from the cyanobacterium Synechocystis sp. PCC6803 was not affected in hydrogenase activity (28).
Hydrogenase activity and nickel uptake assays with R. leguminosarum HupE overexpressed in E. coli further confirmed that this protein mediates Ni transport and promotes hydrogenase activity, indicating a physiological role for this transport. A technical problem, likely related to nonspecific binding of nickel to Rhizobium cells, prevented direct transport experiments in the original genetic background. This nonspecific binding of nickel to cell structures has previously been reported as a major problem interfering with the analysis of nickel transport in different bacteria and in liposome systems (50). Nickel transport assays performed with E. coli cells indicate that HupE is able to mediate a highly efficient nickel uptake system. More interestingly, expression of HupE and HupE2 proteins in the E. coli mutant HYD723 showed different levels of nickel transport. These differences in Ni accumulation rates mediated by HupE and HupE2 were not due to the different growth conditions used for both sets of experiments, since cells grown anaerobically showed a similar pattern of results (data not shown). The different uptake activities might indicate either different levels of active protein or different affinities and/or capacities of these transport proteins.
The site-directed mutagenesis analysis performed for the present report is the first attempt to study functional requirements in a member of the HupE/UreJ family of metal permeases. Data from mutagenesis of histidine residues suggest essential roles associated with two motifs, HX5DH and FHGX[AV]HGXE, conserved in the HupE/UreJ family. Assuming that the identified signal peptide is actually cleaved, we have integrated our data into a hypothetical model for mature HupE topology (Fig. 5), based on previous models proposed for other Ni permeases (19, 24). According to this model, the identified motifs I and II would be located within predicted transmembrane domains I and IV, respectively, and would participate in a membrane channel that allows the passage of nickel through the cytoplasmic membrane. Validation of this hypothesis requires additional experimentation to exclude the possibility that the histidine residues within the identified motifs are required for protein stability or membrane insertion.
FIG. 5.
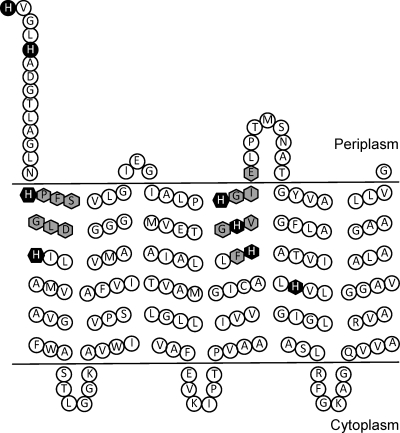
Predicted topology of the Rhizobium leguminosarum HupE protein and localization of amino acid residues involved in activity. Proposed model of HupE topology based on hydropathy predictions and previous models for C. necator HoxN and H. pylori NixA (19, 24). Transmembrane domains are as predicted by DAS and HMMTOP 2.0 bioinformatic programs. Histidine residues are colored black. Residues highlighted in gray correspond to those participating in the conserved domains proposed as essential for nickel transport. Residues corresponding to a predicted cleavable signal peptide have been removed.
The first motif is similar to the motif (HX4DH) identified as essential for nickel uptake in permeases of the NiCoT and UreH families (50). Changes of the histidine residues in this motif suppressed the ability of HupE to provide nickel for hydrogenase synthesis. The second motif (FHGX[AV]HGXE) is likely specific for the HupE/UreJ family and had not previously been identified (Fig. 2). Mutagenesis data indicate that at least one of the histidine residues in the motif (H130) is essential for hydrogenase activity but that alteration of the other residue (H126) leads to reduced levels. A search of nonredundant protein database at NCBI for the sequence FHGX[AV]HGXE revealed that it is present in 142 proteins, all of them members of the HupE/UreJ family. These data suggest that this motif is a specific signature for permeases of this family. A different motif (GX5GHSSVV) has been described for the NiCoT family of transporters; the histidine residue within this motif has been shown as essential for NixA functionality (23). Such a motif is not present in HupE-like proteins. Finally, analysis of histidine residues in the N-terminal part of HupE suggests an interchangeable or additive effect of H22 and H26. Based on their predicted periplasm-exposed location and on the high nickel affinity of histidine residues, we hypothesize a role in ion scavenging and concentration around the transporter (Fig. 5). Then, histidine residues from motifs I and II might bind the ion with higher affinity and introduce it into the cell. Since these residues are well conserved in HupE/UreJ proteins from different microorganisms, an essential role for them in nickel transport in other bacterial backgrounds can be assumed.
