Abstract
The widely conserved second messenger cyclic diguanosine monophosphate (c-di-GMP) plays a key role in quorum-sensing (QS)-dependent production of virulence factors in Xanthomonas campestris pv. campestris. The detection of QS diffusible signal factor (DSF) by the sensor RpfC leads to the activation of response regulator RpfG, which activates virulence gene expression by degrading c-di-GMP. Here, we show that a global regulator in the X. campestris pv. campestris QS regulatory pathway, Clp, is a c-di-GMP effector. c-di-GMP specifically binds to Clp with high affinity and induces allosteric conformational changes that abolish the interaction between Clp and its target gene promoter. Clp is similar to the cyclic AMP (cAMP) binding proteins Crp and Vfr and contains a conserved cyclic nucleotide monophosphate (cNMP) binding domain. Using site-directed mutagenesis, we found that the cNMP binding domain of Clp contains a glutamic acid residue (E99) that is essential for c-di-GMP binding. Substituting the residue with serine (E99S) resulted in decreased sensitivity to changes in the intracellular c-di-GMP level and attenuated bacterial virulence. These data establish the direct role of Clp in the response to fluctuating c-di-GMP levels and depict a novel mechanism by which QS links the second messenger with the X. campestris pv. campestris virulence regulon.
The nucleotide signaling molecule cyclic diguanosine monophosphate (c-di-GMP) has recently emerged as a widely conserved second messenger implicated in the regulation of various biological functions in bacteria, including cellulose biosynthesis (24), bacterial motility (27), biofilm formation (12), and extracellular virulence factor production (25). The intracellular level of c-di-GMP is influenced by the opposite activities of the diguanylate cyclases via GGDEF domain proteins that synthesize c-di-GMP and the phosphodiesterases via EAL or HD-GYP domain proteins that degrade this signaling molecule (15). In accordance with the central role of c-di-GMP in bacterial physiology, the three domains associated with c-di-GMP metabolism (GGDEF, EAL, and HD-GYP) are distributed broadly in a wide range of bacterial species (3, 15, 23). Notably, these domains are commonly linked to various signal sensing or detection domains, suggesting that various environmental cues modulate bacterial physiology by influencing the rate of c-di-GMP synthesis and hydrolysis.
The characterization of proteins that sense and detect changes in the c-di-GMP level is key to understanding how this ubiquitous second messenger modulates such diverse biological functions. Several types of c-di-GMP receptors have been unveiled in recent years, including several enzymes and proteins containing a PilZ domain (1), PleD and PelD containing the GGDEF domain with an RXXD motif (2, 19), and LapD containing a degenerated EAL domain (22). These effectors are not directly involved in transcriptional control because they do not contain DNA binding domains. Recently, the transcription factor FleQ of Pseudomonas aeruginosa, which interacts with c-di-GMP, was reported. FleQ suppresses extracellular polysaccharide (EPS) biosynthesis by binding to the pel promoter, and this binding is inhibited by c-di-GMP (11).
c-di-GMP is implicated in the regulation of virulence in Xanthomonas campestris pv. campestris. The pathogen produces a range of extracellular virulence factors, including protease, cellulose, pectinase, and EPS (3, 10). The production of these virulence factors is regulated by the diffusible signal factor (DSF)-mediated quorum-sensing (QS) mechanism and the RavS/RpsR two-component system. In the QS system, detection of the DSF involves the sensor RpfC, which transduces the QS signal to its cognate response regulator, RpfG, through a conserved phosphorelay mechanism (8). Activated RpfG functions as a phosphodiesterase, degrading c-di-GMP to GMP (25). In the RavS/RpsR system, the sensor RavS contains a PAS domain implicated in hypoxia sensing, whereas the response regulator RavR acts as a c-di-GMP-degrading phosphodiesterase (6). In both systems, c-di-GMP degradation results in enhanced transcriptional expression of the global regulator Clp, which positively regulates virulence factor production (6, 7). However, how Clp detects and responds to the RpfG- and RavR-mediated changes of the steady-state level of c-di-GMP is unclear.
Clp is a global regulator that shares a conserved cyclic nucleotide monophosphate (cNMP) binding domain with the well-characterized cyclic AMP (cAMP) nucleotide receptors Crp of Escherichia coli and Vfr of Pseudomonas aeruginosa (7). Peptide sequence alignment has revealed that Clp differs from Crp and Vfr in only one or two of the six conserved residues implicated in cAMP binding. In this study, we demonstrate that Clp interacts with c-di-GMP with high affinity and that this interaction abolishes the ability of Clp to bind to its target gene promoter. We also demonstrate that the single amino acid variation (E99) in the cNMP binding domain of Clp, which distinguishes Clp from Crp (corresponding residue S83) and Vfr (corresponding residue S88), is the key residue defining the ability of Clp to interact with c-di-GMP. The cNMP binding domain protein from Stenotrophomonas maltophilia, which is homologous to Crp and Vfr but shares the conserved E99 residue with Clp, also binds c-di-GMP but not cAMP. The evidence suggests that a subfamily of cNMP binding domain proteins may be c-d-GMP effectors, with the conserved E99 residue as a signature amino acid residue.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. X. campestris pv. campestris strains were grown at 30°C in YEB medium (29), unless otherwise stated. E. coli strains were grown at 37°C in LB medium. Antibiotics were added at the following concentrations when required: 100 μg/ml kanamycin, 50 μg/ml rifampin, and 10 μg/ml tetracycline. X-Gluc (5-bromo-4-chloro-3-indolyl-β-glucopyranoside) was included in the medium (60 μg/ml) to detect β-glucuronidase (GUS) activity. The synthesis of DSF was described previously (29). The signaling molecule was added to the medium at a final concentration of 5 μM when necessary.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmida | Properties or characteristics | Source or reference |
|---|---|---|
| X. campestris pv. campestris strains | ||
| WT | Wild-type strain 8004, Rifr | Max Dow |
