Abstract
Thermostable direct hemolysin (TDH) and TDH-related hemolysin (TRH) are the major virulence determinants of Vibrio parahaemolyticus. TRH is further differentiated into TRH1 and TRH2 on the basis of genetic and phenotypic differences. We developed a novel and highly specific loop-mediated isothermal amplification (LAMP) assay for sensitive and rapid detection of the tdh, trh1, and trh2 genes of V. parahaemolyticus. The LAMP assay was designed for both combined and individual detection of the tdh, trh1, and trh2 genes and combined detection of the trh1 and trh2 genes. Our results showed that it gave the same results as DNA probes and conventional PCR assays for 125 strains of V. parahaemolyticus, 3 strains of Grimontia hollisae, and 2 strains of Vibrio mimicus carrying the tdh, trh1, and trh2 genes in various combinations. No LAMP products were detected for any of the 20 bacterial strains lacking the tdh, trh1, and trh2 genes. The sensitivities of the LAMP assay for detection of tdh-, trh1-, and trh2-carrying V. parahaemolyticus strains in spiked shrimp samples were 0.8, 21.3, and 5.0 CFU per LAMP reaction tube, respectively. Starting with DNA extraction from a single colony and from spiked shrimp samples, the LAMP assay required only 27 to 60 min and less than 80 min, respectively. This is the first report of a rapid and specific LAMP assay for detection and differentiation of the tdh, trh1, and trh2 genes of V. parahaemolyticus and related Vibrio species.
Vibrio parahaemolyticus, which is widely distributed in estuarine, marine, and coastal environments of tropical and temperate zones, causes seafood-borne gastrointestinal disorders in humans (9). Because most clinical isolates of V. parahaemolyticus produce the thermostable direct hemolysin (TDH), TDH-related hemolysin (TRH), or both (5, 11, 14), these products are considered important virulence markers of V. parahaemolyticus (4, 5, 9, 11, 14). TDH and TRH are encoded by the tdh and trh genes, respectively. Five sequence variants of the tdh gene (tdh1 to tdh5) can be distinguished, which are >97% identical (1, 10). The tdh gene has also been detected in Grimontia (Vibrio) hollisae and some strains of Vibrio mimicus isolated from patients with diarrhea (9). The trh gene shares ca. 68% sequence identity with the tdh gene (5). Although trh gene sequences vary somewhat among strains, the trh variants can be clustered into two subgroups represented by two trh genes (trh1 and trh2), which share 84% sequence identity (5).
Although most clinical isolates carry the tdh and trh genes, either alone or in combination, approximately 99% of environmental isolates do not possess either gene (9). These genes are therefore considered important virulence and epidemiological markers (5, 11, 14). Detection of the tdh and trh genes of V. parahaemolyticus using DNA probe methods is time-consuming and laborious. PCR assays, in contrast, although providing rapid detection of both tdh and trh genes (2, 15), require electrophoresis on an agarose gel, which is time-consuming and tedious. A recent real-time PCR assay for detection of the tdh and trh genes (12) is more rapid than conventional PCR assays but requires sophisticated and expensive equipment.
A recently developed novel nucleic acid amplification method termed loop-mediated isothermal amplification (LAMP) (13) is a promising candidate for rapid and easy detection of the tdh and trh genes. A LAMP assay allows one-step detection of gene amplification by simple turbidity analysis and requires only a simple incubator, such as a heat block or a water bath providing a constant temperature. LAMP assays are faster, easier to perform, and more specific than conventional PCR assays (6, 7). Further, they synthesize a large amount of DNA and its by-product, an insoluble white precipitate of magnesium pyrophosphate, and the by-product can be detected by simple turbidity analysis. The increase in the turbidity of the reaction mixture due to the production of the white precipitate correlates with the amount of DNA synthesized (6, 7, 13). Thus, LAMP assays do not require expensive equipment and are highly precise (3, 18, 19).
Here we describe a rapid and simple LAMP assay for detection of the tdh, trh1, and trh2 genes of V. parahaemolyticus. We also determined the sensitivity of this LAMP assay using spiked shrimp samples.
MATERIALS AND METHODS
Bacterial strains.
Bacterial strains belonging to V. parahaemolyticus, other Vibrio species, and Grimontia (Vibrio) hollisae were used. All cultures were stock cultures from our laboratory which were originally isolated at locations throughout the world over a 30-year period.
Detection of virulence genes.
To detect the tdh and trh genes, a PCR assay was carried out using primers D3 (5′-CCACTACCACTCTCATATGC-3′) and D5 (5′-GGTACTAAATGGCTGACATC-3′) and primers R2 (5′-GGCTCAAAATGGTTAAGCG-3′) and R6 (5′-CATTTCCGCTCTCATATGC-3′), respectively, as previously described (15). trh-positive PCR samples were examined further to determine the presence of the trh1 and trh2 genes by performing a DNA colony hybridization assay as described previously (5, 11). In brief, the DNA probes specific for the trh1 and trh2 genes were 334- and 419-bp DNA fragments, respectively, isolated from recombinant plasmids and were labeled by the random priming method with 32P-labeled dCTP. DNA colony blots were prepared on nitrocellulose membranes, and hybridization was performed under high-stringency conditions (in a solution containing 50% formamide). Hybridization signals on X-ray film were judged visually. Colony blots giving strong and weak hybridization signals with the trh1 and trh2 probes, respectively, were considered positive for the trh1 gene, while colony blots that gave weak and strong hybridization signals with the trh1 and trh2 probes, respectively, were considered positive for the trh2 gene.
