Abstract
Pseudomonas entomophila is an entomopathogenic bacterium that is able to infect and kill Drosophila melanogaster upon ingestion. Its genome sequence suggests that it is a versatile soil bacterium closely related to Pseudomonas putida. The GacS/GacA two-component system plays a key role in P. entomophila pathogenicity, controlling many putative virulence factors and AprA, a secreted protease important to escape the fly immune response. P. entomophila secretes a strong diffusible hemolytic activity. Here, we showed that this activity is linked to the production of a new cyclic lipopeptide containing 14 amino acids and a 3-C10OH fatty acid that we called entolysin. Three nonribosomal peptide synthetases (EtlA, EtlB, EtlC) were identified as responsible for entolysin biosynthesis. Two additional components (EtlR, MacAB) are necessary for its production and secretion. The P. entomophila GacS/GacA two-component system regulates entolysin production, and we demonstrated that its functioning requires two small RNAs and two RsmA-like proteins. Finally, entolysin is required for swarming motility, as described for other lipopeptides, but it does not participate in the virulence of P. entomophila for Drosophila. While investigating the physiological role of entolysin, we also uncovered new phenotypes associated with P. entomophila, including strong biocontrol abilities.
Pseudomonas entomophila is a recently isolated Pseudomonas species that is closely related to the saprophytic soil bacterium Pseudomonas putida. It was initially characterized as a natural pathogen of Drosophila (63). Indeed, P. entomophila was first isolated from flies sampled in Guadeloupe, and it is highly pathogenic for Drosophila larvae and adults. P. entomophila can also effectively kill members of other insect orders (e.g., Bombyx mori, Anopheles gambiae), which makes it a new entomopathogenic bacterium. Its ability to infect and kill Drosophila melanogaster very efficiently after ingestion makes it an appropriate model for the study of host-pathogen interactions (38, 62, 63).
In order to unravel features contributing to the entomopathogenic properties of P. entomophila, its genome was sequenced. The results suggest that this strain is a ubiquitous, metabolically versatile bacterium that may colonize diverse habitats, including soil, rhizosphere, and aquatic systems, as shown for P. putida KT2440 (62). However, in contrast to the P. putida genome, the P. entomophila genome contains many genes that are predicted to be important for virulence toward insects. Notably, P. entomophila could secrete many degradative enzymes (proteases and lipases), putative toxins, and secondary metabolites (62). Similar factors have been shown to play a key role in the virulence of other entomopathogenic bacteria like Photorhabdus and Xenorhabdus sp. (27, 29).
Insertional mutagenesis allowed the identification of several P. entomophila genes required to infect and/or kill Drosophila. This analysis demonstrated that P. entomophila virulence is under the control of the GacS/GacA two-component system (62, 63), a global regulatory system which is known to control secondary metabolite production, protein secretion, and pathogenic abilities in gammaproteobacteria (37, 65). Another study indicates that P. entomophila can counteract the Drosophila gut immune response as a result of the secretion of an abundant protease, AprA, which degrades antimicrobial peptides produced by gut epithelia and thereby promotes bacterial persistence (38). However, an AprA-deficient mutant remains virulent to some extent, indicating that P. entomophila virulence is multifactorial, AprA being one virulence factor among others.
The secretion of virulence factors is a common mechanism employed by pathogens to compromise host defenses. Several entomopathogenic bacteria (e.g., Photorhabdus luminescens) secrete toxins that allow them to impair host function (8). The starting point of this study was the observation that, in contrast to several other Pseudomonas strains, P. entomophila secretes a strong diffusible hemolytic activity (which is also controlled by the Gac system). This raises the possibility of a link between this hemolytic activity and the pathogenicity of P. entomophila for Drosophila. Indeed, bacterial hemolysins are exotoxins that attack blood cell membranes and cause cell rupture by poorly defined mechanisms. It was conceivable that this hemolytic activity could be a readout for the ability of P. entomophila to damage the epithelial cells of the Drosophila gut, which plays a crucial role in its virulence (10, 33, 63).
In this study, the P. entomophila hemolytic factor was identified as a cyclic lipopeptide (CLP) whose structure was elucidated. CLPs are versatile molecules with antimicrobial, cytotoxic, and surfactant properties that are produced by members of the genera Bacillus, Serratia, Burkholderia, and Pseudomonas (31, 41, 43, 50). They are produced by a ribosome-independent mechanism that utilizes multifunctional enzymes called nonribosomal peptide synthetases (NRPSs) (42, 59). These NRPSs are composed of repeated amino acid activation modules containing domains for condensation, aminoacyl adenylation, and thiolation. Modules are responsible for activation and incorporation of amino acids into the growing peptide. A large number of prokaryotic and some eukaryotic organisms synthesize peptide metabolites via this nonribosomal mechanism of biosynthesis (42, 47).
Several genes involved in P. entomophila lipopeptide production were identified, three of them encoding NRPSs. The physiological role of this lipopeptide was also investigated, and it does not seem to play a role in the process of virulence towards Drosophila and Dictyostelium or in the P. entomophila biocontrol activity that was uncovered by this study. This suggests that the lifestyle of this newly identified bacterium is probably quite versatile and that lipopeptide production could be required only under specific circumstances.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
All of the bacterial strains used in this study are listed in Table 1. Escherichia coli DH5α (Invitrogen) was used as the recipient strain for all plasmid constructs, and E. coli strain S17.1 (58) was used to conjugate plasmids into P. entomophila. P. entomophila was grown in LB for all experiments, except for swarming motility assays. When E. coli was grown, antibiotics were used, when necessary, at the following concentrations: G418, 25 μg/ml; tetracycline, 5 μg/ml. When P. entomophila was grown, antibiotics were used, when necessary, at the following concentrations: gentamicin, 50 μg/ml for liquid cultures and 150 μg/ml for solid medium; tetracycline, 40 μg/ml; rifampin, 30 μg/ml.
TABLE 1.
Bacterial strains used in this study
| Strain | Relevant characteristic(s) | Reference or source |
|---|---|---|
| Pseudomonas entomophila | ||
| Pe | Wild type | 63 |
| ΔgacA | gacA deletion mutant | This study |
| IM3332 | pseen3332 (etlA) insertion mutant, Gmr | This study |
| IM3045 | pseen3045 (etlB) insertion mutant, Gmr | This study |
| IM3044 | pseen3044 (etlC) insertion mutant, Gmr | This study |
| IM3043 | pseen3043 (macA) insertion mutant, Gmr | This study |
| IM3046 | pseen3046 insertion mutant, Gmr | This study |
| IM3335 | pseen3335 insertion mutant, Gmr | This study |
| ΔY | rsmY deletion mutant | This study |
| ΔZ | rsmZ deletion mutant | This study |
| ΔYZ | rsmY rsmZ deletion mutant | This study |
| ΔYZ ΔA1 | rsmY rsmZ rsmA1 (pseen3843) deletion mutant | This study |
| ΔYZ ΔA2 | rsmY rsmZ rsmA2 (pseen2282) deletion mutant | This study |
| ΔYZ ΔA3 | rsmY rsmZ rsmA3 (pseen1464) deletion mutant | This study |
| ΔYZ ΔA1 ΔA2 | rsmY rsmZ rsmA1 rsmA2 deletion mutant | This study |
| ΔYZ ΔA1 ΔA3 | rsmY rsmZ rsmA1 rsmA3 deletion mutant | This study |
| ΔYZ ΔA2 ΔA3 | rsmY rsmZ rsmA2 rsmA3 deletion mutant | This study |
| ΔYZ ΔA1 ΔA2 ΔA3 | rsmY rsmZ rsmA1 rsmA2 rsmA3 deletion mutant | This study |
| Pe F3045-lacZ | Pe containing chromosomal translational pseen3045-lacZ reporter | This study |
| ΔYZ F3045-lacZ | ΔYZ mutant containing chromosomal translational pseen3045-lacZ reporter | This study |
| Pseudomonas fluorescens CHA0 | Wild type | 59a |
Construction of strains and plasmids.
Deletion constructs for the gacA, rsmY, rsmZ, rsmA1, rsmA2, and rsmA3 genes were generated by amplifying flanking regions by PCR and then splicing the flanking regions together by overlap extension PCR. The deletions were in frame and contained the linker sequences 5′-GGTACC-5′ (gacA), 5′-AAGCTT-3′ (rsmY, rsmZ, rsmA1, rsmA2), and 5′-GAATTC-3′ (rsmA3). The resulting PCR products were then cloned into plasmid pEXG2 (51), yielding plasmids pEXΔgacA, pEXΔrsmY, pEXΔrsmZ, pEXΔrsmA1, pEXΔrsmA2, and pEXΔrsmA3. These plasmids were then used to create strains ΔgacA, ΔY, ΔZ, ΔYZ, ΔYZ ΔA1, ΔYZ ΔA2, ΔYZ ΔA3, ΔYZ ΔA1 ΔA2, ΔYZ ΔA1 ΔA3, ΔYZ ΔA2 ΔA3, and ΔYZ ΔA1 ΔA2 ΔA3, containing in-frame deletions of the gacA, rsmY, rsmZ, rsmA1, rsmA2, and rsmA3 genes, by allelic exchange. Deletions were confirmed by PCR.
Insertion constructs for the pseen3332, pseen3045, pseen3044, pseen3046, pseen4043, and pseen3335 genes were generated by cloning an internal 500- to 800-bp fragment into the pINT nonreplicative plasmid (Arne Rietsch, unpublished work), generating plasmids pINT3332, pINT3045, pINT3044, pINT3043, pINT3046, and pINT3335. These constructs were then used to create strains IM3332, IM3045, IM3044, IM3043, IM3046, and IM3335 by homologous recombination.
