Abstract
Melamine toxicity in mammals has been attributed to the blockage of kidney tubules by insoluble complexes of melamine with cyanuric acid or uric acid. Bacteria metabolize melamine via three consecutive deamination reactions to generate cyanuric acid. The second deamination reaction, in which ammeline is the substrate, is common to many bacteria, but the genes and enzymes responsible have not been previously identified. Here, we combined bioinformatics and experimental data to identify guanine deaminase as the enzyme responsible for this biotransformation. The ammeline degradation phenotype was demonstrated in wild-type Escherichia coli and Pseudomonas strains, including E. coli K12 and Pseudomonas putida KT2440. Bioinformatics analysis of these and other genomes led to the hypothesis that the ammeline deaminating enzyme was guanine deaminase. An E. coli guanine deaminase deletion mutant was deficient in ammeline deaminase activity, supporting the role of guanine deaminase in this reaction. Two guanine deaminases from disparate sources (Bradyrhizobium japonicum USDA 110 and Homo sapiens) that had available X-ray structures were purified to homogeneity and shown to catalyze ammeline deamination at rates sufficient to support bacterial growth on ammeline as a sole nitrogen source. In silico models of guanine deaminase active sites showed that ammeline could bind to guanine deaminase in a similar orientation to guanine, with a favorable docking score. Other members of the amidohydrolase superfamily that are not guanine deaminases were assayed in vitro, and none had substantial ammeline deaminase activity. The present study indicated that widespread guanine deaminases have a promiscuous activity allowing them to catalyze a key reaction in the bacterial transformation of melamine to cyanuric acid and potentially contribute to the toxicity of melamine.
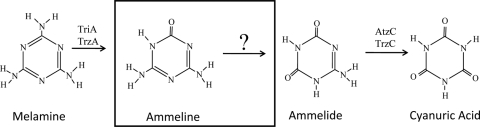
Ammeline is an intermediate in the bacterial metabolism of melamine (Fig. 1). Melamine has become internationally recognized as a chemical adulterant in pet foods and infant formula that caused morbidity and mortality in pets and children (12). In pets, where more than 1,000 deaths have been attributed to melamine poisoning, the composition of the causal kidney precipitate was found to be a 1:1 complex of melamine-cyanuric acid (2, 25). In human babies, melamine-uric acid cocrystals have been identified (11). Feeding animals a mixture of melamine and cyanuric acid or a mixture of melamine, ammeline, ammelide, and cyanuric acid was found to produce acute kidney disease (5). Melamine and cyanuric acid are known to form a highly insoluble, hydrogen-bonded network (33) that can precipitate in the kidneys, causing kidney failure. Since bacterial metabolism of melamine generates cyanuric acid (6, 7, 14), it is possible that bacterial melamine metabolism could contribute to melamine toxicity in some cases.
FIG. 1.
The known metabolic pathway in bacteria for transforming melamine to cyanuric acid.
Bacteria metabolize melamine by sequential deamination (4, 6, 7, 14, 30) to ammeline, ammelide, and cyanuric acid (Fig. 1). The genes and enzymes involved in the deamination of melamine and ammelide are known. Melamine deaminases (TriA and TrzA) have been purified and characterized (20, 28). The enzymes AtzC (34) and TrzC (7) were shown to be capable of ammelide deamination. Although ammeline deamination has been observed in a large number of microbial strains (37), the genes and enzymes involved in bacterial ammeline deamination have remained obscure. Many of the bacteria and fungi that were shown to deaminate ammeline did not deaminate melamine or ammelide (37), indicating that these ammeline deaminating enzymes have not evolved as a component of the melamine degradation pathway.
Enzymes functioning in the metabolism of the s-triazine herbicide atrazine are related to some of the enzymes in the melamine pathway. TriA (melamine deaminase) is related to AtzA (28, 31) and TrzN (35), enzymes that catalyze the dechlorination of atrazine. AtzB, which catalyzes the second step in the atrazine metabolic pathway, was reported to also deaminate ammeline as a side reaction, but the rate of the reaction was not measured in that study (27).
The enzymes involved in melamine and ammelide deamination, along with other enzymes acting on s-triazine herbicides, are all members of the amidohydrolase superfamily (32, 36). These enzymes typically contain one or two metal ions that are involved in activating water for nucleophilic displacement reactions. A significant number of amidohydrolase superfamily members catalyze deamination reactions with nitrogen heterocyclic ring substrates. In this context, we specifically analyzed the amidohydrolase enzymes in ammeline-metabolizing bacteria to identify the enzyme responsible for the activity. Molecular genetic, biochemical, and in silico data support the hypothesis that guanine deaminase functions as the principal ammeline deaminase activity of bacteria. This has implications for enzyme catalytic promiscuity and understanding the bacterial metabolism of melamine. The latter may be relevant to melamine toxicity in humans and animals.
MATERIALS AND METHODS
Bacterial strains and growth media.
