Abstract
Sulfidic, anoxic sediments of the moderately hypersaline Salton Sea contain gradients in salinity and carbon that potentially structure the sedimentary microbial community. We investigated the abundance, community structure, and diversity of Bacteria and Archaea along these gradients to further distinguish the ecologies of these domains outside their established physiological range. Quantitative PCR was used to enumerate 16S rRNA gene abundances of Bacteria, Archaea, and Crenarchaeota. Community structure and diversity were evaluated by terminal restriction fragment length polymorphism (T-RFLP), quantitative analysis of gene (16S rRNA) frequencies of dominant microorganisms, and cloning and sequencing of 16S rRNA. Archaea were numerically dominant at all depths and exhibited a lesser response to environmental gradients than that of Bacteria. The relative abundance of Crenarchaeota was low (0.4 to 22%) at all depths but increased with decreased carbon content and increased salinity. Salinity structured the bacterial community but exerted no significant control on archaeal community structure, which was weakly correlated with total carbon. Partial sequencing of archaeal 16S rRNA genes retrieved from three sediment depths revealed diverse communities of Euryarchaeota and Crenarchaeota, many of which were affiliated with groups previously described from marine sediments. The abundance of these groups across all depths suggests that many putative marine archaeal groups can tolerate elevated salinity (5.0 to 11.8% [wt/vol]) and persist under the anaerobic conditions present in Salton Sea sediments. The differential response of archaeal and bacterial communities to salinity and carbon patterns is consistent with the hypothesis that adaptations to energy stress and availability distinguish the ecologies of these domains.
The vast majority of cultured Archaea isolates are characterized as extremophiles, which thrive under environmental extremes of temperature, pH, salinity, and oxygen availability. Unlike Bacteria, these organisms are well defined by select physiologies or catabolic activities. Cultivated halophilic archaea are obligate aerobes, and with a few exceptions (58), most 16S rRNA gene sequences affiliated with this physiological group have been recovered primarily from environments with oxygen present. Thermophilic archaea, many of which utilize hydrogen-based metabolisms, have temperature requirements that preclude their survival and growth in more moderate environments. Other archaeal physiological groups include acidophiles, which thrive in acidic and mostly high-temperature environments, the obligate anaerobic methanogens, which are capable of competing with Bacteria when more energetically favorable electron acceptors are not available (i.e., sulfate), and methane-oxidizing archaea, which require methane for energy production. Recent work on several Crenarchaeota isolates points to nitrification as their primary energy metabolism, but these organisms have been detected in cold, predominantly aerobic environments, such as open ocean waters and soil (47), and in hyperthermophilic environments (24).
Several archaeal groups identified using only 16S rRNA genes, for which no current isolates exist, have been detected in anaerobic sediments of the marine subsurface (6), estuaries (42), freshwater (46), and salt lakes (29). While their physiology and catabolism remain a source of speculation, the environmental distribution patterns of these mesophilic, presumably anaerobic, groups seemingly exclude the physiological and catabolic types outlined above. That is, the persistence of diverse archaeal populations in anoxic sediments at moderate temperature and salinity and at circumneutral pH with only trace levels of methane strongly suggests that alternative metabolic or physiological activities must characterize these populations.
Saline lakes are ubiquitous and can be found on all continents. Although many saline lakes are labeled “extreme” environments, microbial diversity within their sediments is often equivalent to that reported for studies of freshwater and marine systems (28). Most studies of the microbial ecology within saline lakes have focused on gradients within the water column, with very few studies on patterns within the sediments. Specifically, these studies have examined how changes in water column salinity lead to shifts in microbial productivity and diversity (8). However, particle-associated microbial communities are known to differ fundamentally from water column or free-living populations (1, 18). These observed differences could be explained by the type and strength of environmental gradients that microbial communities in sediments experience, as opposed to those encountered by pelagic communities.
Sediments contain strong environmental gradients, such as time (e.g., sediment age at depth), nutrient and carbon availability, and the dominant terminal electron-accepting process (TEAP) resulting from the sequential use of available oxidants by the microbial community (41). These gradients can lead to changes in the dominant microbial groups (i.e., a shift from sulfate reducers to methanogens with depth and age). Many saline lakes are highly productive and shallow and experience large fluctuations in water level due to climatic changes or to changes in inflows due to urban and agricultural activities. Changes in lake level can lead to dramatic shifts in mixing regimens, nutrient cycling, and water chemistry. Historic fluctuations in water column salinity are often recorded within the sediments in the form of evaporite deposits, which may act as additional sources of ionic loading of the water column (62). These sedimentary salinity gradients may modulate the metabolic activity of some microbial groups. For example, Oren (44) proposed bioenergetic constraints as a possible explanation for the reduced activity or absence of some microbial groups within high-salinity environments. Thus, saline lake sediments are excellent natural laboratories in which to study changes and adaptations of microbial communities due to large-scale changes in environmental gradients.
The Salton Sea is a large (980 km2), eutrophic, moderately hypersaline (48 to 50 g liter−1), terminal lake located 69 m below sea level in the Salton Basin, CA. Several large lakes have formed in the Salton Basin over geologic history, the most recent of which was Lake Cahuilla ca. 300 years ago (7). The current lake was unintentionally created in 1905-1907, when the Colorado River flooded the Salton Basin for a period of 16 months. Profundal sediments are highly sulfidic, and sulfate reduction is suspected to be the dominant TEAP within these sediments (54). Based on elemental analysis (51) and 137Cs activity (37) of sediment layers, a depth of ∼22 cm marks the point when flooding of the Salton Basin occurred. Sediment above this depth represents the ca. 102 years of historical change within the Salton Sea, including a shift from a water column salinity of 35 g liter−1 to the hypersaline conditions that currently exist. Sediments below this depth consist of low-carbon, gypsum-rich evaporite deposits that were present on the older dry lake bed prior to the formation of the current lake. A previous study reported several strong geochemical gradients within pore water across this relatively small depth range (62).
In this work, a suite of cultivation-independent techniques and geochemical analyses was utilized to correlate shifts in abundance, community structure, and diversity of Archaea and Bacteria in Salton Sea sediments with changes in environmental gradients. Large differences in abundance and community structure patterns of Archaea and Bacteria were found along the gradients. In addition, the majority of archaeal sequences retrieved were affiliated with previously described but as yet uncultivated groups identified from various marine sedimentary environments. This indicates that these groups are able to tolerate the higher salinity and anaerobic conditions characteristic of Salton Sea sediments. Fundamental differences between the metabolic capacities and ecologies of Archaea and Bacteria are discussed to explain these patterns.
MATERIALS AND METHODS
Sediment collection and processing.
Intact sediment cores were collected at a site (S-1; 33°25′00"N, 115°55′00"W; 14 m in depth) located in the center of the northern basin of the Salton Sea. This site was one of several reference sampling stations chosen for a reconnaissance study of the biological limnology of the Salton Sea during the period 1997-1999 (63) and is a location where sediment cores representing the complete history of the extant lake can be collected. Bulk sediment was collected using a modified Ekman box corer (15 × 15 × 50 cm) and subsampled using polycarbonate cylinders (7.5 × 50 cm). This method yielded sediment cores with an average length of 35 cm. Sediment from one core was extruded at 1-cm intervals, and the sediment from each interval was then split. Following the method of Wardlaw and Valentine (62), pore water was extracted from one-half of the sediment by centrifugation (12,000 × g, 15 min) and filtered through a 0.2-μm filter. The remaining sediment was stored at −80°C for subsequent molecular analyses. Sediment from a second core was extruded at 1-cm intervals, placed in acid-washed (20% HCl), precombusted (450°C, 4 h) glass vials, and stored at −20°C prior to analysis of bulk sediment chemical properties.
Geochemical analysis.
