Abstract
Bacillus subtilis is a well-established cell factory for efficient secretion of many biotechnologically relevant enzymes that are naturally produced by it or related organisms. However, the use of B. subtilis as a host for production of heterologous secretory proteins can be complicated by problems related to inefficient translocation of the foreign proteins across the plasma membrane or to inefficient release of the exported proteins from the cell surface into the surrounding medium. Therefore, there is a clear need for tools that allow more efficient membrane targeting, translocation, and release during the production of these proteins. In the present study, we investigated the contributions of the pre (prelip) and pro (prolip) sequences of a Staphylococcus hyicus lipase to secretion of a heterologous protein, the alkaline phosphatase PhoA of Escherichia coli, by B. subtilis. The results indicate that the presence of the prolip-peptide, in combination with the lipase signal peptide (prelip), contributes significantly to the efficient secretion of PhoA by B. subtilis and that prelip directs PhoA secretion more efficiently than the authentic signal peptide of PhoA. Genome-wide transcriptional analyses of the host cell responses indicate that, under the conditions tested, no known secretion or membrane-cell wall stress responses were provoked by the production of PhoA with any of the pre- and pro-region sequences used. Our data underscore the view that the pre-pro signals of the S. hyicus lipase are very useful tools for secretion of heterologous proteins in B. subtilis.
Bacillus subtilis is widely appreciated as a cell factory for the production of industrial enzymes that naturally occur in it or closely related species. These enzymes are usually secreted into the growth medium in large and commercially relevant quantities (20, 52, 53). The high secretion capacity is a clear advantage that B. subtilis has over Gram-negative bacteria, such as Escherichia coli, in which secreted proteins are retained in the periplasm. Additionally, the lack of an outer membrane implies that proteins produced with B. subtilis are free of lipopolysaccharide (endotoxin). Other advantages of using B. subtilis as a protein production host are its high genetic amenability, the availability of strains with mutations in nearly all of the ∼4,100 genes, a toolbox containing strains and vectors for gene expression, and the fact that this bacterium is generally recognized as safe (6, 22, 25, 64). However, the use of B. subtilis as a cell factory for production of particular heterologous secretory proteins can also be problematic due to, for example, inefficient translocation of the foreign proteins across the plasma membrane or inefficient release of the exported proteins from the cell envelope into the surrounding medium (4, 33, 39, 47). In addition, premature or dismal protein folding may limit the yield of the desired products and can even be detrimental to the producing host organism.
Proteins that are destined to act outside the cell are targeted from their site of synthesis, the cytoplasm, to the membrane with the aid of so-called signal peptides (or pre-peptides) (57, 58). These molecular Zip codes are usually composed of a positively charged N region, a hydrophobic H region, and a C region with a recognition site for cleavage by signal peptidase during or after membrane translocation. The majority of the secreted proteins are transported in an unfolded state across the cytoplasmic membrane, and this is followed by cleavage of the signal peptide and folding of the protein (52, 56). In addition, some proteins are produced with a pro-peptide that functions as a chaperone in guiding the timely protein folding and, in some cases, also the release from the plasma membrane and cell envelope after translocation (46, 49, 51, 61). Although the general structure of the secretion signals is conserved in all domains of life, particular secretion signals have been optimized during evolution for functioning in the context of a particular host organism and for secretion of a particular protein. Consequently, these targeting signals may perform less efficiently when they are used in heterologous protein production and can thus be important bottlenecks in heterologous protein production. Therefore, the search for efficient secretion signals for interesting heterologous proteins is a useful strategy to improve the production of these proteins by established host organisms, such as B. subtilis (7).
Previously, it was reported that the combined signal peptide (prelip) and pro (prolip) regions of a Staphylococcus hyicus preprolipase can act as a productive secretion signal for general use in Gram-positive bacteria (15, 18, 32, 33, 45, 50). In addition, the pre- and pro-regions have also been used extensively in staphylococcal surface display systems (9, 10, 19, 28-30, 40, 43, 44). The main objective of the present study was to investigate the individual and combined contributions of these regions to the heterologous secretion of the alkaline phosphatase PhoA of E. coli by B. subtilis. For this purpose, we performed a detailed dissection of the subcellular localization and fate of PhoA that was produced with and without the pro-peptide. The results show that both prelip and prolip contributed significantly to the efficient secretion of PhoA by B. subtilis, while the level of secretion of PhoA by its native signal sequence in B. subtilis was marginal. As shown by genome-wide transcription analyses, neither a CssRS-dependent secretion stress response nor a SigW-dependent membrane stress response was observed upon production of PhoA with the different secretion signals tested.
MATERIALS AND METHODS
Plasmids, bacterial strains, media, and growth conditions.
The plasmids and bacterial strains used in this study are listed in Table 1. Strains were grown with agitation at 37°C in Luria-Bertani (LB) medium consisting of 1% tryptone, 0.5% yeast extract, and 1% NaCl (pH 7.4). If appropriate, media were supplemented with chloramphenicol at a concentration of 5 μg/ml.
TABLE 1.
Plasmids, strains, and primers used in this study
| Plasmid, strain, or primer | Relevant propertiesa | Reference or source |
|---|---|---|
| Plasmids | ||
| pJM1 | pUC18 derivative; contains the gene encoding the preprolipase from S. hyicus under control of the regulatory elements of the lac operon; Apr | 32 |
| pJM1-2 | pJM1 derivative with an NaeI site at the position corresponding to the signal peptidase I cleavage site in the preprolipase of S. hyicus; Apr | This study |
| pJM1-87 | pJM1 derivative with an SwaI site between the ribosome-binding site and the ATG start codon of the lipase gene of S. hyicus; Apr | 33 |
| pPA9 | pJM1-87 derivative containing the authentic coding sequence for pre-PhoA of E. coli instead of the preprolipase gene of S. hyicus; Apr | This study |
| pPA10 | pJM1-2 derivative containing a precise fusion between the coding sequence for the signal peptide (prelip) of the preprolipase of S. hyicus and the coding sequence for mature PhoA; Apr | This study |
| pPS2 | pLipPS1 derivative containing the constitutive promoter of the S. hyicus preprolipase gene (pLip) followed by a multiple cloning site; Cmr | 33 |
| pPSPhoA2 | pPS2 derivative carrying the fusion between the coding sequence for the signal peptide (prelip) of the preprolipase of S. hyicus and the mature PhoA coding sequence from pPA10 (prelip-PhoA); Cmr | This study |
| pPSPhoA5 | pPS2 derivative carrying the fusion between the coding sequence for the preprolip part of the preprolipase of S. hyicus and the mature PhoA coding sequence (preprolip-PhoA); Cmr | 13 |
| pPSPhoA6 | pPS2 derivative carrying the authentic coding sequence for pre-PhoA of E. coli from pPA9 (pre-PhoA); Cmr | This study |
| BaSysBio II | Ligation-independent cloning vector based on pDG1727 for promoter-GFP activity analysis; Apr Spr | Stéphane Aymerich, Eric Botella, Kevin Devine, Mark Fogg, Annette Hansen, and Tony Wilkinson, unpublished data |
| E. coli strains | ||
| DH5α | F− φ80dlacZΔM15 endA1 recA1 gyrA96 thi-1 hsdR17(rK− mK+) supE44 relA1 deoR Δ(lacZYA-argF)U169 | Life Technologies, Inc. |
| JM109 | F′ traD36 lacIq Δ(lacZ)M15 proA+B+/e14-(McrA−) Δ(lac-proAB) thi gyrA96 (Nalr) endA1 hsdR17(rk− mk+) relA1 supE44 recA1 | 62 |
| B. subtilis 168 | trpC2 | 25 |
| Primers (5′-3′) | ||
| K2 | TTGTGTTGTCGAATCGCCGGCCTCTGCCACGCCC | This study |
| pho4 | GGGATTTAAATGATATCACGTGTTAACCGGGCTGCTCAGGGCGATAT | 13 |
| pho5 | TTTAAAGCTTGGATCCTTATTTCAGCCCCAGAGCGGC | 13 |
| pho6 | GGGGATTTAAATGAAACAAAGCACTATTGCACTGGC | This study |
| htrA fwd | CCGCGGGCTTTCCCAGCAAAATCAATTGGCACGTATT | This study |
| htrA rev | GTTCCTCCTTCCCACCCGTTAAAAAGACCTCATTCTCATTACC | This study |
| htrB fwd | CCGCGGGCTTTCCCAGCTTCTCTAAATACTTCGTCAGCA | This study |
| htrB rev | GTTCCTCCTTCCCACCTCTGCTGTTCTGTATGTGAAGG | This study |
Underlining in primer sequences indicates restriction sites used for cloning.
DNA techniques.
Procedures for DNA purification, restriction, ligation, agarose gel electrophoresis, and transformation of competent E. coli cells were carried out as previously described (42). Chromosomal DNA of B. subtilis was isolated as described by Bron and Venema (8). PCR was carried out with the Pwo DNA polymerase, using chromosomal DNA as a template. Plasmid DNA from E. coli was isolated using the alkaline lysis method (42) or a High Pure plasmid isolation kit according to the protocol supplied by the manufacturer (Roche Applied Science). B. subtilis was transformed as described by Kunst and Rapoport (26). The nucleotide sequences of primers used for PCR are shown in Table 1. Constructs were first made in E. coli DH5α and then introduced into B. subtilis.