| FE58 | DSF reporter strain | 29 |
| ΔrpfF strain | rpfF in-frame deletion mutant of strain 8004 | 9 |
| Δclp strain | clp in-frame deletion mutant of strain 8004 | Present study |
| ΔrpfG strain | rpfG in-frame deletion mutant of strain 8004 | Present study |
| WT(pDSK) strain | Strain 8004 harboring plasmid pDSK519, Rifr, Kanr | Present study |
| WT(PA5487) strain | Strain 8004 harboring construct pDSK-PA5487, Rifr, Kanr | Present study |
| ΔrpfG(pDSK) strain | ΔrpfG mutant harboring plasmid pDSK519, Rifr, Kanr | Present study |
| ΔrpfG(PA3947) strain | ΔrpfG mutant harboring construct pDSK-PA3947, Rifr, Kanr | Present study |
| ΔrpfG(clp) strain | ΔrpfG mutant harboring construct pDSK-clp, Rifr, Kanr | Present study |
| Δclp[clp(E99S)] strain | Δclp mutant harboring construct pDSK-clp(E99S), Rifr, Kanr | Present study |
| Δclp[clp(T149S)] strain | Δclp mutant harboring construct pDSK-clp(T149S), Rifr, Kanr | Present study |
| Δclp(clp) strain | Δclp mutant harboring construct pDSK-clp, Rifr, Kanr | Present study |
| E99S strain | Strain 8004 derivative with clp replaced in situ by clp(E99S) | Present study |
| T149S strain | Strain 8004 derivative with clp replaced in situ by clp(T149S) | Present study |
| E99S(pDSK) strain | E99S mutant harboring plasmid pDSK519, Rifr, Kanr | Present study |
| E99S(PA5487) strain | E99S mutant harboring vector pDSK-PA5487, Rifr, Kanr | Present study |
| T149S(pDSK) strain | T149S mutant harboring plasmid pDSK519, Rifr, Kanr | Present study |
| T149S(PA5487) strain | T149S mutant harboring vector pDSK-PA5487, Rifr, Kanr | Present study |
| E. coli strains | ||
| DH5α | supE44 ΔlacU169(φ80lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | Laboratory collection |
| BL21 | F−ompT hsdSB(rB− mB−) gal dcm λ(DE3) | Laboratory collection |
| RK2013 | Triparental mating helper strain, Kanr | Laboratory collection |
| Plasmids | ||
| pDSK519 | Broad-host-range cloning vector, Kanr | Laboratory collection |
| pGEX-6P-1 | GST fusion protein expression vector, Ampr | H. W. Song |
| pGEM-4-GUS | Plasmid pGEM-4 containing promoterless gusA, Ampr | Present study |
| pLAFR3 | Broad-host-range cloning vector, Tecr | Laboratory collection |
| pGEMT-easy | Plasmid vector for cloning PCR fragment, Ampr | Promega |
| pK18mobsacB | Sucrose-sensitive suicide plasmid, Kanr | Laboratory collection |
| pL3PengXCA-GUS | DSF sensor with gusA under the control of the engXCA promoter | 29 |
| pDSK-clp | clp placed under the control of the lac promoter in vector pDSK519, Kanr | Present study |
| pDSK-clp(E99S) | clp(E99S) placed under the control of the lac promoter in vector pDSK519, Kanr | Present study |
| pDSK-clp(T149S) | clp(T149S) placed under the control of the lac promoter in vector pDSK519, Kanr | Present study |
| pDSK-PA3947 | PA3947 placed under the control of the lac promoter in vector pDSK519, Kanr | Present study |
| pDSK-PA5487 | PA5487 placed under the control of the lac promoter in vector pDSK519, Kanr | Present study |
| pGEM-clp | clp cloned in pGEMT-easy, Ampr | Present study |
| pGEX-clp | clp cloned in vector pGEX-6P-1 for protein purification, Ampr | Present study |
| pGEX-Smclp | clp homologue from Stenotrophomonas maltophilia cloned in vector pGEX-6P-1 for protein purification, Ampr | Present study |
| pGEX-clp(E99S) | clp(E99S) cloned in vector pGEX-6P-1 for protein purification, Ampr | Present study |
| pGEX-clp(T149S) | clp(T149S) cloned in vector pGEX-6P-1 for protein purification, Ampr | Present study |
| pK-clp(E99S) | clp(E99S) cloned in vector pK18mobsacB, Kanr | Present study |
| pK-clp(T149S) | clp(T149S) cloned in vector pK18mobsacB, Kanr | Present study |
WT, wild type.
Generation of in-frame deletion mutants and in trans expression constructs.
A spontaneous rifampin-resistant derivative of strain 8004 was used as the parental strain for generating deletion mutants. In-frame deletion mutants of rpfG and clp were generated using the primers listed in Table S1 in the supplemental material according to previously described methods (9). For in trans expression, the coding regions of the corresponding proteins were amplified by PCR using the primers listed in Table S1 and cloned separately under the control of the lac promoter in the expression vector pDSK519. The resulting constructs were transferred into X. campestris pv. campestris strains through triparental mating.
Quantitative determination of GUS activity and EPS production.
Bacterial cells were collected by centrifugation at 14,000 rpm for 10 min, and the total soluble protein was prepared by sonication. GUS enzyme activity was determined quantitatively according to the standard protocol (14). The substrate 4-methylumbelliferyl-β-d-gucuronide was used for fluorometric assays on a Hitachi F1000 fluorescence spectrophotometer (Sigma), with excitation at 365 nm and emission at 455 nm. The data are reported as the averages from at least three independent repeats and defined as picomoles of methyl umbelliferone produced per minute per microgram of total soluble protein.
To analyze EPS production, supernatants were collected from overnight bacterial cultures (10 ml, optical density at 600 nm [OD600] = 2.0) after centrifugation, as described above. Two volumes of absolute ethanol were added to the supernatants, and the mixtures were kept at −20°C for a half hour. The precipitated EPS molecules were spun down and dried at 55°C overnight before determining the dry weight.
RNA extraction and RT-PCR analysis.
Total RNA was isolated from fresh bacterial cultures. Briefly, bacterial cells were harvested at an OD600 of 2.0 by centrifugation at 4°C for 4 min at 10,000 rpm. RNA was purified using RNeasy minicolumns (Qiagen) according to the manufacturer's protocol. After digestion with DNase I (Promega) to remove contaminating genomic DNA residues, the RNA samples were repurified using an RNeasy column to remove excess enzyme. The quantity and purity of the RNA were determined by agarose gel electrophoresis and spectrometry. Reverse transcriptase PCR (RT-PCR) analysis was performed using the OneStep RT-PCR kit (Qiagen) according to the manufacturer's instructions. The primers used for RT-PCR analysis are listed in Table S1 in the supplemental material. The density of the PCR band was determined using Image J (http://rsb.info.nih.gov/ij/).
Site-directed mutagenesis of clp and in situ substitution.