The concentration of total dissolved salts in pore water was determined by the refractive index, measured using a handheld refractometer. Samples that had a salinity value of >80 g liter−1 were quantitatively diluted with distilled water prior to measurement. Since the ionic composition of Salton Sea pore water differs from that of seawater, exact values determined using this technique should be interpreted with caution. Pore water sulfate concentrations were determined using a nephelometric method based on the precipitation of barium sulfate (BaSO4) through the addition of excess barium chloride (BaCl2) solution to a known volume of pore water (23). The reported accuracy of this method (∼1 mM) (23) makes it particularly useful for samples with high sulfate concentrations (62). Total carbon (TC), organic carbon (OC; after treatment of sample with 25% HCl), and total nitrogen (TN) contents of bulk sediment were determined from dried (105°C, 48 h), ground sediment samples, using a CHN elemental analyzer (model CEC 440HA; Exeter Analytical). Inorganic carbon (IC) content was determined by subtraction of the OC value from the TC value.
Mineral identification and analysis.
Sediment subsamples (0.2 to 0.5 g) were collected at 2-cm intervals along the 0- to 28-cm sediment depth range and dried onto 5-mm glass slides. Mineral-phase identification and analysis were conducted with powder X-ray diffractometry (XRD), using a Phillips X'Pert diffractometer. The X-ray source was Cu Kα radiation (1.5405 Å) at 40 kV and 50 mA. Diffraction peaks between 2θ values of 2° and 70° were recorded, and peaks were identified using the peak search function of Phillips X'Pert Highscore software (PANalytical). Minerals were identified by comparing the observed mixed-phase diffraction pattern to the ICDD-PDF mineral database. Relative abundances of each mineral phase were determined using the “semiquantitative analysis” function in the X'Pert Highscore software package. This calculation is based on reference intensity ratio values for the mineral assemblage (12). For sediment samples, relative abundances of the following minerals were determined using the “fit profile” command: calcite (ICDD-PDF reference code 00-005-0586), gypsum (ICDD-PDF reference code 99-000-0004), halite (ICDD-PDF reference code 00-005-0628), and silica (ICDD-PDF reference code 00-033-1161).
DNA extraction and T-RFLP analysis.
DNAs were extracted from subsamples (0.3 to 0.5 g) of previously frozen sediment by use of a FastDNA spin kit for soil (Qbiogene, Carlsbad, CA). DNAs from two separate subsamples were extracted, combined, further purified with a Wizard SV gel and PCR cleanup system (Promega, Madison, WI) according to the manufacturer's instructions, and quantified by PicoGreen fluorometry (Molecular Probes, Eugene, OR). Archaeal and bacterial community structure was determined at 2-cm depth intervals, using terminal restriction fragment length polymorphism (T-RFLP) analysis (5). Duplicate PCR (50-μl) reaction mixtures were prepared and contained 50 ng template DNA, 0.2 μM forward primer, 0.2 μM reverse primer, 1× Qiagen PCR buffer, 1.5 mM MgCl2, a 200 μM concentration of each deoxynucleoside triphosphate, 0.2 mM bovine serum albumin (BSA; New England Biolabs [NEB], Ipswich, MA), and 2.5 U Taq DNA polymerase. Partial bacterial and archaeal 16S rRNA genes were amplified using labeled (6-carboxyfluorescein [FAM]; Eurofins MWG Operon) forward and unlabeled reverse primers (Table 1). The specificity of the archaeal reverse primer was confirmed using in silico amplification against databases, using the RDP v10 Probe Match tool (15). Archaeal PCR amplification conditions were as follows: 3-min hot start at 95°C, followed by 30 cycles (45 s of denaturing at 94°C, 1 min of annealing, and 2 min of extension at 72°C) and a final extension step of 5 min at 72°C. Bacterial PCR amplification conditions were as follows: 2-min hot start at 95°C, followed by 30 cycles (45 s of denaturing at 95°C, 45 s of annealing, and 90 s of extension at 72°C) and a final extension step of 7 min at 72°C. Amplicon size was checked by running PCR products on a 0.8% agarose gel for 30 min at 80 V and visualizing them by staining with SYBR gold (Molecular Probes).
TABLE 1.
PCR primers and conditions used in this study
| Primer | Sequence (5′→3′) | Annealing temp (°C) | Targeted group | Reference |
|---|---|---|---|---|
| Primers for T-RFLP | ||||
| 27f | AGAGTTTGATCMTGGCTCAG | 55 | Bacteria | 35 |
| 1392r | ACGGGCGGTGTGTRC | 55 | Universal | 35 |
| Primers for T-RFLP/cloning | ||||
| 519f | CAGCMGCCGCGGTAATWC | 57 | Universal | 36 |
| 1397r | GTGTGCAAGGRGCAGGGA | 57 | Archaea | Unpublished |
| Primers for qPCR | ||||
| Bac331f | TCCTACGGGAGGCAGCAGT | 64.5 | Bacteria | 40 |
| Bac797r | GGACTACCAGGGTCTAATCCTGTT | 64.5 | Bacteria | 40 |
| Arch349f | GYGCASCAGKCGMGAAW | 50 | Archaea | 56 |
| Arch806r | GGACTACVSGGGTATCTAAT | 50 | Archaea | 56 |
| 771f | ACGGTGAGGGATGAAAGCT | 54 | Crenarchaeota | 43 |
| 957r | CGGCGTTGACTCCAATTG | 54 | Crenarchaeota | 43 |
Duplicate PCR products containing amplicons of the correct size were combined, purified, and concentrated using a Wizard SV gel and PCR cleanup system (Promega). Duplicate 40-μl reaction mixtures containing 50 ng PCR product, 5.0 μl 1× enzyme buffer (20×; NEB), 0.5 μl BSA (10 mg ml−1), and 0.5 U of restriction enzyme (NEB) were digested for 8 h. The following restriction enzymes were used: HhaI, DdeI, and DpnI for archaeal PCR products and HhaI, HaeIII, and RsaI for bacterial PCR products. Digestion products were purified and concentrated using a Montage PCR centrifugal device (Millipore, Billerica, MA) and quantified using PicoGreen. Samples were diluted to have equivalent final DNA concentrations and were run in duplicate on an ABI Prism 3100 genetic analyzer (PE Applied Biosystems, Foster City, CA) at the Genomics Technology Support Facility of Michigan State University. Resulting electropherograms were analyzed using GeneScan 3.1 software (Perkin-Elmer).
Community structure analysis.
Terminal restriction fragment (TRF) peaks of ≥50 fluorescence units (above background) and constituting ≥0.5% of the total sample peak area were manually aligned by comparison to an internal size standard. Duplicate T-RFLP profiles were compared, and nonreplicated TRF peaks were removed from further analyses. Replicated TRF peak areas were averaged and divided by the total sample area (based on replicated TRF peaks only) to obtain the individual relative peak area for each TRF.
Microbial community structure patterns were evaluated using hierarchal cluster analysis and nonmetric multidimensional (MDS) analysis (13). Hierarchal cluster analysis was performed on TRF relative abundance and presence-absence data, using the Bray-Curtis (BC) similarity index and flexible-beta linkage (β = −0.25). MDS is an ordination technique that plots samples as points in low-dimensional space while attempting to maintain the relative distances between points as close as possible to the actual rank order of similarities between samples. Thus, sampling stations with similar community structures are plotted closer together in ordination space. A stress factor is calculated for each MDS ordination and indicates how well plotted configurations of sample distances agree with original rank orders calculated from the similarity matrices. MDS analysis of T-RFLP profiles was performed using previously calculated BC indices for TRF relative abundances.
Relationships between microbial community structure and environmental variables were examined using the BEST procedure (14), which calculates modified Spearman rank correlation coefficients (ρs) between MDS configurations and Euclidean similarity matrices of all possible environmental variable combinations. Hydrogen sulfide concentrations were log transformed before calculating Euclidean similarities, whereas dissolved oxygen and temperature values were not transformed. Cluster analysis was done using XLStat (AddinSoft SARL), and all MDS calculations were performed using Primer v6.0 (Primer-E Ltd.).
Phylogenetic assignment of TRFs.