Plasmid pPSPhoA2, which encodes a precise fusion between the signal peptide (prelip) of a lipase from S. hyicus and the mature PhoA protein of E. coli (prelip-PhoA), was constructed as follows. First, primer K2 was employed as previously described (32) to introduce an NaeI site into plasmid pJM1 at the position that corresponds to the signal peptidase I cleavage site of the preprolipase. This resulted in plasmid pJM1-2. Next, the phoA gene of E. coli JM109 was amplified by PCR with primers pho4 and pho5; primer pho4 introduces an HpaI site between the codons for Glu9 and Asn10 of mature PhoA. The amplified phoA fragment was cleaved with HpaI and HindIII and used to replace a 1.9-kb NaeI-HindIII fragment of pJM1-2, which encodes the prolipase. The resulting plasmid was named pPA10. Finally, pPSPhoA2 was obtained by excision of a 1.5-kb SacI-HindIII fragment from pPA10 and ligation of this fragment, which encodes prelip-PhoA, into the SacI-HindIII sites of the expression vector pPS2.
Plasmid pPSPhoA5, which encodes a precise fusion between the signal peptide plus the pro-region of the S. hyicus lipase and mature PhoA of E. coli (preprolip-PhoA), was constructed as described by Darmon et al. (13).
Plasmid pPSPhoA6, which carries the authentic phoA gene of E. coli JM109 fused to the ribosome-binding site of the S. hyicus lipase gene (pre-PhoA), was obtained as follows. First, the phoA gene of E. coli JM109 was PCR amplified with primers pho5 and pho6; primer pho6 introduces an SwaI site directly upstream of the ATG start codon of phoA. Upon cleavage with SwaI and HindIII, the fragment was ligated into the corresponding sites of plasmid pJM1-87, replacing the 2-kb SwaI-HindIII fragment that contains the lipase structural gene. This resulted in plasmid pPA9. Finally, pPSPhoA6 was obtained by insertion of a 1.5-kb SacI-HindIII fragment from pPA9, which encodes the authentic pre-PhoA of E. coli, into the corresponding sites of pPS2.
Localization of PhoA.
The subcellular localization of PhoA was determined using the protocol described by Tjalsma et al. (54). Briefly, cells were grown overnight at 37°C in TY broth and separated from the growth medium by centrifugation. Next, the cells were resuspended in protoplast buffer (20 mM potassium phosphate [pH 7.5], 15 mM MgCl2, 20% sucrose) supplemented with 1 mg/ml lysozyme. After 30 min of incubation at 37°C, proteins released from the cells by protoplasting (i.e., the cell wall fraction) were separated from the protoplasts by centrifugation. The protoplasts were resuspended in protoplast buffer and divided into three aliquots that were supplemented with either 1% trypsin in phosphate-buffered saline (PBS), 1% trypsin plus 1% Triton X-100 in PBS, or PBS alone (PBS contained [per liter] 8 g NaCl, 2.68 g Na2HPO4, 0.2 g KCl, and 0.24 g KH2PO4 and had a pH of 7.4). After 30 min of incubation at 37°C, trypsin was inactivated by adding Complete protease inhibitors (Roche Molecular Biochemicals). All fractions were used for SDS-PAGE and subsequent Western blotting.
SDS-PAGE and Western blotting.
Proteins in the growth medium, the cell wall, protoplasts, and/or cell lysates were separated by SDS-PAGE (using precast bis-Tris NuPAGE gels from Invitrogen) and then semidry blotted (75 min at 1 mA/cm2) onto a nitrocellulose membrane (Protran; Schleicher & Schuell). Subsequently, the PhoA, SipS, BdbD, TrxA, and YolF proteins were detected with specific polyclonal antibodies raised in rabbits as previously described (17). These antibodies were detected with fluorescent IgG secondary antibodies (IRDye 800 CW goat anti-rabbit antibodies obtained from LiCor Biosciences) in combination with the Odyssey infrared imaging system (LiCor Biosciences).
Alkaline phosphatase assays.
Alkaline phosphatase (PhoA) activity assays were carried out essentially as described by Nicholson and Setlow (35), but with minor modifications as described by Darmon et al. (13).
Transcriptome analysis.
Total RNA was isolated from cells in the mid-exponential growth phase using a High Pure RNA isolation kit (catalog no. 1828665; Roche). RNA samples were quantified spectrophotometrically at 260 nm, and the quality was checked by examining the integrity of the 23S and 16S rRNA bands on an agarose gel. Conversion of 20 μg RNA to cDNA and cDNA labeling were performed using a CyScribe postlabeling kit (catalog no. RPN5660X; GE Healthcare) according to the manufacturer's instructions. Probes in 3× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) containing 0.3% SDS and 0.5 μg/μl tRNA were denatured at 90°C for 2 min and cooled on ice for a few seconds, which was followed immediately by application onto the chip.
B. subtilis microarrays were printed on poly-l-lysine-coated glass slides at Novozymes A/S (Bagsvaerd) using a MicroGrid II arrayer (BioRobotics, Huntingdon, United Kingdom), and this was followed by UV cross-linking at 60 mJ with a UV-Stratalinker 1800 (Biocrest B.V., Amsterdam, The Netherlands). Free poly-l-lysine groups were neutralized by washing the slides for 15 min in 1-methyl-2-pyrrolidinone supplemented with 1.7% (wt/vol) succinic anhydride and 4.3% (vol/vol) boric acid (pH 8.0) and then three times in water. Slides were blocked by washing them in 1% (wt/vol) bovine serum albumin, 20% (vol/vol) 20× SSC, 0.5% (wt/vol) SDS for 45 min at 42°C and then five times in water and drying them by centrifugation (500 rpm for 5 min at 20°C). The BACLIB96 OligoLibrary consisting of 65-mer oligonucleotides representing 4,106 identified open reading frames of the B. subtilis 168 genome was purchased from Compugen (United States), diluted to obtain a concentration of 10 μM in 50% dimethyl sulfoxide, and printed three times on each slide. Labeled cDNA was hybridized to B. subtilis chips under a supported coverslip in a humid chamber overnight at 60°C in a dark water bath. After hybridization, the coverslip was gently removed by placing the chip in a buffer consisting of 2× SSC and 0.1% SDS. The chip was washed for 5 min in 2× SSC-0.1% SDS, for 5 min in 1× SSC-0.1% SDS, for 5 min in 0.5× SSC, and for 10 s in deionized water and finally dried by centrifugation (500 rpm for 5 min and 20°C). Probed arrays were scanned at 532 and 635 nm using a GenePix 4000B scanner (Molecular Devices), and image analyses were performed using Imagene 7.5 from BioDiscovery (United States). Data sets were Lowess print tip normalized and merged in GeneSight 4.1 (BioDiscovery, United States). Finally, the data were validated by use of the Confidence Analyzer tool (99% confidence level) in GeneSight 4.1. Two technical replicate hybridizations (dye swap) were performed for each time point in each of two biological replicate experiments. Since each probe was present three times on the DNA chips, this generated two biological replicate data sets, each of which contained six technical replicate values for each gene. Only genes that were found to be significantly regulated at the 99% confidence level in both biological replicate experiments were included in the final list.
Promoter activity assay.
The promoter regions of htrA and htrB were amplified by PCR from genomic DNA prepared from B. subtilis 168 using the primers described in Table 1. The resulting PCR fragments were prepared for ligation-independent cloning using T4 DNA polymerase (Novagen) and dTTP (3). The BaSysBioII cloning vector was digested with SmaI, purified from an agarose gel, and prepared for ligation-independent cloning using T4 DNA polymerase (Novagen) and dATP. One microliter of the treated vector and 3 μl of the treated insert were mixed and left to anneal at room temperature before transformation of E. coli. The resulting constructs PhtrA-GFP and PhtrB-GFP (containing the green fluorescent protein gene) were used to transform B. subtilis 168, and transformants were selected on LB medium plates containing spectinomycin. Strains with htrA-GFP or htrB-GFP fusions were precultured in LB medium containing 1% or 5% NaCl and diluted 1:100 in the same medium 3 h before the start of the growth experiments. Next, the cells were diluted in the appropriate medium to obtain an optical density at 600 nm (OD600) of 0.01. Then the cells were grown in triplicate wells of a 96-well black optical-bottom microtiter plate (Nunc) that was placed in a Biotek Synergy 2 plate reader at 37°C with variable shaking. The OD600 and fluorescence (excitation 485/20 nm, emission 528/20 nm) of the strains were measured for 14 h. Fluorescence measurements were processed essentially as described by Ronen et al. (41). Before the experiment was started, the OD977 and OD900 of the wells were determined to allow light path correction to 1 cm. For each of the recorded fluorescence data points the blank consisting of neat LB medium (average of three wells) was removed, and each point was corrected for a 1-cm light path by multiplying it by 0.18/(OD977 − OD900). The average for the three samples was then calculated. The fluorescence data were further processed by calculating the average for the three samples and subtracting the background fluorescence of parental strain 168 (without GFP) for each data point. The promoter activity was calculated using the following equation: promoter activity = [GFP(t) − GFP(t − 1)]/OD(t), where t is the time point of measurement. Each experiment was repeated at least three times.