The results reported here also indicate that HupE and HupE2 are involved in symbiotic hydrogenase activity, likely by providing nickel to the bacteroid cell inside the nodule. The presence of hupE homologs in Rhizobium genomes is not universal. For instance, this gene is absent in sequenced strains of Sinorhizobium meliloti and B. japonicum. In contrast, genomes from R. etli CE3 and M. loti MAFF303099 contain hupE-like genes (designated RHE_PB00145 and mll4945, respectively) whose predicted proteins show over 40% sequence identity with HupE and ca. 80% and 50%, respectively, with HupE2. These hupE2-like genes are probably responsible for the significant levels of hydrogenase activity induced by the hupE-deficient pALPFE hup cluster in these two strains. In the case of R. tropici CIAT899, the genome sequence is not available but the low levels of symbiotic hydrogenase activity associated with pALPFE suggest that strain CIAT899 does not contain the hupE gene.
The presence of two copies of a hupE gene in R. leguminosarum UPM791 might be related to the plasmid location of the corresponding genes. HupE is encoded by a gene within the hydrogenase gene cluster in the symbiotic plasmid, whereas hupE2 is located in plasmid pRLUPM791b, unrelated to hydrogenase genes. While it is clear that the hupE gene, part of a highly conserved hup cluster (20), is evolutionarily related to hydrogenase genes, the rationale for a hupE2 gene in R. leguminosarum is less clear. The low level of amino acid sequence identity between hupE and hupE2 suggests that these two proteins may play different roles, either providing nickel with a different capacity/affinity or transporting a different ion. In fact, HupE from Synechocystis sp. PCC6803 is likely a cobalt transporter required for the activity of a cobalamin-dependent methionine synthase (28). In that system, as well as in hupE genes from other cyanobacteria, sequences encoding a B12-dependent riboswitch have been identified upstream of hupE, as reported for other cobalt transporters (39). Such elements are not present upstream of the R. leguminosarum hupE and hupE2 coding sequences (data not shown). Further analyses of HupE2 transport capacity and expression will help to elucidate this point.
Analysis of the role of HupE and HupE2 in symbiotic hydrogenase activity revealed differences for the R. leguminosarum symbioses with pea and lentil. The lack of a hydrogenase phenotype of the hupE hupE2 double mutant in pea bacteroids suggests the existence of alternative mechanisms that replace the function of HupE and HupE2 in this symbiosis. Surprisingly, Ni provision by HupE/HupE2 was required in lentil bacteroids, suggesting that these potential alternative mechanisms do not operate in lentils. In symbiosis, bacteroid nickel ions are obtained from the plant cytosol surrounding the bacteroids. In this environment, nickel is likely to be present not as a free ion but rather as complexes with organic compounds (13). Although the possibility that different nickel transporters are induced in pea bacteroids but are absent in cultured cells and lentil bacteroids cannot be excluded, we favor the hypothesis that the observed differences are due to the presence of different nickel complexes in both symbioses (11). We have previously shown that bacteroid hydrogenase activity is reduced in lentil symbiosis due to both transcriptional and posttranscriptional factors (11), and similar limitations were observed when other symbiotic systems were analyzed for heterologous hydrogenase expression (8). One of these factors is apparently related to incorporation of nickel into the hydrogenase. Since the lentil nodule cytosol contains nickel levels similar to those observed in pea nodules (11), it is possible that the observed differences are due to differences in the natures of the nickel forms present. Confirmation of this hypothesis will require further analysis of the composition of nickel complexes in the cytoplasmic fractions of pea and lentil nodules. This analysis is currently under way in our laboratory.
The data presented here represent a first step toward the understanding of nickel provision by HupE, a member of a widespread bacterial nickel transporter family. Further work will allow us to get insight into the mechanism of metal provision in endosymbiotic bacteria in both free-living and symbiotic lifestyles.
Acknowledgments
We are grateful to A. Hidalgo for excellent experimental assistance during her training stay at the laboratory.
This work was supported by projects from the Spanish Ministry of Science and Technology (BIO2007-64147 to J.-M.P.), the U.P.M. (AL09-P[I+D]-06 to B.B.), and the Comunidad Autónoma de Madrid (MICROAMBIENTE-CM to T.R.-A.) and by grants from the Centre National de la Recherche Scientifique (to M.A.M.-B.).