The putative phylogenetic identities and relative abundances of bacterial community members determined from T-RFLP profiles were estimated at selected sediment depths by use of several Web-based programs. First, several 16S rRNA gene sequence databases (Ribosomal Database Project [RDP] v10 [15] and Greengenes [21]) and the In silico PCR and DNA Restriction (ISPaR) tool, a component of the Microbial Community Analysis (MiCA) online resource (53), were used to generate predicted TRF lengths of bacterial 16S rRNA gene sequences. Primers and restriction enzymes used in ISPaR were identical to those used in the sediment T-RFLP analysis. Second, predicted TRF lengths of 16S rRNA gene sequences were imported into PAT (31) and matched to TRF lengths generated from sediment T-RFLP analysis. Third, normalized abundance equations for each TRF peak and its relative peak area in each T-RFLP profile were calculated using the APLAUS tool in MiCA. Abundance equations were then simplified by performing algebraic calculations by hand, further reducing the number of possible phylotypes present in each sample. This procedure resulted in several mathematically valid plausible community compositions for each sample; however, only one plausible community composition was determined in our analysis, and phylogenetic assignments were restricted mostly to the phylum or class level.
In agreement with previous studies, few matches between archaeal T-RFLP patterns and archaeal 16S rRNA gene sequences present in databases were found. Therefore, expected TRFs of archaeal clones obtained in this study were matched to T-RFLP sediment profiles by use of TRiFLe (30). First, partial archaeal 16S rRNA gene sequences of clones from three sediment depth intervals were digested in silico, using PCR primers and the HhaI restriction enzyme that were used for sediment T-RFLP analysis. Second, sediment T-RFLP profiles from each depth interval were matched to expected TRFs from clone sequences by use of TRiFLe. Finally, relative abundances of matched TRFs were used to estimate the relative abundances of Euryarchaeota and Crenarchaeota over depth.
qPCR.
Quantitative real-time PCR (qPCR) was performed using an ABI 7900 HT real-time PCR system (Applied Biosystems) and the primers and annealing conditions listed in Table 1. PCR mixtures (25 μl) contained 12.5 μl of 1× Power SYBR green master mix (Applied Biosystems), 0.9 μM primer (each; final concentration), 1 μl of DNA template (1 ng), and ultrapure water for the balance. Each reaction condition had an initial denaturing step of 10 min at 95°C to activate the AmpliTaq Gold DNA polymerase, followed by 40 cycles of 30 s of denaturing at 95°C, 30 s of primer annealing (Table 1), and 30 s of primer extension at 72°C. Melting curve analysis (0.2°C s−1) and gel electrophoresis (2% agarose) of qPCR products revealed single amplicons of the predicted sizes for all primer pairs. Annealing temperatures used were optimized for each primer pair. Plasmid quantification standards were constructed for each primer pair by cloning PCR products resulting from amplification of environmental DNA. Exact matches between 16S rRNA sequences of PCR product inserts and qPCR primer pairs were confirmed by sequencing. DNA copy numbers of PCR product inserts contained in clones were calculated, and standard curves were generated by serial dilution (3.0 × 103 to 3.0 × 109 DNA copies μl−1). Amplification of environmental samples and standards, including controls containing no DNA template (ultrapure water only), was done in triplicate. Average amplification efficiencies for Bacteria, Archaea, and Crenarchaeota were 89%, 85%, and 97%, respectively, and all amplifications were linear (r2 = 0.999, 0.990, and 0.997, respectively).
Archaeal clone libraries.
Archaeal 16S rRNA gene clone libraries were constructed from the sediment depth intervals 0 to 1 cm, 22 to 23 cm, and 32 to 33 cm. PCR conditions and primers were the same as those used for archaeal T-RFLP analysis (Table 1). Clone libraries were constructed from PCR products by use of a PCR CloningPlus kit (Qiagen, Valencia, CA). Blue-white screening of cloned plasmids was done using LB agar plates containing 100 μg ml−1 ampicillin, 80 μg ml−1 X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside), and 50 μM IPTG (isopropyl-β-d-thiogalactopyranoside; Teknova, Hollister, CA). Picked transformants were grown overnight in 1.6 ml LB broth containing 0.075 mg ml−1 ampicillin, and plasmid DNA was extracted using a QuickClean 5 M miniprep kit (GenScript, Piscataway, NJ) according to the manufacturer's instructions. Approximately 500 ng of DNA from each plasmid containing a PCR product insert was sequenced at the University of California, Berkeley, DNA sequencing facility, using the T7-M13 sequencing primer pair. The average sequence length was 890 nucleotides.
Phylogenetic analysis and clone library comparisons.
Sequence ends were trimmed and edited using Sequencher v4.7 (Gene Codes, Ann Arbor, MI) and were checked for chimeras using Mallard (4) and Bellerophon (26). Suspect sequences were removed prior to further analysis. Unique phylotypes (≥97% sequence similarity) were determined using DOTUR (48). A single representative phylotype sequence was included in the alignments. Edited sequences were compared to previously deposited sequences, using the RDP v10 Seqmatch (15) and NCBI BLASTN (3) online tools. Multiple sequence alignments of the most closely matched isolate and environmental sequences and the clone sequences from this study were performed using CLUSTAL_X v1.81 (57).
Neighbor-joining (NJ) trees of sequences, with the Jukes-Cantor nucleotide substitution correction and 1,000 distance bootstrap replicates, were constructed using PAUP* v.4.0 beta 10 (55). Phylogenetic trees were exported and edited using TreeView v1.6.0 (45). Phylogenetic trees were also constructed using maximum parsimony and maximum likelihood methods, and resulting tree topologies were similar to those generated using NJ methods (data not shown). Clone library diversity was calculated using EstimateS v7.5 (17), richness estimates were calculated using DOTUR (48), and library coverage was calculated using Good's index. Estimated shared richness, community overlap (66), and community structure similarity (65) between clone libraries were calculated using SONS (49).
Nucleotide sequence accession numbers.
Sequence data have been submitted to the GenBank database under accession numbers EU329738 to EU329844.
RESULTS
Bulk sediment and pore water chemistry.
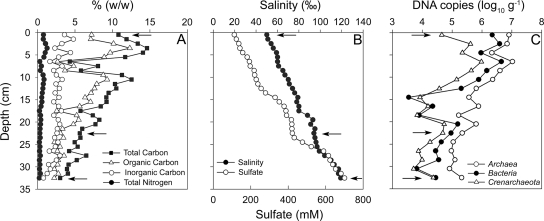
TC, OC, and TN contents (% [wt/wt]) all decreased with increasing depth, whereas IC remained relatively constant (Fig. 1A). The sediment OC content ranged between 12.3 and 2.3%, and IC was <5% over most depths; however, the amount of TC comprised of IC increased from an average of 30% to 40% at depths below 20 cm. Ratios of OC to TN by mass (9.2 to 13.5) were similar to those reported for marine coastal sediments. Pore water salinity (% [wt/vol]) more than doubled over the sampling depth, increasing from 5.0 to 11.8% (50 to 118‰), and sulfate concentrations increased from 112 to 701 mM (Fig. 1B).
FIG. 1.
Depth profiles of bulk sediment (A), pore water chemistry (B), and abundance of major microbial groups determined by qPCR (C). Arrows indicate depth intervals selected for archaeal 16S rRNA gene sequencing.
Powder X-ray diffractometry analysis indicated that calcite, gypsum, halite, and silica were the most abundant minerals present. With the exception of the near-surface (0 to 2 cm) and deeper (23 to 28 cm) sediments, calcite was the most abundant mineral and had an average relative abundance of 73% (wt/wt). Gypsum was detected only in the depth intervals in which calcite abundances were lowest (0 to 2 cm, 67 to 72%; and 23 to 28 cm, 29 to 69%). Average relative abundances of halite and silica were 23% and 6%, respectively, and showed little variation with depth. The high relative abundances of halite are most likely an artifact resulting from the drying process during sample preparation.
Quantification of 16S rRNA genes.
The abundances of major microbial groups, as determined by qPCR, were reported as DNA copy numbers of 16S rRNA genes per g of dry sediment. The abundances of total microbes (Archaea and Bacteria) and of Archaea were 105 to 107 DNA copies g−1, whereas Bacteria and Crenarchaeota abundances were 104 to 106 DNA copies g−1 (Fig. 1C). Archaeal 16S rRNA genes were dominant (64 to 99%) at all sediment depths, and Crenarchaeota abundance, as a percentage of total Archaea, varied from 0.4 to 22%. Changes in the abundances of Crenarchaeota and Bacteria over depth were similar (Fig. 1C).