RESULTS
Secretion and localization of E. coli PhoA by B. subtilis.
It was previously reported that the combined signal peptide (prelip) and pro (prolip) regions of the S. hyicus preprolipase can act as a productive secretion signal for use in Gram-positive bacteria (15, 32, 33, 50) and for efficient secretion of the E. coli alkaline phosphatase PhoA in B. subtilis (5, 13). To investigate to what extent prelip and prolip contribute to efficient secretion of PhoA, this protein was produced in B. subtilis not only with the preprolip region (encoded by plasmid pPSPhoA5) but also with the prelip signal peptide alone (encoded by plasmid pPSPhoA2) or with its own signal peptide (encoded by plasmid pPSPhoA6). Importantly, all three phoA constructs were expressed using the same promoter, ribosome-binding site, and ATG start codon.
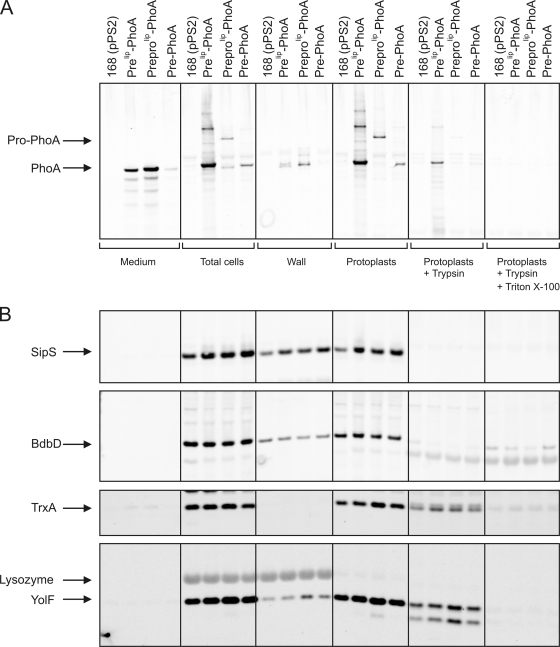
The largest amounts of PhoA were secreted into the growth medium when the protein was synthesized as a preprolip-PhoA hybrid precursor, as shown by Western blotting using growth medium and (sub)cellular fractions (Fig. 1A). Additionally, mature-size PhoA derived from preprolip-PhoA was also detected in the cell wall fraction. Notably, the cell wall fraction also contained larger forms of PhoA (Fig. 1A) representing processing and degradation products of translocated prolip-PhoA. By contrast, the largest forms of PhoA were detected in protoplasts of the preprolip-PhoA-producing cells. These large forms most likely represented prolip-PhoA and degradation products of prolip-PhoA (designated “pro-PhoA” [Fig. 1A]). All pro-PhoA was degraded upon addition of trypsin to intact protoplasts, suggesting that the (pro)protein was effectively translocated across the protoplast membrane.
FIG. 1.
Secretion and subcellular localization of E. coli PhoA in B. subtilis. To determine the localization of the precursor and mature forms of different PhoA fusion proteins, fractionation experiments were performed with cells of B. subtilis 168 transformed with pPSPhoA2 (prelip-PhoA), pPSPhoA5 (preprolip-PhoA), or pPSPhoA6 (pre-PhoA). Parental strain 168 transformed with the “empty” vector pPS2 was used as a negative control. Cells grown in LB medium were protoplasted, and protoplasts were separated from the cell wall fraction by centrifugation. Protoplasts were incubated for 30 min in the presence of 1 mg/ml of trypsin with or without 1% Triton X-100, as indicated. Samples were used for SDS-PAGE and Western blotting, and specific antibodies were used to detect the precursor and mature forms of PhoA (A). Additionally, immunoblotting experiments with marker proteins that resided in the cytoplasm (e.g., TrxA) or in the membrane and were exposed to either the cytoplasmic surface (e.g., YolF) or the extracytoplasmic surface (e.g., BdbD and SipS) were performed (B). The positions of prolip-PhoA (“Pro-PhoA”), mature-size PhoA (“PhoA”), SipS, BdbD, TrxA, and YolF are indicated.
In contrast, the secretion of PhoA was less efficient when the protein was fused to only prelip. The medium contained smaller amounts of mature-size PhoA than the medium of cells producing preprolip-PhoA (Fig. 1A). Consistently, a relatively small amount of mature-size PhoA was present in the cell wall fraction of cells producing prelip-PhoA. In contrast to the results for cells producing preprolip-PhoA, the protoplasts of these cells contained large amounts of mature-size PhoA as well (Fig. 1A). In addition, a different set of forms of PhoA larger than those observed with the preprolip-PhoA precursor was detected in protoplasts of the prelip-PhoA-producing cells. These larger forms could represent aggregated forms of PhoA or PhoA bound to (an)other protein(s). The major part of the mature-size PhoA and the larger PhoA forms were accessible to trypsin in intact protoplasts, indicating that they had been translocated across the protoplast membrane. A minor fraction of PhoA was degraded by trypsin only when 1% Triton X-100 was added to lyse the protoplasts, suggesting that this fraction represented nontranslocated PhoA.
By far the smallest amounts of mature PhoA were secreted into the growth medium when the authentic pre-PhoA of E. coli was expressed in B. subtilis (Fig. 1A). In this case, mature PhoA was also detected in protoplasts, but not in the cell wall fraction. The protoplast-associated PhoA was sensitive to trypsin treatment, suggesting that it had been translocated across the protoplast membrane.
The fractionation data presented here were supported by the results of immunoblotting experiments performed with marker proteins that reside in the cytoplasm (e.g., TrxA), at the cytoplasmic side of the membrane (e.g., YolF), or at the extracytoplasmic membrane surface (e.g., BdbD and SipS). As shown in Fig. 1B, complete degradation of BdbD and SipS occurred upon incubation of protoplasts with trypsin, whereas complete degradation of TrxA and YolF required protoplast lysis with 1% Triton X-100, which is in line with the known subcellular localization of these marker proteins. However, some trypsin-mediated degradation of TrxA and YolF was also observed when protoplasts were incubated with trypsin in the absence of Triton X-100, indicating that some lysis had occurred during protoplasting. This conclusion is supported by the detection of small amounts of BdbD and YolF in the wall fractions. In fact, a limited amount of cell lysis may have occurred prior to protoplasting, as shown by small amounts of TrxA in the growth medium fraction.
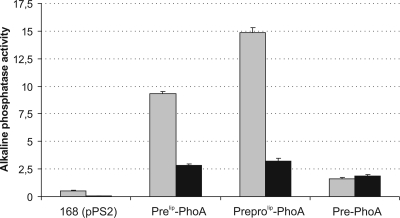
The cellular and secreted amounts of active PhoA were assessed by determining alkaline phosphatase activity in growth medium and cellular fractions of cells producing the three different PhoA precursors (Fig. 2). Analysis of the cellular fractions revealed that cells expressing preprolip-PhoA, prelip-PhoA, and authentic pre-PhoA contained comparable levels of active PhoA. Analysis of the medium fractions showed that, consistent with the results of the Western blotting analyses, the highest PhoA activity was detected in the growth medium of cells expressing preprolip-PhoA, while the levels of PhoA activity were lower in the medium of cells expressing prelip-PhoA. In the medium of cells expressing authentic pre-PhoA, very little PhoA activity was detected, and the level was just above the background levels. Taken together, these observations show that prelip and prolip (in combination with prelip) contribute to efficient export of E. coli PhoA from the cytoplasm and secretion into the growth medium when they are produced in B. subtilis.
FIG. 2.
Secretion and cellular content of active PhoA. Alkaline phosphatase activity was measured in growth media and cells of B. subtilis 168 transformed with pPSPhoA2 (prelip-PhoA), pPSPhoA5 (preprolip-PhoA), or pPSPhoA6 (pre-PhoA). Parental strain 168 transformed with the “empty” plasmid pPS2 was used as a negative control. Cells were grown overnight in LB medium at 37°C, and samples were withdrawn to determine the PhoA activities in the growth medium (gray bars) and cells (black bars). PhoA activities are expressed in U/ml/OD600. Standard deviations are indicated by error bars.
Secretion of PhoA does not cause a secretion stress response in B. subtilis.
The large differences in extracellular accumulation of active PhoA derived from the three different precursor peptides used in our study suggested that at least some of the PhoA produced was degraded. This was most likely due to slow or incorrect PhoA folding upon translocation across the membrane, especially in the case of prelip-PhoA and authentic pre-PhoA. As judged by the observed differences and previous studies of OmpA secretion (32, 33), it is conceivable that the prelip and prolip peptides helped prevent this proteolytic degradation by contributing to effective export and proper folding of PhoA. Thus, cells expressing the different PhoA precursors might be confronted with different amounts of incorrectly folded and degraded precursor proteins, which is potentially stressful. In B. subtilis, this so-called secretion stress response is sensed and combated by the CssRS two-component system (14, 21). Known members of the CssRS regulon are htrA, htrB, cssR, and cssS. In particular, the levels of htrA and htrB transcription are significantly increased in secretion-stressed cells (14). Consequently, the expression of these genes can be used as a reliable indicator of the physiological state of the cells, and it also can be used to assess how well the cells can handle a particular secretory protein that is being produced.