The site-directed mutagenesis of clp was performed using the QuickChange site-directed mutagenesis kit following the manufacturer's protocol (Stratagene). The coding region was amplified using the primers Clp-F2 and Clp-R2 (Table S1 in the supplemental material) and cloned into the vector pGEMT-easy to generate pGEM-clp, which was used for subsequent site-directed mutagenesis. Based on the sequence alignment with Crp and Vfr, residues E99 and T149 of Clp were changed to serine using the primer pairs E99S-FOR/E99S-REV and T149S-FOR/T149S-REV (Table S1), respectively. After verification by DNA sequencing, the mutated clp alleles were ligated with pDSK519 to generate expression constructs pDSK-clp(E99S) and pDSK-clp(T149S), respectively, for in trans expression in X. campestris pv. campestris.
For in situ substitution of wild-type clp in X. campestris pv. campestris, the clp alleles were subcloned into the SmaI site of suicide vector pK18mobsacB. Resulting constructs pK-clp(E99S) and pK-clp(T149S) were separately mobilized into X. campestris pv. campestris wild-type strain 8004 by triparental mating. The transformants were selected on LB medium supplemented with rifampin and kanamycin. A second selection to remove the vector was performed on YEB medium containing 5% (wt/vol) sucrose and rifampin. The corresponding substitutions in the X. campestris pv. campestris genome were confirmed by PCR amplification of the Clp coding region and DNA sequencing.
In trans expression of c-di-GMP metabolic enzymes.
To modulate the c-di-GMP level in X. campestris pv. campestris strains, PA5487 and PA3947, which encode a diguanylate cyclase and a phosphodiesterase (16), respectively, were amplified from Pseudomonas aeruginosa strain PAO1 using the primer pairs PA5487-FOR/PA5487-REV and PA3947-FOR/PA3947-REV (see Table S1 in the supplemental material), respectively. After digestion with XbaI and EcoRI, the PCR products were cloned under the control of the lac promoter in the plasmid vector pDSK519 to generate expression constructs pDSK-PA5487 and pDSK-PA3947, respectively. The constructs were verified by DNA sequencing and mobilized separately into X. campestris pv. campestris strains by triparental mating. The resulting transformants were selected on LB medium supplemented with rifampin and kanamycin.
Protein purification.
To purify Clp; the Clp E99S and Clp T149S variants; and ClpSm, the Clp homologue from S. maltophilia, their coding regions were amplified using the corresponding primer pairs listed in Table S1 in the supplemental material and cloned into the expression vector pGEX-6p-1 (Novagen). The resulting constructs (pGEX-clp, pGEX-clp(E99S), pGEX-clp(T149S), and pGEX-Smclp; Table S1) were transformed into host strain E. coli BL21. After sequencing confirmation, these four proteins were expressed and purified separately using glutathione Sepharose 4B columns according to the procedures recommended by the manufacturer (Amersham). Protein purity was checked by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) followed by Coomassie blue staining. Purified proteins were dialyzed against 20 mM Tris buffer (pH 7.3) containing 50 mM NaCl using 3,000 molecular-weight-cutoff centrifugal filter devices (Millipore, Bedford, MA). The samples were stored as aliquots in 50% (vol/vol) glycerol at −20°C until use.
EMSA.
DNA probes for electrophoretic mobility shift assays (EMSA) were obtained by PCR amplification using the primer pairs listed in Table S1 in the supplemental material. The PCR fragments were purified using the QIAQuick PCR purification kit (Qiagen) and labeled using a digoxigenin (DIG)-end-labeling kit when necessary. The DNA-protein binding reaction was performed according to the manufacturer's instructions (Roche). A 6% polyacrylamide gel was used for separation of the DNA-protein complex. After UV cross-linking, the DIG-labeled probes were detected in the membrane using a DIG luminescent detection kit (Roche).
CD spectroscopy and ITC.
Far-UV circular dichroism (CD) analysis of Clp was carried out on a JASCO J-810 spectropolarimeter as previously described (30). Clp and c-di-GMP were added at a final concentration of 20 μM. The isothermal titration calorimetry (ITC) measurements were obtained using a VP-IPC calorimeter following the manufacturer's protocol (MicroCal, Northampton, MA). In brief, 5-μl aliquots of nucleotide solution (500 μM) were injected at 5-min intervals via a 300-μl syringe into the sample cell containing 1.4 ml of Clp or its homologue (20 μM), with constant stirring at 290 rpm, and the heat changes accompanying these additions were recorded. The protein samples were extensively dialyzed against Tris-HCl buffer (20 mM Tris-HCl, pH 7.3; 50 mM NaCl) before titration. The nucleotide solution was prepared directly in Tris-HCl buffer, and the titrate solutions were degassed under a vacuum prior to loading for ITC. The titration experiment was repeated at least twice, and the data were calibrated with a buffer control and fitted with the one-site model to determine the binding constant (Ka) using the MicroCal ORIGIN version 7.0 software.
Virulence assay.
The virulence assay was conducted by the inoculation of X. campestris pv. campestris strains onto Chinese cabbage following the vacuum infiltration method with minor modifications (4). Briefly, cabbage stem tissue was cut into pieces approximately 2 by 2 cm in size, which were then immersed in an overnight culture of bacterial suspension (OD600 = 2.0) prior to 2-min vacuum exposure to facilitate bacterial infiltration. The treated stem tissues were then transferred to an incubator at 30°C, with humidity at around 90%. Symptom development was monitored daily, and the photographs were taken 4 days later. The experiment was repeated at least three times in duplicate.
RESULTS
c-di-GMP is a specific Clp ligand.