The pseen3045-lacZ translational reporter fusion was constructed according to reference 60. A 499-bp fragment upstream of the pseen3045 gene was cloned into the pSS231 plasmid in such a way that the ATG of the pseen3045 gene was fused in frame to the 10th codon of the lacZ gene. The EcoRI/AatII fragment was then subcloned into plasmid Mini CTX lacZ (30) for the purpose of antibiotic compatibility, generating plasmid Mini CTX F3045-lacZ. This plasmid was then conjugated into the wild-type strain and the ΔYZ mutant, creating strains Pe F3045-lacZ and ΔYZ F3045-lacZ by homologous recombination. These strains carry both the promoter fusion and the wild-type copy of the pseen3045 gene.
The plasmids were made by cloning PCR-amplified DNA fragments containing each of the rsmY, rsmZ, rsmA1, rsmA2, and rsmA3 genes from P. entomophila into the pPSV35 vector (51), generating plasmids pPSVrsmY, pPSVrsmZ, pPSVrsmA1, pPSVrsmA2, and pPSVrsmA3. The sequences of all of the primers used are available upon request.
Bacterial mutagenesis.
Random mutagenesis was performed by biparental mating using P. entomophila and Escherichia coli S17.1-λpir (46) carrying the pUT-Tn5-Tc suicide plasmid as previously described (19).
Lipopeptide extraction and HPLC separation.
Lipopeptide was isolated by liquid-liquid extraction of bacterial culture supernatants, followed by C18 reverse-phase high-performance liquid chromatography (HPLC) separation. Some 10 ml of bacterial culture supernatants was extracted twice with the same volume of ethyl acetate. The extracts were pooled, washed once with the same volume of water, dried, and then taken to a volume of methanol 10 times smaller than the initial bacterial supernatant volume.
Crude extracts obtained from 5 to 10 ml of culture supernatant were dried, taken to 100 μl of eluent A (acetonitrile/water 6:4 [vol/vol], 0.1% trifluoroacetic acid [TFA]), and loaded onto a C18 Hypersil octyldecyl silane column (3 μm, 250 by 4.6 mm; Thermo Fisher Scientific). An acetonitrile/isopropanol mixture (6:4, vol/vol) containing 0.1% TFA was used as eluent B. Elution was performed at a flow rate of 0.6 ml/min with a linear gradient starting at 100/0 (eluent A/eluent B), which subsequently changed to 20/80 in 80 min. The compounds were detected at 206 nm. Fractions corresponding to chromatogram peaks were collected manually and dried under vacuum.
Lipopeptide overall chemical composition analysis.
Amino acid analysis of the HPLC-separated lipopeptide fractions was performed with a Hitachi L-8800 amino acid analyzer equipped with a 2620MSC-PS column (ScienceTec, Les Ulis, France) after hydrolysis of dried HPLC fractions in 6 M HCl (16 h, 95°C). The elution protocol recommended by the manufacturer for the separation of amino acids was used.
Fatty acid analysis was performed by gas chromatography after hydrolysis of dried HPLC fractions in 4 M HCl (2 h, 100°C), fatty acid extraction with ethyl acetate, and esterification (dry methanol/acetyl chloride, 6 h, 85°C). The HP 5890 gas chromatograph used was equipped with an SGE 25QC3/BP10 0.5 capillary column, and a temperature gradient of 130 to 240°C, 2°C/min, was used. The identity of fatty acid isolated from the HPLC fractions was deduced by comparison of its retention time with reference samples (methyl esters of 2-OH and 3-OH C10 to C16 fatty acids, as well as nonhydroxylated C10, C12, C14, and C16 fatty acids).
Matrix-assisted laser desorption ionization (MALDI) mass spectrometry (MS) and tandem MS (MS/MS) analysis.
MALDI MS analysis of crude extracts and lipopeptide HPLC fractions was performed using a Perseptive Voyager STR (PE Biosystems) time-of-flight (TOF) mass spectrometer equipped with an N2 laser (337 nm). Aliquots (0.5 to 2 μl) of the samples were deposited onto the plate, covered with 0.5 to 1 μl of matrix solution (2,5-dihydroxybenzoic acid in methanol at 10 μg/μl), and dried. Different sample-to-matrix ratios were tested to obtain the best spectra. Samples containing trace lipopeptide quantities (extracts from attenuated mutants and minor HPLC fractions) were preconcentrated (5 to 10 times) before deposition. The analysis of positive and negative ions was performed in the reflectron mode.
MALDI MS/MS analysis of major lipopeptide HPLC fractions was performed by MALDI-TOF-TOF (4700 Proteomics analyzer; Applied Biosystems) using an NdYag laser (355 nm, 200 Hz) and a collision energy of 1 keV (gas, N2 at 5 × 10−7 torr). The analyses were performed on both native (cyclic) and open-ring (linear) lipopeptide forms. The lactone cycle was opened by treatment with a 28% ammonium solution at 40°C for 2 h. α-Cyano-4-hydroxycinnamic acid was used as a matrix, and an acetonitrile/water mixture (1:1, vol/vol) containing 0.1% TFA was used as a solvent for both the samples and the matrix. Fragmentation spectra of positive protonated precursor ions were recorded.
Bioinformatic analysis.
The amino acid sequences of the NRPSs involved in putisolvin biosynthesis were compared and analyzed with NRPS-PKS web-based software (http://www.nii.res.in/nrps-pks.html) (3).
Phylogenetic analyses were performed using the Mobyle portal (http://mobyle.pasteur.fr/). Multiple alignments were performed using the clustalw-multalign program. Alignment files were then visualized using the Boxshade program or analyzed using the Quicktree program. Phylogenetic trees were visualized using the Drawtree program.
Surfactant production assay.
To test culture supernatants for biosurfactant activity, overnight cultures were used. After the addition of 5% methylene blue (which is useful for photography but has no influence on droplet surface tension), 20 μl was pipetted as a droplet onto Parafilm. The spreading of the droplet on the Parafilm was observed, the droplet was allowed to dry, and the diameter of the dried droplet was recorded. Measurements of this diameter are in millimeters and represent means of at least five experiments.
Swarming motility assay.
In order to test swarming motility on agar medium, single colonies were spot inoculated onto twofold-diluted M63 minimal medium (45) containing 0.5 mM MgSO4, 0.1% glucose, and 0.25% Casamino Acids and solidified with 0.5% agar. Plates were incubated at 30°C for 24 h.
β-Galactosidase activity.
Cells were grown at 30°C in LB medium supplemented as needed with gentamicin (25 μg/ml) and isopropyl-β-d-thiogalactopyranoside (IPTG) at the concentration indicated. Cells were permeabilized with sodium dodecyl sulfate and CHCl3 and assayed for β-galactosidase activity as described previously (22). Assays were performed at least three times in triplicate on separate occasions. Representative data sets are shown below. The values are averages based on one experiment.
Assay of virulence for Drosophila melanogaster.
Oregonr flies were used as the standard wild-type strain. relishE20 is a recessive mutation that blocks the Imd pathway (28). Drosophila stocks were maintained at 25°C.
Bacterial cultures were pelleted by centrifugation after 24 h of growth, and pellet optical densities at 600 nm were adjusted to 100. A 120-μl sample of the pellet was added to a Whatman paper filter disk that completely covered the agar surface at the bottom of a standard fly culture vial. Thirty 4- to 8-day-old adult female flies were starved for 3 h at 29°C in empty vials prior transfer into these bacterium-containing vials, which were subsequently incubated at 29°C. The flies were monitored for death over 4 days. Virulence assays were performed at least three times in triplicate on separate occasions. Representative data sets are shown. The values are averages based on one experiment.
Assay of virulence for Dictyostelium discoideum.
Virulence assays were performed essentially as previously described (1, 12, 26). Briefly, 50 μl of an overnight culture of P. entomophila was deposited into an individual well of a 24-well plate containing 2 ml of SM-Agar (peptone at 10 g/liter, yeast extract at 1 g/liter, KH2PO4 at 2.2 g/liter, K2HPO4 at 1 g/liter, MgSO4 · 7H2O at 1 g/liter, agar at 20 g/liter) and dried. A 5-μl droplet containing 100 Dictyostelium cells was then added to the middle of the well, and the cells were allowed to grow for 6 days at 21°C. Robust growth of Dictyostelium was seen on gacA mutant cells, indicating that this strain has lost its virulence. No growth was observed on wild-type P. entomophila or entolysin-deficient mutants, indicating that these mutants are still virulent.
Biocontrol assay.
Cucumber root rot biocontrol assays were carried out essentially as described previously (4). Briefly, 200-ml Erlenmeyer flasks were partially filled with 60 g of a natural sandy loam soil. When appropriate, the soil was infested with 2.5 g/kg of a 5-day-old millet seed inoculum of Pythium ultimum. Three sterilely grown 4-day-old cucumber (Cucumis sativus cv. Chinese Snake) seedlings were then placed in each flask. Pseudomonas strains were added to soil as a suspension (5 ml per flask) of cells washed twice in sterile distilled water to give 1 × 107 CFU/g of soil. Control flasks received the same amount of sterile water. The microcosms were incubated in a growth chamber containing 80% relative humidity at 22°C with light (200 μmol s−1 m−2; 1.37 ratio of 660-nm light to 730-nm light) for 16 h, followed by an 8-h dark period at 15°C. After 7 days of incubation, the number of surviving plants was recorded. Plants were then removed from the flasks, washed, briefly dried with paper towels, and weighed.
