Abstract
Swimming in ocean water, including ocean water at beaches not impacted by known point sources of pollution, is an increasing health concern. This study was an initial evaluation of the presence of indicator microbes and pathogens and the association among the indicator microbes, pathogens, and environmental conditions at a subtropical, recreational marine beach in south Florida impacted by non-point sources of pollution. Twelve water and eight sand samples were collected during four sampling events at high or low tide under elevated or reduced solar insolation conditions. The analyses performed included analyses of fecal indicator bacteria (FIB) (fecal coliforms, Escherichia coli, enterococci, and Clostridium perfringens), human-associated microbial source tracking (MST) markers (human polyomaviruses [HPyVs] and Enterococcus faecium esp gene), and pathogens (Vibrio vulnificus, Staphylococcus aureus, enterovirus, norovirus, hepatitis A virus, Cryptosporidium spp., and Giardia spp.). The enterococcus concentrations in water and sand determined by quantitative PCR were greater than the concentrations determined by membrane filtration measurement. The FIB concentrations in water were below the recreational water quality standards for three of the four sampling events, when pathogens and MST markers were also generally undetectable. The FIB levels exceeded regulatory guidelines during one event, and this was accompanied by detection of HPyVs and pathogens, including detection of the autochthonous bacterium V. vulnificus in sand and water, detection of the allochthonous protozoans Giardia spp. in water, and detection of Cryptosporidium spp. in sand samples. The elevated microbial levels were detected at high tide and under low-solar-insolation conditions. Additional sampling should be conducted to further explore the relationships between tidal and solar insolation conditions and between indicator microbes and pathogens in subtropical recreational marine waters impacted by non-point source pollution.
Global estimates indicate that each year more than 120 million cases of gastrointestinal disease and 50 million cases of severe respiratory diseases are caused by swimming and bathing in wastewater-polluted coastal waters (42). Swimming-related illness is attributed predominantly to exposure to microbial pathogens, which enter the water through point sources, such as sewage outfalls. Water quality at beaches may also be impacted by non-point sources, such as storm water runoff, sand resuspension, animal fecal inputs, and human bather shedding (8, 12, 22, 47, 59).
The concentration of indicator microorganisms in a body of recreational water is used to estimate the health risk to bathers. These microbes serve as surrogates for microbial pathogens. Studies show that the U.S. Environmental Protection Agency (EPA)-recommended indicator microbe for marine beaches, enterococci, shows a significant correlation with illness in marine beaches impacted by point source pollution (38, 54). However, a similar correlation has not been identified at beaches impacted by non-point source pollution or subtropical marine beaches (17, 29, 38, 54).
The failure to consistently demonstrate an association between enterococci and illness at non-point source beaches calls into question the ability of indicator microbes to predict the presence of pathogens. Studies conducted on the west coast of the United States have shown that indicators are often not correlated with measured pathogens at non-point source beaches (31, 32, 33, 37). Additional studies conducted in a subtropical environment, such as that of South Florida, where this study was conducted, have repeatedly shown the limited accuracy of indicator microbe standards for determining the presence of pathogens (27, 35). This lack of correlation is understandable since an indicator microbe, such as enterococci, may come from relatively low-risk sources of fecal pollution and therefore may not be related to human or other high-risk sources of fecal pollution and pathogens (9). It has also been shown in both subtropical and temperate climates that indicator bacteria can multiply in the environment, resulting in a false impression of increased microbial pollution and pathogen presence (4, 7, 19, 24, 41, 45, 57, 58). Environmental factors, such as tide, rain, and solar insolation, can also have significant and varying effects on the levels of indicator and pathogenic microbes (21, 24, 33).
The lack of correlation between pathogens and indicator microbes at non-point source beaches can result in two problematic scenarios. If indicator microbes are absent and pathogens are present (false-negative scenario), regulatory monitoring may fail to identify the potential adverse health effects on bathers due to the pathogens. This problem is likely to occur since indicator bacteria are less resistant to environmental stresses and disinfection at wastewater treatment plants than certain pathogens (6, 15). However, if indicator microbes are present and pathogens are absent (false-positive scenario), there can be unnecessary economic losses due to recreational beach advisories and/or closures. A 4-month closure of Huntington Beach in 1999 due to microbial standard violations resulted in the loss of millions of dollars in tourism income to the business community and almost 2 million dollars in beach closure investigation fees (55; for a review, see reference 27).