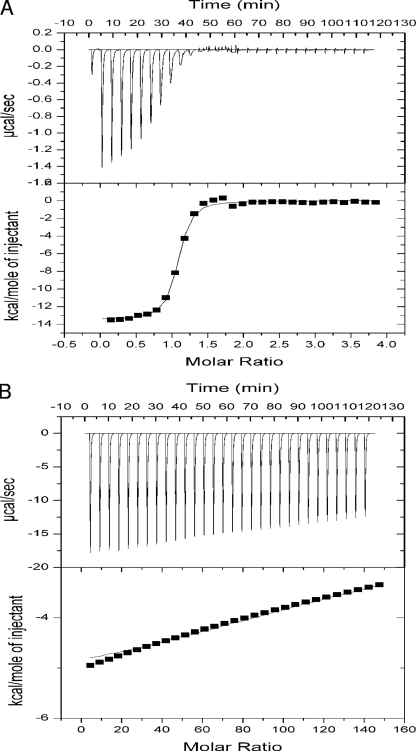
The available evidence suggests that Clp is a global transcriptional regulator located downstream of the RpfC/RpfG two-component system in the DSF pathway (7). Based on this model, the intracellular level of DSF is under a threshold level at low cell density, which favors c-di-GMP accumulation, as its degradation enzyme RpfG exists in its inactive form. On the other hand, at high cell density, the accumulated DSF activates RpfG through the sensor RpfC, resulting in c-di-GMP degradation and the accumulation of GMP. To understand the mechanism by which Clp and c-di-GMP act in the DSF signaling pathway, we tested whether Clp can detect c-di-GMP or its related nucleotides as ligands using ITC. Clp was expressed as a glutathione S-transferase (GST) fusion protein, which was used in subsequent analysis because the solubility of recombinant Clp that is separated from GST is poor. In the ITC analysis, the Clp fusion protein solution was titrated with c-di-GMP and its derivatives at 20°C, and the heat released upon binding measured. The results showed a strong interaction between the Clp fusion protein and c-di-GMP (Fig. 1A). As expected, no physical interaction was observed between the control protein, GST, and c-di-GMP (data not shown). The binding isotherm data suggested that Clp tightly binds c-di-GMP in 1:1 stoichiometry, with an estimated dissociation constant (Kd) of 1.61 ± 0.22−7 M. In contrast, the addition of cAMP, even at a level 20-fold higher than that of the Clp fusion protein, did not cause a heat release similar to that of c-di-GMP (Fig. 1B). Similarly, no molecular interaction was observed between Clp and GMP or GTP (see Fig. S1A and S1B in the supplemental material), indicating that they are not the cognate signal ligands of Clp.
FIG. 1.
ITC analysis of c-di-GMP (A) and cAMP (B) binding to Clp. The top of each panel shows the ITC titration of 20 μM Clp with 5-μl aliquots of 500 μM c-di-GMP or 10 mM cAMP in Tris-HCl buffer at 20°C. The bottom of each panel shows the binding isotherm for the titration represented in the top of the panel; the solid line is the best fit of the data to a one-site binding model.
c-di-GMP inhibits the formation of the protein-promoter complex consisting of Clp and its target promoter PengXCA.
Clp is a global regulator with a DNA binding domain at its C terminus (7). To further analyze the impact of c-di-GMP on Clp functionality, we amplified the promoter region of engXCA and performed an EMSA. engXCA encodes an extracellular endoglucanase (5) that contributes to X. campestris pv. campestris virulence through its cellulolytic activity. Previous analyses showed that engXCA belongs to the DSF and Clp regulon (7, 9) and is under the direct control of Clp (13). When purified Clp fusion protein was incubated with a DIG-labeled engXCA promoter (PengXCA) fragment (382 bp), protein-DNA complexes formed in a Clp dosage-dependent manner (see Fig. S2A in the supplemental material). In contrast, no band shift was observed with the negative control-lacking protein or with GST (Fig. S2A in the supplemental material). The specificity of Clp fusion protein binding to PengXCA was further verified using competitor DNA. The intensity of the shifted band was diminished in the presence of unlabeled PengXCA (see Fig. S2B in the supplemental material) but not in the presence of the unlabeled prt1 promoter (Fig. S2B). A previous study showed that prt1, which encodes a protease, is not directly regulated by Clp (13). Under the same experimental conditions, we tested whether the ligand c-di-GMP influences the formation of the protein-promoter complex consisting of Clp and PengXCA. The addition of c-di-GMP, at either 10 μM or 100 μM, abolished the ability of Clp to form a complex with its target promoter (Fig. 2A). Consistent with the results of the ITC analysis, the addition of GMP and GTP, up to 100 μM, had no effect on the Clp-promoter interaction (Fig. 2A).
FIG. 2.
The effect of c-di-GMP on the conformational structure and promoter binding activity of Clp. (A) EMSA analysis of the impact of c-di-GMP on Clp binding to the engXCA promoter. DIG-labeled promoter fragments were incubated with purified Clp proteins in the presence or absence of nucleotide, as indicated. The final concentration of nucleotide in the reaction mixture was 10 μM or 100 μM. (B and C) Far-UV CD spectra of Clp and the control, glutathione-S-transferase (GST), in the presence or absence of c-di-GMP, as indicated. deg, degrees; com2, cm2.
c-di-GMP acts as an allosteric regulator of Clp.
To understand how c-di-GMP might inhibit the formation of the Clp-promoter complex, we used CD spectroscopy to determine the effect of c-di-GMP on the conformational structure of Clp. The CD spectrum of the Clp protein showed intensive negative ellipticity from 200 nm to 230 nm (Fig. 2B), suggesting a large unordered contribution and a small, but detectable, contribution of the α-helical structure (32). In contrast, the addition of c-di-GMP resulted in notable conformational changes in Clp, including decreased intensity of the negative ellipticity from 200 nm to 230 nm and a shifted peak of positive ellipticity (Fig. 2B), indicating reduced content of the α-helical structure and increased β-sheet content, respectively (32). As a negative control, the addition of c-di-GMP did not alter the CD spectra of the GST protein (Fig. 2C). Taken together, the results demonstrate that c-di-GMP binding induces allosteric conformational changes in Clp, which might consequently affect the affinity of the transcription factor to its target promoter. In addition, the findings suggest that c-di-GMP shares a mechanism of action with cAMP, which is known as the allosteric regulator of Crp (31).
Modulation of intracellular c-di-GMP levels alters the expression pattern of engXCA.
The above data suggest that c-di-GMP acts as an inhibitor of Clp-dependent virulence gene expression. To test this possibility, we fused the GUS coding region to the engXCA promoter, generating reporter construct pL3PengXCA-GUS. First, we tested the expression pattern of engXCA in the presence and absence of the functional Clp gene. The reporter construct pL3PengXCA-GUS was introduced into the clp in-frame deletion mutant, the Δclp mutant, and its wild-type strain 8004. The mutation of clp did not affect X. campestris pv. campestris growth (data not shown) but diminished the expression of engXCA (Fig. 3A). In agreement with the previous report (13), the data indicate that Clp is a key transcriptional factor in the regulation of engXCA expression under the experimental conditions used in this study.
FIG. 3.