Recently, it was reported based on the use of lacZ fusions that production of the preprolip-PhoA precursor in B. subtilis htrB mutant cells triggered a mild htrB-specific secretion stress response. Remarkably, however, htrA expression was not affected by preprolip-PhoA production (13). To assess whether the production of either prelip-PhoA, preprolip-PhoA, or pre-PhoA provoked a secretion or other stress response in B. subtilis, we performed genome-wide transcriptional analyses with strains producing these precursor proteins. As a control, we analyzed the effects of the empty pPS2 plasmid vector. The first interesting finding was that the transcription levels of the htrA, htrB, cssR, and cssS genes in cells producing either prelip-PhoA, preprolip-PhoA, or pre-PhoA did not differ from the transcription levels of these genes in parental strain 168 (data not shown). This indicated that the production of the three different PhoA precursors did not cause a CssRS-dependent secretion stress response in B. subtilis under the conditions tested. To verify these transcriptome data, we studied the activity of the htrA and htrB promoters in cells producing the different PhoA precursors in more detail using transcriptional fusions between the promoters of htrA or htrB and the GFP gene. The results of this analysis show that the htrA and htrB promoters in cells producing the different PhoA precursors displayed average activities similar to those of the htrA and htrB promoters in parental 168 strain or the parental strain harboring the empty pPS2 vector (results not shown). For comparison, we also analyzed the activity of the htrA and htrB promoters in cells containing plasmid pKTH10. This plasmid directs high-level production of the α-amylase AmyQ from Bacillus amyloliquefaciens (38), which is a known inducer of htrA and htrB expression (1, 31). It is noteworthy that the average activities of the htrA and htrB promoters in this strain were about 10-fold higher than those in the strains harboring the PhoA precursors, showing that the different PhoA precursors indeed did not cause a CssRS-dependent secretion stress response in B. subtilis.
To investigate whether production of the three PhoA precursors led to a different, CssRS-independent stress response, the expression levels of different sigma factors in the array analysis were inspected. The results showed that the expression levels of the sigma factors were not significantly altered, although the pre-PhoA-producing cells did show a slight increase (1.4-fold) in the expression of sigW (data not shown). SigW is a transcription factor which regulates the expression of genes in response to membrane and cell wall stress (11, 12). Thus, increased levels of SigW could indicate potential membrane or cell wall stress, like that in potentia caused by inefficient PhoA translocation and export. However, the results of the microarray analyses revealed that in response to the production of prelip-PhoA, preprolip-PhoA, or pre-PhoA, there is no altered expression of any gene that is known to be under the control of SigW. Taken together, these (negative) results show that production of the prelip-PhoA, preprolip-PhoA, or pre-PhoA precursors does not provoke a secretion or membrane-cell wall stress response.
Interestingly, production of the prelip-PhoA, preprolip-PhoA, or pre-PhoA precursors did induce increased expression of a limited set of genes, including trxA, tagC, dinB, clpE, yvaV, vyqH, yqbO, ydcL, yvaN, yvaO, yomZ, yonD, yonE, yonH, and yonN (Table 2). It should be noted, however, that most of the apparently PhoA-specific induction was relatively moderate (2- to 3.4-fold). Higher levels of induction were observed only in pre-PhoA-producing cells for the yvaN (8.2-fold increase) and yvaO (5.4-fold increase) genes, which encode phage-related proteins. Overall, production of pre-PhoA seems to cause the most pronounced changes in transcription of the trxA, clpE, yvaV, vyqH, yqbO, yvaN, and yvaO genes, while prelip-PhoA production had the greatest impact on transcription of the tagC, dinB, clpE, ydcL, yomZ, yonD, yonE, yonH, and yonN genes. Production of preprolip-PhoA had the mildest effects.
TABLE 2.
Microarray analysesa
| Accession no.b | pPS2c | pPSPhoA2c | pPSPhoA5c | pPSPhoA6c | PBSX | SPβ | Phage related | Gene | Comments |
|---|---|---|---|---|---|---|---|---|---|
| BG10348 | 3,4 | trxA | Thioredoxin | ||||||
| BG10453 | 2,0 | tagC | Possibly involved in polyglycerol phosphate teichoic acid biosynthesis | ||||||
| BG10539 | 2,4 | dinB | Nuclease inhibitor | ||||||
| BG12578 | 2,4 | clpE | ATP-dependent Clp protease-like (class III stress gene) | ||||||
| BG12736 | 3,9 | 4,5 | 2,9 | 3,3 | ybfG | Unknown; similar to unknown proteins | |||
| BG14073 | 2,3 | yvaV | Unknown; similar to unknown proteins | ||||||
| BG14137 | 3,0 | yvqH | Unknown; similar to unknown proteins from B. subtilis | ||||||
| BG11286 | 2,7 | Phage related | yqbO | Unknown; similar to phage-related protein | |||||
| BG12099 | 2,4 | Phage related | ydcL | Unknown; similar to integrase | |||||
| BG14069 | 8,2 | Phage related | yvaN | Unknown; similar to immunity repressor protein | |||||
| BG14070 | 5,4 | Phage related | yvaO | Unknown; similar to immunity repressor protein | |||||
| BG13613 | 2,0 | SPβ | yomZ | Unknown | |||||
| BG13617 | 2,6 | SPβ | yonD | Unknown | |||||
| BG13618 | 2,5 | SPβ | yonE | Unknown | |||||
| BG13621 | 2,6 | 2,5 | SPβ | yonH | Unknown | ||||
| BG13625 | 2,9 | SPβ | yonN | Unknown; similar to HU-related DNA-binding protein | |||||
| BG10959 | 3,5 | 3,2 | PBSX | xepA | PBSX prophage lytic exoenzyme | ||||
| BG10960 | 2,8 | 3,6 | 2,6 | PBSX | xhlA | Involved in cell lysis upon induction of PBSX | |||
| BG10961 | 4,3 | 5,7 | 3,1 | 4,6 | PBSX | xhlB | Hydrolysis of 5-bromo-4-chloroindolyl phosphate upon induction of PBSX | ||
| BG10962 | 3,9 | 2,6 | 2,9 | PBSX | xlyA | N-Acetylmuramoyl-l-alanine amidase (PBSX prophage-mediated lysis) | |||
| BG10997 | 2,9 | 3,5 | PBSX | xkdD | PBSX prophage | ||||
| BG10998 | 2,0 | PBSX | xpf | RNA polymerase PBSX sigma factor-like | |||||
| BG10999 | 2,8 | 3,9 | 3,5 | 3,1 | PBSX | xtmA | PBSX terminase (small subunit) | ||
| BG11000 | 2,3 | 3,3 | 2,4 | 2,4 | PBSX | xtmB | PBSX terminase (large subunit) | ||
| BG11540 | 3,0 | 4,1 | 2,9 | PBSX | xkdE | PBSX prophage | |||
| BG11541 | 3,7 | 5,7 | 2,9 | 4,0 | PBSX | xkdF | PBSX prophage | ||
| BG11542 | 3,1 | 3,7 | 2,8 | PBSX | xkdG | PBSX prophage | |||
| BG11543 | 3,5 | 5,2 | 3,0 | 3,7 | PBSX | xkdH | PBSX prophage | ||
| BG11544 | 3,5 | 3,9 | 2,6 | PBSX | xkdI | PBSX prophage | |||
| BG11545 | 4,9 | 9,0 | 4,3 | 4,8 | PBSX | xkdJ | PBSX prophage | ||
| BG11546 | 3,6 | 6,5 | 3,2 | 4,1 | PBSX | xkdK | PBSX prophage | ||
| BG11547 | 4,6 | 6,5 | 3,0 | 4,6 | PBSX | xkdM | PBSX prophage | ||
| BG11548 | 5,3 | 4,8 | 3,3 | PBSX | xkdN | PBSX prophage | |||
| BG11549 | 3,1 | 4,9 | 2,8 | 5,1 | PBSX | xkdO | PBSX prophage | ||
| BG11550 | 2,4 | 2,1 | PBSX | xkdP | PBSX prophage | ||||
| BG11551 | 2,2 | 2,8 | PBSX | xkdQ | PBSX prophage | ||||
| BG11552 | 2,7 | 3,0 | 2,2 | 2,5 | PBSX | xkdR | PBSX prophage | ||
| BG11553 | 3,0 | 4,2 | 3,4 | 3,2 | PBSX | xkdS | PBSX prophage | ||
| BG11554 | 2,6 | 2,6 | PBSX | xkdT | PBSX prophage | ||||
| BG11555 | 4,0 | 5,8 | 4,0 | 4,1 | PBSX | xkdU | PBSX prophage | ||
| BG11556 | 2,6 | PBSX | xkdV | PBSX prophage | |||||
| BG11557 | 3,2 | 5,0 | 3,2 | 3,4 | PBSX | xkdW | PBSX prophage | ||
| BG11558 | 3,3 | PBSX | xkdX | PBSX prophage | |||||
| BG11559 | 2,1 | 2,5 | 2,6 | PBSX | xtrA | PBSX prophage | |||
| BG12699 | 3,7 | 5,0 | 2,5 | 3,6 | PBSX | xlyB | N-Acetylmuramoyl-l-alanine amidase (PBSX prophage-mediated lysis) |
To monitor the effects of PhoA precursor production on genome-wide transcriptional activity, microarray analyses were performed. Preparation of total RNA, cDNA synthesis, labeling, and DNA microarray hybridization and analyses were performed as described in Materials and Methods. The RNA samples were obtained from independent cultures of parental 168 strain and strain 168 harboring the following plasmids: pPSPhoA2 for production of prelip-PhoA, pPSPhoA5 for production of preprolip-PhoA, pPSPhoA6 for production of pre-PhoA, and the “empty” cloning vector pPS2. RNA samples were used for independent cDNA synthesis and subsequent competitive DNA array hybridization with samples of the plasmid-containing strains and the parental 168 strain. Genes were considered significantly upregulated when the Cyber-T Bayesian P values for differences in the mRNA abundance between the parental strain and the corresponding plasmid-containing strain were <0.001 and the individual differences for two biological replicates were at least 2-fold. Notably, no downregulated genes were identified. Genes belonging to prophage regions are indicated.