RESULTS
Identification of mutants deficient for hemolytic activity.
P. entomophila secretes a strong diffusible hemolytic activity that can be easily detected as a large lysis zone on blood plates (see Fig. 2A and reference 62). In order to identify factors responsible for the hemolytic activity of P. entomophila, we generated a Tn5-derived library of mutants that were individually screened for the ability to produce a lysis zone on blood plates. We isolated 3 clones (out of 500) that were deficient for hemolytic activity.
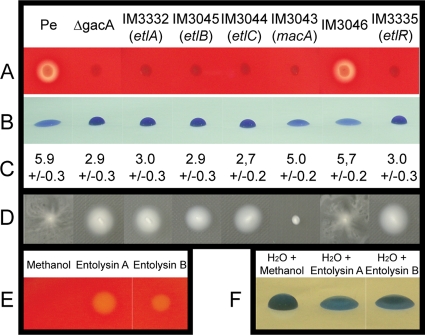
FIG. 2.
Hemolytic activity, biosurfactant production, and swarming motility of wild-type P. entomophila and mutants affected in different genes of the entolysin biosynthesis gene clusters. (A) Hemolytic activity as visualized on blood plates. Single colonies were patched onto sheep blood plates and allowed to grow for 24 h at 30°C. (B) Biosurfactant activity was visualized by the drop-collapsing assay, in which 20-μl droplets of each culture supernatant were deposited onto a hydrophobic (Parafilm) substrate. Bromophenol blue was added to stain the supernatants for photographic purposes and had no influence on the shape of the droplets. (C) After drying, the size (in millimeters) of each supernatant droplet was measured. Each value is the mean of five experiments. (D) Swarming motility was measured on minimal medium plates containing 0.5% agar. Plates were inoculated with a toothpick, and bacteria were allowed to grow for 24 h at 30°C. (E) Hemolytic activity of purified entolysins A and B. Five microliters of concentrated products was deposited onto a blood plate, and activity was allowed to develop for 24 h. (F) Surface tension activity of purified entolysins A and B. Two microliters of concentrated product was added to 20 μl of water, which was deposited onto Parafilm.
Subsequent cloning and sequencing of the region flanking the transposon revealed that it was integrated into different genes of the three hemolysin-deficient mutants. One mutant was a gacA mutant. GacA is the response regulator of the GacS/GacA two-component system that controls P. entomophila virulence, and it was already known that P. entomophila gac mutants are defective in the secretion of protease and hemolysin (62). The other two mutants were affected in the gene pseen3045 and the gene pseen3042, encoding an NRPS and an ABC transporter component, respectively (Fig. 1). A BLAST analysis showed that the pseen3045 gene product is 62% identical and 72% similar (across 88% of the protein) to PsoB, an NRPS involved in putisolvin biosynthesis. Putisolvins are CLPs produced by Pseudomonas putida whose production requires three NRPS proteins encoded by the psoA, psoB, and psoC genes (23). Analogs of these genes, pseen3332, pseen3045 (which was previously identified as involved in hemolysin production), and pseen3044, respectively, were found in the P. entomophila genome. In order to determine the contribution of these genes to the hemolytic activity of P. entomophila, insertion mutants were constructed using single homologous recombination, resulting in strains IM3332, IM3045, and IM3044 (inactivated for the genes pseen3332, pseen3045, and pseen3044, respectively). These strains were tested on blood plates, which revealed that they were indeed deprived of hemolytic activity (Fig. 2A).
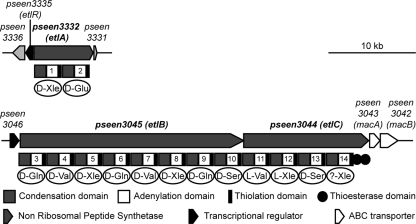
FIG. 1.
Schematic representation of the entolysin biosynthesis gene clusters and surrounding open reading frames in the P. entomophila genome. Below the genes is the module and domain organization of EtlA, EtlB, and EtlC. Predicted amino acid specificity is indicated below each module. All amino acids are identified by standard three-letter biochemical notation, with D and L referring to their stereochemistry.
Surfactant production by P. entomophila and its involvement in swarming motility.
CLPs are versatile surface-active molecules composed of a fatty acid tail linked to a short oligopeptide which is cyclized to form a lactone ring between two amino acids in the peptide chain. In order to investigate CLP production by P. entomophila, we used the so-called “drop-collapsing assay” (see Materials and Methods), which consists of looking at the shape of a culture supernatant droplet on Parafilm. When no surfactant is produced, the droplet stays round, but when surfactant is present in the spent medium, the droplet flattens out. Figure 2B shows that P. entomophila produces a molecule able to strongly decrease the surface tension of the culture medium. This molecule is not produced by the gacA mutant or by the IM3332, IM3045, and IM3044 mutants (Fig. 2B and C), which indicates that the hemolytic activity of P. entomophila is linked to surfactant production.
Surfactant production is known to be linked to swarming motility (7, 14, 21, 34). The swarming motility of P. entomophila was thus tested (see Materials and Methods) and compared to the motility of the gacA mutant and the IM3044 mutant (Fig. 2D). In contrast to what is observed with the wild-type strain, both of these mutants are unable to swarm on minimal medium plates, which indicates that P. entomophila lipopeptide seems to be involved in swarming motility, as are the other lipopeptides produced by Pseudomonas species (50).
Lipopeptide isolation.
In order to purify and isolate the putative lipopeptide produced by P. entomophila, culture supernatants of the P. entomophila wild-type strain (Pe), the gacA mutant (ΔgacA), and the IM3044 strain were subjected to ethyl acetate extraction and the crude extracts were fractionated by reversed-phase HPLC using a C18 column as described in Materials and Methods.
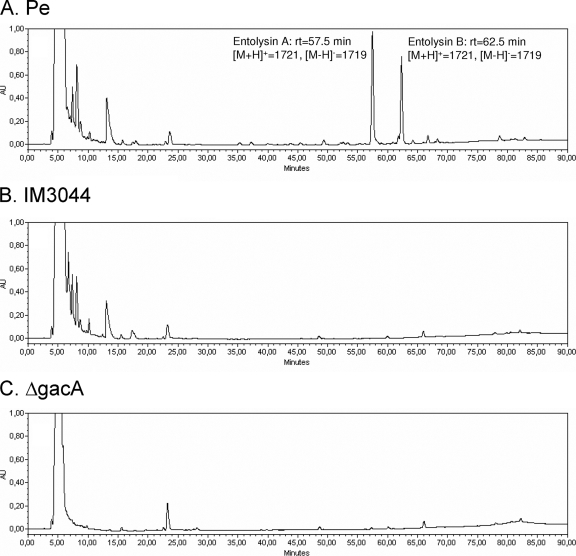
Chromatograms obtained through sample analysis are presented in Fig. 3. Two major peaks observed in the wild-type P. entomophila chromatogram at retention times of 57.5 and 62.5 min (Fig. 3A) were totally absent from the chromatogram obtained with strain IM3044 (Fig. 3B). Comparison with the gacA mutant chromatogram shows that these peaks, together with some other peaks, are also lacking (Fig. 3C). This correlates with the fact that the Gac system is known to be involved in global secondary metabolite regulation in other Pseudomonas species (37).
FIG. 3.
HPLC analysis of the surface active compound(s) of P. entomophila. C18 reverse-phase HPLC profiles of ethyl acetate extracts derived from 6 ml of culture supernatants of wild-type P. entomophila (A), hemolysin-deficient mutant IM3045 (B), and a gacA mutant (C). Chromatograms were obtained through sample analysis at a wavelength of 206 nm. The two main peaks represent entolysin isoforms A and B. AU, arbitrary units.
HPLC fractions corresponding to each of the two major peaks (Fig. 3A) were collected and analyzed using MALDI MS in the positive and negative ion modes. Both fractions gave rise to molecular ions with the same m/z values in the positive ion mode [M + H]+ at m/z = 1,721 and in the negative ion mode [M − H]− at m/z = 1,719, suggesting that these two fractions probably contain isoforms of the same product. We thus postulated that this product is a CLP produced by P. entomophila and linked to its hemolytic activity. This lipopeptide was named entolysin, and its putative isoforms were named entolysin A and entolysin B.
HPLC fractions containing these isoforms were dried, and entolysins A and B were dissolved in methanol in order to test their biological activity. Both entolysins A and B are able to decrease surface tension and to lyse blood cells, as illustrated in Fig. 2E and F.
Lipopeptide structural characterization.
HPLC fractions containing the two putative isoforms entolysin A and entolysin B were first subjected to amino acid and fatty acid analyses as described in Materials and Methods. Similar results were obtained. The amino acid analysis demonstrated that Ser, Gln/Glu, Val, Ile, and Leu residues are present in both fractions in a molar ratio of approximately 2:4:2.2:1:3.5 (see Table S1 in the supplemental material). Gas chromatography of methyl esters of fatty acids isolated from entolysin A and entolysin B by acid hydrolysis revealed the presence of a 3-C10OH fatty acid in both of them. This fatty acid was identified by comparison of its retention time with external standards by gas chromatographic analysis.