Modulation of the intracellular level of c-di-GMP changes the expression pattern of engXCA. (A) The effect of rpfG and clp mutations on engXCA transcription in X. campestris pv. campestris (Xcc). (B) Effect of c-di-GMP synthase (PA5487) and degradation enzyme (PA3947) on engXCA expression. The genes for in trans expression were cloned under the control of the lac promoter in expression vector pDSK519 (pDSK); the empty vector was introduced into wild-type strain 8004 (WT) and its ΔrpfG deletion mutant as controls. (C) RT-PCR analysis of engXCA with the strains described above. The relative signal intensity for each strain was derived after normalization against the corresponding 16S rRNA loading control. In panels A and B, the data are the means of three repeats, and the error bars indicate standard deviation.
Next, we determined whether a fluctuation in the intracellular level of c-di-GMP affects the transcriptional expression of the engXCA operon in X. campestris pv. campestris. For this purpose, we cloned the genes PA5487 and PA3947 from Pseudomonas aeruginosa for in trans expression in X. campestris pv. campestris. Previously, PA5487 was shown to encode a diguanylate cyclase that synthesizes c-di-GMP, and PA3947 encodes a phosphodiesterase that degrades the second messenger (16). In the wild-type X. campestris pv. campestris carrying the vector pDSK519 alone, roughly 2,500 units of β-glucuronidase was detected. In contrast, in trans expression of the c-di-GMP synthase encoded by PA5487 resulted in an approximately 10-fold reduction in engXCA transcription in X. campestris pv. campestris (Fig. 3B), which is consistent with the notion that high cellular c-di-GMP levels suppress virulence gene expression. To test the impact of decreased c-di-GMP levels, we used the RpfG null mutant, the ΔrpfG mutant, which should have an intracellular c-di-GMP level higher than that of wild-type X. campestris pv. campestris (25). Consistent with this expectation, increased engXCA expression was observed with the ΔrpfG mutant expressing the c-di-GMP degradation enzyme encoded by PA3947 compared to the control strain, the ΔrpfG mutant containing the vector (Fig. 3B). RT-PCR analysis confirmed that in trans expression of PA3947 in the strain carrying ΔrpfG resulted in increased engXCA transcription (Fig. 3C).
The conserved amino acid residues implicated in the cNMP binding domain are required for the full activity of Clp.
Peptide sequence alignment revealed that Clp shares approximately 45% and 48% of its amino acids with Crp and Vfr, respectively. Notably, of the six amino acid residues implicated in Crp binding to cAMP (18, 30), four are conserved in Clp (G87, E88, R98, and T148; the varied residues are E99 and T149) (7). Compared to Vfr, Clp differs in only one of these key residues (E99) (7). To test the impact of this variation in the cAMP binding motif on the transcription factor activity of Clp, we generated two clp alleles [clp(E99S) and clp(T149S)] by site-directed mutagenesis. Wild-type clp and its alleles were cloned under the control of the lac promoter in the plasmid vector pDSK519 for in trans expression. In trans expression of wild-type clp in the Δclp deletion mutant resulted in increased engXCA expression compared to the wild-type control, whereas expression of the clp variants in the same Δclp mutant only partially restored engXCA expression (Fig. 4A). In addition, a similar pattern was observed when we tested the impact of these Clp variants on EPS production (Fig. 4B), which is another trait of X. campestris pv. campestris regulated by the DSF and Clp regulon (7, 9). The data suggest that the key residues in the cNMP binding domain are associated with the full activity of Clp. Given that the transcription factor activity of Clp does not require the presence of c-di-GMP (13) and that the residues involved in cAMP binding are located at the interface of two subunits of the Crp dimer (20, 21), we speculate that the E99S and T149S substitutions may influence Clp dimer association or stability, which consequently affects its activity as a transcription factor.
FIG. 4.
The role of conserved amino acid residues implicated in cAMP binding in the functionality of Clp. (A) The effect of substituting the conserved amino acid residues on the regulatory activity of Clp in modulating engXCA expression. (B) The effect of substituting the conserved amino acid residues on the regulatory activity of Clp in modulating EPS production. The data are the means of three repeats, and the error bars indicate standard deviation.
Substitution of a key amino acid residue (E99) in the cNMP domain of Clp diminishes its response to c-di-GMP.
To determine the effect of a key amino acid variation in the cNMP domain on ligand binding, we purified the E99S and T149S variant proteins using the same method as that for Clp purification. In the absence of the c-di-GMP ligand, the E99S and T149S variants bound to the engXCA promoter in a manner similar to that of wild-type Clp; the T149S substitution did not seem to influence its response to the c-di-GMP ligand, as the addition of 5 μM or 50 μM c-di-GMP diminished the promoter binding activity of both Clp and the T149S variant (Fig. 5A). However, the E99S variant still formed a protein-DNA complex in the presence of c-di-GMP (Fig. 5A). The size of the E99S variant-DNA complex formed in the presence of c-di-GMP was smaller than that formed in the absence of a second messenger (Fig. 5A). One plausible explanation is that addition of c-di-GMP to the E99S variant substantially reduced its affinity for one of the two Clp binding sites present in the engXCA promoter (see Fig. S3 in the supplemental material) (13).
FIG. 5.
The impact of amino acid substitution on the sensitivity of Clp to its inhibitory ligand c-di-GMP. (A) EMSA analysis of Clp and its derivatives (25 nM) in the presence or absence of c-di-GMP. (B) Effect of in trans expression of c-di-GMP synthase (PA5487) on engXCA expression in X. campestris pv. campestris strains expressing wild-type Clp or substituted alleles. (C) The effect of in trans expression of c-di-GMP synthase (PA5487) on the virulence of X. campestris pv. campestris strains expressing wild-type Clp or substituted alleles against cabbage. The experiment was repeated three times, and the photograph, taken 4 days after inoculation, shows a representative set of results.