Microbial community structure patterns.
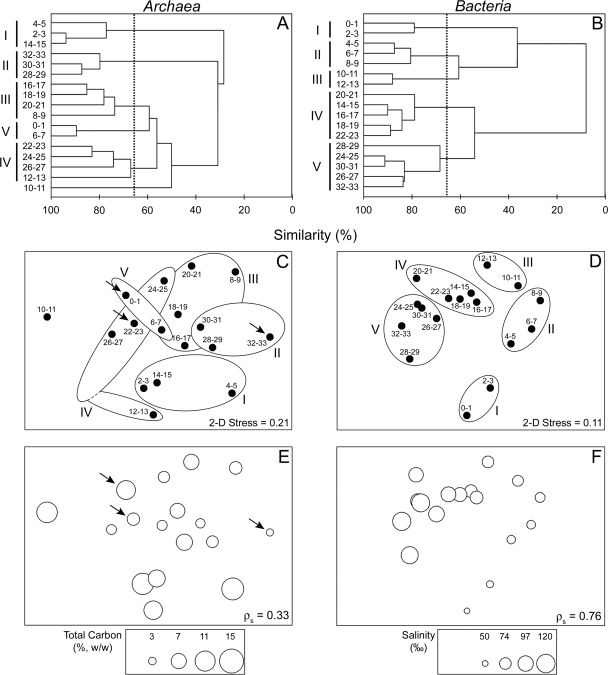
Three different restriction enzymes were used to generate TRFs for Archaea and Bacteria. Although the number of TRFs generated with each restriction enzyme was variable, community structure patterns determined by multivariate techniques were similar. Therefore, only results for the HhaI enzyme are presented. Relative TRF peak areas were used in the analysis of community structure, but similar patterns were found using presence-absence data. Cluster analysis revealed five distinct subgroups (I to V) with ≥65% similarity for both microbial groups (Fig. 2A and B). Two major bacterial subgroups with <10% similarity were observed (Fig. 2B). Additionally, an overall trend with sediment depth was found for the bacterial community, where adjacent sediment depths were most similar to each other. No such trend with depth was apparent for the archaeal community.
FIG. 2.
Hierarchical cluster analyses (A and B) and nonmetric MDS plots (C to F) of arcsine square root-transformed relative T-RFLP peak areas. Distance matrices used for both analyses were calculated using the Bray-Curtis similarity index. Sediment depth intervals (cm) are listed on the y axes, and dotted lines (A and B), ellipses (C and D), and Roman numerals (A to D) designate subgroups with ≥65% similarity. Corresponding concentrations of environmental variables most correlated with community structure are plotted as open circles (E and F). Arrows indicate depth intervals selected for archaeal 16S rRNA gene sequencing (C and E). 2-D, two-dimensional; ρs, Spearman rank correlation coefficient. Only results of clustering and MDS analyses obtained using data from HhaI restriction are presented. MDS topology and resulting stress values obtained using other restriction enzymes were similar.
MDS was used to perform ordination of T-RFLP profiles (Fig. 2C and D). An examination of the stress values, which represent unexplained information, determined that two dimensions were sufficient for explaining community patterns. Bacterial community patterns from MDS analysis closely matched those obtained by hierarchal clustering (Fig. 2B and D), further validating the overall trend, and were correlated with pore water salinity (ρs = 0.76) (Fig. 2F). Archaeal community structure patterns revealed by MDS analysis also matched those from hierarchal clustering (Fig. 2A and C). Archaeal community structure patterns were weakly correlated with TC (ρs = 0.33) and pore water salinity (ρs = 0.11) (Fig. 2E).
Bacterial community composition patterns.
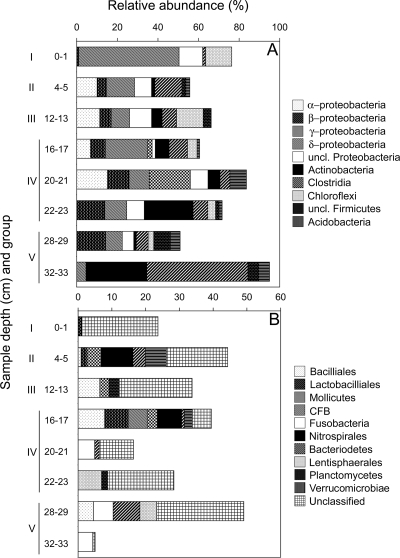
Bacterial 16S rRNA gene sequences representing a total of 16 bacterial phylogenetic groups were matched to T-RFLP profiles by use of APLAUS (Fig. 3). Abundance trends for several bacterial groups were evident. Within the Proteobacteria, the abundance of Betaproteobacteria increased with depth (and therefore salinity). Gammaproteobacteria abundance decreased with depth, and Alphaproteobacteria abundance showed little change with depth. The abundance of Firmicutes members (Clostridia, Bacilliales, Lactobacilliales, and Mollicutes) and Actinobacteria also generally increased with depth. Members of the Deltaproteobacteria, which include dissimilatory sulfate reducers, were detected only within the 16- to 17-cm and 20- to 21-cm depth intervals. Matches to members of the class Dehalococcoides, which are members of the phylum Chloroflexi, were found in the 16- to 17-cm and 28- to 29-cm depth intervals. The presence of this group was also detected only within sediments from depths of >8 cm by use of Dehalococcoides-specific PCR primers (data not shown).
FIG. 3.
Relative abundances of major bacterial groups identified by APLAUS at selected sediment depth intervals. CFB, Cytophaga-Flavobacterium-Bacteriodes group; uncl. and unclassified, unknown taxon. Roman numerals correspond to major subgroups identified by hierarchical cluster and MDS analyses, shown in Fig. 2. Note the different x axis scales in panels A and B.
Archaeal clone library richness and diversity.
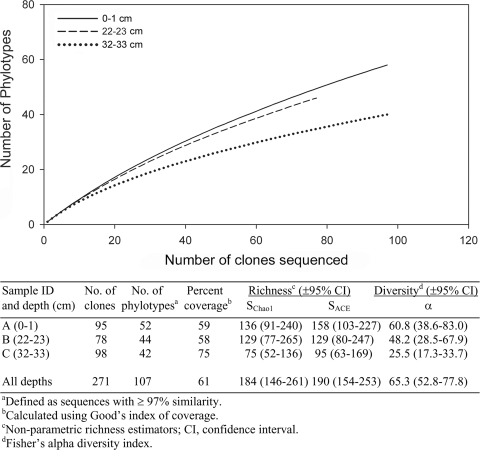
A total of 271 sequences representing 107 phylotypes was identified from clone libraries (Fig. 4). Rarefaction curves, coverage estimates, and two estimators of species richness (SChao1 and SACE [9, 10]) indicated that sediment depth intervals were not completely sampled to capture the total estimated richness at each depth (Fig. 4). Estimates of phylotype richness and diversity for each clone library were high, although any significant differences in these measures between libraries were not supported due to large confidence intervals (Fig. 4). The number of archaeal phylotypes recovered from clone libraries was also relatively large compared to estimates from other sediment environments (2).
FIG. 4.
Rarefaction curves showing phylotype (≥97% similarity) richness of partial archaeal 16S rRNA gene clone libraries from three sediment depth intervals. The table lists characteristics of individual and combined clone libraries.
Phylogenetic analysis of Archaea.