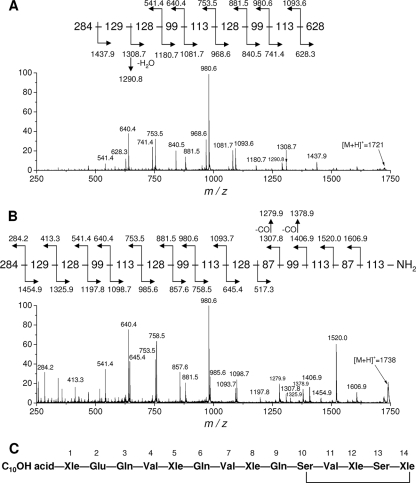
MALDI MS/MS analysis of entolysin A and entolysin B was performed before and after ammonium hydroxide treatment. After treatment, the major ions were observed in the mass spectra at 17 U higher than before, i.e., at m/z = 1,738. This mass increase corresponds to the opening of the lactone ring, present in many other lipopeptides, by the addition of ammonia across the ester bond (36, 50). Collision-induced dissociation (CID) MS/MS spectra of the [M + H]+ precursor ions from native (cyclic) and open-ring (linear) samples were again found to be almost identical for entolysin A and entolysin B. Fragmentation spectra obtained for cyclic and open-ring entolysin A are presented in Fig. 4A and B. In both spectra, b and y sequence ions were observed. As expected, the C-terminal region of the untreated lipopeptide failed to yield fragment ions (Fig. 4A) because of the cycling between the C terminus and one of the Ser residues. Only the linear part of the structure including the fatty acid linked to the N terminus and the first eight amino acid residues was covered by the fragmentation. The linear, open-ring lipopeptide yielded fragment ions covering the complete sequence (Fig. 4B). Comparison of the cyclic and open-ring forms showed that fragmentation of the cyclic form ceased right before the Ser residue at position 10, suggesting that this residue was involved in cycling with the C-terminal Leu/Ile. Under these experimental conditions, no ions of the w series were observed, making discrimination between Leu and Ile impossible. Taken together, these data suggested the entolysin structure presented in Fig. 4C. It was not possible to establish a structural difference between entolysin A and entolysin B; this will be the subject of a further study. However, amino acid analysis showed the presence of one isoleucine and four leucine residues, whose positions were not determined. This observation suggests that these isoforms could differ in the positioning of the isoleucine residue, as observed earlier for viscosin and massetolide L synthesized by P. fluorescens SS101 (15).
FIG. 4.
CID tandem mass spectra and fragmentation schemes of the cyclic form of entolysin A (the m/z 1,721 species) (A) and the open-ring form (m/z 1,738 [1,721 + 17] species) (B). (C) Proposed structure of entolysin, based on MALDI MS/MS, fatty acid, and amino acid analyses. Ser, serine; Glu, glutamic acid; Gln, glutamine; Val, valine; Xle, isoleucine or leucine.
Genetic analysis of the entolysin synthetases.
The genes involved in entolysin synthesis were renamed etlA, etlB, and etlC, corresponding to the genes pseen3332, pseen3045, and pseen3044, respectively (Fig. 1). These three genes encode NRPSs. Analysis of the deduced amino acid sequences revealed that EtlA, EtlB, and EtlC comprise two, eight, and four modules, respectively, which corresponds to the number of amino acid residues found in entolysin. These modules are composed of a condensation domain, an adenylation domain, and a thiolation domain. Two additional thioesterization domains are found at the end of EtlC.
Using NRPS-PKS web-based software (see Materials and Methods), the primary structures of the 14 adenylation domains were analyzed. The signature sequence consists of 10 selectivity-conferring amino acid residues embedded in the substrate-binding pocket. For the predicted selectivity of the 14 A domains of EtlA, EtlB, and EtlC, see Table S2 in the supplemental material. These prediction results correlate well with the proposed entolysin structure, but they give no information about the putative position of the isoleucine residue. A phylogenetic analysis of the amino acid sequences of A domains identified in NRPSs involved in lipopeptide synthesis in different Pseudomonas species was also performed (see Fig. S1 in the supplemental material). Clustering of A domains with the same specificity suggests that the isoleucine residue could be preferentially assembled by the last domain (EtlC A14).
In order to gain insight into the stereochemistry of the assembled amino acids, a phylogenetic analysis of the amino acid sequences of C domains was also performed. It is known that dual condensation/epimerization (C/E) domains could be responsible for incorporating D residues into several Pseudomonas-produced lipopeptides (5). These dual domains are known to cluster together (5, 15). Figure S2 in the supplemental material shows that, according to the phylogenetic analysis, all of the C domains involved in entolysin production but two (C12 and C13) could function as dual C/E domains. Subsequent analysis of the primary sequence of the proposed C/E domains showed that they contain an N-terminal sequence described as typical of the C/E domains (5). It includes an elongated His motif (HHL/IxxxxGD), which is only found as perfect in the C3, C5, and C8 domains. In the other putative C/E domains, the Gly residue in front of the Asp residue is either missing (C2) or replaced by an Ala (C4, C14) or a His (C7, C11, C9, C10, C6) (see Fig. S3 in the supplemental material). It is worth noting that the two His residues and the Asp residue are the ones predicted to have a catalytic function (5).
Moreover, it was suggested that T domains interacting with dual C/E domains present highly conserved specific amino acids (23). Analysis of the primary sequence of T domains involved in entolysin production showed that these amino acids are found in all but T11, T12, and T14. T11 and T12 present a conserved amino acid sequence typical of T domains interacting with classical C domains (23). Interestingly, the last T domain (EtlC T14) lacks most of these conserved amino acids, like the last T domains involved in putisolvin (PsoC T11) and arthrofactin (ArfC T11) synthesis (see Fig. S4 in the supplemental material).
Overall, these data suggest that the amino acids in positions 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, and 13 could be in the D configuration (see Fig. 1).
Finally, the close clustering of the first condensation domain of EtlA with C1 domains of other NRPSs involved in CLP biosynthesis (see Fig. S2 in the supplemental material) confirmed its role in N acylation of the first amino acid of the CLP molecule (16, 54).
Involvement of other genes in entolysin production.
Two genes encoding an ABC transporter similar to the MacAB macrolide transporter (35) are localized immediately downstream of etlB and etlC, and two genes encoding transcriptional regulators are found upstream of etlA and etlB (Fig. 1). In other systems, it has been suggested that similar genes were involved in lipopeptide secretion and regulation (17, 23).
In order to investigate whether these genes are also involved in entolysin production, pseen3043 (macA), pseen3046, and pseen3335 insertion mutants were constructed by single homologous recombination. The abilities of these mutants (IM3043, IM3046, and IM3335, respectively) to lyse blood cells, to produce biosurfactant, and to swarm were checked. The results presented in Fig. 2 show that pseen3043 and pseen3335, but not pseen3046, are involved in lipopeptide production. The LuxR-like regulator encoded by pseen3335 (which we thus renamed etlR) presents similarity with PsoR, another LuxR-like regulator involved in putisolvin regulation (23). A mutant affected in the etlR gene is not able to synthesize entolysin any more, as it is deprived of hemolytic activity. Comparison to what is known about other systems suggests that EtlR controls the transcription of entolysin biosynthesis genes. Interestingly, the IM3043 mutant (affected in the macA gene) is completely deprived of hemolytic activity and swarming ability (similar results were obtained with the macB transposon mutant [data not shown]). However, the drop-collapsing assay indicates that some amount of surfactant is still produced in the culture supernatant, as the surface tension of the droplet is only partially recovered. It was proposed that the macAB genes may encode an ABC transporter involved in lipopeptide export (35, 50). Thus, one hypothesis that could explain this slight discrepancy is that entolysin might be produced by a macA mutant but not exported. During growth in liquid medium, some level of cell lysis could cause the produced entolysin to leak into the growth medium. Consistent with this hypothesis, we observed that if the macA mutant was allowed to grow on blood plates for several days, some hemolytic activity could be detected (data not shown). It was also noted that a macA mutant seems to be affected in its growth in minimal medium.
Overall, this genetic analysis indicates that three NRPSs (distributed in two loci separated by 355 kb), a transcription factor, and an ABC transporter are involved in entolysin production by P. entomophila.
Entolysin production is directly regulated by the GacS/GacA two-component system.
It was shown that the GacS/GacA two-component system regulates the hemolytic activity of P. entomophila, as well as protease production and virulence. We wondered if this regulation was due to a direct effect of the Gac system on the entolysin biosynthesis genes or was part of a more global, indirect mechanism. The functioning of the Gac system, a conserved two-component system, is well known and happens mainly at a posttranscriptional level (37). Upon phosphorylation by the GacS sensor, the GacA transcriptional regulator positively controls the expression of genes specifying small noncoding RNAs, which are characterized by repeated GGA motifs. These motifs are essential for binding small RNA-binding proteins called RsmA proteins that act as translational repressors of target mRNAs.