Given these two possible scenarios, the relationship between indicator microbes and pathogens under different environmental conditions at non-point source beaches representing different geographic and climatic settings should be assessed further. Investigation of this relationship would require a large sample size in order to establish possible significant associations between the various factors and targets. The objective of this study was to conduct a preliminary evaluation of the presence of indicator microbes and pathogens and the possible association between indicator microbes, pathogen measurements, and environmental conditions at a subtropical recreational marine beach in South Florida. Because of cost limitations when multiple targets, including pathogens, are screened, this study was not intended to establish a conclusive relationship between the various factors and targets but was intended to provide insight into both the presence of organisms and possible associations which should be investigated further. Such information would be useful for understanding the potential health risks to bathers from non-point sources of microbes and would also contribute to determining the appropriateness of using indicator microbes to monitor the water quality at non-point source beaches. Although previous studies have assessed the presence of either viral, protozoan, or bacterial pathogens along with indicator microbes in point or non-point source recreational beach waters (27, 33, 37, 47), to our knowledge, this is the first study to assess the presence of all three classes of pathogens (viral, protozoan, and bacterial) as well as indicator microbes at a non-point source recreational beach. This study is also the first study to sample for all these microbes in both water and sand at a non-point source recreational beach. Through analysis of the various microbes under different targeted environmental conditions, this study also included a preliminary evaluation of the sources of microbial contaminants and pathogens and the effectiveness of various analytical methods for microbe detection. The latter part of the study included a comparison of three different methods for enterococcus enumeration, as well as an innovative method for simultaneously concentrating protozoans and viruses from water samples.
MATERIALS AND METHODS
Site description.
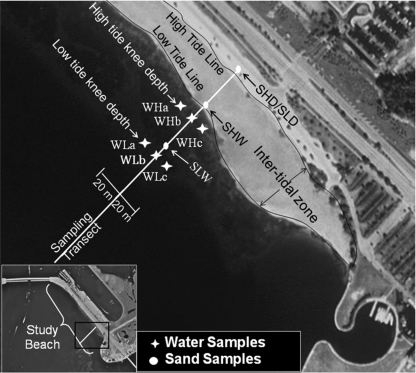
Samples were collected from a study beach on Virginia Key in Miami-Dade County, Florida (Fig. 1). The climate of Miami is classified as “subtropical” because of its geographic location and its average ambient temperature, 24.8°C. The beach studied is an irregularly narrow beach with an average distance between the mean water line and the outer edge of the sand of approximately 5 m. The beach is 1.6 km long, relatively shallow, and characterized by weak water circulation (41). This beach is the only beach in Miami-Dade County where visitors can bring their pets, particularly dogs. Admission to the beach is free, and many bathers frequent the beach, particularly during the summer months. Extensive evaluation of the vicinity around the beach did not find point sources of pollution for it, such as sewage outfalls, failing lift stations, cross-connections of sewage with storm drains, or less obvious non-point sources, such as septic tanks (41). This beach is usually in compliance with regulatory monitoring criteria, but periodically (4.5 days per year [average for 2002 to 2007]) it has been placed under an advisory due to microbial water quality violations (http://esetappsdoh.doh.state.fl.us/irm00beachwater/default.aspx).
FIG. 1.
Water and sand sampling locations at high and low tide along the sampling transect at the study beach in Miami, FL. Photo courtesy of the U.S. Geological Survey.
Sample collection and measurement of physical-chemical parameters.
Four sampling events were conducted during the summer of 2007, two on 18 July (day 1) and two on 8 August (day 2). Each event was timed to coincide with either high or low tide. Samples were collected within 1 h before peak high tide and within 1 h before peak low tide. On day 1, low tide was at dawn (6:30 a.m.) with a solar insolation level of −1.7 W/m2, while high tide occurred at noon (12:30 p.m.) with a solar insolation level of 861 W/m2. On day 2, high tide occurred before sunrise (5:45 a.m.) with a solar insolation level of −1.8 W/m2, while low tide occurred at noon (12:00 p.m.) with a solar insolation level of 402 W/m2. The lower solar insolation level at noon on day 2 was due to overcast conditions. The values for solar insolation, defined as the amount of electromagnetic energy (solar radiation) incident on the surface of the earth, were obtained with a precision spectral pyranometer located within 1 km of the sampling site (http://www.rsmas.miami.edu/etc/download-weatherpak.cgi). During each event, three water samples (WLa, WLb, and WLc or WHa, WHb, and WHc) and two sand samples (SLW and SLD or SHW and SHD) were collected; in the sample designations “H” and “L” indicate high and low tide, respectively, “a,” “b,” and “c” indicate the lateral locations with respect to the sampling transect, and “W” and “D” indicate wet and dry sand, respectively (Fig. 1). Therefore, a total of 12 water samples and 8 sand samples were collected during this study.