Footnotes
Published ahead of print on 18 December 2009.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baginsky, C., B. Brito, J. Imperial, J. M. Palacios, and T. Ruiz-Argüeso. 2002. Diversity and evolution of hydrogenase systems in rhizobia. Appl. Environ. Microbiol. 68:4915-4924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baginsky, C., J. M. Palacios, J. Imperial, T. Ruiz-Argüeso, and B. Brito. 2004. Molecular and functional characterization of the Azorhizobium caulinodans ORS571 hydrogenase gene cluster. FEMS Microbiol. Lett. 237:399-405. [DOI] [PubMed] [Google Scholar]
- 4.Beringer, J. 1974. R factor transfer in Rhizobium leguminosarum. J. Gen. Microbiol. 84:188-198. [DOI] [PubMed] [Google Scholar]
- 5.Blokesch, M., M. Rohrmoser, S. Rode, and A. Bock. 2004. HybF, a zinc-containing protein involved in NiFe hydrogenase maturation. J. Bacteriol. 186:2603-2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Böck, A., P. W. King, M. Blokesch, and M. C. Posewitz. 2006. Maturation of hydrogenases. Adv. Microb. Physiol. 51:1-71. [DOI] [PubMed] [Google Scholar]
- 7.Brito, B., M. Martínez, D. Fernández, L. Rey, E. Cabrera, J. M. Palacios, J. Imperial, and T. Ruiz-Argüeso. 1997. Hydrogenase genes from Rhizobium leguminosarum bv. viciae are controlled by the nitrogen fixation regulatory protein NifA. Proc. Natl. Acad. Sci. U. S. A. 94:6019-6024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brito, B., J. Monza, J. Imperial, T. Ruiz-Argüeso, and J. M. Palacios. 2000. Nickel availability and hupSL activation by heterologous regulators limit symbiotic expression of the Rhizobium leguminosarum bv viciae hydrogenase system in Hup− rhizobia. Appl. Environ. Microbiol. 66:937-942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brito, B., J. M. Palacios, E. Hidalgo, J. Imperial, and T. Ruiz-Argüeso. 1994. Nickel availability to pea (Pisum sativum L.) plants limits hydrogenase activity of Rhizobium leguminosarum bv. viciae bacteroids by affecting the processing of the hydrogenase structural subunits. J. Bacteriol. 176:5297-5303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brito, B., J. M. Palacios, J. Imperial, and T. Ruiz-Argüeso. 2002. Engineering the Rhizobium leguminosarum bv. viciae hydrogenase system for expression in free-living microaerobic cells and increased symbiotic hydrogenase activity. Appl. Environ. Microbiol. 68:2461-2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brito, B., A. Toffanin, R. I. Prieto, J. Imperial, T. Ruiz-Argüeso, and J. M. Palacios. 2008. Host-dependent expression of Rhizobium leguminosarum bv. viciae hydrogenase is controlled at transcriptional and post-transcriptional levels in legume nodules. Mol. Plant Microbe Interact. 21:597-604. [DOI] [PubMed] [Google Scholar]
- 12.Casadaban, M. J., and S. N. Cohen. 1979. Lactose genes fused to exogenous promoters in one step using a Mu-lac bacteriophage: in vivo probe for transcriptional control sequences. Proc. Natl. Acad. Sci. U. S. A. 76:4530-4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cataldo, D. A., T. R. Garland, and R. E. Wildung. 1978. Nickel in plants: II. Distribution and chemical form in soybean plants. Plant Physiol. 62:566-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen, C., D. Huang, and J. Liu. 2009. Functions and toxicity of nickel in plants: recent advances and future prospects. Clean 37:304-313. [Google Scholar]
- 15.Cserzö, M., E. Wallin, I. Simon, G. von Heijne, and A. Elofsson. 1997. Prediction of transmembrane alpha-helices in prokaryotic membrane proteins: the dense alignment surface method. Protein Eng. 10:673-676. [DOI] [PubMed] [Google Scholar]
- 16.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Degen, O., and T. Eitinger. 2002. Substrate specificity of nickel/cobalt permeases: insights from mutants altered in transmembrane domains I and II. J. Bacteriol. 184:3569-3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eitinger, T., J. Suhr, L. Moore, and J. A. Smith. 2005. Secondary transporters for nickel and cobalt ions: theme and variations. Biometals 18:399-405. [DOI] [PubMed] [Google Scholar]