E. coli JW5466-3 (ΔguaD::kan mutant) (1) was obtained from the E. coli Genetic Stock Center, Yale University. A human guaD clone was obtained already cloned into the pET-derived expression vector PNIC-Bsa4, courtesy of Martin Hammarström at the Structural Genomics Consortium (Stockholm, Sweden), and was used to transform Rosetta 2 competent cells (Merck, Darmstadt, Germany). P. putida KT2440 (ATCC 47054) was obtained from the American Type Culture Collection (Manassas, VA). Bacterial strains were grown in Luria-Bertani (LB) medium with the appropriate antibiotics at 37°C, unless otherwise noted. Bacteria were grown using 2 mM ammeline or ammelide as the sole source of nitrogen on HM salts minimal medium (3) containing 0.1% sodium gluconate and 0.1% l-arabinose as carbon sources.
Analytical methods and standards.
High-pressure liquid chromatography (HPLC) analysis was performed on a Hewlett-Packard HP 1100 series chromatographic system equipped with a diode array detector and interfaced with an HP Chemstation. Compounds were separated on a Varian Microsorb-MV 100-8 CN column (250 by 4.6 mm) with a 1 ml/min flow rate and an isocratic mobile phase of 5 mM phosphate buffer (pH 6.7) with the detector absorbance set at 224 nm. Retention times were the following: melamine, 4.7 min; ammeline, 3.5 min; and ammelide, 2.8 min. Product identities were further confirmed by UV spectroscopy of the eluting materials. Ammonia concentrations were determined using the Berthelot method as previously described (22). Standard melamine was obtained from Sigma-Aldrich (St. Louis, MO), and cyanuric acid was from Fluka (Buchs, Switzerland). Ammeline and ammelide were synthesized as previously described (28).
Deaminase enzyme assays.
Deamination rates were measured by determining ammonia released as described above. When rates were low, activities were confirmed using HPLC. Uniform suspensions (25 to 50 mM) of potential substrates were prepared by sonication in 25 mM sodium hydroxide (guanine) or 50% ethanol plus 25 mM sodium hydroxide (s-triazines) and stored at −20°C. All compounds were observed to be stable with minimal hydrolysis when stored under these conditions for 1 month. Substrate-only controls were run with all assays to test for nonenzymatic hydrolysis. Guanine deaminases were assayed in 100 mM potassium phosphate buffer, pH 7.0, with 2 to 180 μg of protein and 300 μM substrate. Assays with all other amidohydrolases (creatinine deaminase [MP Biomedicals, Solon, OH], adenosine deaminase [Roche Applied Science, Mannheim, Germany], and d-aminoacylase and urease [Sigma-Aldrich, St. Louis, MO]) were done as described above with 50 μg of protein in 50 mM potassium phosphate buffer, pH 7.5 (except urease, for which the buffer was pH 7.8).
Cloning the Bradyrhizobium japonicum USDA 110 guanine deaminase.
Genomic DNA from B. japonicum USDA 110 was isolated as previously described (26). Guanine deaminase (guaD) from B. japonicum USDA 110, encoded by open reading frame (ORF) blr3880, was PCR amplified using primers 14FNdeI (5′-GAAGGAACCAGCATATGACCACCGTCGG-3′) and 1561compHindIII (5′-GCAAAGCTTTCGTCGGCGATGCGGC-3′) and cloned into the NdeI/HindIII sites of vector pET28b+ (Novagen, Madison, WI). The resulting plasmid was used to transform E. coli BL21(DE3) pLysS cells.
Proteins, expression, and purification.
Expression strains were grown to an optical density at 600 nm (OD600) of 0.5 to 1 at 37°C. B. japonicum USDA 110 guaD expression was induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at 37°C; human guanine deaminase gene expression was induced with 0.1 mM IPTG at 15°C for 19 h. After induction, the cells were harvested and resuspended in the assay buffer plus Roche Complete Mini EDTA-free protease inhibitor and then lysed with two cycles in a French pressure cell at 124 MPa. The His-tagged GuaD was purified on a HiTrap IMAC HP column (GE Healthcare, Piscataway, NJ), as per the manufacturer's instructions, using washes of 0.05 M, 0.1 M, 0.25 M, and 0.5 M imidazole in 100 mM potassium phosphate buffer (pH 7.0). Guanine deaminase activity eluted with 0.5 M imidazole. The protein was dialyzed against 100 mM potassium phosphate buffer (pH 7.0), followed by one change of buffer with 0.2 mM zinc sulfate added and then a final change of buffer without zinc. Purified enzyme aliquots were flash frozen in liquid nitrogen and stored at −80°C. Creatinine deaminase was purchased from MP Biomedicals (Solon, OH), adenosine deaminase was purchased from Roche Applied Science (Mannheim, Germany), and d-aminoacylase and urease were purchased from Sigma-Aldrich (St. Louis, MO). The TriA (28), AtzB (27), and TrzN (35) proteins were purified as described elsewhere.
Computational methods.