Water samples were collected in 20-liter sterile containers along the sampling transect at knee depth. Sand samples were collected from the upper 2.5 cm in a 0.25-m2 area and placed into sterile Whirlpak bags. Wet sand samples (900 g) were collected from the intertidal zone, and dry sand samples (500 g) were collected along the sampling transect above the high-tide line (Fig. 1). The sand samples were then transported to the lab, and each sample was transferred into a carboy with 10 liters of phosphate-buffered saline. Each sand-water mixture was mixed vigorously for 10 min and then allowed to settle for 10 min. This method was utilized to remove the “washable” microbes from the sand and is based on a method suggested by Van Elsas and Smalla (53). The supernatants of the sand-water mixtures (sand eluates) along with the 20-liter water samples were used for analyses.
Physical-chemical parameters, including pH, temperature, salinity (YSI model 650-01m environmental monitoring systems; YSI, Yellow Springs, OH), and turbidity, were measured for water samples; volatile organic compounds and moisture were measured for the sand. Rainfall data were obtained from seven tipping-bucket rain gauges located within 1 km of the site (http://www.rsmas.miami.edu/etc/download-weatherpak.cgi). Wind speed and beach conditions (numbers of bathers, dogs, and birds within 100 m of the sampling transect) were also recorded.
Sample concentration and analysis.
Water and sand eluate samples were split for analysis of the EPA-recommended marine bacterial indicator, enterococci. Three different methods were used to enumerate enterococci: the membrane filtration (MF), chromogenic substrate, and quantitative PCR (qPCR) methods. Additional microbial indicators (fecal coliforms, Escherichia coli, and Clostridium perfringens) were evaluated using traditional MF methods. Human-associated microbial source tracking (MST) markers were evaluated using PCR and included human polyomaviruses (HPyVs) and the esp gene of Enterococcus faecium. The pathogens evaluated included bacteria (Staphylococcus aureus evaluated by MF and Vibrio vulnificus evaluated by MF with confirmation by PCR), protozoans (Cryptosporidium spp. and Giardia spp. evaluated by microscopy and qPCR), and viruses (enterovirus, norovirus, and hepatitis A evaluated by qPCR) (Table 1). See the supplemental material for a detailed description of the methods used.
TABLE 1.
Methods used to detect target organisms
| Target(s) | Methoda | Reference(s) |
|---|---|---|
| Fecal coliforms | Standard method 9222 D | 5 |
| E. coli | U.S. EPA method 1603 | 49 |
| C. perfringens | U.S. EPA method 600-R-95/030 | 48 |
| Enterococci (membrane filtration) | U.S. EPA method 1600 | 50 |
| Enterococci (chromogenic substrate) | IDEXX method | |
| Enterococci (qPCR) | Method of Haughland et al. | 30 |
| E. faecium esp | Method of Scott et al. | 40 |
| Human polyomaviruses | Method of McQuaig et al. | 36 |
| V. vulnificus | Method of Kaysner and DePaola and Gordon et al. | 26, 34 |
| Cryptosporidium spp. and Giardia spp. | Traditional filtration (IDEXX Filtamax) followed by traditional microscopy using method 1623 | 52 |
| Cryptosporidium spp. and Giardia spp. | Bilayer filtration system followed by qPCR as described by Guy et al. | 1, 3, 28 |
| Enterovirus, norovirus, and hepatitis A virus | Bilayer filtration system followed by qPCR as described by DeLeon et al. | 1, 3, 18 |
Details concerning the methods used are provided in the supplemental material.
One-way analysis of variance was used to determine significant differences among mean concentrations of fecal indicator bacteria (FIB). Pearson correlation analysis was used to determine correlations between microbe concentrations. Data sets which did not fit a normal distribution were log transformed prior to analysis.
RESULTS
Environmental parameters.