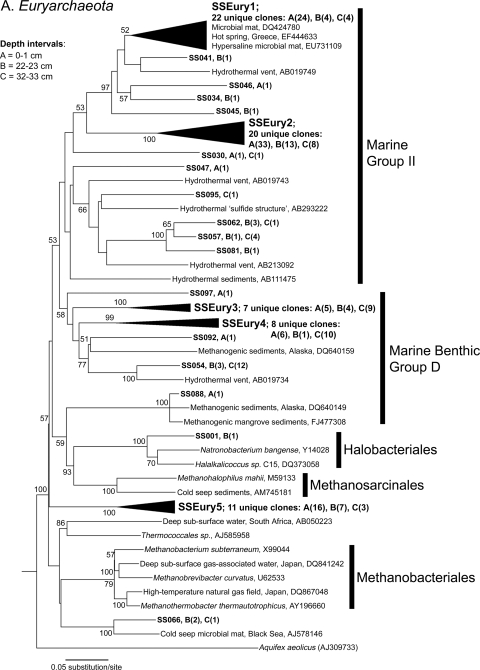
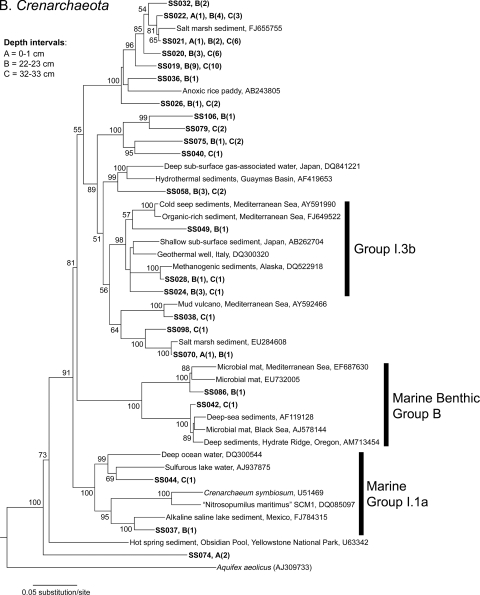
All 107 archaeal phylotypes were classified within the Euryarchaeota or Crenarchaeota phyla (Fig. 5A and B). The majority of phylotypes (79% [84 of 107 phylotypes]) were classified within the Euryarchaeota, whereas 21% (23 of 107 phylotypes) were classified within the Crenarchaeota. Only one euryarchaeotal phylotype (clone SS001) was classified within a group that included cultured representatives, Halobacteriales, and it was most similar (90%) to Halalkalicoccus sp. strain C15 (Fig. 5A). The majority of Euryarchaeota phylotypes grouped within several previously recognized archaeal groups, including marine group II (MG-II) and marine benthic group D (MBG-D), and a few were distantly related to the Methanosarcinales and Methanobacteriales. Approximately one-third (7 of 23 phylotypes) of the Crenarchaeota phylotypes were grouped within the uncultured environmental clades MG-I.1a (20), MG-I.3b, and MBG-B. Remaining crenarchaeotal phylotypes were not affiliated with any known clades and grouped with uncultured organism sequences recovered from marine, estuarine, and anoxic rice paddy sediments.
FIG. 5.
Phylogenetic relationships of Euryarchaeota (A) and Crenarchaeota (B) Salton Sea clone sequences (in bold) from three sediment depth intervals, as well as previously reported environmental sequences and associated GenBank accession numbers. Trees were constructed using the NJ algorithm with Jukes-Cantor-corrected DNA distances. Bootstrap (1,000 replicates) values of >50 are indicated at the nodes. The number of sequences within each clone group is shown in parentheses, and the depth interval in which each clone was found is indicated by a letter. The scale bars represent a 5% sequence difference, and Aquifex aeolicus was used as an outgroup.
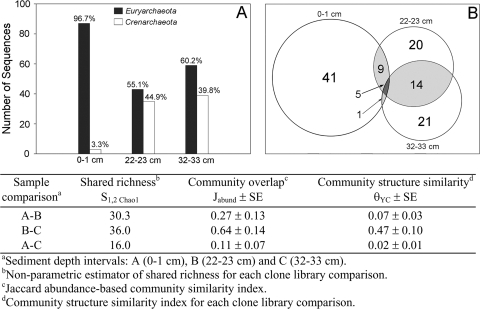
The distributions of Euryarchaeota and Crenarchaeota over depth were investigated by comparing numbers of total sequences and phylotypes recovered, as well as numbers of phylotypes of each group shared between depth intervals. Comparisons between the numbers of sequences affiliated with Crenarchaeota and Euryarchaeota indicated that Crenarchaeota sequences were more abundant in the deeper sediment layers (Fig. 6A). The largest number of shared phylotypes was between the 22- to 23-cm and 32- to 33-cm clone libraries, whereas the 0- to 1-cm clone library shared few phylotypes with those from deeper sediment (Fig. 6B). These trends were also reflected in the results from several statistical pairwise comparisons of clone library structure and composition (Fig. 6). The estimated number of shared phylotypes (S1,2 Chao1) was lowest between the 0- to 1-cm and 32- to 33-cm depth intervals. Trends in estimates of the probability that a phylotype from one of two libraries being compared is found in both (Jabund) and the similarity of relative abundance estimates of phylotypes shared between libraries (θYC) further support the observed compositional differences between clone libraries.
FIG. 6.
Total number of Euryarchaeota and Crenarchaeota 16S rRNA gene sequences identified in each sediment depth interval (A) and number of archaeal phylotypes shared between sediment depth intervals (B). The table lists community richness and similarity measures for each clone library comparison.
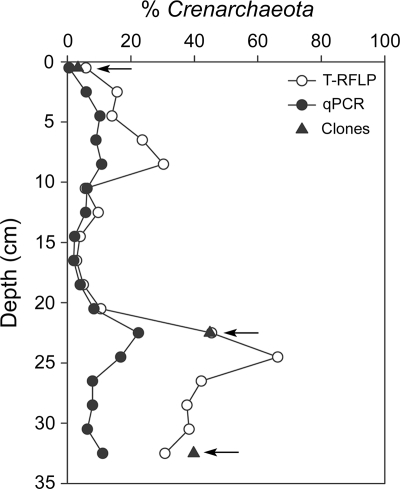
The relative abundance of Crenarchaeota as a function of total archaeal abundance (qPCR) or total archaeal 16S rRNA gene sequences recovered (T-RFLP and clone library composition analyses) increased with sediment depth in two distinct regions: within the 0- to 8-cm and 20- to 26-cm depth intervals (Fig. 7). The largest percentage of Crenarchaeota organisms was found in the deepest sediment layers (>20 cm). Although estimates of Crenarchaeota abundance were dependent on the methodology used, the general trends with sediment depth were similar between methods.
FIG. 7.
Relative abundance of Crenarchaeota as a function of total archaeal abundance, determined by three culture-independent 16S rRNA gene-based techniques.
DISCUSSION
Microbial abundance patterns along environmental gradients.
qPCR estimates of microbial group abundances revealed that the anoxic sediments of the Salton Sea were dominated by Archaea at all depths. Percent abundance values equaled or exceeded (64 to 99%) those measured from marine subsurface sediments (6) and other saline lakes (29). On average, the abundance of Bacteria was an order of magnitude lower than that of Archaea and showed a more marked decrease with depth. The largest decline in bacterial 16S rRNA gene abundance occurred in the 14- to 18-cm depth interval, just above the depth (∼22 cm) when the extant lake was formed. It is tempting to speculate that this decline was due to the lower productivity, and therefore less carbon export to the sediments, of the newly formed lake. However, no limnological information exists to adequately support this hypothesis. Also, it is difficult to explain why there was no concomitant decrease in archaeal abundance. Except in the near-surface layers, the abundance of Crenarchaeota closely followed the pattern of Bacteria; however, the percentage of Crenarchaeota organisms as a function of total Archaea abundance slightly increased with depth. It is important that because extracted DNA served as the template for qPCR, an unknown fraction of quantified 16S rRNA genes belong to potentially inactive cells. Average values of 3.6 to 4.1 16S rRNA gene copies per cell, obtained from a bacterial genome database (33), are often used to estimate cell numbers in natural environments. Although bacterial 16S rRNA gene copy numbers of growing cells can vary widely, estimates for Archaea are typically 1 to 1.5 16S rRNA gene copies per cell. Therefore, it is unlikely that the greater abundance of archaeal 16S rRNA gene copies in Salton Sea sediments was due to a larger number of gene copies per cell than that for Bacteria.
Presence of marine archaeal groups in the Salton Sea.