Extraction of DNA from pure bacterial cultures for the LAMP assay.
Bacterial DNA for the LAMP assay was extracted as described previously (18, 19). In brief, a loopful (1 μl) of bacterial growth on an appropriate agar medium was inoculated into 50 μl of an NaOH solution (25 mmol liter−1) in a 1.5-ml microcentrifuge tube using a disposable inoculating loop (diameter, ca. 1 mm; Nunc Ltd., Roskilde, Denmark), and the cell mixture was heated at 95°C for 5 min. After neutralization with 4 μl of Tris-HCl buffer (1 mol liter−1, pH 7.5), cell debris was pelleted by centrifugation at 20,000 × g and 4°C for 5 min, and the supernatant was used as the template DNA for the LAMP assay.
Primer design for the LAMP assay.
All primers were designed using Primer Explorer V4 software (Fujitsu System Solutions Ltd., Tokyo, Japan) and were synthesized using sequence-grade purification by Hokkaido System Science Co., Ltd. (Sapporo, Japan). Nucleotide sequences specific for the tdh, trh1, and trh2 genes were detected by multiple alignments of 22 tdh sequences, 6 trh1 sequences, and 12 trh2 sequences available from the DDBJ/EMBL/GenBank database. The sequences selected were used as LAMP primers (Table 1).
TABLE 1.
LAMP primers used in this studya
| Primer | Sequence (5′ to 3′) | Gene location |
|---|---|---|
| Tdh-FIP | GTACCTGACGTTGTGAATACTGATTGTCTCTGACTTTTGGACAAAC | 353-327, 268-288 |
| Tdh-BIP | TGACATCCTACATGACTGTGAACACTTATAGCCAGACACCGC | 362-385, 429-412 |
| Tdh-F3 | AGATATTGTTTGTTGTTCGAGAT | 209-231 |
| Tdh-B3 | AACACAGCAGAATGACCG | 449-432 |
| Tdh-LF | GTACGGTTTTCTTTTTACATTACG | 312-289 |
| Tdh-LB | AAGACTATACAATGGCAGCG | 395-414 |
| Trh1-FIP | AGGCTTGTTTTTTCTGATTTTGTGACTACACAATGGCTGCTCT | 418-394, 343-360 |
| Trh1-BIP | TCTTCTGTTAGTGATTTCGTTGGTTTTCATCCAAATACGTTACACT | 432-455, 498-477 |
| Trh1-F3 | GCGCCTATATGACGGTAA | 310-327 |
| Trh1-B3 | ACATTGACGAAATATTCTGGC | 520-500 |
| Trh1-LF | AGACCGTTGARAGGCC | 393-378 |
| Trh2-FIP | CCGATTGACCGTATACATCTTTGTTGTGGAGGACTATTGGACAA | 493-470, 426-445 |
| Trh2-BIP | TCAAAGTGGTTAAGCGCCTATATGCCATSTTTATAACCAGAAAGAGC | 511-534, 593-571 |
| Trh2-F3 | CATCAATACCTTTTCCTTCTCC | 335-356 |
| Trh2-B3 | GCTTGTTTTCTCTGATTTTGTG | 630-609 |
| Trh2-LF | TGGTTTTCTTTTTATGKTTCGGT | 468-446 |
| Trh2-LB | ATGGTCAYAACTATACRATGGC | 548-569 |
LAMP assay.
The LAMP assay was performed using a Loopamp DNA amplification kit (Eiken Chemical Co. Ltd., Tokyo, Japan) as previously described (18, 19). Individual LAMP assays, which targeted the tdh, trh1, and trh2 genes individually, were performed using 25-μl 1× reaction mixtures (Eiken Chemical) containing 1 μl of template DNA, 1 μl of Bst DNA polymerase (Eiken Chemical), 1.6 μmol liter−1 each of inner primers FIP and BIP, 0.2 μmol liter−1 each of outer primers F3 and B3, and 0.8 μmol liter−1 each of loop primers LF and LB. The reaction mixtures were incubated in a Loopamp real-time turbidimeter (LA-320; Teramecs Co. Ltd., Kyoto, Japan) at 65°C for 60 min and then at 80°C for 2 min to complete the reaction. LAMP was detected in real time by determining the turbidity at 650 nm using an LA-320 turbidimeter. A reaction was considered positive when the turbidity reached 0.1 within 60 min. White precipitation visible with the unaided eye was also considered an indication of a successful LAMP procedure (6).
LAMP assays using a mixture of the tdh, trh1, and trh2 primer sets and a mixture of the trh1 and trh2 primer sets were also performed to evaluate if multiplex assays were possible under the same conditions.