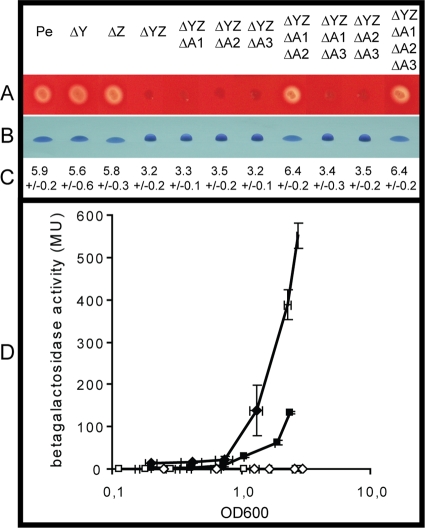
The functioning of the GacS/GacA two-component system in P. entomophila and its impact on entolysin production were investigated in further detail. P. entomophila genome annotation revealed that two genes corresponding to small RNAs, rsmY and rsmZ, are present (62). Typical GacA-binding boxes were found upstream of these genes, and we showed, by using transcriptional fusions, that expression of these genes is indeed under the control of the Gac system (data not shown). Deletion of either one of these genes does not strongly affect entolysin production, as shown in Fig. 5, in contrast to the deletion of both genes. The rsmY rsmZ double mutant (ΔYΔZ) is deprived of hemolytic activity (Fig. 5) and of virulence (data not shown), like the gacA mutant (ΔgacA).
FIG. 5.
Regulation of lipopeptide production by the Gac system. (A) Hemolytic activities of mutants affected in the different constituents of the Gac system. (B) Biosurfactant production by these mutants visualized by the drop-collapsing effect and quantified by measuring droplet size (C). (D) Quantification of the β-galactosidase activity of the translational fusion etlB-lacZ (Pseen3045-lacZ) (see Materials and Methods) as a function of bacterial growth in different genetic contexts. Diamonds represent expression of the fusion in the wild-type strain carrying the empty vector pPSV35 (closed diamonds) or overexpressing the gene encoding the translational repressor RsmA2 from the same vector (pPSVrsmA2, open diamonds). Squares represent expression of the fusion in the rsmY rsmZ double mutant carrying the empty vector pPSV35 (opened squares) or overexpressing the rsmY gene from the same vector (pPSVrsmY, closed squares). Cultures were grown with gentamicin and 1 mM IPTG to allow full expression of the genes from the plasmids. Each value is the average of three different cultures, and error bars represent standard deviations. MU, Miller units; OD600, optical density at 600 nm.
Similarly, three genes encoding RsmA-like proteins are found in the P. entomophila genome: rsmA1 (pseen3843), rsmA2 (pseen2282), and rsmA3 (pseen1464). Deletion of any of these genes in the wild-type strain does not lead to any visible phenotype regarding entolysin production. As was reported for other species (37), deletion of both rsmA1 and rsmA2 strongly impairs bacterial growth, which is why we chose to investigate the role of these RsmA proteins in an rsmY rsmZ background. Deletion of any of the rsmA genes in the rsmY rsmZ mutant does not restore hemolytic activity. However, when both rsmA1 and rsmA2 are deleted in the rsmY rsmZ mutant, entolysin production is restored, whereas no additional effect can be seen upon rsmA3 deletion (Fig. 5). Moreover, overexpression of either rsmA1 or rsmA2, but not rsmA3, from a plasmid in the wild-type P. entomophila strain abolishes hemolytic activity and protease production, as well as virulence (data not shown). We concluded that the P. entomophila Gac system implicates two small RNAs, rsmY and rsmZ, and two small RNA-binding proteins, RsmA1 and RsmA2. We cannot exclude the possibility that RsmA3 plays a specific role under certain conditions; however, such a role could not be seen in our experimental settings.
In order to determine if the effect of the Gac system on entolysin production is exerted at the biosynthesis gene level, we constructed translational fusions of the etlA (pseen3332) and etlB (pseen3045) genes and the lacZ gene (see Materials and Methods). These fusions were introduced into the chromosome of the wild-type P. entomophila strain and the rsmY rsmZ double mutant. β-Galactosidase production was measured during the growth of these strains, showing that these translational fusions are produced in wild-type P. entomophila. However, they are not produced in the absence of rsmY and rsmZ (Fig. 5D and data not shown). Synthesis of either one of the small RNAs from a plasmid in the rsmY rsmZ double mutant restored production of the translational fusions (Fig. 5D and data not shown). In a reciprocal way, overexpression of either rsmA1 or rsmA2 from a plasmid in the wild-type strain abolishes the production of the etlA and etlB translational fusions (Fig. 5D and data not shown). These results correlate with the fact that the Gac system could exert its effect on entolysin production through the control of the etl genes.
Role of entolysin in P. entomophila virulence.
In order to determine if the hemolytic activity of P. entomophila is linked to its ability to infect and kill Drosophila, we investigated whether an entolysin-deficient mutant is affected in its virulence for Drosophila. Two different Drosophila lines were tested, the wild-type Oregonr line and the relish mutant line, in which the Imd pathway, which plays a important role in defense against infection by Gram-negative bacteria, is inactivated (see Materials and Methods). Adult female flies were fed with wild-type P. entomophila, the avirulent gacA mutant (ΔgacA), and an entolysin-deficient mutant (IM3044) (see Materials and Methods). Flies deprived of a functional immune system die more quickly than wild-type flies upon infection by P. entomophila, as previously described (38), but in either case, no difference in survival between P. entomophila and the etlC mutant (IM3044) could be detected (Fig. 6). Similar results were obtained with third-instar larvae. This demonstrates that entolysin does not play a major role in the virulence of P. entomophila for Drosophila.
FIG. 6.
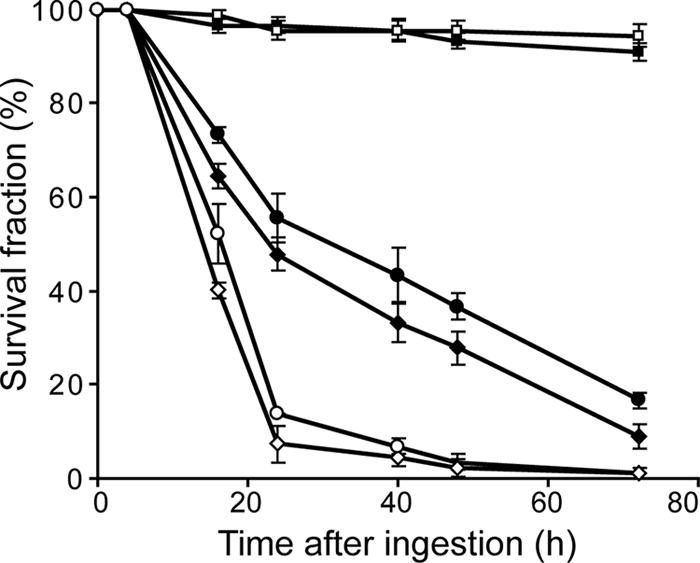
Entolysin is not involved in the virulence of P. entomophila for Drosophila. Wild-type Oregonr female flies (closed symbol) and relish mutant female flies (opened symbol) were naturally infected by the P. entomophila wild-type strain (Pe, diamonds), a gacA mutant (ΔgacA, squares), and an entolysin-deficient mutant (IM3045, circles).
We also wondered if the ability to lyse blood cells could be involved in virulence for other organisms. Dictyostelium discoideum is an amoeba that is used as a model for phagocytosis. The virulence of many bacteria, including Pseudomonas aeruginosa, has been studied by using Dictyostelium as a model of host-pathogen interactions (9, 11). Wild-type P. entomophila (Pe), a gacA mutant, and an entolysin-deficient mutant (IM3044) were tested in the Dictyostelium model (see Materials and Methods). We observed that the wild-type strain and the entolysin-deficient mutant do not allow the growth of Dictyostelium, in contrast to the gacA mutant (data not shown). This indicates that P. entomophila, which was first isolated as virulent for Drosophila, could also be virulent for Dictyostelium. Interestingly, the Gac system that controls virulence for Drosophila also controls virulence for Dictyostelium. However, entolysin does not play a role in either type of virulence.
Biocontrol abilities of P. entomophila do not rely on entolysin production.
Many CLPs are produced by plant-associated Pseudomonas species (50) like P. syringae (a plant pathogen) and P. fluorescens (a bacterium beneficial to plants). It was already known that P. entomophila is not a plant pathogen (62), but its biocontrol activity was never tested.
In order to determine whether or not P. entomophila is able to promote plant growth and to protect plants from root diseases caused by pathogenic fungi, plant assays were set up in natural soil microcosms involving cucumber as the host plant and Pythium ultimum as a root pathogen (see Materials and Methods). Addition of the well-characterized P. fluorescens biocontrol strain CHA0 (4) to pathogen-infested soil considerably enhanced plant survival (by about 70%). Root and shoot fresh weights were increased severalfold compared with those of a control without biocontrol bacteria (see Table S3 in the supplemental material). Interestingly, addition of P. entomophila gave similar results, showing that P. entomophila is very efficient as a biocontrol agent in this plant assay. A gacA mutant is impaired in the ability to protect cucumber from the root disease, as revealed by shoot and root fresh weights that were about 40% lower than those of plants protected by wild-type P. entomophila. However, the gacA mutant retains some biocontrol ability, as the plant survival rates and plant fresh weights observed after the addition of this mutant were higher than those of pathogen-treated control plants (see Table S3 in the supplemental material). The etlC mutant (IM3044), deficient in entolysin production, presents the same level of plant protection as the wild-type strain, indicating that entolysin is not involved in P. entomophila biocontrol activity.
DISCUSSION
Entolysin is a new CLP.