To verify the EMSA findings, we tested the effect of overexpression of the c-di-GMP synthase encoded by PA5487 of P. aeruginosa. We transferred the altered clp alleles [clp(E99S) and clp(T149S)] to the chromosome of strain 8004 to replace wild-type clp by in situ substitution and used the resulting X. campestris pv. campestris derivatives for further analysis. Increased c-di-GMP content due to the expression of PA5487 decreased engXCA expression in strains with wild-type clp or its allele clp(T149S) roughly 10-fold, whereas expression of PA5487 in the strain containing the clp(E99S) allele reduced engXCA expression only by less than 20% (Fig. 5B). Consistently, the virulence assay showed that production of c-di-GMP due to the expression of PA5487 in the strain with wild-type clp or its altered allele clp(T149S) resulted in a loss of virulence in cabbage, whereas the expression of PA5487 in the strain with the clp(E99S) allele failed to stop bacterial infection; the maceration symptom was less extensive without PA5487 compared to the E99S parental strain (Fig. 5C). Taken together, these data suggest that E99 is a key amino acid residue that specifies the ability of Clp to respond to c-di-GMP, whereas the substitution of T149 with serine affects only the transcription factor activity of Clp and has no influence on its response to the inhibitory ligand c-di-GMP.
To further confirm the interaction between the Clp E99S mutant and c-di-GMP, ITC was carried out. The Clp E99S fusion protein solution was titrated with c-di-GMP at 20°C, and the heat released upon binding was measured. However, no physical interaction was observed between the Clp E99S mutant and c-di-GMP (see Fig. S4 in the supplemental material).
The Clp homologue from Stenotrophomonas maltophilia also interacts with c-di-GMP.
Interestingly, the alignment of the amino acid sequences of Clp homologues revealed that these regulatory proteins can be grouped into two categories based on the amino acid aligned with E99 of Clp; one group shares the conserved residue serine (S), represented by Crp and Vfr, and the other shares the conserved residue glutamic acid (E), including a range of Clp homologues from various Xanthomonas species and Stenotrophomonas maltophilia (Fig. 6A). To test whether other proteins containing the conserved E99 residue also interact with c-di-GMP, we purified the Clp homologue (ClpSm) from S. maltophilia for ITC. ClpSm did not interact with cAMP but bound strongly to c-di-GMP, with a Kd of 2.16 ± 0.54−7 M (Fig. 6B and C).
FIG. 6.
The E99 residue of Clp may be a signature residue of c-di-GMP effectors with a cNMP binding domain. (A) Clp homologues can be grouped into two categories based on the signature amino acid E99. Colored shading indicates the conserved residues corresponding to E99 and T149 of Clp. XCV0519, Xanthomonas campestris pv. vesicatoria (NCBI accession no. CAJ22150); XAC0483, Xanthomonas axonopodis pv. citri (NCBI accession no. AAM35374); XOO_3933, Xanthomonas oryzae pv. oryzae MAFF 311018 (NCBI accession no. BAE70688); Clp, Xanthomonas campestris pv. campestris strain 8004 (NCBI accession no. AAY47667); XfasM23_0792, Xylella fastidiosa M23 (NCBI accession no. ACB92231); ClpSm, Stenotrophomonas maltophilia K279a (NCBI accession no. YP_001973974); PaerPA_01000634, Pseudomonas aeruginosa (NCBI accession no. ZP_01363536.1); Vfr, Pseudomonas aeruginosa PAO1 (NCBI accession no. AAG04041); Avin_46100, Azotobacter vinelandii DJ (NCBI accession no. ACO80719); PSPTOT1_1546, Pseudomonas syringae pv. tomato T1 (NCBI accession no. ZP_03395776); Crp, Escherichia coli strain K-12 substrain MG1655 (NCBI accession no. AAC76382). (B) ITC analysis of c-di-GMP (500 μM) binding to ClpSm. The dissociation constant (Kd) was estimated as 2.16 ± 5.4−7 M. (C) ITC analysis of ClpSm binding cAMP (1 mM).
DISCUSSION
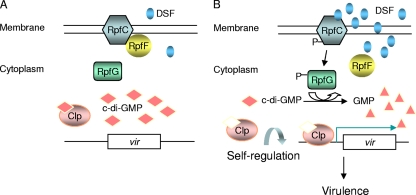
One of the intriguing puzzles in the X. campestris pv. campestris DSF-dependent QS system is the mechanism by which QS signal sensing is coupled to intracellular regulatory networks through the second messenger c-di-GMP and global regulator Clp (10). In this study, we present several lines of evidence that Clp is a novel regulator of the c-di-GMP response. First, Clp specifically binds c-di-GMP with a high affinity (Kd = 1.61 ± 0.22−7 M). Second, c-di-GMP inhibits the binding of Clp to the engXCA promoter. Third, c-di-GMP induces intensive allosteric conformational changes in Clp. Fourth, increasing the intercellular content of c-di-GMP by overexpressing c-di-GMP synthase downregulates virulence gene expression and attenuates bacterial virulence. These data, together with previous findings on the DSF-dependent QS, including the transcriptional self-regulation of Clp (7), depict a detailed DSF signaling model of the regulation of bacterial virulence (Fig. 7). In this model, a basal level of DSF is produced at a low cell density because RpfF is bound to RpfC (8); RpfG is in its inactive state, and c-di-GMP is maintained at a relatively high level that inactivates Clp by direct binding (Fig. 7A). When bacterial cells reach a high population density, the detection of DSF by RpfC results in the release of RpfF and phosphorylation of RpfG, which leads to increased DSF production and the degradation of c-di-GMP (7, 25). Consequently, the cellular level of free Clp increases, and the regulator acts as a positive transcription factor to induce its own gene transcription and virulence gene expression (Fig. 7B).
FIG. 7.
Schematic representation of the role of Clp and c-di-GMP in the DSF-mediated QS regulation of virulence at low (A) and high (B) population density. The Clp-dependent virulence regulon is represented by vir, which includes engXCA and other virulence genes, as depicted in our previous study (7).
While the manuscript was being prepared, a paper was published ahead of print that shows the inhibitory effect of c-di-GMP on the binding of the Clp homologue from Xanthomonas axonopodis pv. citri to the synthetic DNA fragment with the conserved Crp binding site (17). In addition to confirming that Clp of X. campestris pv. campestris is also an effector protein of c-di-GMP, this study has demonstrated the role of a key residue associated with ligand binding in the DSF-dependent modulation of virulence factor production under both in vitro and in vivo conditions. The identification of Clp as a c-di-GMP-responsive regulator has added a new member to the expanding superfamily of c-di-GMP effectors. Several types of c-di-GMP effectors have been identified in recent years, including PilZ, PleD, PelD, LapD, and FleQ (1, 11, 19, 26). Clp shares little homology with these known effector proteins at the amino acid level and differs from others in domain structure (data not shown). In contrast to the previously known c-di-GMP effectors, which require c-di-GMP for their functionality, Clp expands the regulatory mechanism of c-di-GMP by acting as a positive global regulator with c-di-GMP as an inhibitory ligand in the modulation of a large regulon, which includes over 300 genes, that encodes various biological functions (7). The identification of Clp as a c-di-GMP responsive global regulator provides new insight into how X. campestris pv. campestris can couple the signal inputs from various c-di-GMP metabolic mechanisms, such as RpfC/RpfG and RavS/RavR (6, 9, 28), to its virulence regulon.