Sequences of the amidohydrolase superfamily were obtained from listings in the Structure Function Linkage Database (23). Sequence clustering was performed by combining functional information with network clusters obtained from the program structureViz (19) in a method described by Seffernick et al. (29). Connectivity is depicted at a BLASTP score cutoff of e−5. Docking studies were performed using Maestro, version 8.5, and XP Glide, version 5.0 (Schrodinger Suite 2008, Update 1; Schrodinger, Inc.) (9). Because of different substrate/product orientations in human (Protein Data Bank [PDB] code 2uz9), B. japonicum USDA 110 (PDB 2ood), and Clostridium acetobutylicum (PDB 2i9u) structures, substrates/products were removed from the PDB files prior to docking.
RESULTS
Computational studies for identifying ammeline deaminase.
A previous study by Zeyer and coworkers (37) suggested that many bacteria are able to catalyze the deamination of ammeline to ammelide. We tested for this activity in E. coli and Pseudomonas strains, including E. coli strain K-12 MG1655 and P. putida KT2440, for which complete genome sequences are available. The E. coli and P. putida strains metabolized ammeline to ammelide but did not metabolize ammelide or melamine. The gene(s) underlying this phenotype was then sought by comparative genomic analysis.
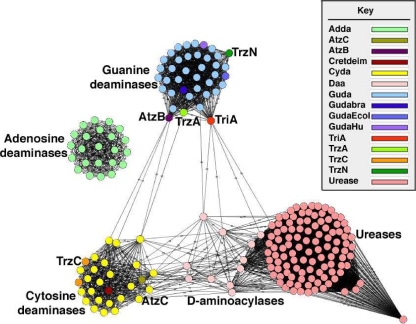
A detailed analysis of the known enzymes acting on s-triazine substrates including melamine and ammelide indicated that they clustered into one of two subfamilies within the amidohydrolase superfamily: the guanine deaminase or cytosine deaminase subfamilies (Fig. 2). Figure 2 shows a cluster network diagram where the nodes represent protein sequences, and the edges represent BLASTP linkages at an e-score cutoff of e−5. The enzymes previously shown to act on melamine, TriA (28) and TrzA (20), and hydroxyatrazine, AtzB (27), clustered most closely with the guanine deaminases. TrzN, an enzyme that hydrolytically displaces a chlorine substituent from the s-triazine herbicide atrazine, also clusters with the guanine deaminases. The two naturally occurring enzymes shown to react with ammelide (AtzC and TrzC) cluster with the cytosine deaminases (Fig. 2). Several other extensively characterized amidohydrolase superfamily members are also shown in Fig. 2.
FIG. 2.
A clustering network diagram of a subset of the amidohydrolase superfamily, representing enzymes tested in this study for activity with melamine, ammeline, and ammelide. The node colors represent different functions, as indicated on the figure. The distance between nodes represents the closeness of sequence relatedness. Adda, adenosine deaminases; Cretdeim, creatinine deaminase; Cyda, cytosine deaminases; Daa, d-aminoacylases; Guda, guanine deaminases; Gudabra, guanine deaminase from B. japonicum USDA 110; GudaEcol, guanine deaminase from E. coli strain K-12 MG16655; GudaHu, guanine deaminase from humans.
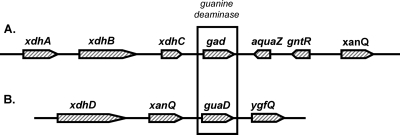
Ammeline deaminase was suspected to be a member of the amidohydrolase superfamily that contained one active-site metal (monometallic) since cytosine and guanine deaminase subfamily enzymes are monometallic. Eighteen completed genomes from various Pseudomonas and E. coli strains were searched for monometallic amidohydrolase superfamily members using BLASTP. Three candidates were identified as being present in all Pseudomonas genomes (locus names are given for P. putida KT2440): PP_2584 (annotated as hydroxydechloro-s-trazine ethylaminohydrolase), PP_3209 (annotated as amidohydrolase), and PP_4281 (annotated as guanine deaminase). Two candidates were identified in the E. coli strains (locus names are given for E. coli K-12 substrain MG1655): b2879 (annotated as a predicted chlorohydrolase/amidohydrolase) and b2883 (guanine deaminase). The common enzyme was annotated as a guanine deaminase, and genome context in both P. putida KT2440 and E. coli MG1655 indicated that these enzymes are likely to be genuine guanine deaminases (Fig. 3). Genes linked to the annotated guanine deaminase genes gudA and guaD are involved in metabolism of xanthine, the product of the guanine deaminase reaction. For example, in P. putida (Fig. 3A), the genes xdhA and xdhB encode the two subunits of xanthine dehydrogenase, and xdhC encodes a xanthine dehydrogenase accessory protein. In E. coli, XdhD is a xanthine dehydrogenase fusion protein of both subunits (Fig. 3B). The XanQ and YgfQ are purine permeases. Lastly, the GuaD protein of E. coli has been purified and shown to have guanine deaminase activity (18). It was not tested for activity with ammeline.