The majority of archaeal phylotypes recovered from Salton Sea sediments belong to phylogenetic groups previously described from marine sediments (i.e., MG-II and MBG-B and -D). A few previous studies have also reported the presence of archaeal marine groups in saline lakes (28, 29). Jiang et al. (29) suggested that similarities in pH (e.g., alkaline) and salinity composition (e.g., NaCl dominant) in marine and Tibetan saline lake sediments could explain the presence and apparent dominance of these groups in these two seemingly disparate environments. The Salton Sea is NaCl-salinity dominant, alkaline, and located close (<240 km) to both the Pacific Ocean and the Gulf of California. The lake also serves as critical wetland habitat and functions as a major stopover along the Pacific Flyway for millions of migratory birds. The potential for exchange of material (sediment and water) from nearby marine environments provides adequate means for the introduction of marine Archaea into the Salton Sea.
Linking community structure and composition to environmental gradients.
We employed the whole-community fingerprint T-RFLP method to examine changes in relative microbial community structure at a high resolution and to indicate where community compositional differences occurred. TRF peak data were also used to quantify the relative gene frequencies of bacterial and archaeal taxa. While the utility of this technique has been demonstrated previously (52), it is important to recognize the limitations of such an approach. The effectiveness of T-RFLP and clone library analyses in estimating community structure and diversity, respectively, can be hindered by artifacts in the form of preferential DNA extraction, PCR and cloning biases (50), and a lack of primer specificity. Also, individual TRF peaks generated with one restriction enzyme often belong to more than one taxon. We addressed the last caveat by utilizing data from multiple restriction enzyme digests for TRF-matching protocols (e.g., APLAUS and TRiFLe). In addition, the majority (>80% of the relative peak area) of archaeal TRFs were identifiable in our clone libraries.
Shifts in bacterial community structure were strongly depth dependent and correlated with pore water salinity. Groupings of sediment depths with similar community structures, revealed by multivariate analysis of T-RFLP patterns, were further supported by differences in phylogenetic composition. The bacterial community composition was dominated by Proteobacteria, Actinobacteria, and Chloroflexi. Among the Proteobacteria, an increase in Betaproteobacteria and decreases in Alphaproteobacteria and Gammaproteobacteria sequences were found with salinity. This is opposite to the trend observed by Wu et al. (64) in studying the diversity of water column microbes in lakes varying in salinity, further highlighting the fundamental differences between sediment and water column microbial communities. The majority of described Gammaproteobacteria are heterotrophic, and this group is characterized as being physiologically diverse. Most of the Gammaproteobacteria sequences detected in the Salton Sea belonged to sulfide oxidizers or matched sequences retrieved from sulfide-rich environments. Although no direct physiological link could be made, Clostridia and Actinobacteria sequences were detected at almost every depth, a pattern commonly found in reducing marine sediment environments (16, 38).
Changes in archaeal community structure based on T-RFLP analysis were not a simple function of depth and were weakly correlated with measured environmental gradients. However, the three depth intervals from which clone libraries were constructed belonged to distinct sample groupings determined by T-RFLP analysis. The weaker community structure patterns of Archaea could be explained by the relatively constant percentage of Euryarchaeota abundance over depth and the overlapping TRFs between both groups. Although Crenarchaeota abundance determined by qPCR showed a slight increase in deeper sediments, Euryarchaeota species were dominant at all depths. Also, many Crenarchaeota sequences had identical TRFs to those of the Euryarchaeota. This problem was minimized during the clone-matching procedure by utilizing data from all three restriction enzyme digests simultaneously with the TRiFLe program. T-RFLP and clone library frequencies gave almost identical estimates of Crenarchaeota abundance. The discrepancy observed between these estimates and abundances determined using qPCR can be explained by cloning bias or differences in primer specificity for this group.
Archaeal diversity as a function of environmental gradients.
As a whole, the level of 16S rRNA gene diversity of the Salton Sea archaeal community was relatively high compared to that in other archaeal clone libraries constructed from aquatic sediments (2) and, specifically, compared to that in other saline lakes (22, 28). Due to incomplete sampling, direct comparisons of richness estimates between our clone libraries cannot be made; however, large differences in clone library composition were apparent. Both the number of total Crenarchaeota sequences and the number of phylotypes increased at deeper depths (22 to 23 and 32 to 33 cm), with a concomitant decrease in Euryarchaeota sequences. In contrast, Jiang et al. (28) found a predominance of crenarchaeotal sequences in sediments associated with past freshwater conditions in Lake Chaka, with euryarchaeotal sequences being found almost exclusively in saline and hypersaline sediments. Although Salton Sea pore water salinity increased threefold along the depth range investigated, the salinity range was much lower than that reported by Jiang et al. (28). While salinity values reached near-saturation levels (40%) in their study, salinities of 5.0 to 11.8% (wt/vol) were measured in Salton Sea sediments. This range is more similar to that found by Walsh et al. (61), who also reported an increase in crenarchaeotal sequences within higher-salinity soils. It is possible that Euryarchaeota are better adapted than Crenarchaeota to higher salinities present in other saline lake sediments.
In addition to salinity, other environmental differences may help to explain the observed archaeal diversity patterns across sediment depth. Carbon supply, sediment redox conditions, and the dominant TEAP at depth are expected to be important drivers of microbial communities. Profundal sediments in the Salton Sea are sulfidic and remain permanently anoxic, even during times of deep-water column mixing (54, 63), and sulfate reduction is the suspected TEAP. Carbon content decreases (e.g., TC decreasing from 14.6 to 3.0% [wt/wt]) and the number of evaporite deposits increases with depth. Little is known about the physiology and metabolic requirements of Archaea detected in marine water and sediment, despite their fairly extensive distribution in anoxic environments (25, 46, 60). It has been hypothesized that some anaerobic, mesophilic archaea may carry out some form of hydrogen metabolism (19), sulfate reduction (34), or methane production/oxidation (27). Kiene et al. (32) reported on methylated reduced sulfur compounds (i.e., dimethylsulfide) being utilized for methane formation in several anoxic sediment environments, including several alkaline saline lakes. Salton Sea sediment and water contain <1 μM methane (D. L. Valentine, unpublished data), and methanogenic archaea must compete with sulfate reducers for hydrogen and acetate or must utilize other compounds (e.g., methylotrophy). In addition, no clone sequences recovered matched closely to known methanogens or methane oxidizers, although this is not definitive proof of their absence. It remains most likely that some form of sulfur-based or syntrophic metabolism (i.e., interspecies hydrogen transfer) plays a key role in the success of these particle-associated archaea in the Salton Sea and other anoxic sediment environments.
Energetic stress and the structuring of microbial communities.
The results of this study indicate that salinity and carbon gradients play a significant role in driving microbial abundance and diversity patterns in Salton Sea sediments. Increasing salinity and decreasing carbon content over depth represent conditions of increased energetic stress for the microbial communities present. Valentine (59) proposed a framework in which the success of Archaea over Bacteria in any environment may be explained by the amount of energetic stress experienced by microbes within these environments and by the physiological adaptations that each group possesses to deal with such stress. Several studies have suggested that Archaea are dominant within the deeper sediments of the marine subsurface (6, 11, 39). We hypothesize that the high-salinity and low-carbon conditions that exist in the deeper sediments of the Salton Sea provide an environmental niche in which mesophilic, presumably anaerobic archaea are able to thrive, though their exact biogeochemical roles remain unknown. Future studies of the metabolic activities and physiologies of the microbial communities described in this study will be useful for providing a more complete understanding of how energetic stress may dictate the ecologies of both domains.
Acknowledgments
Funding for this work was provided by NSF grant MCB-0604191 (D.L.V.) and by a CMIS California NASA space grant (B.K.S.). Additional support was provided by a Philip and Aida Siff Education Foundation graduate fellowship awarded to B.K.S.
We thank Ian Hewson and Jed A. Fuhrman for advice and assistance with sediment DNA extractions, PCR primer selection, and T-RFLP analysis. We also thank Christine N. Desautels for assistance with T-RFLP analysis and sequencing and Bryan Brinegar of Environmental Recovery Solutions for providing boat logistics.