Crystal structure analysis of the Crp-cAMP complex revealed that serine 83 (S83, the corresponding residue of E99 in Clp) within the conserved cNMP binding domain of Crp may play a key role in protein-ligand interactions by forming a hydrogen bond with the phosphate oxygen of cAMP through its functional hydroxyl group (20). Substitution of S83 in Crp with a similar amino acid, threonine (S83T), to retain a hydroxyl group for hydrogen bonding does not significantly affect the ability of Crp to respond to cAMP, whereas replacement of S83 with alanine, which has a methyl functional group and is unable to form a hydrogen bond with the ligand, abolishes Crp activity (21). Intriguingly, wild-type Clp contains a glutamic acid (E99) at the position corresponding to residue S83 of Crp. Glutamic acid is different from serine in that it contains a carbonyl functional group instead of a hydroxyl group and thus is unable to donate a hydrogen atom to hydrogen bond formation with phosphate oxygen. In this study, we found that the replacement of E99 with serine (E99S) in Clp deprives it of its ability to bind to c-di-GMP in vitro and results in a loss of sensitivity to c-di-GMP inhibition under in vivo conditions. These results establish the role of the conserved cNMP binding domain, which contains the residue E99, in the Clp and c-di-GMP interaction. The results also suggest that the role of residue E99 in ligand binding should be different from its counterpart Crp in cAMP binding due to the different physicochemical properties of their functional groups.
Furthermore, we provide evidence that the Clp-type c-di-GMP receptors are likely conserved widely in other bacterial species. The intriguing findings that Crp is the receptor for cAMP and Clp is the receptor for c-di-GMP encouraged us to conduct a sequence alignment of the proteins with cNMP binding domains (Fig. 6A) and biochemical analysis of the Clp homologue (ClpSm) from S. maltophilia (Fig. 6B). The results indicate that the amino acid residue at the position corresponding to E99 in Clp may play a key role in determining ligand specificity. The cAMP receptors, represented by Crp and Vfr, share the conserved residue serine (S), whereas the c-di-GMP receptors Clp and ClpSm share the conserved residue glutamic acid (E) (Fig. 6A). These findings suggest that the conserved residue E99 in Clp represents a signature residue of the c-di-GMP effectors with cNMP binding domains. The notion is further strengthened by the finding that the Clp homologue of Xanthomonas axonopodis pv. citri, which was recently shown to be a c-di-GMP effector (17), also contains the conserved E99 residue (Fig. 6A).
Supplementary Material
Acknowledgments
We thank Zhi-Hong Cheng, Chao Wang, and Sharon-Hee-Ming Ling for providing excellent technique assistance in the ITC analysis, as well as Qi-Hui Seet for help with EMSA.
This work was financially supported by the Biomedical Research Council, the Agency of Science, Technology, and Research (A*Star), Singapore.
Footnotes
Published ahead of print on 11 December 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1.Amikam, D., and M. Y. Galperin. 2006. PilZ domain is part of the bacterial c-di-GMP binding protein. Bioinformatics 22:3-6. [DOI] [PubMed] [Google Scholar]
- 2.Christen, B., M. Christen, R. Paul, F. Schmid, M. Folcher, P. Jenoe, M. Meuwly, and U. Jenal. 2006. Allosteric control of cyclic di-GMP signaling. J. Biol. Chem. 281:32015-32024. [DOI] [PubMed] [Google Scholar]
- 3.Dow, J. M., Y. Fouhy, J. F. Lucey, and R. P. Ryan. 2006. The HD-GYP domain, cyclic di-GMP signaling, and bacterial virulence to plants. Mol. Plant Microbe Interact. 19:1378-1384. [DOI] [PubMed] [Google Scholar]
- 4.Fett, W. F., and L. Sequeira. 1980. A new bacterial agglutinin from soybean. I. Isolation, partial purification, and characterization. Plant Physiol. 66:847-852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gough, C. L., J. M. Dow, J. Keen, B. Henrissat, and M. J. Daniels. 1990. Nucleotide sequence of the engXCA gene encoding the major endoglucanase of Xanthomonas campestris pv. campestris. Gene 89:53-59. [DOI] [PubMed] [Google Scholar]
- 6.He, Y. W., C. Boon, L. Zhou, and L. H. Zhang. 2009. Co-regulation of Xanthomonas campestris virulence by quorum sensing and a novel two-component regulatory system RavS/RavR. Mol. Microbiol. 71:1464-1476. [DOI] [PubMed] [Google Scholar]
- 7.He, Y. W., A. Y. Ng, M. Xu, K. Lin, L. H. Wang, Y. H. Dong, and L. H. Zhang. 2007. Xanthomonas campestris cell-cell communication involves a putative nucleotide receptor protein Clp and a hierarchical signalling network. Mol. Microbiol. 64:281-292. [DOI] [PubMed] [Google Scholar]
- 8.He, Y. W., C. Wang, L. Zhou, H. Song, J. M. Dow, and L. H. Zhang. 2006. Dual signaling functions of the hybrid sensor kinase RpfC of Xanthomonas campestris involve either phosphorelay or receiver domain-protein interaction. J. Biol. Chem. 281:33414-33421. [DOI] [PubMed] [Google Scholar]
- 9.He, Y. W., M. Xu, K. Lin, Y. J. Ng, C. M. Wen, L. H. Wang, Z. D. Liu, H. B. Zhang, Y. H. Dong, J. M. Dow, and L. H. Zhang. 2006. Genome scale analysis of diffusible signal factor regulon in Xanthomonas campestris pv. campestris: identification of novel cell-cell communication-dependent genes and functions. Mol. Microbiol. 59:610-622. [DOI] [PubMed] [Google Scholar]
- 10.He, Y. W., and L. H. Zhang. 2008. Quorum sensing and virulence regulation in Xanthomonas campestris. FEMS Microbiol. Rev. 32:842-857. [DOI] [PubMed] [Google Scholar]