- 19.Eitinger, T., L. Wolfram, O. Degen, and C. Anthon. 1997. A Ni2+ binding motif is the basis of high affinity transport of the Alcaligenes eutrophus nickel permease. J. Biol. Chem. 272:17139-17144. [DOI] [PubMed] [Google Scholar]
- 20.Fernández, D., A. Toffanin, J. M. Palacios, T. Ruiz-Argüeso, and J. Imperial. 2005. Hydrogenase genes are uncommon and highly conserved in Rhizobium leguminosarum bv. viciae. FEMS Microbiol. Lett. 253:83-88. [DOI] [PubMed] [Google Scholar]
- 21.Fontecilla-Camps, J. C., A. Volbeda, C. Cavazza, and Y. Nicolet. 2007. Structure/function relationships of [NiFe]- and [FeFe]-hydrogenases. Chem. Rev. 107:4273-4303. [DOI] [PubMed] [Google Scholar]
- 22.Fu, C., S. Javedan, F. Moshiri, and R. J. Maier. 1994. Bacterial genes involved in incorporation of nickel into a hydrogenase enzyme. Proc. Natl. Acad. Sci. U. S. A. 91:5099-5103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fulkerson, J. F., Jr., R. M. Garner, and H. L. Mobley. 1998. Conserved residues and motifs in the NixA protein of Helicobacter pylori are critical for the high affinity transport of nickel ions. J. Biol. Chem. 273:235-241. [DOI] [PubMed] [Google Scholar]
- 24.Fulkerson, J. F., Jr., and H. L. Mobley. 2000. Membrane topology of the NixA nickel transporter of Helicobacter pylori: two nickel transport-specific motifs within transmembrane helices II and III. J. Bacteriol. 182:1722-1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Furia, T. E. 1972. Sequestrants in food, p. 271-294. In T. E. Furia (ed.), CRC handbook of food additives. CRC Press, Boca Raton, FL.
- 26.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 28.Hoffmann, D., K. Gutekunst, M. Klissenbauer, R. Schulz-Friedrich, and J. Appel. 2006. Mutagenesis of hydrogenase accessory genes of Synechocystis sp. PCC 6803. Additional homologues of hypA and hypB are not active in hydrogenase maturation. FEBS J. 273:4516-4527. [DOI] [PubMed] [Google Scholar]
- 29.Leach, M. R., J. W. Zhang, and D. B. Zamble. 2007. The role of complex formation between the Escherichia coli hydrogenase accessory factors HypB and SlyD. J. Biol. Chem. 282:16177-16186. [DOI] [PubMed] [Google Scholar]
- 30.Leyva, A., J. M. Palacios, T. Mozo, and T. Ruiz-Argüeso. 1987. Cloning and characterization of hydrogen uptake genes from Rhizobium leguminosarum. J. Bacteriol. 169:4929-4934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leyva, A., J. M. Palacios, J. Murillo, and T. Ruiz-Argüeso. 1990. Genetic organization of the hydrogen uptake (hup) cluster from Rhizobium leguminosarum. J. Bacteriol. 172:1647-1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.López, M., V. Carbonero, E. Cabrera, and T. Ruiz-Argüeso. 1983. Effects of host on the expression of the H2-uptake hydrogenase of Rhizobium in legume nodules. Plant Sci. Lett. 29:191-199. [Google Scholar]
- 33.McMillan, D., M. Mau, and M. Walker. 1998. Characterisation of the urease gene cluster in Bordetella bronchiseptica. Gene 208:243-251. [DOI] [PubMed] [Google Scholar]
- 34.Mulrooney, S. B., and R. P. Hausinger. 2003. Nickel uptake and utilization by microorganisms. FEMS Microbiol. Rev. 27:239-261. [DOI] [PubMed] [Google Scholar]
- 35.Navarro, C., L. F. Wu, and M. A. Mandrand-Berthelot. 1993. The nik operon of Escherichia coli encodes a periplasmic binding-protein-dependent transport system for nickel. Mol. Microbiol. 9:1181-1191. [DOI] [PubMed] [Google Scholar]
- 36.Noel, K., A. Sánchez, L. Fernández, J. Leemans, and M. Cevallos. 1984. Rhizobium phaseoli symbiotic mutants with transposon Tn5 insertions. J. Bacteriol. 158:148-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O'Gara, F., and K. T. Shanmugam. 1976. Regulation of nitrogen fixation by rhizobia: export of fixed nitrogen as NH4+. Biochim. Biophys. Acta 437:313-321. [DOI] [PubMed] [Google Scholar]