Accession numbers were derived from the subtilist database (http://genolist.pasteur.fr/SubtiList).
Numbers indicate the fold factor by which transcription of an indicated gene was increased due to the presence of pPS2, pPSPhoA2, pPSPhoA5, or pPSPhoA6.
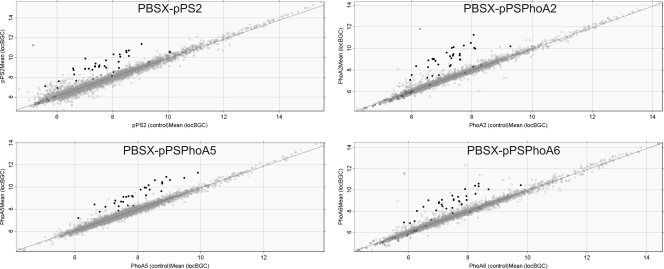
Plasmid pPS2 induces the PBSX prophage in B. subtilis.
A remarkable observation from the array analyses was that a significant number of genes that showed increased expression in prelip-PhoA-, preprolip-PhoA-, or pre-PhoA-producing cells belong to the PBSX prophage region. Notably, similar induction of these genes was observed in the strain containing the empty pPS2 vector, showing that pPS2-borne sequences are responsible for this effect. The PBSX region contains genes from a phage that inserted itself into the B. subtilis 168 genome during the evolution of this strain (60, 63). Unlike most other prophage regions of the B. subtilis genome, the PBSX prophage can still be induced, specifically under DNA-damaging conditions (27, 37). To address the overall PBSX induction, we monitored the expression of all PBSX prophage genes in the B. subtilis cells producing the three different PhoA precursors or containing the empty pPS2 vector. For this purpose, the expression levels of all genes included in the array analyses were plotted in individual scatter plots (Fig. 3). The scatter plots clearly show that the PBSX prophage is induced in the strains containing the empty pPS2 vector or one of the three pPS2-derived plasmids that encode prelip-PhoA, preprolip-PhoA, or pre-PhoA (Fig. 3). The average levels of induction of PBSX genes were found to be similar for all four different plasmids used in our analyses compared to the parental plasmid-free strain B. subtilis 168. This implies that the pPS2 plasmid generates a stimulus that is responsible for induction of the PBSX prophage. Only one other gene, ybfG, was clearly induced by the presence of pPS2.
FIG. 3.
Induction of PBSX genes. Scatter plots were used to visualize the results of transcriptome analyses performed with B. subtilis 168 carrying pPSPhoA2 (prelip-PhoA), pPSPhoA5 (preprolip-PhoA), pPSPhoA6 (pre-PhoA), or the empty vector pPS2. The plots show the gene transcription levels in a culture of bacteria carrying a plasmid (y axis) relative to the gene transcription levels in a plasmid-free control culture (x axis). Filled circles indicate genes that are located on the PBSX prophage, and open circles indicate all remaining genes.
DISCUSSION
The present study was aimed at dissection of the contributions of the pre and pro sequences of a secreted S. hyicus lipase to the secretion of a heterologous model protein by B. subtilis. The alkaline phosphatase PhoA from E. coli was the model protein used for this, because it was previously shown to be secreted at relatively high levels when it was fused to the combined preprolip regions (5, 13) and because it contains two disulfide bonds. As a consequence of these disulfide bonds, PhoA has folding requirements that are significantly different from those of proteins that do not contain disulfide bonds, and thus it can act as a model for secretion of heterologous proteins that contain disulfide bonds (23, 24). Although use of the prepro-region of the S. hyicus lipase has been described in previous studies (15, 18, 32, 33, 45, 50), these studies did not include an in-depth examination of the roles of the pre- and pro-regions in the subcellular localization and fate of exported “cargo” proteins like the examination that we performed here for E. coli PhoA using cell fractionation and protease accessibility assays. Several novel and unexpected findings were obtained. First, it was shown that the presence of the prolip-peptide, in combination with the lipase signal peptide (prelip), contributes significantly to efficient secretion of PhoA by B. subtilis and that prelip directs PhoA secretion more efficiently than the authentic signal peptide of PhoA. Second, while there were clear differences in the efficiency with which B. subtilis was able to secrete processed products of the preprolip-PhoA, prelip-PhoA, and pre-PhoA precursors, these differences could not be correlated with a general secretion stress response or with another known stress response in the producing cells. Third, our genome-wide analysis of transcriptional responses in the producing cells revealed that the pPS2 vector, which was used for expression of E. coli phoA, triggered induction of genes of the PBSX prophage in B. subtilis.
The observation that the signal peptide (prelip) of the S. hyicus lipase directs the secretion of E. coli PhoA by B. subtilis with higher efficiency than the authentic PhoA signal peptide probably is related to the fact that prelip conforms better to the requirements for productive interactions with the secretion machinery of B. subtilis than the PhoA signal peptide. It was previously shown that the average length of the signal peptides of 47 extracellular proteins of B. subtilis is 30 residues, including an N region with an average of two positively charged residues, a hydrophobic H region consisting of ∼20 residues, and a C region with a preferred Ala-X-Ala signal peptidase I recognition site (2). Accordingly, the total length of prelip is 38 residues, including an N region with at least four positively charged residues (indicated by bold type in the sequence MKETKHQHTFSIRKS), an H region consisting of 20 residues, and a predicted Ala-Glu-Ala cleavage site. In contrast, the authentic signal peptide of PhoA (total length, 21 residues) includes an N region with one positively charged Lys residue, an H region consisting of 10 residues, and a Thr-Lys-Ala signal peptidase I recognition site and seems to be suboptimal for the B. subtilis secretion machinery.
The observation that the pro-peptide of the S. hyicus lipase (prolip) also contributes significantly to efficient secretion of PhoA is consistent with the previously reported observation that preprolip supports efficient secretion of E. coli OmpA by both B. subtilis and Staphylococcus carnosus (32, 33). It was proposed that prolip could help translocated and unfolded OmpA escape from proteases at the membrane-cell wall interface of these two Gram-positive bacteria by accelerating the release of OmpA from the plasma membrane and/or passage through the cell wall (32, 33). prolip might play a similar role in the secretion of E. coli PhoA by B. subtilis. Indeed, our Western blotting experiments showed that the preprolip-PhoA-expressing strain contains more PhoA in the medium, while in the prelip-PhoA-expressing strain a much larger fraction of the PhoA protein remains attached to the membrane. It is also possible that PhoA may be exported more efficiently from the cytoplasm of B. subtilis with preprolip than with prelip because the junction between prelip and prolip was optimized during evolution, whereas the junction between prelip and mature PhoA is artificial and therefore perhaps suboptimal for secretion. Indeed, the present study indicates that the presence of prolip also results in clearly improved export of PhoA from the cytoplasm, at least when this process is directed by the prelip signal peptide. This finding is interesting since pro-peptides are generally not thought to be involved in protein translocation across the membrane, which is initiated by the signal peptide. Rather, pro-peptides are generally known to be involved in the posttranslocational folding process to achieve an active and stable form of the secretory protein (46, 55). Although this has not been previously documented for B. subtilis, there is some precedence for a potential role of prolip in protein export from the cytoplasm of S. carnosus, and it has been proposed that the prolip peptide can keep precursor proteins in a translocation-competent state (50). Whether this is also the case in B. subtilis is worth further investigation. In any case, the present data show that great increases in heterologous protein secretion can probably be achieved by modulating the pre- and pro-peptides of the protein of interest.