Sensitivity of the LAMP assay with spiked shrimp samples. (i) Preparation of shrimp samples.
The sensitivity of the LAMP assay with spiked shrimp (Trachypenaeus curvirostris) samples was determined as previously described (17, 18), with slight modifications. Shrimp samples were purchased at a supermarket in Osaka, Japan, in 2009, and whole bodies were used without peeling the shells. For preparation of the template DNA, 25 g of a shrimp sample was added to 225 ml of alkaline peptone water (APW) (Eiken Chemical) in a plastic stomacher bag and stomached for 30 s. After overnight incubation at 36°C, APW enrichment cultures were confirmed to be V. parahaemolyticus negative by plating onto thiosulfate-citrate-bile salts-sucrose (TCBS) agar and examination of the APW enrichment cultures using a V. parahaemolyticus species-specific LAMP assay (18). V. parahaemolyticus-negative APW enrichment cultures were spiked with positive and negative control strains for the sensitivity test.
(ii) Preparation of bacterial cells.
In parallel, a small amount of bacterial growth on TCBS agar was inoculated into 4 ml of tryptic soy broth (TSB) (Becton Dickinson) and incubated overnight at 36°C. Then 40 μl of an enriched TSB culture was transferred to 4 ml of TSB and incubated for 4 to 5 h with shaking at 150 rpm at 36°C in a water bath to obtain mid-log-phase cells. Serial 10-fold dilutions of the test strain cultures were prepared in phosphate-buffered saline (pH 7.5).
(iii) Preparation of template DNA.
In a parallel experiment, a 100-μl mid-log-phase APW culture of a negative control strain, V. parahaemolyticus VP02 (tdh negative, trh negative; 5.7 × 107 CFU/ml), and 100 μl each of serial dilutions of positive control strains in the mid-log phase were inoculated into 900 μl of APW enrichment broth in 1.5-ml microcentrifuge tubes. After thoroughly mixed using a Vortex-Genie 2 (Scientific Industries Inc., NY), the spiked samples were centrifuged at 900 × g for 1 min to remove larger debris. Each resulting supernatant was transferred to a new 1.5-ml microcentrifuge tube and centrifuged at 10,000 × g for 5 min. After removal of the supernatant, the pellets were resuspended in 100 μl of NaOH (25 mmol liter−1), thoroughly mixed using a Vortex-Genie 2, and then heated at 95°C for 5 min. After neutralization with 8 μl of Tris-HCl buffer (1 mol liter−1, pH 7.5), debris was removed by centrifugation at 20,000 × g at 4°C for 5 min. Supernatants were collected, and 1- or 2-μl portions were used as the template DNAs for LAMP assay.
(iv) Determination of sensitivity.
Using the template DNAs prepared as described above, the sensitivities of the individual LAMP assays for the tdh, trh1, and trh2 genes were determined in triplicate, and the detection limits were defined as the highest dilutions at which all three samples tested positive. The sensitivities of the multiplex LAMP assays (combined detection of the tdh, trh1, and trh2 genes and combined detection of the trh1 and trh2 genes) were also determined as described above, except that primers for the three and two genes were used simultaneously. In parallel, to determine the inoculum size, 100 μl of each dilution was plated onto tryptic soy agar (TSA) (Nissui, Tokyo, Japan) supplemented with 2.5% NaCl in duplicate and incubated overnight at 36°C. Further, the sensitivity of the LAMP assay for spiked APW enrichment cultures with different ratios of the bacteria (AQ3815/AQ4037/AT4/VP02 ratios of 1:1:1:1, 2:2:2:1, 4:4:4:1, and 8:8:8:1, which corresponded to CFU ratios of 0.8:21.3:5.0:105,000, 1.7:42.6:10.0:105,000, 3.4:85.2:20.0:105,000, and 6.7:170.4:40.0:105,000, respectively, in LAMP reaction tubes) was determined.
RESULTS
LAMP products were detected for all 125 strains of V. parahaemolyticus, 3 strains of G. hollisae, and 2 strains of V. mimicus carrying the tdh, trh1, and trh2 genes in various combinations (Table 2). No LAMP products were detected for any of the 20 bacterial strains not carrying the tdh, trh1, and trh2 genes. These results were perfectly consistent with those obtained by PCR and DNA colony hybridization assays.
TABLE 2.