FIG. 3.
Gene regions of the annotated guanine deaminase genes in P. putida KT2440 (gad) (A) and E. coli K-12 strain MG1655 (guaD) (B).
Genetic evidence for guanine deaminase having ammeline deaminase activity.
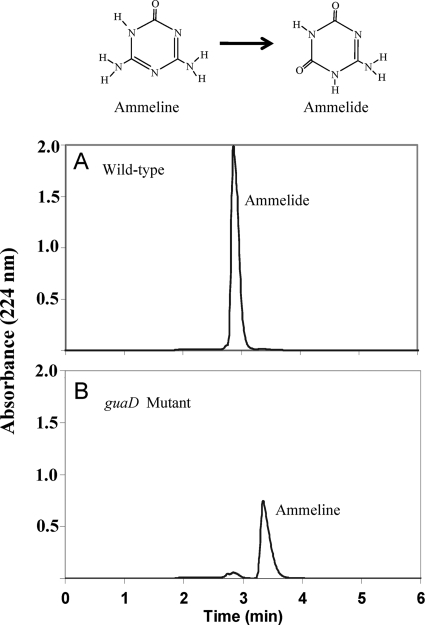
To genetically test the hypothesis that guanine deaminase was involved in ammeline deamination, an E. coli K-12 guanine deaminase mutant (ΔguaD) from the Keio gene knockout collection, strain JW5466-3 (1), was tested and found to lack ammeline deamination activity in vivo that was present in the wild type (Fig. 4). The small amount of material eluting at 2.9 min when the guanine deaminase mutant was tested was also present in a control incubation without cells. Thus, there was no discernible ammeline deamination with the mutant following 14 h of incubation. This result not only suggested the involvement of guanine deaminase in deamination of ammeline, but it also indicated that GuaD was solely responsible for ammeline deamination in E. coli.
FIG. 4.
HPLC traces of E. coli wild type and a GuaD guanine deaminase mutant after incubation for 14 h with ammeline. The small amount of material eluting at 2.9 min with the guanine deaminase mutant is also present in a control incubation without cells.
Purified guanine deaminases have ammeline deaminase activity.
Guanine deaminases from B. japonicum USDA 110 and Homo sapiens, for which X-ray structures are available, were purified and tested in vitro for ammeline deaminase activity. The enzymes were expressed as His-tagged proteins and purified to homogeneity using a nickel affinity column. The activities of the enzymes with guanine were comparable to previously reported values (18, 21). Measured activities with ammeline were 73 and 57 nmol per min per mg for the B. japonicum USDA 110 and human guanine deaminases, respectively (Table 1).
TABLE 1.
Specific activities of amidohydrolase superfamily members with melamine, ammeline, and ammelide
| Enzyme type and specific enzymea | Enzyme source | Specific activity with the indicated substrate (nmol per min per mg of protein) |
||
|---|---|---|---|---|
| Melamine | Ammeline | Ammelide | ||
| Guanine deaminases | ||||
| Guanine deaminase | B. japonicum | <0.3b | 73 ± 11 | <0.3 |
| Guanine deaminase | Human | <0.3 | 57 ± 7 | <0.3 |
| Triazine hydrolases | ||||
| Melamine deaminase (TriA) | Pseudomonas sp. NRRL B-12227 | 12,000 | 0.7 ± 0.1 | <0.3 |
| Triazine hydrolase (TrzA)c | Rhodococcus corralinus | 270,000 | 0 | NRd |
| Hydroxyatrazine hydrolase (AtzB) | Pseudomonas sp. ADP | 0.3 | <0.3e | <0.3 |
| N-Isopropylammelide hydrolase (AtzC)f | Pseudomonas sp. ADP | NRd | 0 | 2010 |
| Triazine hydrolase (TrzN) | Arthrobacter aurescens TC1 | <0.3 | <0.3 | <0.3 |
| Other amidohydrolases | ||||
| Adenosine deaminase | Bovine | <0.3 | <0.3 | <0.3 |
| Creatinine deaminase | Bacteriag | <0.3 | <0.3 | <0.3 |
| d-Aminoacylase | E. coli | <0.3 | <0.3 | <0.3 |
| Urease | Jack bean | <0.3 | <0.3 | <0.3 |
Based on known substrate(s).
No detectable activity at limit of detection indicated.
Activities as reported in reference 20.
Activities were not reported for these substrates in the reference indicated.
Activity below the detection limit in a standard assay, but ammelide was detected in 24-h incubations.
Activities as reported in reference 34.
The manufacturer obtains enzyme from different bacterial sources.
Structural examination of ammeline deamination by guanine deaminase.