During the day 1 and 2 morning sampling events there were no bathers or animals near the sampling area; however, in the afternoon there were five bathers and two dogs on day 1 and two bathers and one dog on day 2. The turbidity of the water samples ranged from 3 to 9 nephelometric turbidity units. The water temperature varied from approximately 30°C in the morning to 32 to 34°C in the afternoon on both sampling days. The salinity and pH for all water samples ranged from 31.5 to 32.8 practical salinity units and from 7.4 to 8.2, respectively. The volatile organic compound levels for sand ranged from 0.75 to 1.54% of the dry sand weight, while the moisture content levels for dry sand samples (samples SHD and SLD) ranged from 0.3 to 4% and the moisture content levels for wet sand samples (samples SHW and SLW) ranged from 18 to 22% for all four sampling events. The wind speed during all sampling events ranged from 3 to 3.85 m/s. No rain was recorded 6 h prior to any of the four sampling events, while less than 0.5 cm of rain was recorded 24 h prior to each of the four sampling events.
Microbial indicators.
The enterococcus levels in water samples for all four sampling events ranged from <2 to 110 CFU 100 ml−1 when MF methods were used, from <10 to 100 most probable numbers (MPN) 100 ml−1 when the chromogenic substrate method was used, and from 13 to 157 MPN 100 ml−1 when qPCR was used (Table 2). The concentrations measured by qPCR were greater (P = 0.008) than the concentrations measured by MF. No significant differences (P > 0.1) were found between the concentrations determined by the MF and chromogenic substrate methods or between the concentrations determined by the qPCR and chromogenic substrate methods for water samples. The results of qPCR correlated with MF results (r = 0.86; P < 0.01) for water samples. The measured concentrations determined by the chromogenic substrate method did not significantly correlate with the MF results or the qPCR results (r = 0.78; P = 0.07). For sand, the enterococcus levels for all four sampling events ranged from 4 to 1,088 CFU g (dry weight)−1 when the MF method was used, from 1 to 1,006 MPN g (dry weight)−1 when the chromogenic substrate method was used, and from 74 to 22,100 MPN g (dry weight)−1 when qPCR was used. The enterococcus levels determined by qPCR were significantly higher than those determined by the MF method (P = 0.002) and the chromogenic substrate method (P = 0.0001), and there were no significant differences (P > 0.1) between the MF and chromogenic substrate data for sand (Table 2). The concentrations determined by the chromogenic substrate method correlated with the concentrations determined by the MF method (r = 0.92; P < 0.004); however, there were no significant correlations (P > 0.2) between the results obtained with the other methods for sand samples.
TABLE 2.
Levels of indicator and pathogenic bacteria in water during the four sampling eventsa
| Indicator or pathogenic bacteria (detection method) | Sample location | Concnb |
|||
|---|---|---|---|---|---|
| Day 1 |
Day 2 |
||||
| Low tide, morning | High tide, afternoon | Low tide, afternoon | High tide, morning | ||
| Enterococci (MF) | a | 1 (1) | 2 (3) | <2 | 110 (12) |
| b | 3 (4) | 1 (1) | <2 | 77 (25) | |
| c | 3 (3) | 10 (6) | 8 (6) | 37 (10) | |
| Wet | 4 (6) | 4 (3) | 8 (11) | 8 (12) | |
| Dry | 152 (52) | 45 (87) | 1,088 (381) | 63 (51) | |
| Enterococci (chromogenic substrate) | a | <10 | <10 | <50 | 100 |
| b | 10 | 30 | <50 | 50 | |
| c | 31 | 40 | <50 | <50 | |
| Wet | 3 | 1 | 3 | 1 | |
| Dry | 39 | 6 | 1,006 | 34 | |
| Enterococci (qPCR) | a | 13 | 37 | 40 | 144 |
| b | 17 | 55 | 42 | 157 | |
| c | 18 | 48 | 36 | 128 | |
| Wet | 1,033 | 680 | 276 | 140 | |
| Dry | 1,383 | 22,100 | Inhibited | 74 | |
| E. coli (MF) | a | 2 | <2 | <2 | 218 |
| b | <2 | <2 | <2 | 147 | |
| c | <2 | 4 | <2 | 27 | |
| Wet | <5 | <5 | 2 (1)c | 2 | |
| Dry | <5 | <5 | 4 (1) | 29 (35) | |
| Fecal coliforms (MF) | a | 2 | <2 | <2 | 275 |
| b | 4 | 4 | 2 | 213 | |
| c | 4 | 22 | <2 | 107 | |
| Wet | <5 | 13 (21) | 3 (1) | 26 (40) | |
| Dry | 387 (165) | 26 (10) | 20 (12) | 1,421 | |