Footnotes
Published ahead of print on 30 November 2009.
REFERENCES
- 1.Acinas, S. G., J. Antón, and F. Rodriguez-Valera. 1999. Diversity of free-living and attached bacteria in offshore western Mediterranean waters as depicted by analysis of genes encoding 16S rRNA. Appl. Environ. Microbiol. 65:514-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aller, J. Y., and P. F. Kemp. 2008. Are Archaea inherently less diverse than Bacteria in the same environments? FEMS Microbiol. Ecol. 65:74-87. [DOI] [PubMed] [Google Scholar]
- 3.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 4.Ashelford, K. E., N. A. Chuzhanova, J. C. Fry, A. J. Jones, and A. J. Weightman. 2006. New screening software shows that most recent large 16S rRNA gene clone libraries contain chimeras. Appl. Environ. Microbiol. 72:5734-5741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Avaniss-Aghajani, E., K. Jones, D. Chapman, and C. Brunk. 1994. A molecular technique for identification of bacteria using small subunit ribosomal RNA sequences. Biotechniques 17:144-149. [PubMed] [Google Scholar]
- 6.Biddle, J. F., J. S. Lipp, M. A. Lever, K. G. Lloyd, K. B. Sørensen, R. Anderson, H. F. Fredricks, M. Elvert, T. J. Kelly, D. P. Schrag, M. L. Sogin, J. E. Brenchley, A. Teske, C. H. House, and K.-U. Hinrichs. 2006. Heterotrophic Archaea dominate sedimentary subsurface ecosystems off Peru. Proc. Natl. Acad. Sci. USA 103:3846-3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buckles, J. E., K. Kashiwase, and T. Krantz. 2002. Reconstruction of prehistoric Lake Cahuilla in the Salton Sea Basin using GIS and GPS. Hydrobiologia 473:55-57. [Google Scholar]
- 8.Casamayor, E. O., R. Massana, S. Benlloch, L. Øvreås, B. Díez, V. J. Goddard, J. M. Gasol, I. Joint, F. Rodríguez-Valera, and C. Pedrós-Alió. 2002. Changes in archaeal, bacterial and eukaryal assemblages along a salinity gradient by comparison of genetic fingerprinting methods in a multipond solar saltern. Environ. Microbiol. 4:338-348. [DOI] [PubMed] [Google Scholar]
- 9.Chao, A. 1987. Estimating the population size for capture-recapture data with unequal catchability. Biometrics 43:783-791. [PubMed] [Google Scholar]
- 10.Chao, A., and M. C. K. Yang. 1993. Stopping rules and estimation for recapture debugging with unequal failure rates. Biometrika 80:193-201. [Google Scholar]
- 11.Chapelle, F. H., K. O'Neill, P. M. Bradley, B. A. Methe, S. A. Ciufo, L. L. Knobel, and D. R. Lovley. 2002. A hydrogen-based subsurface microbial community dominated by methanogens. Nature 415:312-315. [DOI] [PubMed] [Google Scholar]
- 12.Chung, F. H. 1974. Quantitative interpretation of X-ray diffraction patterns of mixtures. I. Matrix-flushing method for quantitative multicomponent analysis. J. Appl. Crystallogr. 7:519-525. [Google Scholar]
- 13.Clarke, K. R., and M. Ainsworth. 1993. A method of linking multivariate community structure to environmental variables. Mar. Ecol. Prog. Ser. 92:205-219. [Google Scholar]
- 14.Clarke, K. R., and R. M. Warwick. 2001. Change in marine communities: an approach to statistical analysis and interpretation, 2nd ed. PRIMER-E, Plymouth, United Kingdom.
- 15.Cole, J. R., Q. Wang, E. Cardenas, J. Fish, B. Chai, R. J. Farris, A. S. Kulam-Syed-Mohideen, D. M. McGarrell, T. Marsh, G. M. Garrity, and J. M. Tiedje. 2009. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37:D141-D145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Colwell, F., R. Matsumoto, and D. Reed. 2004. A review of the gas hydrates, geology, and biology of the Nankai Trough. Chem. Geol. 205:391-404. [Google Scholar]
- 17.Colwell, R. K. 2005. EstimateS: statistical estimation of species richness and shared species from samples, version 7.5. http://purl.oclc.org/estimates.
- 18.Crump, B. C., E. V. Armbrust, and J. A. Baross. 1999. Phylogenetic analysis of particle-attached and free-living bacterial communities in the Columbia River, its estuary, and the adjacent coastal ocean. Appl. Environ. Microbiol. 65:3192-3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dawson, S. C., E. F. Delong, and N. R. Pace. 2006. Phylogenetic and ecological perspectives on uncultured Crenarchaeota and Korarchaeota, p. 281-289. In M. Dworkin, S. Falkow, E. Rosenberg, K.-H. Schleifer, and E. Stackebrandt (ed.), The prokaryotes, volume 3. Archaea. Bacteria: Firmicutes, Actinomycetes. Springer-Verlag, New York, NY. [Google Scholar]
- 20.DeLong, E. F. 1992. Archaea in coastal marine environments. Proc. Natl. Acad. Sci. USA 89:5685-5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DeSantis, T. Z., P. Hugenholtz, N. Larsen, M. Rojas, E. L. Brodie, K. Keller, T. Huber, D. Dalevi, P. Hu, and G. L. Andersen. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72:5069-5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dong, H., G. Zhang, H. Jiang, B. Yu, L. Chapman, C. Lucas, and M. Fields. 2006. Microbial diversity in sediments of saline Qinghai Lake, China: linking geochemical controls to microbial ecology. Microb. Ecol. 51:65-82. [DOI] [PubMed] [Google Scholar]
- 23.Gieskes, J. M., T. Gamo, and H. Brumsack. 1991. Chemical methods for interstitial water analysis aboard Joides Resolution. Ocean drilling program, technical note 15. Texas A&M University, College Station, TX.
- 24.Hatzenpichler, R., E. V. Lebedeva, E. Spieck, K. Stoecker, A. Richter, H. Daims, and M. Wagner. 2008. A moderately thermophilic ammonia-oxidizing crenarchaeote from a hot spring. Proc. Natl. Acad. Sci. USA 105:2134-2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hershberger, K. L., S. M. Barns, A.-L. Reysenbach, S. C. Dawson, and N. R. Pace. 1996. Wide diversity of Crenarchaeota. Nature 384:420. [DOI] [PubMed] [Google Scholar]
- 26.Huber, T., G. Faulkner, and P. Hugenholtz. 2004. Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20:2317-2319. [DOI] [PubMed] [Google Scholar]
- 27.Inagaki, F., T. Nunoura, S. Nakagawa, A. Teske, M. Lever, A. Lauer, M. Suzuki, K. Takai, M. Delwiche, F. S. Colwell, K. H. Nealson, K. Horikoshi, S. D'Hondt, and B. B. Jørgensen. 2006. Biogeographical distribution and diversity of microbes in methane hydrate-bearing deep marine sediments on the Pacific Ocean Margin. Proc. Natl. Acad. Sci. USA 103:2815-2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang, H., H. Dong, B. Yu, X. Liu, Y. Li, S. Ji, and C. L. Zhang. 2007. Microbial response to salinity change in Lake Chaka, a hypersaline lake on Tibetan plateau. Environ. Microbiol. 9:2603-2621. [DOI] [PubMed] [Google Scholar]
- 29.Jiang, H., H. Dong, B. Yu, Q. Ye, J. Shen, H. Rowe, and C. Zhang. 2008. Dominance of putative marine benthic Archaea in Qinghai Lake, north-western China. Environ. Microbiol. 10:2355-2367. [DOI] [PubMed] [Google Scholar]
- 30.Junier, P., T. Junier, and K.-P. Witzel. 2008. TRiFLe, a program for in silico terminal restriction fragment length polymorphism analysis with user-defined sequence sets. Appl. Environ. Microbiol. 74:6452-6456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kent, A. D., D. J. Smith, B. J. Benson, and E. W. Triplett. 2003. Web-based phylogenetic assignment tool for analysis of terminal restriction fragment length polymorphism profiles of microbial communities. Appl. Environ. Microbiol. 69:6768-6776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kiene, R. P., R. S. Oremland, A. Catena, L. G. Miller, and D. G. Capone. 1986. Metabolism of reduced methylated sulfur compounds in anaerobic sediments and by a pure culture of an estuarine methanogen. Appl. Environ. Microbiol. 52:1037-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klappenbach, J. A., P. R. Saxman, J. R. Cole, and T. M. Schmidt. 2001. rrndb: the ribosomal RNA operon copy number database. Nucleic Acids Res. 29:181-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Knittel, K., T. Losekann, A. Boetius, R. Kort, and R. Amann. 2005. Diversity and distribution of methanotrophic Archaea at cold seeps. Appl. Environ. Microbiol. 71:467-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, Chichester, United Kingdom.