Results of LAMP assays for detection of the tdh, trh1, and trh2 genes
| Species | Genotypea | No. of strains tested | No. of LAMP-positive strains using a primer set(s) for: |
||||
|---|---|---|---|---|---|---|---|
| tdh | trh1 | trh2 | tdh, trh1, and trh2 | trh1 and trh2 | |||
| V. parahaemolyticus | tdh+, trh negative | 30 | 30 | 0 | 0 | 30 | 0 |
| V. parahaemolyticus | tdh+, trh1+ | 32 | 32 | 32 | 0 | 32 | 32 |
| V. parahaemolyticus | tdh negative, trh1+ | 13 | 0 | 13 | 0 | 13 | 13 |
| V. parahaemolyticus | tdh+, trh2+ | 13 | 13 | 0 | 13 | 13 | 13 |
| V. parahaemolyticus | tdh negative, trh2+ | 37 | 0 | 0 | 37 | 37 | 37 |
| V. parahaemolyticus | tdh negative, trh negative | 10 | 0 | 0 | 0 | 0 | 0 |
| G. hollisae | tdh+, trh negative | 3 | 3 | 0 | 0 | 3 | 0 |
| V. mimicus | tdh+, trh negative | 2 | 2 | 0 | 0 | 2 | 0 |
| V. alginolyticus | tdh negative, trh negative | 3 | 0 | 0 | 0 | 0 | 0 |
| V. cholerae | tdh negative, trh negative | 1 | 0 | 0 | 0 | 0 | 0 |
| V. fluvialis | tdh negative, trh negative | 2 | 0 | 0 | 0 | 0 | 0 |
| V. furnissii | tdh negative, trh negative | 1 | 0 | 0 | 0 | 0 | 0 |
| V. metschnikovii | tdh negative, trh negative | 1 | 0 | 0 | 0 | 0 | 0 |
| V. mimicus | tdh negative, trh negative | 1 | 0 | 0 | 0 | 0 | 0 |
| V. vulnificus | tdh negative, trh negative | 1 | 0 | 0 | 0 | 0 | 0 |
Determined by PCR and DNA colony hybridization assays.
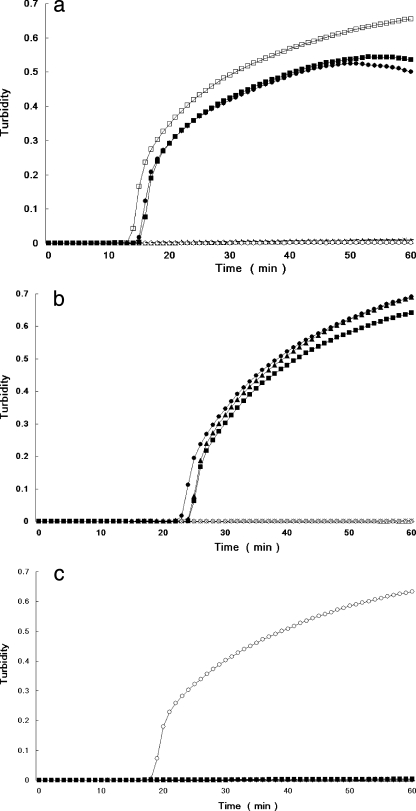
The PCR assay took more than 4 h, but the LAMP assay using DNA extracted from a single colony on TCBS agar was markedly faster, requiring only 14 to 22 min for amplification of tdh, 24 to 38 min for amplification of trh1, and 18 to 28 min for amplification of trh2. Further, the multiplex LAMP assay for comprehensive detection of the hemolysin genes required 14 to 36 min for amplification of tdh, trh1 and trh2 and 20 to 47 min for amplification of trh1 and trh2. Representative results for the individual tdh, trh1, and trh2 LAMP assays obtained using an LA-320 turbidimeter are shown in Fig. 1a, 1b, and 1c, respectively. Results for combined detection of tdh, trh1, and trh2 and for combined detection of trh1 and trh2 are shown in Fig. 1d and 1e, respectively. When tdh-bearing G. hollisae and V. mimicus strains were used, the times for the LAMP assay were 20 to 23 min and 16 to 18 min, respectively, for tdh detection and 24 to 25 min and 16 to 18 min, respectively, for tdh detection by the multiplex (tdh, trh1, and trh2) system using DNA extracted from a single colony on TCBS agar or TSA (data not shown). All LAMP detection assays, including the DNA extraction step, were completed in 27 to 60 min. An increase in turbidity due to the LAMP reaction was confirmed both by using the LA-320 turbidimeter and by visual inspection, and the results of the two methods agreed perfectly.
FIG. 1.
Real-time monitoring of LAMP reactions with V. parahaemolyticus control strains for multiplex and individual detection of the tdh, trh1, and trh2 genes. Profiles for individual detection of tdh (a), trh1 (b), and trh2 (c) and for multiplex detection of tdh, trh1, and trh2 (d) and of trh1 and trh2 (e) are shown. Total DNAs extracted from AQ3815 (tdh1+, tdh2+, trh1 negative, trh2 negative) (□), AQ3776 (tdh3+, tdh4+, trh1+, trh2 negative) (▪), AQ3860 (tdh5+, trh1+, trh2 negative) (•), AQ4037 (tdh negative, trh1+, trh2 negative) (▴), AT4 (tdh negative, trh1 negative, trh2+) (○), and Vp02 (tdh negative, trh1 negative, trh2 negative [negative control]) (×) were examined.