| Presumptive S. aureus (MF)c | a | <2 | 11 | 11 | 21 |
| b | <2 | <2 | <2 | 32 | |
| c | 2 | 4 | 12 | 12 | |
| Wet | 14 (6) | 24 (6) | 20 (7) | 13 (14) | |
| Dry | 188 | 31 (51) | 1,048 | 132 (14) | |
| C. perfringens (MF) | a | <2 | 2 | 2 | 4 |
| b | 8 | 6 | <2 | 13 | |
| c | <2 | 4 | <2 | 8 | |
| Wet | 7 (2) | 6 (2) | 6 (3) | 3 (2) | |
| Dry | 38 (19) | 64 (24) | 11 (3) | 44 (5) | |
Sample locations a, b, and c are water sample locations at different distances from the sampling transect, and the wet and dry locations are sand sample locations in and above the intertidal zone, respectively, as shown in Fig. 1. For water, three samples were obtained for each enterococcus method and one or two samples were obtained for the other indicator and pathogenic bacteria. For sand, 5 to 10 samples were obtained for enterococci, 2 to 5 samples were obtained for presumptive S. aureus and C. perfringens, and 3 or 4 samples were obtained for E. coli and fecal coliforms.
The values obtained by the MF, chromogenic substrate, and qPCR methods are expressed in CFU, MPN, and genome equivalents, respectively, per 100 ml for water and per gram for sand. The values for each type of sample were averaged to obtain the results reported. The numbers in parentheses are standard deviations.
None of the presumptive S. aureus results were confirmed. No organisms were determined to be cocci by Gram staining, and none of the organisms grew on sheep blood agar of ORSAB media.
The enterococcus levels for the day 1 low- and high-tide water samples and for the day 2 low-tide water samples were less than 25 CFU 100 ml−1 based on only MF results. The average level for day 2 high-tide samples was 65 CFU ml−1 based on only MF results, and the average level for the WHa sample on day 2 was 110 CFU 100 ml−1 (standard deviation = 12) (Table 2). The day 2 high-tide water samples also had the highest E. coli, fecal coliform, and C. perfringens levels (averages, 131, 198, and 8.3 CFU 100 ml−1, respectively) (Table 2). For the water samples, strong correlations were found between enterococci and E. coli (r = 0.98; P < 0.01), between enterococci and fecal coliforms (r = 0.98; P < 0.01), and between E. coli and fecal coliforms (r = 0.97; P < 0.06).
For sand, strong correlations were found between E. coli and C. perfringens (r = 0.96; P = 0.04). All indicator microbe levels were higher for the dry sand than for the wet sand for all samples except the day 2 high-tide sample analyzed for enterococci by qPCR (Table 2).
MST markers.
The human polyomavirus marker was detected in samples WHa, WHc, WLc, SLW, and SHW on day 2, while the levels in all other samples were below the detection limit. The E. faecium esp gene was not detected in any of the samples collected during the four sampling events (Table 3).
TABLE 3.
Results for MST markers and pathogens for all four sampling events
| Sample type | Marker or pathogen | Detection ona: |
|||
|---|---|---|---|---|---|
| Day 1 |
Day 2 |
||||
| Low tide, morning | High tide, afternoon | Low tide, afternoon | High tide, morning | ||
| Water | Polyomavirus | − | − | c | a, c |
| E. faecium gene | − | − | − | − | |
| V. vulnificus | − | − | − | b | |
| Giardia spp. (PCR) | − | − | − | a, b | |
| Giardia spp. (microscopy)b | NA | − | NA | b | |
| Cryptosporidium spp. (PCR) | − | − | − | − | |
| Cryptosporidium spp. (microscopy)b | NA | − | NA | − | |
| Enterovirus | − | − | − | − | |
| Norovirus | − | − | − | − | |
| Hepatitis A virus | − | − | − | − | |
| Sand | Polyomavirus | − | − | Wet | Dry |
| E. faecium gene | − | − | − | − | |
| V. vulnificus | − | − | Wet, dry | Dry | |
| Giardia spp. (PCR) | − | − | − | − | |
| Giardia spp. (microscopy) | NA | − | NA | − | |
| Cryptosporidium spp. (PCR) | − | − | Dry | Wet | |
| Cryptosporidium spp. (microscopy) | NA | − | NA | − | |
| Enterovirus | − | − | − | − | |
| Norovirus | − | − | − | − | |
| Hepatitis A virus | − | − | − | − | |
−, negative (the signal was below the detection limit); a, b, and c, positive for water at locations a, b, or c, respectively (see Fig. 1); wet and dry, positive for wet and dry sand, respectively; NA, not analyzed.