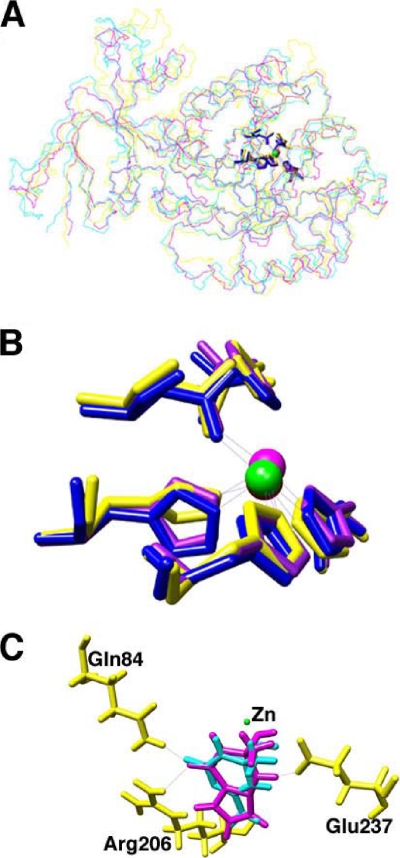
Figure 5A compares the X-ray structures of guanine deaminases derived from humans and bacteria. The overlaid backbone tracings reveal a striking degree of similarity, considering that human and bacterial guanine deaminase genes diverged more than 1 billion years ago and share only ∼30 to 40% amino acid sequence identity. The ligands to the active-site metal are nearly superimposable, and several residues that might interact with substrate are in comparable positions (Fig. 5B).
FIG. 5.
Crystal structure and docking analysis of guanine deaminases. (A) Overlay of X-ray structure backbones of guanine deaminases from B. japonicum USDA 110 (PDB 2ood; yellow), human (PDB 2uz9; blue), and C. acetobutylicum (PDB 2i9u; purple). (B) Catalytically significant residues in the guanine deaminase active sites. The spheres represent the active-site zinc atoms. (C) Docking results of guanine (purple) and ammeline (cyan) tetrahedral intermediates within B. japonicum USDA 110. Residues that interact with both substrates include Arg206, Gln84, and Glu237.
Potential protein-substrate interactions were modeled using Glide (9) to dock a series of potential substrates, substrate analogs, tetrahedral intermediates, and products, into the B. japonicum USDA 110 guanine deaminase structure (PDB code 2ood). Guanine deaminase showed similar binding interactions with both guanine and ammeline. The N-H groups of Arg206 and Gln84 appear to be within hydrogen bonding distance to the keto group on each of the substrates. In addition, Glu237 appears to be within hydrogen bonding distance from the ring nitrogen adjacent to the leaving group, an interaction conserved in many of the mononuclear members of the amidohydrolase superfamily (Fig. 5C). In all cases, the best docking scores were obtained from tetrahedral reaction intermediates, especially those for guanine and ammeline, which is consistent with these compounds being substrates.
Do other amidohydrolase superfamily members show activity with ammeline?
All of the enzymes known to catalyze the reactions shown in Fig. 1 are members of the amidohydrolase superfamily, prompting us to test other amidohydrolase enzymes with each substrate within the pathway of melamine transformation to cyanuric acid. Amidohydrolases that were tested here were known to act on (i) guanine, (ii) s-triazine substrates, or (iii) miscellaneous substrates (Table 1). Previously, melamine deaminase (TriA) and hydroxyatrazine hydrolase (AtzB) were reported to have some activity with ammeline. However, as observed here, those activities were very low, less than 1 nmol per min per mg. Other amidohydrolases tested had no detectable activity with ammeline. The last category of amidohydrolases (adenosine deaminase, creatinine deaminase, d-aminoacylase, and urease) had no activity with melamine, ammeline, or ammelide. The results shown in Table 1 are consistent with the hypothesis that ammeline deamination is not an activity that is common throughout the amidohydrolase superfamily and suggest that guanine deaminase alone might be responsible for this activity in many bacteria.
Distribution and classification of guanine deaminases.
Enzymes annotated as guanine deaminase derive from two groups, the amidohydrolase-type and cytidine deaminase-type superfamilies. Both superfamilies use a metal to activate water to catalyze hydrolytic nucleophilic substitution reactions. However, the backbone structures and catalytic residues of each are significantly different, indicating different ancestral origins. The amidohydrolase superfamily consists of a (βα)8 barrel with three histidine and one aspartate metal ligand while the cytidine deaminase superfamily has an alpha-beta-alpha structure with cysteine metal ligands. Of the 185 annotated guanine deaminase genes in the KEGG database as of 10 April 2009, 162 (88%) were found to be in the amidohydrolase superfamily. Amidohydrolase-type guanine deaminases are broadly dispersed throughout the tree of life and are known in human, insects, protozoa, fungi, bacteria, and archaea. It remains to be tested if guanine deaminases from the cytidine deaminase superfamily also show significant activity with ammeline as a substrate. However, the genera of bacteria shown to grow and metabolize ammeline in this and in a previous study (37) are now known from our genomic analysis to largely express the amidohydrolase-type guanine deaminase. This suggests that the amidohydrolase guanine deaminase may be largely responsible for the widespread ability of bacteria to metabolize ammeline.