In this study, the hemolytic activity of P. entomophila was investigated, which led to the isolation and characterization of a new CLP called entolysin. Compared to other Pseudomonas-produced CLPs, entolysin harbors some specific features, including a peptide moiety of 14 amino acids and a cyclization different from that of other CLPs. Indeed, the lactone ring is formed between the C-terminal carboxyl group (isoleucine) and the 10th amino acid (serine) instead of one of the first amino acids (50). Another example of such a different way of cyclization is found in the putisolvin structures, where it occurs between the 12th and last C-terminal carboxy group (serine) and the 9th amino acid (serine) (36). Interestingly, the putisolvin biosynthesis enzymes are the closest phylogenetically to the NRPSs involved in entolysin synthesis. However, the fatty acid moiety (3-hydroxydecanoic acid) of entolysin is similar to the fatty acid moiety of CLPs belonging to the viscosin or amphisin group (50).
Entolysin was purified as two isoforms, entolysins A and B, indistinguishable from one another by MS/MS analysis. This suggests that the isoleucine position could differ in these isoforms. Phylogenetic analysis of the A domains suggests that isoleucine could be preferentially assembled as the last amino acid of entolysin, but some level of flexibility in A domain specificity has been reported (20, 49).
Three nonribosomal peptide synthetases participate in entolysin biosynthesis.
Using a genetic approach, we showed that the biosynthesis of entolysin requires three NRPSs encoded in two different loci of the P. entomophila genome. Although CLP biosynthesis genes are often part of the same transcriptional unit, such a case has been reported for massetolide A synthesis by P. fluorescens SS101 (15) and viscosin synthesis by P. fluorescens SBW25 (16).
The genetic organization of these two loci reveals that several genes flanking the etl genes are conserved among other Pseudomonas CLP biosynthesis clusters, including two genes encoding a putative macrolide transporter that are located downstream of etlB and etlC, called macA and macB. Similar genes have been identified in arthrofactin (53), syringopeptin and syringomycin (25, 56), massetolide A (15), and putisolvin (23) biosynthesis gene clusters. Inactivation of either of these genes leads to a defect in hemolytic activity, suggesting that the products of these genes could be involved in entolysin secretion.
Another conserved gene found upstream of etlA, called etlR (pseen3335), encodes a transcriptional regulator that is also involved in entolysin production. EtlR contains a C-terminal DNA-binding helix-turn-helix domain typical of the LuxR-type transcriptional regulators but lacks typical N-acylhomoserine lactone-binding or response regulator domains. Interestingly, similar transcriptional regulators, also involved in CLP biosynthesis, have been identified in many other Pseudomonas strains (17).
Finally, another regulator-encoding gene was found upstream of etlB, pseen3046. This gene is not conserved in other Pseudomonas CLP biosynthesis clusters and is not involved in entolysin regulation. This regulator contains an N-terminal DNA-binding domain specific of the LysR-type family, as well as a LysR-like substrate-binding domain. Even if no role for this regulator in entolysin production was observed in our experimental settings, this regulator might play a role under specific environmental conditions.
Analysis of the entolysin biosynthetic enzymes shows that each module of EtlA, EtlB, and EtlC comprises a C domain, an A domain, and a T domain. No additional domain was found, like an internal epimerase domain, for instance, which is characteristic of many Pseudomonas CLP biosynthetic enzymes (50). However, phylogenetic analysis and primary sequence analysis suggest that some of the C domain could have dual catalytic roles for condensation and epimerization, as was described for other CLPs produced by Pseudomonas (5, 15).
Strikingly, the in silico analysis of A domain specificity correlates perfectly with the experimentally determined entolysin structure. Two thioesterase (TE) domains are found at the end of EtlC. TE domains have also been called peptide cyclases, as the cleavage of the linear peptide that they catalyze is often accompanied by an intramolecular cyclization reaction (57). These cyclases display a high level of specificity by selecting a particular residue of the substrate for cyclization, which possibly explains the structural diversity of the peptide ring sizes described previously for the various CLPs (50). Tandem TE domains have been reported for several Pseudomonas CLP biosynthetic enzymes, and it was shown that both of them contribute to macrocyclization of arthrofactin (55). We can hypothesize that the same is true for entolysin.
The GacS/GacA two-component system regulates entolysin production at the biosynthetic gene level.
The global regulatory system GacS/GacA is known to be involved in regulating secondary metabolite production (37). Here, the core components of the GacS/GacA two-component system of P. entomophila were identified. In addition to the GacS sensor and the GacA transcriptional regulator, this system involves the two small RNAs rsmY and rsmZ and two of the three small RNA-binding proteins encoded in the P. entomophila genome, RsmA1 and RsmA2. In addition, we provide evidence that this GacS/GacA two-component system controls entolysin production at the biosynthesis gene level. Analysis of the etlA and etlB putative 5′ leaders revealed the presence of the A/UCANGGANGU/A consensus binding site for RsmA-like proteins, in which the central GGA motif is part of a loop placed on variable short stems. However, no additional GGA motif could be found, in contrast to what was described for the hcnA 5′ leader, a well-studied RsmA target (37). Interestingly, the RsmA consensus binding site is also found upstream of the etlR gene, encoding a LuxR-like regulator also involved in entolysin production. This is consistent with another report that showed that GacS has an effect on massetolide A biosynthetic gene expression through a LuxR-like regulator (18). Here, we can hypothesize that the Gac system exerts a double control on the entolysin biosynthesis genes, directly through RsmAs binding of the etlA and etlB transcripts and indirectly through EltR.
Entolysin is required for swarming motility and hemolytic activity.
Several natural roles for CLPs and other biosurfactants were proposed, including their function in motility, antimicrobial activity, biofilm formation, and pathogenicity (52).
We did not investigate antimicrobial activity and biofilm formation in detail, as no obvious effect of entolysin on these processes could be detected. It was known that P. entomophila inhibits Bacillus sp. growth when they are cocultivated on plates (N. Vodovar, unpublished data), and an entolysin-deficient mutant behaves exactly the same (data not shown). Similarly, P. entomophila is able to attach strongly to plastic, like an entolysin-deficient mutant (I. Vallet-Gely, unpublished data). This makes entolysin different in its function from putisolvins and massetolide A, which influence biofilm formation either positively (massetolide A) (15) or negatively (putisolvins) (36). As we did not look in detail at biofilm structure, the possibility of a role for entolysin in biofilm maturation and structure, as was reported for arthrofactin (53), cannot be excluded. However, an increase in the bacterial biomass of the biofilm of an arthrofactin-deficient mutant was observed, which was not detected in an entolysin-deficient mutant (data not shown).
Reduction of surface tension is essential for surface motility (2). Entolysin appears to be essential for swarming motility, as was observed with many other Pseudomonas-produced CLPs. Interestingly, this surface motility can facilitate movements of plant-pathogenic Pseudomonas spp. on the phylloplane (39) or efficient colonization of the rhizosphere by antagonistic Pseudomonas spp. (40, 64). However, entolysin is not involved in P. entomophila biocontrol activity in our experimental settings. Interestingly, CLPs are not always involved in biocontrol abilities. For instance, massetolide A plays a role in protecting tomato plant roots against Phytophthora infestans (61) but is not required for suppression of complex Pythium sp. populations resident in orchard soils (44). We cannot exclude the possibilities that P. entomophila possesses extended biocontrol activity and that entolysin could play a role under conditions other than those of Pythium infection of cucumber plant roots.
The mode of action of CLPs produced by plant-pathogenic pseudomonads involves the formation of ion channels in the host plasma membrane, leading to cytolysis (13, 31, 32, 48). It was reported that several Pseudomonas sp.-produced CLPs are able to cause erythrocyte hemolysis (6). Recent studies involving surfactin (produced by Bacillus subtilis) showed that the cyclic character of the peptide moiety is important for erythrocyte hemolysis, as linear products of surfactin failed to cause lysis (24). Therefore, it was not surprising to find that the hemolytic activity of P. entomophila is due to the production of a CLP.
Our initial hypothesis was that this hemolytic activity could be linked to the pathogenicity of P. entomophila for Drosophila, especially as this pathogenicity is linked to its ability to cause irreversible damage of the gut epithelium. This hypothesis was not verified in our experimental setup, as an entolysin-deficient mutant was not affected in its pathogenicity. We cannot exclude the possibility that entolysin contributes in a redundant way to the pathogenicity of P. entomophila. Altogether, our study revealed a clear role for entolysin in swarming motility and hemolytic activity but did not identify a physiological role. Nevertheless, it is very likely that this ability to lyse cells is necessary for the bacterium to survive in one of its environments.
Concluding remarks.
There is an increasing scientific and commercial interest in biologically produced surfactants, as they have potential environmental, biotechnological, and pharmacological applications. Entolysin is a new CLP that presents characteristics different from those of previously identified CLPs and is therefore potentially a new molecule of interest.
Even without giving a definitive statement concerning the main physiological role of entolysin, this study clearly provided important information about the lifestyle of P. entomophila. It showed that, according to what was revealed by its genome sequence, this bacterium can be a soil inhabitant. Moreover, considering the high level of biocontrol that P. entomophila provides against Pythium damping-off and root rot, its use may provide an attractive alternative strategy to control this economically important oomycete pathogen.
Supplementary Material
Acknowledgments
We thank Sang-Jin Suh (Auburn, AL) for providing plasmids and Frederic Boccard and Onya Opota for critical reading of the manuscript. We are grateful to Audrey Boniface, Didier Blanot (Orsay, France), and Olivier Binggeli for technical help and insightful discussions.
The laboratories of B.L. were funded, respectively, by CNRS and the Agence Nationale de la Recherche (MIME2005) and by the Swiss National Fund (project 31003A_120709). We acknowledge financial support to C.K. from the Swiss National Science Foundation (project 31003A-120121). The laboratory of P.C. is supported by the Fonds National Suisse pour la Recherche Scientifique. P.C. holds the Doerenkamp-Naef-Zbinden chair at the Geneva Faculty of Medicine. A.N. is a recipient of a young researcher grant from INSERM (France). Teams 1 and 2 were also supported by the Group of Research CNRS GDR 3048.