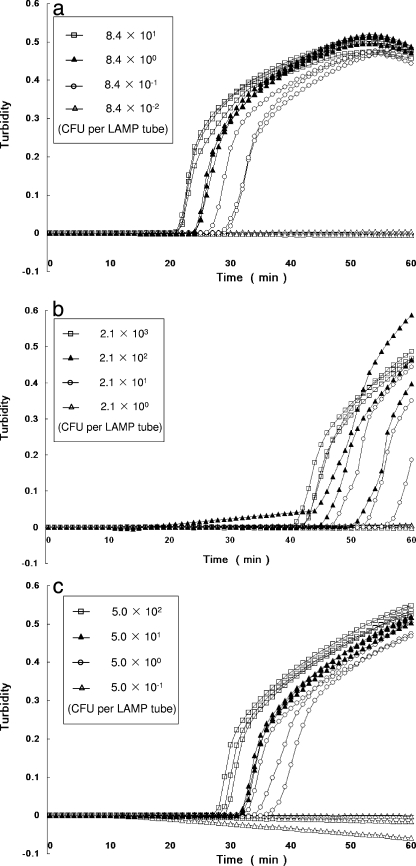
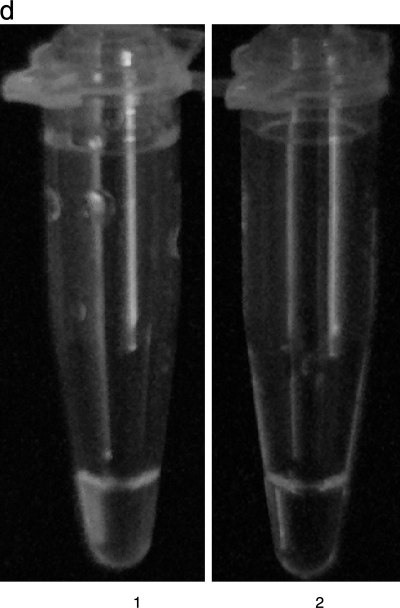
As shown in Fig. 2 and Table 3, the sensitivities of the individual LAMP assays for V. parahaemolyticus AQ3815 (tdh+, trh negative), AQ4037 (tdh negative, trh1+) and AT4 (tdh negative, trh2+) in spiked shrimp samples were 0.8, 21.3, and 5.0 CFU per LAMP tube, respectively. Dilutions yielding 84.4 to 0.8 CFU per LAMP tube (Fig. 2d, tube 1), 2,130 to 21.3 CFU per LAMP tube, and 500 to 5.0 CFU per LAMP tube showed an increase in turbidity, whereas dilutions yielding 0.1 CFU per LAMP tube (Fig. 2d, tube 2), 2.1 CFU per LAMP tube, and 0.5 CFU per LAMP tube did not show an increase. DNA amplification in the LAMP assay could also be judged using the white precipitate by visual assessment with the unaided eye. Representative LAMP results used for visual assessment with the unaided eye are shown in Fig. 2d. The sensitivities of the two methods were the same.
FIG. 2.
Sensitivity test for detection of the tdh, trh1, trh2 genes in V. parahaemolyticus strains in spiked shrimp samples by real-time turbidimetry. The curves from left to right indicate decreasing concentrations of CFU from bacterial colonies. (a) Detection of the tdh gene of V. parahaemolyticus AQ3815. (b) Detection of the trh1 gene of V. parahaemolyticus AQ4037. (c) Detection of the trh2 gene of V. parahaemolyticus AT4. (d) Visual detection of the tdh gene by observation of precipitation. Tube 1, 84.4 CFU of AQ3815; tube 2, 0.1 CFU of AQ3815.
TABLE 3.
Sensitivities of the LAMP assays for multiplex and individual detection of the tdh, trh1, and trh2 genes
| Strain | Genotype | Sensitivity of the LAMP assay (CFU/LAMP tube) using a primer set(s) fora: |
||||
|---|---|---|---|---|---|---|
| tdh | trh1 | trh2 | tdh, trh1, and trh2 | trh1 and trh2 | ||
| AQ3815 | tdh+ | 0.8 | NDb | ND | 1.7 | ND |
| AQ4037 | trh1+ | ND | 21.3 | ND | 85.2 | 21.3 |
| AT4 | trh2+ | ND | ND | 5.0 | 5.0 | 5.0 |
The values are the lower limits of the number of viable cells detectable by the LAMP assay.
ND, not determined.
When sensitivity tests with shrimp inoculated with known amounts of positive control strains (AQ3815 [tdh+], AQ4037 [trh1+], and AT4 [trh2+]) were performed using the two multiplex LAMP assays with mixtures of the tdh, trh1, and trh2 primer sets and the trh1 and trh2 primer sets, the sensitivities were slightly lower than those obtained with the LAMP assay for individual gene detection (Table 3). Further, when sensitivity tests were carried out using shrimp samples inoculated with various amounts of the positive control strains and Vp02, a negative control strain, the two multiplex LAMP assays exhibited slightly higher sensitivities than the LAMP assays for individual gene detection (Table 4). In these sensitivity tests, all of the results obtained with the two multiplex LAMP assays were in agreement (Table 4).
TABLE 4.