DISCUSSION
Guanine deaminase activity is widespread in microbes, where it functions in the assimilation of nitrogen from guanine and the regulation of cellular guanine nucleotide pools (18). In the human brain, proteins with guanine deaminase activity play additional roles in dendritic growth and neuronal development (8). Guanine deaminases are found in plants, fungi, and protozoa. The large majority of enzymes that function physiologically in guanine deamination are structurally placed within the aminohydrolase superfamily of proteins.
The conclusion here that guanine deaminase is the likely enzyme used by many bacteria for ammeline deamination was initially surprising but completely consistent with previous observations. First, it was previously shown that ammeline deamination activity is widespread in bacteria (37) and that guanine deaminase is also widespread. Second, analysis of gene regions identified genes for melamine and ammelide deamination (6, 7) but not ammeline deaminase. Third, previous studies showed significant accumulation of ammeline during bacterial melamine metabolism (14), consistent with metabolism by a set of enzymes that are not tightly controlled and coregulated. It had previously been considered possible that the melamine deaminase was the main ammeline-deaminating enzyme (6, 7, 14, 28). However, none of these studies used enzymes purified to homogeneity as was done in the present study. The new data presented here revealed that the specific activity of the melamine deaminase TriA with ammeline is quite minimal, 0.7 nmol per min per mg of protein, and 2 orders of magnitude less than the specific activity of guanine deaminase with ammeline (Table 1). Some bacteria, like E. coli, lack melamine-deaminating activity completely and only catalyze the transformation of ammeline to ammelide, which they do stoichiometrically (Fig. 4). These observations showing that ammeline deamination is widespread and separate from melamine and ammelide deamination reactions are completely consistent with the idea that ammeline is transformed by a housekeeping enzyme such as guanine deaminase.
The activity of guanine deaminase with ammeline was measured in the range of 57 to 73 nmol per min per mg of protein. However, B. japonicum USDA 110, from which the guanine deaminase catalyzes ammeline deamination in vitro, grew on ammeline as the sole nitrogen source (data not shown). Human guanine deaminase was found to be inactive with ammeline but was engineered for activity with this new substrate (21). The engineered human guanine deaminase had a reported specific activity, at saturating ammelide concentration, of 0.25 nmol per min per mg of protein, which is 2 orders of magnitude less than the activity of the wild-type human enzyme shown here with ammeline.
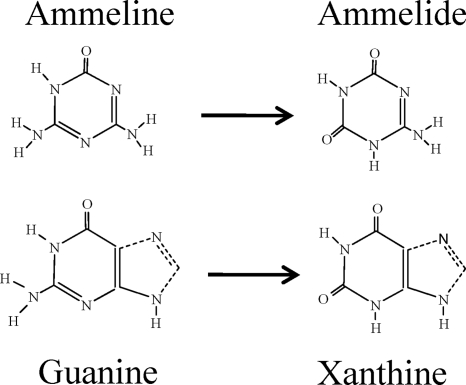
To compare the thermodynamic feasibility of guanine deaminase deaminating guanine or ammeline, one must consider the proper tautomeric form of each compound at physiological pH (Fig. 6). Ammeline had previously been considered to be a hydroxy-triazine (24), but more recent studies suggest that the keto tautomer, as depicted in Fig. 6, is the energetically favored form (15). The keto tautomer of guanine has been known to be the energetically favorable form in water at near neutral pH (10). In this context, the Gibbs free energy for deamination of the corresponding keto tautomers of guanine and ammeline can be analyzed computationally using the group contribution method pioneered by Mavrovouniotis (16, 17) and subsequently expanded upon by Jankowski et al. (13). The consensus calculation was that both deamination reactions had a calculated Gibbs free energy change of −5.7 kcal/mole. The comparative energetics of these two reactions, along with the structural similarities of the substrates (Fig. 6), illustrates how an enzyme evolved for activity with guanine could have a substantial promiscuous activity with ammeline.
FIG. 6.
Comparative tautomeric structures for ammeline and guanine and their reaction products, ammelide and xanthine, respectively. The ammeline (15) and guanine (10) tautomers shown are the predominant forms in aqueous solution at neutral pH.
The present study identified the unknown enzyme in the catabolic pathway from melamine to cyanuric acid (Fig. 1) and provided the basis for identifying genes encoding this enzyme. These data now enable screening for the presence of the genes in different environments. Large-scale metagenomic studies of microorganisms present in the mammalian intestine are currently under way, and these data could help to determine if melamine is transformed to cyanuric acid in the human and animal intestine, thus contributing to melamine toxicity. Regardless of this outcome, it is clear that guanine deaminases, which also act as ammeline deaminases, are widespread in the biological world. In this context, guanine deaminase was a preexisting enzyme that, through industrial activity and food adulteration, has become relevant to food toxicology and human health.
Acknowledgments
We thank Misha Mehta for help with phylogenetic analyses and tree construction and Naomi Kreamer and Erik Reynolds for supplying purified TriA and TrzN, respectively. We also acknowledge Martin Hammarström for supplying cloned human guaD and Nicholas P. Labelo for assisting with docking studies. Andrew Vail assisted with cloning B. japonicum USDA 110 guaD.