To minimize differences in the synthesis of the preprolip-PhoA, prelip-PhoA, and pre-PhoA precursors, all three precursors were produced using the same promoter, ribosomal binding site, and start codon. Nevertheless, clear differences were observed in the yields of cell-bound and extracellular active PhoA, indicating that there were important differences in the efficiency with which B. subtilis was able to export the PhoA precursors and to release processed forms of PhoA into the growth medium. The best “outcome” was achieved with cultures producing preprolip-PhoA, which accumulated the largest amounts of active mature PhoA in the growth medium and relatively small amounts in the cells. Judged by the results obtained with preprolip-PhoA, substantial amounts of the PhoA that was produced as prelip-PhoA or pre-PhoA were apparently not correctly translocated, folded, and/or released. In particular, the accumulation of cell-associated PhoA forms with a molecular weight that was higher than that of mature PhoA suggests that prelip-PhoA production is associated with a PhoA folding problem that gives rise to aggregates or complexes with other, as-yet-unidentified proteins or cell wall compounds. Apparently, the pro-peptide can prevent the occurrence of such high-molecular-weight forms. In the case of authentic pre-PhoA synthesis, the overall yields of PhoA were very low compared to the yields of the other two constructs tested. This suggests that substantial degradation of the synthesized pre-PhoA or the resulting mature PhoA occurred. In view of these findings, it was quite remarkable that in our transcriptome analyses, no CssRS-dependent secretion stress response was observed for any of the precursor-producing cells. This would have been a predicted result in view of the detection of PhoA processing and degradation products and even aggregates in the B. subtilis cell envelope. Indeed, mild htrB induction was observed previously when preprolip-PhoA was produced in the B. subtilis IhtrB strain (13). However, in the IhtrB strain, the secretion stress responses are strongly enhanced due to inactivation of the htrB gene for construction of the htrB-lacZ transcriptional gene fusion (36). Importantly, consistent with our present results, the previous studies with a transcriptional htrA-lacZ gene fusion did not reveal induction of htrA transcription in response to preprolip-PhoA production. The very low and/or undetectable secretion stress response in the PhoA-producing B. subtilis strains contrasts strongly with the severe secretion stress response that was previously observed for cells producing the α-amylase AmyQ of Bacillus amyloliquefaciens (1, 31). The difference is all the more remarkable because, at least in the case of preprolip-PhoA-expressing cells, the PhoA is produced at even higher levels than AmyQ (1). This may indicate that, compared to PhoA, higher levels of malfolded AmyQ are present at critical locations in the B. subtilis cell envelope, at least under the conditions tested. Alternatively, the CssRS system might be more sensitive to malfolded AmyQ than to malfolded PhoA so that the current production levels of PhoA are simply too low to trigger a secretion stress response. If this is true, increasing the production of PhoA to significantly higher levels may establish possible differences in the secretion stress responses by the host cells producing the different PhoA precursors.
The genome-wide transcriptome analysis confirmed that production of the different PhoA precursors provoked no clear overall stress response, although it was evident that production of the pre-PhoA precursor caused somewhat more pronounced transcriptional changes in the producing cells than production of the preprolip-PhoA precursor caused. This in fact confirms the notion that substantial amounts of the pre-PhoA protein were synthesized, but that the synthesis remained largely undetectable due to rapid proteolysis. Furthermore, this finding is consistent with the observation that preprolip-PhoA is more effectively exported to the medium than the pre-PhoA variant. The function of some genes with altered expression in the pre-PhoA-producing strain further supports the idea that pre-PhoA is inefficiently exported from the cytoplasm and degraded. The ATP-dependent Clp protease subunit ClpE, for example, is known to be involved in the breakdown of misfolded proteins (16, 34). The upregulated expression of this gene in the pre-PhoA-producing strain, therefore, suggests that there is incorrect cytoplasmic folding of some of this protein. Furthermore, the gene for the thiol-disulfide oxidoreductase TrxA is known to be upregulated in response to oxidative stress (48). Since folded PhoA has two disulfide bonds, the presence of cytoplasmic PhoA may impose some sort of oxidative stress on the producing cells. The latter view is consistent with the detection of cytoplasmic PhoA in cells producing prelip-PhoA, which also display increased transcription of trxA. Importantly, irrespective of the precise origin of the few stress responses observed, the prolip peptide, at least in combination with the prelip signal peptide, seems to prevent most of these responses from occurring.
The observed induction of PBSX prophage genes by the cloning vector pPS2 explains the previously documented accumulation of the PBSX-specific proteins XkdG, XkdK, and XkdM in the growth medium of B. subtilis cells producing preprolip-PhoA with plasmid pPSPhoA5 (13). At present, it is unclear which sequences of the vector are responsible for this prophage induction. To our knowledge, the only reported stimulus for PBSX prophage induction is DNA damage (27, 37). One possibility is that prophage induction is triggered by accumulating replication intermediates of pPS2, but other explanations are also conceivable since our data do not indicate that pPS2 triggers a strong SOS response in B. subtilis. It is nevertheless possible that pPS2 causes a very mild SOS stimulus, which is sufficient only for induction of PBSX. This view is supported by the upregulated expression of the DNA damage-inducible genes tagC and dinB and a few SPβ genes in prelip-PhoA-producing cells. However, these genes were not significantly induced in the other strains with pPS2-derived plasmids. To our knowledge, prophage induction in B. subtilis 168 in response to the presence of plasmids has not been reported previously. Thus, we do not know whether this is a pPS2-specific phenomenon or a more general phenomenon that so far has escaped attention. Clearly, prophage induction causes cell lysis, and the low levels of the cytoplasmic TrxA protein observed in growth medium fractions may actually be related to this phenomenon. Since cell lysis decreases the production capacity of B. subtilis as a cell factory, it may be relevant to test plasmid vectors for prophage-inducing properties prior to their use in an industrial context. Alternatively, a prophage-free production strain, like the previously constructed B. subtilis Δ6 strain that lacks all prophages (59), might be an appropriate host if there is a reason to use plasmids for bioproduction purposes.
In conclusion, the work presented here shows that combined use of the pre- and pro-region sequences of the S. hyicus lipase can contribute significantly to productive secretion of a heterologous protein by B. subtilis. At least in our experiments with E. coli PhoA fused to the preprolip sequences, no secretion stress response could be detected, which supports the view that these sequences represent a very promising tool for the production of heterologous secretory proteins in B. subtilis.
Acknowledgments
This study was supported in part by CEU projects LSHG-CT-2004-503468, LSHG-CT-2004-005257, LSHM-CT-2006-019064, LSHG-CT-2006-037469, and PITN-GA-2008-215524, by the transnational SysMO initiative through project BACELL SysMO, by the European Science Foundation under the EUROCORES Programme EuroSCOPE, and by grant 04-EScope 01-011 from the Research Council for Earth and Life Sciences of the Netherlands Organization for Scientific Research.
We acknowledge the collaborative effort with the teams of Stéphane Aymerich, Kevin Devine, Vincent Fromion, and Tony Wilkinson in standardizing the fluorescence measurements.
Footnotes
Published ahead of print on 30 November 2009.