Giardia spp. and Cryptosporidium spp. in water were analyzed by microscopy only for sample location b.
Bacterial pathogens.
V. vulnificus was detected on day 2 in samples WHb, SHD, SLW, and SLD by culture, and the results were confirmed by PCR. The levels in all other samples were below the detection limit (Table 3). Presumptive S. aureus isolates were also detected (Table 2). The levels of presumptive S. aureus were generally low in water samples when indicator levels were low. The highest levels of presumptive S. aureus were obtained for the day 2 high-tide sampling event (average, 22 CFU 100 ml−1). Confirmation tests were conducted for 27 presumptive S. aureus isolates obtained from day 1 samples and for 30 presumptive S. aureus isolates obtained from day 2 samples. Two isolates from each day were positive for mannitol fermentation, catalase, clumping factor, and protein A latex agglutination. However, no isolate was determined to be a Gram-positive coccus by Gram staining, and none of the isolates grew on sheep blood agar of ORSAB media (trypticase soy agar with 5% sheep blood in an oxacillin resistance-selective agar base). Therefore, none of the presumptive S. aureus in the collections were confirmed to be S. aureus.
Protozoan and viral pathogens.
The levels of Cryptosporidium spp. and Giardia spp. in samples (collected by traditional filtration of water and sand on days 1 and 2, respectively) were generally below the detection limit. The one exception was day 2 water sample WHb, which was positive for Giardia spp. as determined by microscopy, and the concentration was 1 cyst per 144 liters or (0.7 cyst/100 liter). A matrix spike analysis for the positive Giardia sp. sample showed that the level of recovery was 39% for Giardia spp. Concentration with the bilayer filtration system followed by qPCR analysis gave positive results for samples WHa and WHb on day 2 for Giardia spp. The estimated Giardia sp. concentrations were 11 and 51 cysts per liter, which confirmed detection by traditional methods, although the qPCR values were greater than the values obtained by the standard method. Samples SLD and SHW were positive on day 2 for Cryptosporidium spp. (12 and 6 oocysts per 100 g of dry and wet sand, respectively) when the bilayer filtration system followed by qPCR analysis was used.
The results of confirmation tests further supported the positive Giardia sp. and Cryptosporidium sp. results. The first round of confirmation involved sequencing the qPCR amplicons (after cloning) and resulted in a 100% match with the target beta-giardin gene and COWP gene sequences, respectively. However, as these sequences were the gene fragments used to generate the standard curves, a second primer set was used for each organism, and the gene targets were amplified, cloned, and sequenced in a nonquantitative experiment. For Giardia spp. a larger fragment (513 bp) of the beta-giardin gene was amplified by qPCR. For Cryptosporidium spp. a 232-bp fragment of the 18S rRNA gene was amplified. A BLAST (Basic Local Alignment Search Tool) search of the GenBank sequence database yielded 100% matches with Giardia intestinalis (Giardia lamblia) strain ATCC 50803 (accession number XM_001705373) and Cryptosporidium hominis (accession number L16996). The levels in all other samples were below the detection limits for protozoans (Table 3).
An initial positive result for norovirus II in samples WHa and WHb on day 1 was obtained using the bilayer filtration system followed by qPCR analysis. However, further analysis did not confirm the presence of norovirus, demonstrating the importance of confirming qPCR data for complex matrices. All other samples were negative for norovirus, enterovirus, and hepatitis A (Table 3).
DISCUSSION
The primary objective of this study was to conduct a preliminary evaluation of the presence of indicator microbes and possible associations between these microbes, pathogen measurements, and environmental conditions at a subtropical recreational marine beach. The secondary objectives included evaluation of multiple enterococcus analysis methods, investigation of the presence of microbial source tracking markers, and determination of the applicability of using an innovative low-volume concentration system for pathogens.
Indicator bacteria.