- 38.Palacios, J. M., H. Manyani, M. Martinez, A. C. Ureta, B. Brito, E. Bascones, L. Rey, J. Imperial, and T. Ruiz-Argüeso. 2005. Genetics and biotechnology of the H(2)-uptake [NiFe] hydrogenase from Rhizobium leguminosarum bv. viciae, a legume endosymbiotic bacterium. Biochem. Soc. Trans. 33:94-96. [DOI] [PubMed] [Google Scholar]
- 39.Rodionov, D. A., P. Hebbeln, M. S. Gelfand, and T. Eitinger. 2006. Comparative and functional genomic analysis of prokaryotic nickel and cobalt uptake transporters: evidence for a novel group of ATP-binding cassette transporters. J. Bacteriol. 188:317-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rossmann, R., T. Maier, F. Lottspeich, and A. Böck. 1995. Characterization of a protease from Escherichia coli involved in hydrogenase maturation. Eur. J. Biochem. 227:545-550. [DOI] [PubMed] [Google Scholar]
- 41.Rossmann, R., G. Sawers, and A. Böck. 1991. Regulation of the formate-hydrogenlyase pathway by oxygen, nitrate and pH: definition of the formate regulon. Mol. Microbiol. 5:2807-2814. [DOI] [PubMed] [Google Scholar]
- 42.Ruiz-Argüeso, T., F. J. Hanus, and H. J. Evans. 1978. Hydrogen production and uptake by pea nodules as affected by strains of Rhizobium leguminosarum. Arch. Microbiol. 116:113-118. [Google Scholar]
- 43.Ruiz-Argüeso, T., J. Imperial, and J. M. Palacios. 2000. Uptake hydrogenases in root nodule bacteria, p. 489-507. In E. W. Triplett (ed.), Prokaryotic nitrogen fixation: a model system for analysis of a biological process. Horizon Scientific Press, Wymondham, United Kingdom.
- 44.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. CSHL Press, Cold Spring Harbor, NY.
- 45.Schäfer, A., A. Tauch, W. Jäger, J. Kalinowski, G. Thierbach, and A. Pühler. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69-73. [DOI] [PubMed] [Google Scholar]
- 46.Simon, R., U. Priefer, and A. Pühler. 1983. Vector plasmids for in-vivo and in-vitro manipulations of Gram-negative bacteria, p. 98-106. In A. Pühler (ed.), Molecular genetics of the bacteria-plant interactions. Springer-Verlag, Berlin, Germany.
- 47.Smith, P. K., R. I. Krohn, G. T. Hermanson, A. K. Mallia, F. H. Gartner, M. D. Provenzano, E. K. Fujimoto, N. M. Goeke, B. J. Olson, and D. C. Klenk. 1985. Measurement of protein using bicinchoninic acid. Anal. Biochem. 150:76-85. [DOI] [PubMed] [Google Scholar]
- 48.Toffanin, A., E. Cadahia, J. Imperial, T. Ruiz-Argüeso, and J. M. Palacios. 2002. Characterization of the urease gene cluster from Rhizobium leguminosarum bv. viciae. Arch. Microbiol. 177:290-298. [DOI] [PubMed] [Google Scholar]
- 49.Vincent, J. M. 1970. A manual for the practical study of root-nodule bacteria. Blackwell Scientific Publications, Ltd., Oxford, United Kingdom.
- 50.Wolfram, L., B. Friedrich, and T. Eitinger. 1995. The Alcaligenes eutrophus protein HoxN mediates nickel transport in Escherichia coli. J. Bacteriol. 177:1840-1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu, L. F., M. A. Mandrand-Berthelot, R. Waugh, C. J. Edmons, S. E. Holt, and D. H. Boxer. 1989. Nickel deficiency gives rise to the defective hydrogenase phenotype of hydC and fnr mutants in Escherichia coli. Mol. Microbiol. 3:1709-1718. [DOI] [PubMed] [Google Scholar]
- 52.Young, J. P., L. C. Crossman, A. W. Johnston, N. R. Thomson, Z. F. Ghazoui, K. H. Hull, M. Wexler, A. R. Curson, J. D. Todd, P. S. Poole, T. H. Mauchline, A. K. East, M. A. Quail, C. Churcher, C. Arrowsmith, I. Cherevach, T. Chillingworth, K. Clarke, A. Cronin, P. Davis, A. Fraser, Z. Hance, H. Hauser, K. Jagels, S. Moule, K. Mungall, H. Norbertczak, E. Rabbinowitsch, M. Sanders, M. Simmonds, S. Whitehead, and J. Parkhill. 2006. The genome of Rhizobium leguminosarum has recognizable core and accessory components. Genome Biol. 7:R34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang, Y., D. A. Rodionov, M. S. Gelfand, and V. N. Gladyshev. 2009. Comparative genomic analyses of nickel, cobalt and vitamin B12 utilization. BMC Genomics 10:78. [DOI] [PMC free article] [PubMed] [Google Scholar]