Sensitivity of the LAMP assay using DNA template mixtures obtained from small amounts of bacterial cells
| No. of bacterial cells (CFU/LAMP tube) |
LAMP results (no. positive/no. tested) using a primer set(s) fora: |
|||||||
|---|---|---|---|---|---|---|---|---|
| AQ3815 | AQ4037 | AT4 | Vp02 | tdh | trh1 | trh2 | tdh, trh1, and trh2 | trh1 and trh2 |
| 6.7 | 170.4 | 40.0 | 105,000 | ND | + (3/3) | ND | ND | ND |
| 3.4 | 85.2 | 20.0 | 105,000 | ND | ± (2/3) | ND | ND | ND |
| 1.7 | 42.6 | 10.0 | 105,000 | + (3/3) | ± (2/3) | + (3/3) | ND | ND |
| 0.8 | 21.3 | 5.0 | 105,000 | ± (2/3) | − (0/3) | ± (2/3) | + (3/3) | + (3/3) |
+, triplicate assays were all positive. ±, triplicate assays were both positive and negative; −, triplicate assays were all negative; ND, not determined.
DISCUSSION
Here we report a novel and highly specific LAMP assay for detection of the tdh, trh1, and trh2 genes. This assay provides markedly simpler and more rapid detection of the tdh, trh1, and trh2 genes than conventional PCR and DNA colony hybridization assays.
While our study was in progress, Nemoto and colleagues described a tdh LAMP assay performed with 42 strains of V. parahaemolyticus (8). However, this assay did not include a system to detect the trh1 and trh2 genes, which are also important virulence factors of V. parahaemolyticus. In contrast, our LAMP assay detects and differentiates the tdh, trh1, and trh2 genes and was evaluated using 135 strains of V. parahaemolyticus carrying these genes in various combinations. These strains represent isolates obtained from clinical cases in various locations around the world collected over a 30-year period. These assays should therefore prove to be useful for facilitating clinical diagnosis and for epidemiological investigations of outbreaks of food-borne V. parahaemolyticus infection.
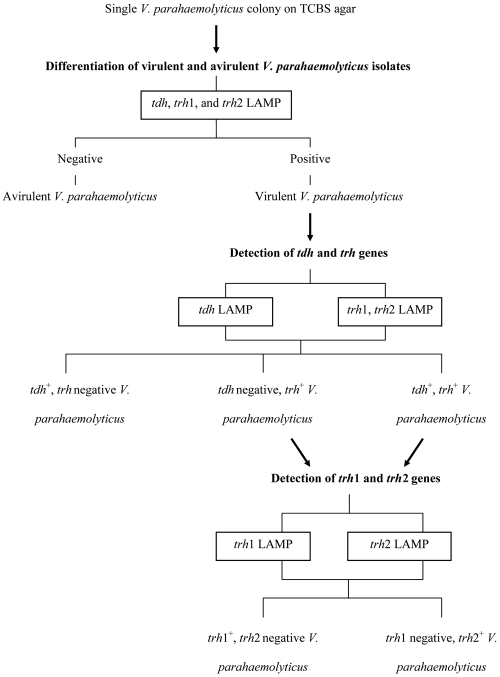
Figure 3 shows a scheme for a suggested practical application of our LAMP system. The multiplex LAMP system, which allows detection of any of the tdh, trh1, and trh2 genes, is first system used to distinguish virulent and avirulent strains. Virulent strains are then distinguished into groups of strains with the tdh and trh genes by the tdh-specific and trh-specific LAMP assays, and the latter group is subsequently separated into trh1 and trh2 subtypes by the trh1- and trh2-specific LAMP assays.
FIG. 3.
Schematic representation of detection and differentiation of the tdh, trh1, and trh2 genes from a single V. parahaemolyticus colony on TCBS agar.
All strains of G. hollisae and some strains of V. mimicus and V. cholerae isolated from patients with diarrhea are known to possess the tdh gene (9). Three G. hollisae and two V. mimicus tdh-carrying strains examined in this study were positive for the tdh gene as determined by our individual tdh LAMP assay. If this LAMP assay and the V. parahaemolyticus species-specific LAMP assay that we described previously (18) are combined, tdh-bearing non-V. parahaemolyticus species can be identified quickly and easily.
In our preliminary tests, at least 10 primer sets each were designed for the tdh, trh1, and trh2 genes and were evaluated for their specificity and speed of amplification (data not shown). The best sets were then selected (Table 1) and proven to be highly successful for the individual LAMP systems, as well as both of the multiplex LAMP systems (Tables 2, 3, and 4 and Fig. 2). One of the preliminary primer sets for the trh1 gene required only 14 to 25 min for amplification (data not shown). When a preliminary set of mixed primers comprising the trh1 primers, as well as the tdh and trh2 primers, that was finally adopted in the present study was used, the amplification time was conspicuously extended (35 to 70 min), and this set was thus considered unsuitable for rapid LAMP system detection. A possible explanation for this extension of the amplification time was the influence of competitive overlapping of the three primer sets in the coding region. The coding regions of the tdh, trh1, and trh2 genes are 567 bp long and have high levels of sequence identity (around 68% for the tdh and trh genes and 84% for the trh1 and trh2 genes) (5). A trh1 primer set designed with another region was used as a second choice. We attempted to design a Trh1-LB primer for the sequence between two segments of the newly designed Trh1-BIP primer, corresponding to the gene location between positions 456 and 476 in S67850. However, a Trh1-LB primer could not be designed due to the insufficient G+C content of the region that the primer was intended to locate. This in turn decreased the sensitivity of the trh1 LAMP assay due to the lack of an LB primer to accelerate the LAMP reaction. As representative results in Fig. 1d show, comprehensive detection of the tdh, trh1, and trh2 genes was obtained in 14 to 36 min using the combination of the three primer sets, and therefore, we used these primer sets in the final assay.