Footnotes
Published ahead of print on 18 December 2009.
REFERENCES
- 1.Baba, T., T. Ara, M. Hasegawa, Y. Takai, Y. Okumura, M. Baba, K. A. Datsenko, M. Tomita, B. L. Wanner, and H. Mori. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown, C. A., K. S. Jeong, R. H. Poppenga, B. Puschner, D. M. Miller, A. E. Ellis, K. Kang, S. Sum, A. M. Cistola, and S. A. Brown. 2007. Outbreaks of renal failure associated with melamine and cyanuric acid in dogs and cats in 2004 and 2007. J. Vet. Diagn. Invest. 19:525-531. [DOI] [PubMed] [Google Scholar]
- 3.Cole, M. A., and G. H. Elkan. 1973. Transmissible resistance to penicillin G, neomycin, and chloramphenicol in Rhizobium japonicum. Antimicrobiol. Agents Chemother. 4:248-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cook, A. M., H. Grossenbacher, and R. Hütter. 1984. Bacterial degradation of N-cyclopropylmelamine. The steps to ring cleavage. Biochem. J. 222:315-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dobson, R., S. Motlagh, M. Quijano, T. Cambron, A. Pullen, B. Regg, A. Gigalow-Kern, T. Vennard, A. Fix, R. Reimschuessel, G. Overmann, Y. Shan, and G. Daston. 2008. Identification and characterization of toxicity of contaminants in pet food leading to an outbreak of renal toxicity in cats and dogs. Toxicol. Sci. 106:251-262. [DOI] [PubMed] [Google Scholar]
- 6.Eaton, R. W., and J. S. Karns. 1991a. Cloning and analysis of s-triazine catabolic genes from Pseudomonas sp strain NRRLB-12227. J. Bacteriol. 173:1215-1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eaton, R. W., and J. S. Karns. 1991b. Cloning and comparison of the DNA encoding ammelide aminohydrolase and cyanuric acid amidohydrolase from three s-triazine-degrading bacterial strains. J. Bacteriol. 173:1363-1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fernández, J. R., W. J. Welsh, and B. L. Firestein. 2008. Structural characterization of the zinc binding domain in cytosolic PSD-95 interactor (cypin): role of zinc binding in guanine deamination and dendrite branching. Proteins 70:873-881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Friesner, R. A., J. L. Banks, R. B. Murphy, T. A. Halgren, J. J. Klicic, D. T. Mainz, M. P. Repasky, E. H. Knoll, M. Shelley, J. K. Perry, D. E. Shaw, P. Francis, and P. S. Shenkin. 2004. Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 47:1739-1749. [DOI] [PubMed] [Google Scholar]
- 10.Gorb, L., A. Kaczmarek, A. Gorb, A. Sadlej, and J. Leszczynski. 2005. Thermodynamics and kinetics of intramolecular proton transfer in guanine. Post Hartree-Fock Study. J. Phys. Chem. B 109:13770-13776. [DOI] [PubMed] [Google Scholar]
- 11.Grases, F., A. Costa-Bauzá, I. Gomila, S. Serra-Trespalle, F. Alonso-Sainz, and J. M. del Valle. 2009. Melamine urinary bladder stone. Urology 73:1262-1263. [DOI] [PubMed] [Google Scholar]
- 12.Ingelfinger, J. R. 2008. Melamine and the global implications of food contamination. N. Engl. J. Med. 2008:2745-2748. [DOI] [PubMed] [Google Scholar]
- 13.Jankowski, M. D., C. S. Henry, L. J. Broadbelt, and V. Hatzimanikas. 2008. Group contribution method for thermodynamic analysis of complex metabolic networks. Biophys. J. 95:1487-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jutzi, K., A. M. Cook, and R. Hütter. 1982. The degradative pathway of the s-triazine melamine Biochem. J. 208:679-684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lotsch, B. V., and W. Schnick. 2006. Synthesis and structural characterization of the ammelinium salts [C3H6N5O]Cl, [C3H6N5O]Br, and [C3H6N5O]NO3. Z. Anorg. Allg. Chem. 632:1457-1464. [Google Scholar]
- 16.Mavrovouniotis, M. 1990. Group contributions for estimating standard Gibbs energies of formation of biochemical compounds in aqueous solution. Biotechnol. Bioeng. 36:1070-1082. [DOI] [PubMed] [Google Scholar]
- 17.Mavrovouniotis, M. 1991. Estimation of standard Gibbs energy changes of biotransformations. J. Biol. Chem. 266:1444-1445. [PubMed] [Google Scholar]
- 18.Maynes, J. T., R. G. Yuan, and F. F. Snyder. 2000. Identification, expression, and characterization of Escherichia coli guanine deaminase. J. Bacteriol. 182:4658-4660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morris, J. H., C. C. Huang, P. C. Babbitt, and T. E. Ferrin. 2007. structureViz: linking Cytoscape and UCSF Chimera. Bioinformatics 23:2345-2347. [DOI] [PubMed] [Google Scholar]