Footnotes
Published ahead of print on 18 December 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Alibaud, L., T. Kohler, A. Coudray, C. Prigent-Combaret, E. Bergeret, J. Perrin, M. Benghezal, C. Reimmann, Y. Gauthier, C. van Delden, I. Attree, M. O. Fauvarque, and P. Cosson. 2008. Pseudomonas aeruginosa virulence genes identified in a Dictyostelium host model. Cell. Microbiol. 10:729-740. [DOI] [PubMed] [Google Scholar]
- 2.Andersen, J. B., B. Koch, T. H. Nielsen, D. Sorensen, M. Hansen, O. Nybroe, C. Christophersen, J. Sorensen, S. Molin, and M. Givskov. 2003. Surface motility in Pseudomonas sp. DSS73 is required for efficient biological containment of the root-pathogenic microfungi Rhizoctonia solani and Pythium ultimum. Microbiology 149:37-46. [DOI] [PubMed] [Google Scholar]
- 3.Ansari, M. Z., G. Yadav, R. S. Gokhale, and D. Mohanty. 2004. NRPS-PKS: a knowledge-based resource for analysis of NRPS/PKS megasynthases. Nucleic Acids Res. 32:W405-W413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baehler, E., P. de Werra, L. Y. Wick, M. Pechy-Tarr, S. Mathys, M. Maurhofer, and C. Keel. 2006. Two novel MvaT-like global regulators control exoproduct formation and biocontrol activity in root-associated Pseudomonas fluorescens CHA0. Mol. Plant-Microbe Interact. 19:313-329. [DOI] [PubMed] [Google Scholar]
- 5.Balibar, C. J., F. H. Vaillancourt, and C. T. Walsh. 2005. Generation of D amino acid residues in assembly of arthrofactin by dual condensation/epimerization domains. Chem. Biol. 12:1189-1200. [DOI] [PubMed] [Google Scholar]
- 6.Bender, C. L., F. Alarcon-Chaidez, and D. C. Gross. 1999. Pseudomonas syringae phytotoxins: mode of action, regulation, and biosynthesis by peptide and polyketide synthetases. Microbiol. Mol. Biol. Rev. 63:266-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berti, A. D., N. J. Greve, Q. H. Christensen, and M. G. Thomas. 2007. Identification of a biosynthetic gene cluster and the six associated lipopeptides involved in swarming motility of Pseudomonas syringae pv. tomato DC3000. J. Bacteriol. 189:6312-6323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bode, H. B. 2009. Entomopathogenic bacteria as a source of secondary metabolites. Curr. Opin. Chem. Biol. 13:224-230. [DOI] [PubMed] [Google Scholar]
- 9.Bozzaro, S., C. Bucci, and M. Steinert. 2008. Phagocytosis and host-pathogen interactions in Dictyostelium with a look at macrophages. Int. Rev. Cell Mol. Biol. 271:253-300. [DOI] [PubMed] [Google Scholar]
- 10.Buchon, N., N. A. Broderick, M. Poidevin, S. Pradervand, and B. Lemaitre. 2009. Drosophila intestinal response to bacterial infection: activation of host defense and stem cell proliferation. Cell Host Microbe 5:200-211. [DOI] [PubMed] [Google Scholar]
- 11.Cosson, P., and T. Soldati. 2008. Eat, kill or die: when amoeba meets bacteria. Curr. Opin. Microbiol. 11:271-276. [DOI] [PubMed] [Google Scholar]
- 12.Cosson, P., L. Zulianello, O. Join-Lambert, F. Faurisson, L. Gebbie, M. Benghezal, C. Van Delden, L. K. Curty, and T. Kohler. 2002. Pseudomonas aeruginosa virulence analyzed in a Dictyostelium discoideum host system. J. Bacteriol. 184:3027-3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dalla Serra, M., G. Fagiuoli, P. Nordera, I. Bernhart, C. Della Volpe, D. Di Giorgio, A. Ballio, and G. Menestrina. 1999. The interaction of lipodepsipeptide toxins from Pseudomonas syringae pv. syringae with biological and model membranes: a comparison of syringotoxin, syringomycin, and two syringopeptins. Mol. Plant-Microbe Interact. 12:391-400. [DOI] [PubMed] [Google Scholar]
- 14.Daniels, R., S. Reynaert, H. Hoekstra, C. Verreth, J. Janssens, K. Braeken, M. Fauvart, S. Beullens, C. Heusdens, I. Lambrichts, D. E. De Vos, J. Vanderleyden, J. Vermant, and J. Michiels. 2006. Quorum signal molecules as biosurfactants affecting swarming in Rhizobium etli. Proc. Natl. Acad. Sci. U. S. A. 103:14965-14970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Bruijn, I., M. J. de Kock, P. de Waard, T. A. van Beek, and J. M. Raaijmakers. 2008. Massetolide A biosynthesis in Pseudomonas fluorescens. J. Bacteriol. 190:2777-2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Bruijn, I., M. J. de Kock, M. Yang, P. de Waard, T. A. van Beek, and J. M. Raaijmakers. 2007. Genome-based discovery, structure prediction and functional analysis of cyclic lipopeptide antibiotics in Pseudomonas species. Mol. Microbiol. 63:417-428. [DOI] [PubMed] [Google Scholar]
- 17.de Bruijn, I., and J. M. Raaijmakers. 2009. Diversity and functional analysis of LuxR-type transcriptional regulators in cyclic lipopeptide biosynthesis in Pseudomonas fluorescens. Appl. Environ. Microbiol. 75:4753-4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Bruijn, I., and J. M. Raaijmakers. 2009. Regulation of cyclic lipopeptide biosynthesis in Pseudomonas fluorescens by the ClpP protease. J. Bacteriol. 191:1910-1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Lorenzo, V., M. Herrero, U. Jakubzik, and K. N. Timmis. 1990. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J. Bacteriol. 172:6568-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De Souza, J. T., M. De Boer, P. De Waard, T. A. Van Beek, and J. M. Raaijmakers. 2003. Biochemical, genetic, and zoosporicidal properties of cyclic lipopeptide surfactants produced by Pseudomonas fluorescens. Appl. Environ. Microbiol. 69:7161-7172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Déziel, E., F. Lepine, S. Milot, and R. Villemur. 2003. rhlA is required for the production of a novel biosurfactant promoting swarming motility in Pseudomonas aeruginosa: 3-(3-hydroxyalkanoyloxy)alkanoic acids (HAAs), the precursors of rhamnolipids. Microbiology 149:2005-2013. [DOI] [PubMed] [Google Scholar]
- 22.Dove, S. L., and A. Hochschild. 2004. A bacterial two-hybrid system based on transcription activation. Methods Mol. Biol. 261:231-246. [DOI] [PubMed] [Google Scholar]
- 23.Dubern, J. F., E. R. Coppoolse, W. J. Stiekema, and G. V. Bloemberg. 2008. Genetic and functional characterization of the gene cluster directing the biosynthesis of putisolvin I and II in Pseudomonas putida strain PCL1445. Microbiology 154:2070-2083. [DOI] [PubMed] [Google Scholar]
- 24.Dufour, S., M. Deleu, K. Nott, B. Wathelet, P. Thonart, and M. Paquot. 2005. Hemolytic activity of new linear surfactin analogs in relation to their physico-chemical properties. Biochim. Biophys. Acta 1726:87-95. [DOI] [PubMed] [Google Scholar]
- 25.Feil, H., W. S. Feil, P. Chain, F. Larimer, G. DiBartolo, A. Copeland, A. Lykidis, S. Trong, M. Nolan, E. Goltsman, J. Thiel, S. Malfatti, J. E. Loper, A. Lapidus, J. C. Detter, M. Land, P. M. Richardson, N. C. Kyrpides, N. Ivanova, and S. E. Lindow. 2005. Comparison of the complete genome sequences of Pseudomonas syringae pv. syringae B728a and pv. tomato DC3000. Proc. Natl. Acad. Sci. U. S. A. 102:11064-11069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Froquet, R., E. Lelong, A. Marchetti, and P. Cosson. 2009. Dictyostelium discoideum: a model host to measure bacterial virulence. Nat. Protoc. 4:25-30. [DOI] [PubMed] [Google Scholar]
- 27.Goodrich-Blair, H., and D. J. Clarke. 2007. Mutualism and pathogenesis in Xenorhabdus and Photorhabdus: two roads to the same destination. Mol. Microbiol. 64:260-268. [DOI] [PubMed] [Google Scholar]
- 28.Hedengren, M., B. Asling, M. S. Dushay, I. Ando, S. Ekengren, M. Wihlborg, and D. Hultmark. 1999. Relish, a central factor in the control of humoral but not cellular immunity in Drosophila. Mol. Cell 4:827-837. [DOI] [PubMed] [Google Scholar]
- 29.Herbert, E. E., and H. Goodrich-Blair. 2007. Friend and foe: the two faces of Xenorhabdus nematophila. Nat. Rev. Microbiol. 5:634-646. [DOI] [PubMed] [Google Scholar]
- 30.Hoang, T. T., A. J. Kutchma, A. Becher, and H. P. Schweizer. 2000. Integration-proficient plasmids for Pseudomonas aeruginosa: site-specific integration and use for engineering of reporter and expression strains. Plasmid 43:59-72. [DOI] [PubMed] [Google Scholar]