The sensitivity of the tdh LAMP assay with spiked shrimp samples shown in Table 3 and Fig. 2 (0.8 CFU per LAMP reaction tube in triplicate) may appear to be somewhat high. However, previous studies have demonstrated that optimized LAMP assays performed with both LF and LB primers to accelerate the reaction can detect the target cells at concentrations on the order of 100 CFU per LAMP reaction tube (17, 20). On this basis, the sensitivity of our LAMP assays appears to be reasonable. As shown in Fig. 2c, the turbidity for 5.0 × 10−1 CFU per LAMP tube decreased to less than zero. A possible explanation for this phenomenon is the gradual sedimentation of the extracted APW enrichment broth cultures containing food components during LAMP because this phenomenon was also observed in our previous studies using DNA templates from fecal and food samples (17-19). In contrast, this phenomenon was never observed in our previous studies (17-19) or the present study when DNA templates from pure bacterial cultures were used.
When dilutions of AQ3815, AQ4037, AT4, and Vp02 cells in mixtures containing different amounts of the bacteria were used, the sensitivities of the two sets of multiplex LAMP assays (one detecting the tdh, trh1, and trh2 genes and the other detecting the trh1 and trh2 genes) were not decreased. The sensitivities of the individual tdh, trh1, and trh2 LAMP assays were affected, however, with 2-, 8-, and 2-fold decreases, respectively (Table 4), possibly due to genetic similarities of the three target sequences. On this basis, the multiplex LAMP assays would be useful for detecting virulent strains in seafood samples containing tdh, trh1, and trh2 at concentrations of 1.7, 170.4, and 10.0 CFU per LAMP reaction tube, corresponding to 9.2 × 101, 9.2 × 103, and 5.4 × 102 CFU per ml in APW enrichment cultures, respectively.
Risk assessment for V. parahaemolyticus in seafood is an increasingly important issue worldwide. However, current methods for quantitative detection of virulent strains are inconvenient and time-consuming (16). A method combining a three-tube most-probable-number culture method and PCR examination for virulent strains is useful but cumbersome (16). The sensitivity of the LAMP assay is at least 10-fold higher than that of a conventional PCR assay and typically equivalent to that of a real-time PCR assay (17-20). The high sensitivity of our LAMP assay and the fact that the amplification reaction results can be judged by simple visual assessment (Fig. 2) make it a more attractive tool for risk assessment, and future studies based on this characteristic would likely make a valuable contribution. Worldwide, outbreaks caused by virulent V. parahaemolyticus are frequent, highlighting the need for effective control of contaminants in seafood. The LAMP assay is a rapid, simple, and practical method which may facilitate surveillance for virulent V. parahaemolyticus contamination in seafood, screening of contaminated seafood samples before consumption, and investigations to determine the causative agents of food poisoning, as well as ecological studies associated with environmental factors, seasons, regions, and practices.
Acknowledgments
This work was supported in part by the Project for Zoonoses Education and Research, University of Miyazaki, by a grant-in-aid from the Ministry of Health, Labor and Welfare, Japan, and by a grant-in-aid for scientific research (KAKENHI 19101010) from the Japan Society for the Promotion of Sciences.
Footnotes
Published ahead of print on 4 December 2009.