- 36.Lane, D. J., B. Pace, G. J. Olsen, D. A. Stahl, M. L. Sogin, and N. R. Pace. 1985. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc. Natl. Acad. Sci. USA 82:6955-6959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li, H. C., T. L. Ku, X. M. Xu, R. Peters, and H. P. Buchheim. 2003. Hydrologic and climatic variations in the Salton Basin, California, between 1,400 and 16,500 14C yr BP as read from geochemical signals in lake deposits, p. 162-163. Abstr. 3rd Int. Limnogeol. Cong., Tucson, AZ.
- 38.Li, Y., F. Li, X. Zhang, S. Qin, Z. Zeng, H. Dang, and Y. Qin. 2008. Vertical distribution of bacterial and archaeal communities along discrete layers of a deep-sea cold sediment sample at the East Pacific Rise (∼13°N). Extremophiles 12:573-585. [DOI] [PubMed] [Google Scholar]
- 39.Lipp, J. S., Y. Morono, F. Inagaki, and K.-U. Hinrichs. 2008. Significant contribution of Archaea to extant biomass in marine subsurface sediments. Nature 454:991-994. [DOI] [PubMed] [Google Scholar]
- 40.Nadkarni, M. A., F. E. Martin, N. A. Jacques, and N. Hunter. 2002. Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set. Microbiology 148:257-266. [DOI] [PubMed] [Google Scholar]
- 41.Nealson, K., and W. Berelson. 2003. Layered microbial communities and the search for life in the universe. Geomicrobiol. J. 20:451-462. [Google Scholar]
- 42.Nelson, K. A., N. S. Moin, and A. E. Bernhard. 2009. Archaeal diversity and the prevalence of Crenarchaeota in salt marsh sediments. Appl. Environ. Microbiol. 75:4211-4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ochsenreiter, T., D. Selezi, A. Quaiser, L. Bonch-Osmolovskaya, and C. Schleper. 2003. Diversity and abundance of Crenarchaeota in terrestrial habitats studied by 16S RNA surveys and real time PCR. Environ. Microbiol. 5:787-797. [DOI] [PubMed] [Google Scholar]
- 44.Oren, A. 2001. The bioenergetic basis for the decrease in metabolic diversity at increasing salt concentrations: implications for the functioning of salt lake ecosystems. Hydrobiologia 466(Dev. Hydrobiol. 162):61-72. [Google Scholar]
- 45.Page, R. D. M. 1996. Tree View: an application to display phylogenetic trees on personal computers. Bioinformatics 12:357-358. [DOI] [PubMed] [Google Scholar]
- 46.Schleper, C., W. Holben, and H.-P. Klenk. 1997. Recovery of crenarchaeotal ribosomal DNA sequences from freshwater-lake sediments. Appl. Environ. Microbiol. 63:321-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schleper, C., G. Jurgens, and M. Jonuscheit. 2005. Genomic studies of uncultivated archaea. Nat. Rev. Microbiol. 3:479-488. [DOI] [PubMed] [Google Scholar]
- 48.Schloss, P. D., and J. Handelsman. 2005. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71:1501-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schloss, P. D., and J. Handelsman. 2006. Introducing SONS, a tool for operational taxonomic unit-based comparisons of microbial community memberships and structures. Appl. Environ. Microbiol. 72:6773-6779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schmalenberger, A., F. Schwieger, and C. C. Tebbe. 2001. Effect of primers hybridizing to different evolutionarily conserved regions of the small-subunit rRNA gene in PCR-based microbial community analyses and genetic profiling. Appl. Environ. Microbiol. 67:3557-3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schroeder, R. A., W. H. Orem, and Y. K. Kharaka. 2002. Chemical evolution of the Salton Sea, California: nutrient and selenium dynamics. Hydrobiologia 473(Dev. Hydrobiol. 161):23-45. [Google Scholar]
- 52.Schütte, U., Z. Abdo, S. Bent, C. Shyu, C. Williams, J. Pierson, and L. Forney. 2008. Advances in the use of terminal restriction fragment length polymorphism (T-RFLP) analysis of 16S rRNA genes to characterize microbial communities. Appl. Microbiol. Biotechnol. 80:365-380. [DOI] [PubMed] [Google Scholar]
- 53.Shyu, C., T. Soule, S. Bent, J. Foster, and L. Forney. 2007. MiCA: a web-based tool for the analysis of microbial communities based on terminal-restriction fragment length polymorphisms of 16S and 18S rRNA genes. Microb. Ecol. 53:562-570. [DOI] [PubMed] [Google Scholar]
- 54.Swan, B. K., J. M. Watts, K. M. Reifel, and S. H. Hurlbert. 2007. Role of the polychaete Neanthes succinea in phosphorus regeneration from sediments in the Salton Sea, California. Hydrobiologia 576:111-125. [Google Scholar]
- 55.Swofford, D. L. 2003. PAUP*. Phylogenetic analysis using parsimony (*and other methods), version 4.0. Sinauer Associates, Sunderland, MA.
- 56.Takai, K., and K. Horikoshi. 2000. Rapid detection and quantification of members of the archaeal community by quantitative PCR using fluorogenic probes. Appl. Environ. Microbiol. 66:5066-5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tindall, B. J., and H. G. Trüper. 1986. Ecophysiology of the aerobic halophilic archaebacteria. Syst. Appl. Microbiol. 7:202-212. [Google Scholar]
- 59.Valentine, D. L. 2007. Adaptations to energy stress dictate the ecology and evolution of the Archaea. Nat. Rev. Microbiol. 5:316-323. [DOI] [PubMed] [Google Scholar]
- 60.Vetriani, C., A.-L. Reysenbach, and J. Doré. 1998. Recovery and phylogenetic analysis of archaeal rRNA sequences from continental shelf sediments. FEMS Microbiol. Ecol. 161:83-88. [DOI] [PubMed] [Google Scholar]
- 61.Walsh, D. A., R. T. Papke, and W. F. Doolittle. 2005. Archaeal diversity along a soil salinity gradient prone to disturbance. Environ. Microbiol. 7:1655-1666. [DOI] [PubMed] [Google Scholar]
- 62.Wardlaw, G. D., and D. L. Valentine. 2005. Evidence for salt diffusion from sediments contributing to increasing salinity in the Salton Sea, California. Hydrobiologia 533:77-85. [Google Scholar]
- 63.Watts, J. M., B. K. Swan, M. A. Tiffany, and S. H. Hurlbert. 2001. Thermal, mixing, and oxygen regimes of the Salton Sea, California, 1997-1999. Hydrobiologia 466:159-176. [Google Scholar]
- 64.Wu, Q. L., G. Zwart, M. Schauer, M. P. Kamst-van Agterveld, and M. W. Hahn. 2006. Bacterioplankton community composition along a salinity gradient of sixteen high-mountain lakes located on the Tibetan Plateau, China. Appl. Environ. Microbiol. 72:5478-5485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yue, J. C., and M. K. Clayton. 2005. A similarity measure based on species proportions. Commun. Stat. Theor. Methods 34:2123-2131. [Google Scholar]
- 66.Yue, J. C., M. K. Clayton, and F.-C. Lin. 2001. A nonparametric estimator of species overlap. Biometrics 57:743-749. [DOI] [PubMed] [Google Scholar]