The analytical methods used for enterococci may be a significant source of variability in results and hence an important factor to consider in beach monitoring and research studies. Comparison of the different methods showed that the values obtained from MF enumeration were generally less than the values determined by qPCR. This is consistent with results of other studies (30, 43) and might be explained by the fact that qPCR can also detect target DNA from nonviable and nonculturable cells, whereas the culture-based method does not.
Spatial and temporal variations are also an important factor in beach monitoring. For the water, the average enterococcus levels determined by MF by two separate labs were below the regulatory guideline levels (104 CFU 100 ml−1 for single samples) for all samples for three of the four sampling events. The average enterococcus levels for the day 2 high-tide sampling event were higher than the levels for the three other sampling events, and the level for one water sample (sample WHa) (110 CFU 100 ml−1; standard deviation, 12 CFU 100 ml−1) exceeded the regulatory guideline level for single-sample analysis. This is important since regulators may have opted to close the beach or place warning signs for bathers if they had processed the one (of three) high-tide samples in which the level exceeded the single-sample enterococcus guideline level. However, this would not have been the case if a different sample had been processed. Thus, these results emphasize that the spatial and temporal variability of indicator levels in recreational marine water could result in different management decisions at a beach site (8, 21). The results for the supplemental fecal indicator bacteria (fecal coliforms, E. coli, and C. perfringens) were predominantly consistent with the enterococcus results for both the water and sand samples, indicating that the environmental factors and source conditions affecting enterococci may also affect these indicators. Significant correlations were also found between results for enterococci, fecal coliforms, and E. coli in water samples.
The results for sand eluate samples support the notion that there was indicator microbe wash-in from the shoreline. The levels of washable bacteria observed in the pore water of the wet sand (samples SLW and SHW) collected from the intertidal zone and in dry sand (samples SLD and SHD) collected within a few feet above the intertidal zone (on a per milliliter basis) were 2 to 4 orders of magnitude higher than the levels observed in the water column, and the levels for dry sand were 1 to 3 orders of magnitude higher than the levels for wet sand. Essentially, the sand appeared to serve as a reservoir in which microbes were captured, persisted, and then released into the water column as the shoreline was subjected to periodic wetting, such as the wetting which occurs through tidal action. The initial source of the indicators in the sand is unknown, but these organisms could have come from direct fecal deposits from animals and humans (59) and from potential regrowth in the sand (10, 13).
Pathogens.
The occurrence of pathogens impacting water is highly intermittent depending on the illnesses afflicting, or the pathogens carried by, the human and animal populations contributing to the pollution source entering the beach. In this study, pathogens were detected predominantly during the same sampling event that resulted in the highest indicator microbe levels. However, because of the low number of sampling events, a significant relationship could not be established between indicator microbes and pathogens. The potential bacterial pathogens evaluated included both S. aureus and V. vulnificus. The only detection of V. vulnificus in the water occurred during the day 2 high-tide sampling event, indicating that further analysis should be conducted to determine whether there is an association between the indicator microbes and this opportunistic pathogen. This relationship should be studied further, especially since it would support the use of these indicator microbes as surrogates for bacterial pathogen contamination in non-point source subtropical marine recreational waters.
Although S. aureus was evaluated, none of the isolates were confirmed to be S. aureus isolates. This may have been because the chrome agar plates used to test for S. aureus allowed growth of organisms other than S. aureus. When S. aureus is abundant relative to the background microbial population, as it is in clinical samples (25, 39) or in captured marine waters for which an abundance of people is a potential source (22), chrome agar has been shown to be useful for isolation. However, in environmental samples, such as those collected during the sampling events in this study, S. aureus is not an abundant species and shares its niche with other organisms capable of producing changes in colony color on the agar used in this study. The number of isolates reported to be positive based solely on colorimetric changes of the indicator agar represents all of the microorganisms capable of causing these changes, not specifically S. aureus. These findings could be explained either if there was no S. aureus in the samples or if other environmental microbes, some capable of causing colorimetric changes, overwhelmed the growth of S. aureus (23). In future studies an alternate selective medium more suitable for isolation of S. aureus from general environmental samples should be used (16).