- 11.Hickman, J. W., and C. S. Harwood. 2008. Identification of FleQ from Pseudomonas aeruginosa as a c-di-GMP-responsive transcription factor. Mol. Microbiol. 69:376-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hickman, J. W., D. F. Tifrea, and C. S. Harwood. 2005. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc. Natl. Acad. Sci. U. S. A. 102:14422-14427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hsiao, Y. M., H. Y. Liao, M. C. Lee, T. C. Yang, and Y. H. Tseng. 2005. Clp upregulates transcription of engA gene encoding a virulence factor in Xanthomonas campestris by direct binding to the upstream tandem Clp sites. FEBS Lett. 579:3525-3533. [DOI] [PubMed] [Google Scholar]
- 14.Jefferson, R. A., T. A. Kavanagh, and M. W. Bevan. 1987. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6:3901-3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jenal, U., and J. Malone. 2006. Mechanisms of cyclic-di-GMP signaling in bacteria. Annu. Rev. Genet. 40:385-407. [DOI] [PubMed] [Google Scholar]
- 16.Kulasakara, H., V. Lee, A. Brencic, N. Liberati, J. Urbach, S. Miyata, D. G. Lee, A. N. Neely, M. Hyodo, Y. Hayakawa, F. M. Ausubel, and S. Lory. 2006. Analysis of Pseudomonas aeruginosa diguanylate cyclases and phosphodiesterases reveals a role for bis-(3′-5′)-cyclic-GMP in virulence. Proc. Natl. Acad. Sci. U. S. A. 103:2839-2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leduc, J. L., and G. P. Roberts. 2009. Cyclic di-GMP allosterically inhibits the CRP-like protein (Clp) of Xanthomonas axonopodis pv. citri. J. Bacteriol. 191:7121-7122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee, E. J., J. Glasgow, S. F. Leu, A. O. Belduz, and J. G. Harman. 1994. Mutagenesis of the cyclic AMP receptor protein of Escherichia coli: targeting positions 83, 127 and 128 of the cyclic nucleotide binding pocket. Nucleic Acids Res. 22:2894-2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee, V. T., J. M. Matewish, J. L. Kessler, M. Hyodo, Y. Hayakawa, and S. Lory. 2007. A cyclic-di-GMP receptor required for bacterial exopolysaccharide production. Mol. Microbiol. 65:1474-1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McKay, D. B., and T. A. Steitz. 1981. Structure of catabolite gene activator protein at 2.9 A resolution suggests binding to left-handed B-DNA. Nature 290:744-749. [DOI] [PubMed] [Google Scholar]
- 21.Moore, J., M. Kantorow, D. Vanderzwaag, and K. McKenney. 1992. Escherichia coli cyclic AMP receptor protein mutants provide evidence for ligand contacts important in activation. J. Bacteriol. 174:8030-8035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Newell, P. D., R. D. Monds, and G. A. O'Toole. 2009. LapD is a bis-(3′,5′)-cyclic dimeric GMP-binding protein that regulates surface attachment by Pseudomonas fluorescens Pf0-1. Proc. Natl. Acad. Sci. U. S. A. 106:3461-3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Römling, U., and D. Amikam. 2006. Cyclic di-GMP as a second messenger. Curr. Opin. Microbiol. 9:218-228. [DOI] [PubMed] [Google Scholar]
- 24.Ross, P., H. Weinhouse, Y. Aloni, D. Michaeli, P. Weinberger-Ohana, R. Mayer, S. Braun, E. de Vroom, G. A. van der Marel, J. H. van Boom, and M. Benziman. 1987. Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid. Nature 325:279-281. [DOI] [PubMed] [Google Scholar]
- 25.Ryan, R. P., Y. Fouhy, J. F. Lucey, L. C. Crossman, S. Spiro, Y. W. He, L. H. Zhang, S. Heeb, M. Camara, P. Williams, and J. M. Dow. 2006. Cell-cell signaling in Xanthomonas campestris involves an HD-GYP domain protein that functions in cyclic di-GMP turnover. Proc. Natl. Acad. Sci. U. S. A. 103:6712-6717. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26.Ryjenkov, D. A., R. Simm, U. Römling, and M. Gomelsky. 2006. The PilZ domain is a receptor for the second messenger c-di-GMP: the PilZ domain protein YcgR controls motility in enterobacteria. J. Biol. Chem. 281:30310-30314. [DOI] [PubMed] [Google Scholar]
- 27.Simm, R., M. Morr, A. Kader, M. Nimtz, and U. Römling. 2004. GGDEF and EAL domains inversely regulate cyclic di-GMP levels and transition from sessility to motility. Mol. Microbiol. 53:1123-1134. [DOI] [PubMed] [Google Scholar]
- 28.Slater, H., A. Alvarez-Morales, C. E. Barber, M. J. Daniels, and J. M. Dow. 2000. A two-component system involving an HD-GYP domain protein links cell-cell signalling to pathogenicity gene expression in Xanthomonas campestris. Mol. Microbiol. 38:986-1003. [DOI] [PubMed] [Google Scholar]
- 29.Wang, L. H., Y. He, Y. Gao, J. E. Wu, Y. H. Dong, C. He, S. X. Wang, L. X. Weng, J. L. Xu, L. Tay, R. X. Fang, and L. H. Zhang. 2004. A bacterial cell-cell communication signal with cross-kingdom structural analogues. Mol. Microbiol. 51:903-912. [DOI] [PubMed] [Google Scholar]
- 30.Wang, L. H., L. X. Weng, Y. H. Dong, and L. H. Zhang. 2004. Specificity and enzyme kinetics of the quorum-quenching N-acyl homoserine lactone lactonase (AHL-lactonase). J. Biol. Chem. 279:13645-13651. [DOI] [PubMed] [Google Scholar]
- 31.Weber, I. T., T. A. Steitz, J. Bubis, and S. S. Taylor. 1987. Predicted structures of cAMP binding domains of type I and II regulatory subunits of cAMP-dependent protein kinase. Biochemistry 26:343-351. [DOI] [PubMed] [Google Scholar]
- 32.Won, H. S., T. W. Lee, S. H. Park, and B. J. Lee. 2002. Stoichiometry and structural effect of the cyclic nucleotide binding to cyclic AMP receptor protein. J. Biol. Chem. 277:11450-11455. [DOI] [PubMed] [Google Scholar]
- 33.Yang, J. T., C. S. Wu, and H. M. Martinez. 1986. Calculation of protein conformation from circular dichroism. Methods Enzymol. 130:208-269. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.