REFERENCES
- 1.Baba, K., H. Shirai, A. Terai, Y. Takeda, and M. Nishibuchi. 1991. Analysis of the tdh gene cloned from a tdh gene- and trh gene-positive strain of Vibrio parahaemolyticus. Microbiol. Immunol. 35:253-258. [DOI] [PubMed] [Google Scholar]
- 2.Bej, A. K., D. P. Patterson, C. W. Brasher, M. C. L. Vickery, D. D. Jones, and C. A. Kaysner. 1999. Detection of total and hemolysin-producing Vibrio parahaemolyticus in shellfish using multiplex PCR amplification of tl, tdh and trh. J. Microbiol. Methods 36:215-225. [DOI] [PubMed] [Google Scholar]
- 3.Hill, J., S. Beriwal, I. Chandra, V. K. Paul, A. Kapil, T. Singh, R. M. Wadowsky, V. Singh, A. Goyal, T. Jahnukainen, J. R. Johnson, P. I. Tarr, and A. Vats. 2008. Loop-mediated isothermal amplification assay for rapid detection of common strains of Escherichia coli. J. Clin. Microbiol. 46:2800-2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Honda, S., I. Goto, I. Minematsu, N. Ikeda, N. Asano, M. Ishibashi, Y. Kinoshita, N. Nishibuchi, T. Honda, and T. Miwatani. 1987. Gastroenteritis due to Kanagawa negative Vibrio parahaemolyticus. Lancet i:331-332. [DOI] [PubMed] [Google Scholar]
- 5.Kishishita, M., N. Matsuoka, K. Kumagai, S. Yamasaki, Y. Takeda, and M. Nishibuchi. 1992. Sequence variation in the thermostable direct hemolysin-related hemolysin (trh) gene of Vibrio parahaemolyticus. Appl. Environ. Microbiol. 58:2449-2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mori, Y., K. Nagamine, N. Tomita, and T. Notomi. 2001. Detection of loop-mediated isothermal amplification reaction by turbidity derived from magnesium pyrophosphate formation. Biochem. Biophys. Res. Commun. 289:150-154. [DOI] [PubMed] [Google Scholar]
- 7.Nagamine, K., T. Hase, and T. Notomi. 2002. Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol. Cell. Probes 16:223-229. [DOI] [PubMed] [Google Scholar]
- 8.Nemoto, J., C. Sugawara, K. Akahane, K. Hashimoto, T. Kojima, M. Ikedo, H. Konuma, and Y. Hara-Kudo. 2009. Rapid and specific detection of the thermostable direct hemolysin gene in Vibrio parahaemolyticus by loop-mediated isothermal amplification. J. Food Prot. 72:748-754. [DOI] [PubMed] [Google Scholar]
- 9.Nishibuchi, M., and J. B. Kaper. 1995. Thermostable direct hemolysin gene of Vibrio parahaemolyticus: a virulence gene acquired by a marine bacterium. Infect. Immun. 63:2093-2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nishibuchi, M., and J. B. Kaper. 1990. Duplication and variation of the thermostable direct haemolysin (tdh) gene in Vibrio parahaemolyticus. Mol. Microbiol. 4:87-99. [DOI] [PubMed] [Google Scholar]
- 11.Nishibuchi, M., and J. B. Kaper. 1985. Nucleotide sequence of the thermostable direct hemolysin gene of Vibrio parahaemolyticus. J. Bacteriol. 162:558-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nordstrom, J. L., M. C. Vickery, G. M. Blackstone, S. L. Murray, and A. DePaola. 2007. Development of a multiplex real-time PCR assay with an internal amplification control for the detection of total and pathogenic Vibrio parahaemolyticus bacteria in oysters. Appl. Environ. Microbiol. 73:5840-5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Notomi, T., H. Okayama, H. Masubuchi, T. Yonekawa, K. Watanabe, N. Amino, and T. Hase. 2000. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 28:E63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shirai, H., H. Ito, T. Hirayama, Y. Nakamoto, N. Nakabayashi, K. Kumagai, Y. Takeda, and M. Nishibuchi. 1990. Molecular epidemiologic evidence for association of thermostable direct hemolysin (TDH) and TDH-related hemolysin of Vibrio parahaemolyticus with gastroenteritis. Infect. Immun. 58:3568-3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tada, J., T. Ohashi, N. Nishimura, Y. Shirasaki, H. Ozaki, S. Fukushima, J. Takano, M. Nishibuchi, and Y. Takeda. 1992. Detection of the thermostable direct hemolysin gene (tdh) and the thermostable direct hemolysin-related hemolysin gene (trh) of Vibrio parahaemolyticus by polymerase chain reaction. Mol. Cell. Probes 6:477-487. [DOI] [PubMed] [Google Scholar]
- 16.Yamamoto, A., J. Iwahori, V. Vuddhakul, W. Charernjiratragulc, D. Vose, K. Osaka, M. Shigematsu, H. Toyofuku, S. Yamamoto, M. Nishibuchi, and F. Kasuga. 2008. Quantitative modeling for risk assessment of Vibrio parahaemolyticus in bloody clams in southern Thailand. Int. J. Food Microbiol. 124:70-78. [DOI] [PubMed] [Google Scholar]
- 17.Yamazaki, W., M. Taguchi, T. Kawai, K. Kawatsu, J. Sakata, K. Inoue, and N. Misawa. 2009. Comparison of loop-mediated isothermal amplification assay and conventional culture methods for detection of Campylobacter jejuni and Campylobacter coli in naturally contaminated chicken meat samples. Appl. Environ. Microbiol. 75:1597-1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamazaki, W., M. Ishibashi, R. Kawahara, and K. Inoue. 2008. Development of a loop-mediated isothermal amplification assay for sensitive and rapid detection of Vibrio parahaemolyticus. BMC Microbiol. 8:163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamazaki, W., K. Seto, M. Taguchi, M. Ishibashi, and K. Inoue. 2008. Sensitive and rapid detection of cholera toxin-producing Vibrio cholerae using a loop-mediated isothermal amplification. BMC Microbiol. 8:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yano, A., R. Ishimaru, and R. Hujikata. 2007. Rapid and sensitive detection of heat-labile I and heat-stable I enterotoxin genes of enterotoxigenic Escherichia coli by loop-mediated isothermal amplification. J. Microbiol. Methods 68:414-420. [DOI] [PubMed] [Google Scholar]