- 20.Mulbry, W. W. 1994. Purification and characterization of an inducible s-triazine hydrolase from Rhodococcus corallinus NRRL B-15444R. Appl. Environ. Microbiol. 60:613-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murphy, P. M., J. M. Bolduc, J. L. Gallaher, B. L. Stoddard, and D. Baker. 2009. Alteration of enzyme specificity by computational loop remodeling and design. Proc. Natl. Acad. Sci. U. S. A. 106:9215-9220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Okamura, T., and K. Kigasawa. 1994. Re-evalution of the colorimetric assay for cytidine deaminase activity. Prenatal Diagn. 14:213-218. [DOI] [PubMed] [Google Scholar]
- 23.Pegg, S. C., S. Brown, S. Ojha, J. Seffernick, E. C. Meng, J. H. Morris, P. J. Chang, C. C. Huang, T. E. Ferrin, and P. C. Babbitt. 2006. Leveraging enzyme structure-function relationships for functional inference and experimental design: the structure-function linkage database. Biochemistry 45:2545-2555. [DOI] [PubMed] [Google Scholar]
- 24.Plass, M., A. Kristl, and M. H. Abraham. 1999. Spectroscopic investigations of the tautomeric equilibria in guanine derivatives of acyclovir. J. Chem. Soc. Perkin Trans. I 2:2641-2646. [Google Scholar]
- 25.Puschner, B., R. Poppenga, L. Lowenstine, M. Filigenzi, and P. Pesavento. 2007. Assessment of melamine and cyanuric acid in cats. J. Vet. Diagn. Invest. 19:616-624. [DOI] [PubMed] [Google Scholar]
- 26.Sadowsky, M., R. Tully, P. Cregan, and H. Keyser. 1987. Genetic diversity in Bradyrhizobium japonicum serogroup 123 and its relation to genotype-specific nodulation of soybeans. Appl. Environ. Microbiol. 53:2624-2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seffernick, J. L., A. Aleem, J. P. Osborne, G. Johnson, M. J. Sadowsky, and L. P. Wackett. 2007. Hydroxyatrazine N-ethylaminohydrolase (AtzB): an amidohydrolase superfamily enzyme catalyzing deamination and dechlorination. J. Bacteriol. 189:6989-6997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seffernick, J. L., M. L. de Souza, M. J. Sadowsky, and L. P. Wackett. 2001. Melamine deaminase and atrazine chlorohydrolase: 98 percent identical but functionally different J. Bacteriol. 183:2405-2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seffernick, J. L., S. K. Samanta, T. M. Louie, L. P. Wackett, and M. Subramanian. 2009. Investigative mining of sequence data for novel enzymes: a case study with nitrilases J. Biotechnol. 143:17-26. [DOI] [PubMed] [Google Scholar]
- 30.Seffernick, J. L., N. Shapir, M. Schoeb, G. Johnson, M. J. Sadowsky, and L. P. Wackett. 2002. Enzymatic degradation of chlorodiamino-s-triazine. Appl. Environ. Microbiol. 68:4672-4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seffernick, J. L., and L. P. Wackett. 2001. Rapid evolution of bacterial catabolic enzymes: a case study with atrazine chlorohydrolase. Biochemistry 40:12747-12753. [DOI] [PubMed] [Google Scholar]
- 32.Seibert, C. M., and F. M. Raushel. 2005. Structural and catalytic diversity within the amidohydrolase superfamily. Biochemistry 44:6383-6391. [DOI] [PubMed] [Google Scholar]
- 33.Seto, C. T., and G. M. Whitesides. 1990. Self-assembly based on the cyanuric acid-melamine lattice. J. Am. Chem. Soc. 112:6409-6411. [Google Scholar]
- 34.Shapir, N., J. P. Osborne, G. Johnson, M. J. Sadowsky, and L. P. Wackett. 2002. Purification, substrate range, and metal center of AtzC: the N-isopropylammelide aminohydrolase involved in bacterial atrazine metabolism. J. Bacteriol. 184:5376-5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shapir, N., C. Pedersen, O. Gil, L. Strong, J. Seffernick, M. J. Sadowsky, and L. P. Wackett. 2006. TrzN from Arthrobacter aurescens TC1 is a zinc amidohydrolase. J. Bacteriol. 188:5859-5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wackett, L. P., M. J. Sadowsky, B. Martinez, and N. Shapir. 2002. Biodegradation of atrazine and related triazine compounds: From enzymes to field studies. Appl. Microbiol. Biotechnol. 58:39-45. [DOI] [PubMed] [Google Scholar]
- 37.Zeyer, J., J. Bodmer, and R. Hütter. 1981. Microbial degradation of ammeline. Zentralbl. Bakteriol. Mikrobiol. Hyg. A 2:289-298. [Google Scholar]