- 31.Hutchison, M. L., and D. C. Gross. 1997. Lipopeptide phytotoxins produced by Pseudomonas syringae pv. syringae: comparison of the biosurfactant and ion channel-forming activities of syringopeptin and syringomycin. Mol. Plant-Microbe Interact. 10:347-354. [DOI] [PubMed] [Google Scholar]
- 32.Hutchison, M. L., M. A. Tester, and D. C. Gross. 1995. Role of biosurfactant and ion channel-forming activities of syringomycin in transmembrane ion flux: a model for the mechanism of action in the plant-pathogen interaction. Mol. Plant-Microbe Interact. 8:610-620. [DOI] [PubMed] [Google Scholar]
- 33.Jiang, H., P. H. Patel, A. Kohlmaier, M. O. Grenley, D. G. McEwen, and B. A. Edgar. 2009. Cytokine/Jak/Stat signaling mediates regeneration and homeostasis in the Drosophila midgut. Cell 137:1343-1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kearns, D. B., and R. Losick. 2003. Swarming motility in undomesticated Bacillus subtilis. Mol. Microbiol. 49:581-590. [DOI] [PubMed] [Google Scholar]
- 35.Kobayashi, N., K. Nishino, and A. Yamaguchi. 2001. Novel macrolide-specific ABC-type efflux transporter in Escherichia coli. J. Bacteriol. 183:5639-5644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuiper, I., E. L. Lagendijk, R. Pickford, J. P. Derrick, G. E. Lamers, J. E. Thomas-Oates, B. J. Lugtenberg, and G. V. Bloemberg. 2004. Characterization of two Pseudomonas putida lipopeptide biosurfactants, putisolvin I and II, which inhibit biofilm formation and break down existing biofilms. Mol. Microbiol. 51:97-113. [DOI] [PubMed] [Google Scholar]
- 37.Lapouge, K., M. Schubert, F. H. Allain, and D. Haas. 2008. Gac/Rsm signal transduction pathway of gamma-proteobacteria: from RNA recognition to regulation of social behaviour. Mol. Microbiol. 67:241-253. [DOI] [PubMed] [Google Scholar]
- 38.Liehl, P., M. Blight, N. Vodovar, F. Boccard, and B. Lemaitre. 2006. Prevalence of local immune response against oral infection in a Drosophila/Pseudomonas infection model. PLoS Pathog. 2:e56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lindow, S. E., and M. T. Brandl. 2003. Microbiology of the phyllosphere. Appl. Environ. Microbiol. 69:1875-1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lugtenberg, B. J., L. Dekkers, and G. V. Bloemberg. 2001. Molecular determinants of rhizosphere colonization by Pseudomonas. Annu. Rev. Phytopathol. 39:461-490. [DOI] [PubMed] [Google Scholar]
- 41.Maget-Dana, R., and F. Peypoux. 1994. Iturins, a special class of pore-forming lipopeptides: biological and physicochemical properties. Toxicology 87:151-174. [DOI] [PubMed] [Google Scholar]
- 42.Marahiel, M. A., T. Stachelhaus, and H. D. Mootz. 1997. Modular peptide synthetases involved in nonribosomal peptide synthesis. Chem. Rev. 97:2651-2674. [DOI] [PubMed] [Google Scholar]
- 43.Matsuyama, T., K. Kaneda, Y. Nakagawa, K. Isa, H. Hara-Hotta, and I. Yano. 1992. A novel extracellular cyclic lipopeptide which promotes flagellum-dependent and -independent spreading growth of Serratia marcescens. J. Bacteriol. 174:1769-1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mazzola, M., X. Zhao, M. F. Cohen, and J. M. Raaijmakers. 2007. Cyclic lipopeptide surfactant production by Pseudomonas fluorescens SS101 is not required for suppression of complex Pythium spp. populations. Phytopathology 97:1348-1355. [DOI] [PubMed] [Google Scholar]
- 45.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 46.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mootz, H. D., D. Schwarzer, and M. A. Marahiel. 2002. Ways of assembling complex natural products on modular nonribosomal peptide synthetases. Chembiochem 3:490-504. [DOI] [PubMed] [Google Scholar]
- 48.Mott, K. A., and J. Y. Takemoto. 1989. Syringomycin, a bacterial phytotoxin, closes stomata. Plant Physiol. 90:1435-1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nielsen, T. H., D. Sorensen, C. Tobiasen, J. B. Andersen, C. Christophersen, M. Givskov, and J. Sorensen. 2002. Antibiotic and biosurfactant properties of cyclic lipopeptides produced by fluorescent Pseudomonas spp. from the sugar beet rhizosphere. Appl. Environ. Microbiol. 68:3416-3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Raaijmakers, J. M., I. de Bruijn, and M. J. de Kock. 2006. Cyclic lipopeptide production by plant-associated Pseudomonas spp.: diversity, activity, biosynthesis, and regulation. Mol. Plant-Microbe Interact. 19:699-710. [DOI] [PubMed] [Google Scholar]
- 51.Rietsch, A., I. Vallet-Gely, S. L. Dove, and J. J. Mekalanos. 2005. ExsE, a secreted regulator of type III secretion genes in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 102:8006-8011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ron, E. Z., and E. Rosenberg. 2001. Natural roles of biosurfactants. Environ. Microbiol. 3:229-236. [DOI] [PubMed] [Google Scholar]
- 53.Roongsawang, N., K. Hase, M. Haruki, T. Imanaka, M. Morikawa, and S. Kanaya. 2003. Cloning and characterization of the gene cluster encoding arthrofactin synthetase from Pseudomonas sp. MIS38. Chem. Biol. 10:869-880. [DOI] [PubMed] [Google Scholar]
- 54.Roongsawang, N., S. P. Lim, K. Washio, K. Takano, S. Kanaya, and M. Morikawa. 2005. Phylogenetic analysis of condensation domains in the nonribosomal peptide synthetases. FEMS Microbiol. Lett. 252:143-151. [DOI] [PubMed] [Google Scholar]
- 55.Roongsawang, N., K. Washio, and M. Morikawa. 2007. In vivo characterization of tandem C-terminal thioesterase domains in arthrofactin synthetase. Chembiochem 8:501-512. [DOI] [PubMed] [Google Scholar]
- 56.Scholz-Schroeder, B. K., J. D. Soule, S. E. Lu, I. Grgurina, and D. C. Gross. 2001. A physical map of the syringomycin and syringopeptin gene clusters localized to an approximately 145-kb DNA region of Pseudomonas syringae pv. syringae strain B301D. Mol. Plant-Microbe Interact. 14:1426-1435. [DOI] [PubMed] [Google Scholar]
- 57.Sieber, S. A., and M. A. Marahiel. 2005. Molecular mechanisms underlying nonribosomal peptide synthesis: approaches to new antibiotics. Chem. Rev. 105:715-738. [DOI] [PubMed] [Google Scholar]
- 58.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 59.Stachelhaus, T., H. D. Mootz, and M. A. Marahiel. 1999. The specificity-conferring code of adenylation domains in nonribosomal peptide synthetases. Chem. Biol. 6:493-505. [DOI] [PubMed] [Google Scholar]
- 59a.Stutz, E., G. Défago, and H. Kern. 1986. Naturally occurring fluorescent pseudomonads involved in suppression of black root rot of tobacco. Phytopathology 76:181-185. [Google Scholar]
- 60.Suh, S. J., L. A. Silo-Suh, and D. E. Ohman. 2004. Development of tools for the genetic manipulation of Pseudomonas aeruginosa. J. Microbiol. Methods 58:203-212. [DOI] [PubMed] [Google Scholar]
- 61.Tran, H., A. Ficke, T. Asiimwe, M. Hofte, and J. M. Raaijmakers. 2007. Role of the cyclic lipopeptide massetolide A in biological control of Phytophthora infestans and in colonization of tomato plants by Pseudomonas fluorescens. New Phytol. 175:731-742. [DOI] [PubMed] [Google Scholar]
- 62.Vodovar, N., D. Vallenet, S. Cruveiller, Z. Rouy, V. Barbe, C. Acosta, L. Cattolico, C. Jubin, A. Lajus, B. Segurens, B. Vacherie, P. Wincker, J. Weissenbach, B. Lemaitre, C. Medigue, and F. Boccard. 2006. Complete genome sequence of the entomopathogenic and metabolically versatile soil bacterium Pseudomonas entomophila. Nat. Biotechnol. 24:673-679. [DOI] [PubMed] [Google Scholar]
- 63.Vodovar, N., M. Vinals, P. Liehl, A. Basset, J. Degrouard, P. Spellman, F. Boccard, and B. Lemaitre. 2005. Drosophila host defense after oral infection by an entomopathogenic Pseudomonas species. Proc. Natl. Acad. Sci. U. S. A. 102:11414-11419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Whipps, J. M. 2001. Microbial interactions and biocontrol in the rhizosphere. J. Exp. Bot. 52:487-511. [DOI] [PubMed] [Google Scholar]
- 65.Yang, S., Q. Peng, Q. Zhang, X. Yi, C. J. Choi, R. M. Reedy, A. O. Charkowski, and C. H. Yang. 2008. Dynamic regulation of GacA in type III secretion, pectinase gene expression, pellicle formation, and pathogenicity of Dickeya dadantii (Erwinia chrysanthemi 3937). Mol. Plant-Microbe Interact. 21:133-142. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.