REFERENCES
- 1.Antelmann, H., E. Darmon, D. Noone, J. W. Veening, H. Westers, S. Bron, O. P. Kuipers, K. M. Devine, M. Hecker, and J. M. van Dijl. 2003. The extracellular proteome of Bacillus subtilis under secretion stress conditions. Mol. Microbiol. 49:143-156. [DOI] [PubMed] [Google Scholar]
- 2.Antelmann, H., H. Tjalsma, B. Voigt, S. Ohlmeier, S. Bron, J. M. van Dijl, and M. Hecker. 2001. A proteomic view on genome-based signal peptide predictions. Genome Res. 11:1484-1502. [DOI] [PubMed] [Google Scholar]
- 3.Aslanidis, C., and P. J. de Jong. 1990. Ligation-independent cloning of PCR products (LIC-PCR). Nucleic Acids Res. 18:6069-6074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bolhuis, A., H. Tjalsma, H. E. Smith, A. de Jong, R. Meima, G. Venema, S. Bron, and J. M. van Dijl. 1999. Evaluation of bottlenecks in the late stages of protein secretion in Bacillus subtilis. Appl. Environ. Microbiol. 65:2934-2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bolhuis, A., G. Venema, W. J. Quax, S. Bron, and J. M. van Dijl. 1999. Functional analysis of paralogous thiol-disulfide oxidoreductases in Bacillus subtilis. J. Biol. Chem. 274:24531-24538. [DOI] [PubMed] [Google Scholar]
- 6.Braun, P., G. Gerritse, J. M. van Dijl, and W. J. Quax. 1999. Improving protein secretion by engineering components of the bacterial translocation machinery. Curr. Opin. Biotechnol. 10:376-381. [DOI] [PubMed] [Google Scholar]
- 7.Brockmeier, U., M. Caspers, R. Freudl, A. Jockwer, T. Noll, and T. Eggert. 2006. Systematic screening of all signal peptides from Bacillus subtilis: a powerful strategy in optimizing heterologous protein secretion in Gram-positive bacteria. J. Mol. Biol. 362:393-402. [DOI] [PubMed] [Google Scholar]
- 8.Bron, S., and G. Venema. 1972. Ultraviolet inactivation and excision-repair in Bacillus subtilis. I. Construction and characterization of a transformable eightfold auxotrophic strain and two ultraviolet-sensitive derivatives. Mutat. Res. 15:1-10. [DOI] [PubMed] [Google Scholar]
- 9.Cano, F., S. Liljeqvist, T. N. Nguyen, P. Samuelson, J. Y. Bonnefoy, S. Stahl, and A. Robert. 1999. A surface-displayed cholera toxin B peptide improves antibody responses using food-grade staphylococci for mucosal subunit vaccine delivery. FEMS Immunol. Med. Microbiol. 25:289-298. [DOI] [PubMed] [Google Scholar]
- 10.Cano, F., H. Plotnicky-Gilquin, T. N. Nguyen, S. Liljeqvist, P. Samuelson, J. Bonnefoy, S. Stahl, and A. Robert. 2000. Partial protection to respiratory syncytial virus (RSV) elicited in mice by intranasal immunization using live staphylococci with surface-displayed RSV-peptides. Vaccine 18:2743-2752. [DOI] [PubMed] [Google Scholar]
- 11.Cao, M., P. A. Kobel, M. M. Morshedi, M. F. Wu, C. Paddon, and J. D. Helmann. 2002. Defining the Bacillus subtilis sigma(W) regulon: a comparative analysis of promoter consensus search, run-off transcription/macroarray analysis (ROMA), and transcriptional profiling approaches. J. Mol. Biol. 316:443-457. [DOI] [PubMed] [Google Scholar]
- 12.Cao, M., T. Wang, R. Ye, and J. D. Helmann. 2002. Antibiotics that inhibit cell wall biosynthesis induce expression of the Bacillus subtilis sigma(W) and sigma(M) regulons. Mol. Microbiol. 45:1267-1276. [DOI] [PubMed] [Google Scholar]
- 13.Darmon, E., R. Dorenbos, J. Meens, R. Freudl, H. Antelmann, M. Hecker, O. P. Kuipers, S. Bron, W. J. Quax, J. Y. Dubois, and J. M. van Dijl. 2006. A disulfide bond-containing alkaline phosphatase triggers a BdbC-dependent secretion stress response in Bacillus subtilis. Appl. Environ. Microbiol. 72:6876-6885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Darmon, E., D. Noone, A. Masson, S. Bron, O. P. Kuipers, K. M. Devine, and J. M. van Dijl. 2002. A novel class of heat and secretion stress-responsive genes is controlled by the autoregulated CssRS two-component system of Bacillus subtilis. J. Bacteriol. 184:5661-5671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Demleitner, G., and F. Gotz. 1994. Evidence for importance of the Staphylococcus hyicus lipase pro-peptide in lipase secretion, stability and activity. FEMS Microbiol. Lett. 121:189-197. [DOI] [PubMed] [Google Scholar]
- 16.Derre, I., G. Rapoport, K. Devine, M. Rose, and T. Msadek. 1999. ClpE, a novel type of HSP100 ATPase, is part of the CtsR heat shock regulon of Bacillus subtilis. Mol. Microbiol. 32:581-593. [DOI] [PubMed] [Google Scholar]
- 17.Dubois, J. Y., T. R. Kouwen, A. K. Schurich, C. R. Reis, H. T. Ensing, E. N. Trip, J. C. Zweers, and J. M. van Dijl. 2009. Immunity to the bacteriocin sublancin 168 is determined by the SunI (YolF) protein of Bacillus subtilis. Antimicrob. Agents Chemother. 53:651-661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gotz, F., H. M. Verheij, and R. Rosenstein. 1998. Staphylococcal lipases: molecular characterisation, secretion, and processing. Chem. Phys. Lipids 93:15-25. [DOI] [PubMed] [Google Scholar]
- 19.Gunneriusson, E., P. Samuelson, M. Uhlen, P. A. Nygren, and S. Stahl. 1996. Surface display of a functional single-chain Fv antibody on staphylococci. J. Bacteriol. 178:1341-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harwood, C. R., and R. Cranenburgh. 2008. Bacillus protein secretion: an unfolding story. Trends Microbiol. 16:73-79. [DOI] [PubMed] [Google Scholar]
- 21.Hyyrylainen, H. L., A. Bolhuis, E. Darmon, L. Muukkonen, P. Koski, M. Vitikainen, M. Sarvas, Z. Pragai, S. Bron, J. M. van Dijl, and V. P. Kontinen. 2001. A novel two-component regulatory system in Bacillus subtilis for the survival of severe secretion stress. Mol. Microbiol. 41:1159-1172. [DOI] [PubMed] [Google Scholar]
- 22.Kobayashi, K., S. D. Ehrlich, A. Albertini, G. Amati, K. K. Andersen, M. Arnaud, K. Asai, S. Ashikaga, S. Aymerich, P. Bessieres, F. Boland, S. C. Brignell, S. Bron, K. Bunai, J. Chapuis, L. C. Christiansen, A. Danchin, M. Debarbouille, E. Dervyn, E. Deuerling, K. Devine, S. K. Devine, O. Dreesen, J. Errington, S. Fillinger, S. J. Foster, Y. Fujita, A. Galizzi, R. Gardan, C. Eschevins, T. Fukushima, K. Haga, C. R. Harwood, M. Hecker, D. Hosoya, M. F. Hullo, H. Kakeshita, D. Karamata, Y. Kasahara, F. Kawamura, K. Koga, P. Koski, R. Kuwana, D. Imamura, M. Ishimaru, S. Ishikawa, I. Ishio, D. Le Coq, A. Masson, C. Mauel, R. Meima, R. P. Mellado, A. Moir, S. Moriya, E. Nagakawa, H. Nanamiya, S. Nakai, P. Nygaard, M. Ogura, T. Ohanan, M. O'Reilly, M. O'Rourke, Z. Pragai, H. M. Pooley, G. Rapoport, J. P. Rawlins, L. A. Rivas, C. Rivolta, A. Sadaie, Y. Sadaie, M. Sarvas, T. Sato, H. H. Saxild, E. Scanlan, W. Schumann, J. F. Seegers, J. Sekiguchi, A. Sekowska, S. J. Seror, M. Simon, P. Stragier, R. Studer, H. Takamatsu, T. Tanaka, M. Takeuchi, H. B. Thomaides, V. Vagner, J. M. van Dijl, K. Watabe, A. Wipat, H. Yamamoto, M. Yamamoto, Y. Yamamoto, K. Yamane, K. Yata, K. Yoshida, H. Yoshikawa, U. Zuber, and N. Ogasawara. 2003. Essential Bacillus subtilis genes. Proc. Natl. Acad. Sci. U. S. A. 100:4678-4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kouwen, T. R., J. Y. Dubois, R. Freudl, W. J. Quax, and J. M. van Dijl. 2008. Modulation of thiol-disulfide oxidoreductases for increased production of disulfide-bond-containing proteins in Bacillus subtilis. Appl. Environ. Microbiol. 74:7536-7545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kouwen, T. R., A. van der Goot, R. Dorenbos, T. Winter, H. Antelmann, M. C. Plaisier, W. J. Quax, J. M. van Dijl, and J. Y. Dubois. 2007. Thiol-disulphide oxidoreductase modules in the low-GC Gram-positive bacteria. Mol. Microbiol. 64:984-999. [DOI] [PubMed] [Google Scholar]
- 25.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, M. G. Bertero, P. Bessieres, A. Bolotin, S. Borchert, R. Borriss, L. Boursier, A. Brans, M. Braun, S. C. Brignell, S. Bron, S. Brouillet, C. V. Bruschi, B. Caldwell, V. Capuano, N. M. Carter, S. K. Choi, J. J. Codani, I. F. Connerton, and A. Danchin, and. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 26.Kunst, F., and G. Rapoport. 1995. Salt stress is an environmental signal affecting degradative enzyme synthesis in Bacillus subtilis. J. Bacteriol. 177:2403-2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lazarevic, V., A. Dusterhoft, B. Soldo, H. Hilbert, C. Mauel, and D. Karamata. 1999. Nucleotide sequence of the Bacillus subtilis temperate bacteriophage SPbetac2. Microbiology 145:1055-1067. [DOI] [PubMed] [Google Scholar]
- 28.Lehtio, J., H. Wernerus, P. Samuelson, T. T. Teeri, and S. Stahl. 2001. Directed immobilization of recombinant staphylococci on cotton fibers by functional display of a fungal cellulose-binding domain. FEMS Microbiol. Lett. 195:197-204. [DOI] [PubMed] [Google Scholar]
- 29.Liljeqvist, S., F. Cano, T. N. Nguyen, M. Uhlen, A. Robert, and S. Stahl. 1999. Surface display of functional fibronectin-binding domains on Staphylococcus carnosus. FEBS Lett. 446:299-304. [DOI] [PubMed] [Google Scholar]
- 30.Liljeqvist, S., P. Samuelson, M. Hansson, T. N. Nguyen, H. Binz, and S. Stahl. 1997. Surface display of the cholera toxin B subunit on Staphylococcus xylosus and Staphylococcus carnosus. Appl. Environ. Microbiol. 63:2481-2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lulko, A. T., J. W. Veening, G. Buist, W. K. Smits, E. J. Blom, A. C. Beekman, S. Bron, and O. P. Kuipers. 2007. Production and secretion stress caused by overexpression of heterologous alpha-amylase leads to inhibition of sporulation and a prolonged motile phase in Bacillus subtilis. Appl. Environ. Microbiol. 73:5354-5362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meens, J., E. Frings, M. Klose, and R. Freudl. 1993. An outer membrane protein (OmpA) of Escherichia coli can be translocated across the cytoplasmic membrane of Bacillus subtilis. Mol. Microbiol. 9:847-855. [DOI] [PubMed] [Google Scholar]
- 33.Meens, J., M. Herbort, M. Klein, and R. Freudl. 1997. Use of the pre-pro part of Staphylococcus hyicus lipase as a carrier for secretion of Escherichia coli outer membrane protein A (OmpA) prevents proteolytic degradation of OmpA by cell-associated protease(s) in two different gram-positive bacteria. Appl. Environ. Microbiol. 63:2814-2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miethke, M., M. Hecker, and U. Gerth. 2006. Involvement of Bacillus subtilis ClpE in CtsR degradation and protein quality control. J. Bacteriol. 188:4610-4619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nicholson, W. L., and P. Setlow. 1990. Sporulation, germination and outgrowth, p. 391-450. In C. R. Harwood and S. M. Cutting (ed.), Molecular biological methods for Bacillus. Wiley, Chichester, United Kingdom.
- 36.Noone, D., A. Howell, R. Collery, and K. M. Devine. 2001. YkdA and YvtA, HtrA-like serine proteases in Bacillus subtilis, engage in negative autoregulation and reciprocal cross-regulation of ykdA and yvtA gene expression. J. Bacteriol. 183:654-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Okamoto, K., J. A. Mudd, and J. Marmur. 1968. Conversion of Bacillus subtilis DNA to phage DNA following mitomycin C induction. J. Mol. Biol. 34:429-437. [DOI] [PubMed] [Google Scholar]
- 38.Palva, I. 1982. Molecular cloning of alpha-amylase gene from Bacillus amyloliquefaciens and its expression in B. subtilis. Gene 19:81-87. [DOI] [PubMed] [Google Scholar]
- 39.Puohiniemi, R., M. Simonen, S. Muttilainen, J. P. Himanen, and M. Sarvas. 1992. Secretion of the Escherichia coli outer membrane proteins OmpA and OmpF in Bacillus subtilis is blocked at an early intracellular step. Mol. Microbiol. 6:981-990. [DOI] [PubMed] [Google Scholar]
- 40.Robert, A., P. Samuelson, C. Andreoni, T. Bachi, M. Uhlen, H. Binz, T. N. Nguyen, and S. Stahl. 1996. Surface display on staphylococci: a comparative study. FEBS Lett. 390:327-333. [DOI] [PubMed] [Google Scholar]
- 41.Ronen, M., R. Rosenberg, B. I. Shraiman, and U. Alon. 2002. Assigning numbers to the arrows: parameterizing a gene regulation network by using accurate expression kinetics. Proc. Natl. Acad. Sci. U. S. A. 99:10555-10560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 43.Samuelson, P., F. Cano, A. Robert, and S. Stahl. 1999. Engineering of a Staphylococcus carnosus surface display system by substitution or deletion of a Staphylococcus hyicus lipase propeptide. FEMS Microbiol. Lett. 179:131-139. [DOI] [PubMed] [Google Scholar]
- 44.Samuelson, P., M. Hansson, N. Ahlborg, C. Andreoni, F. Gotz, T. Bachi, T. N. Nguyen, H. Binz, M. Uhlen, and S. Stahl. 1995. Cell surface display of recombinant proteins on Staphylococcus carnosus. J. Bacteriol. 177:1470-1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sandgathe, A., D. Tippe, S. Dilsen, J. Meens, M. Halfar, D. Weuster-Botz, R. Freudl, J. Thommes, and M. R. Kula. 2003. Production of a human calcitonin precursor with Staphylococcus carnosus: secretory expression and single-step recovery by expanded bed adsorption. Process Biochem. 38:1351-1363. [Google Scholar]
- 46.Sarvas, M., C. R. Harwood, S. Bron, and J. M. van Dijl. 2004. Post-translocational folding of secretory proteins in Gram-positive bacteria. Biochim. Biophys. Acta 1694:311-327. [DOI] [PubMed] [Google Scholar]
- 47.Saunders, C. W., B. J. Schmidt, R. L. Mallonee, and M. S. Guyer. 1987. Secretion of human serum albumin from Bacillus subtilis. J. Bacteriol. 169:2917-2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scharf, C., S. Riethdorf, H. Ernst, S. Engelmann, U. Volker, and M. Hecker. 1998. Thioredoxin is an essential protein induced by multiple stresses in Bacillus subtilis. J. Bacteriol. 180:1869-1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shinde, U., and M. Inouye. 2000. Intramolecular chaperones: polypeptide extensions that modulate protein folding. Semin. Cell Dev. Biol. 11:35-44. [DOI] [PubMed] [Google Scholar]
- 50.Sturmfels, A., F. Gotz, and A. Peschel. 2001. Secretion of human growth hormone by the food-grade bacterium Staphylococcus carnosus requires a propeptide irrespective of the signal peptide used. Arch. Microbiol. 175:295-300. [DOI] [PubMed] [Google Scholar]
- 51.Takagi, H., M. Koga, S. Katsurada, Y. Yabuta, U. Shinde, M. Inouye, and S. Nakamori. 2001. Functional analysis of the propeptides of subtilisin E and aqualysin I as intramolecular chaperones. FEBS Lett. 508:210-214. [DOI] [PubMed] [Google Scholar]
- 52.Tjalsma, H., H. Antelmann, J. D. Jongbloed, P. G. Braun, E. Darmon, R. Dorenbos, J. Y. Dubois, H. Westers, G. Zanen, W. J. Quax, O. P. Kuipers, S. Bron, M. Hecker, and J. M. van Dijl. 2004. Proteomics of protein secretion by Bacillus subtilis: separating the “secrets” of the secretome. Microbiol. Mol. Biol. Rev. 68:207-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tjalsma, H., A. Bolhuis, J. D. Jongbloed, S. Bron, and J. M. van Dijl. 2000. Signal peptide-dependent protein transport in Bacillus subtilis: a genome-based survey of the secretome. Microbiol. Mol. Biol. Rev. 64:515-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tjalsma, H., S. Bron, and J. M. van Dijl. 2003. Complementary impact of paralogous OxaI-like proteins of Bacillus subtilis on post-translocational stages in protein secretion. J. Biol. Chem. 278:15622-15632. [DOI] [PubMed] [Google Scholar]
- 55.van Dijl, J. M., A. Bolhuis, H. Tjalsma, J. D. Jongbloed, A. de Jong, and S. Bron. 2001. Protein transport pathways in Bacillus subtilis: a genome-based road map, p. 337-355. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, DC.
- 56.van Wely, K. H., J. Swaving, R. Freudl, and A. J. Driessen. 2001. Translocation of proteins across the cell envelope of Gram-positive bacteria. FEMS Microbiol. Rev. 25:437-454. [DOI] [PubMed] [Google Scholar]
- 57.von Heijne, G. 1990. Protein targeting signals. Curr. Opin. Cell Biol. 2:604-608. [DOI] [PubMed] [Google Scholar]
- 58.von Heijne, G. 1990. The signal peptide. J. Membr. Biol. 115:195-201. [DOI] [PubMed] [Google Scholar]
- 59.Westers, H., R. Dorenbos, J. M. van Dijl, J. Kabel, T. Flanagan, K. M. Devine, F. Jude, S. J. Seror, A. C. Beekman, E. Darmon, C. Eschevins, A. de Jong, S. Bron, O. P. Kuipers, A. M. Albertini, H. Antelmann, M. Hecker, N. Zamboni, U. Sauer, C. Bruand, D. S. Ehrlich, J. C. Alonso, M. Salas, and W. J. Quax. 2003. Genome engineering reveals large dispensable regions in Bacillus subtilis. Mol. Biol. Evol. 20:2076-2090. [DOI] [PubMed] [Google Scholar]
- 60.Wood, H. E., M. T. Dawson, K. M. Devine, and D. J. McConnell. 1990. Characterization of PBSX, a defective prophage of Bacillus subtilis. J. Bacteriol. 172:2667-2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yabuta, Y., H. Takagi, M. Inouye, and U. Shinde. 2001. Folding pathway mediated by an intramolecular chaperone: propeptide release modulates activation precision of pro-subtilisin. J. Biol. Chem. 276:44427-44434. [DOI] [PubMed] [Google Scholar]
- 62.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 63.Zahler, S. A., R. Z. Korman, R. Rosenthal, and H. E. Hemphill. 1977. Bacillus subtilis bacteriophage SPbeta: localization of the prophage attachment site, and specialized transduction. J. Bacteriol. 129:556-558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zeigler, D. R., and J. B. Perkins. 2008. The genus Bacillus, p. 309-338. In E. Goldman and L. Green (ed.), Practical handbook of microbiology. CRC Press, Boca Raton, FL.