Giardia spp. were detected in the water both by traditional filtration followed by microscopy and by bilayer filtration followed by qPCR only for the day 2 high-tide sampling event, when the highest levels of indicator microbes were detected. The detection of Giardia spp. by the bilayer filtration method indicates the potential of this concentration method since a much smaller volume (5 liters) of sample water may be used for detection (2, 3). Since the risk posed by these pathogens at a given concentration is less in bathing water than in drinking water, this volume may prove to be adequate to detect an acceptable risk. However, the obvious difference in the measured concentration of Giardia spp. between the traditional and innovative methods may be due to the fact that qPCR detects any intact DNA, while microscopy detects only intact cysts. During the same sampling event, Cryptosporidium spp. were also found in the sand eluate by bilayer filtration followed by qPCR analysis.
In terms of the health effects resulting from the presence of these pathogens, a thorough quantitative microbial risk assessment and/or epidemiological study should be conducted. A simple risk calculation is presented here to shed some light on the potential risk due to Giardia spp., which were detected at our study site. Levels as low as 10 G. lamblia cysts have been shown to cause infection in humans (46), and the typical volume of water ingested per swimmer during one swimming event is approximately 16 ml for adults and 37 ml for children (20). Fifty milliliters ingested may be a more conservative estimate. Combining these values with the Giardia sp. cyst values (assuming that the Giardia strain detected was in fact G. lamblia) detected by traditional concentration-microscopy and bilayer concentration-qPCR methods (0.007 and 51 cysts per liter) yields risks of infection to bathers of 3.5/106 (0.00035%) and 25/103 (2.5%), respectively. The concentrations of cysts detected by the bilayer concentration-qPCR method were similar to those detected in another study of G. lamblia (0 to 33 cysts per liter) in recreational beach waters (47). Given that EPA's standard acceptable risk for gastrointestinal illness-causing agents in marine waters is 19 organisms per 1,000 bathers, the first risk value may be acceptable, while the second may not be (51). However, the risk value accepted by the EPA was based on an epidemiologic study performed with a known point source in temperate climates comparing bathers to nonbathers and therefore may not be comparable to the simple calculated risk described here (14). A similar risk calculation may be conducted for the other presumptive pathogens detected at the beach, assuming that their infectivity is well documented, to gain insight into the potential threat to bathers.
Environmental conditions.
The elevated levels of indicator and pathogenic microbes observed for the day 2 morning high-tide sampling event are hypothesized to be due to a combination of factors, including lack of sunlight during and immediately before sample collection (solar insolation, <1 W/m2) and sample collection during high tide. However, given the limited number of samples, such a relationship, although scientifically plausible, needs to be assessed with more samples. Sunlight and UV light are known to inactivate microbes in water (24, 44). The sampling on day 1 during high tide was characterized by elevated solar insolation levels (861 W/m2), which is typical for summer afternoons without rain in Florida. The increased solar insolation levels raised the water temperature to >32°C. Viruses specifically can be vulnerable to high water temperatures (56), and this may be a reason why they were not detected during the low-tide afternoon sampling event, while other MST markers and pathogens were detected in both water and sand. However, the day 2 high-tide sample was collected before sunrise so inactivation from solar insolation was minimal.
Tidal height can also impact indicator microbe levels in water, as sand has been identified as a possible reservoir of indicator microbes at our study beach (10, 11, 21, 41). During high tide, the intertidal zone is submerged, which permits transfer of indicator microbes from the sand and pore water between the sand particles into the water column. Therefore, based on environmental parameters, the day 2 sampling event represents the ideal case for increased indicator microbe levels as a sample was collected during high tide in the early morning before inactivation by solar insolation.
Microbial source tracking markers.
The human source tracking markers HPyVs (human urine and sewage source) were detected during the high-tide and low-tide events on day 2, supporting the hypothesis that the site was impacted by human waste (36) during or prior to the morning high-tide sampling event. Since there are no known point sources or other sewage sources for the beach, such as sewage outfalls or septic systems (41), these results indicate that there are fecal sources which originate either near shore from people and/or animals or offshore from boats that dump their waste (compared to sewage from a large community). Although intersections between canals or rivers and the ocean are not close to the sampling site, it is possible that persistent pathogens originating from canals and rivers are transported to the beach. However, more samples, as well as a comprehensive sanitary survey, are needed in order to determine the sources of these microbial source tracking markers.
Recommendations.
The data obtained in this study, which were limited due to cost limitations during sampling for a large suite of microbes, indicate that additional sampling should be conducted to assess the possible relationships alluded to above between indicator microbes, pathogens, and environmental conditions. The levels of microbes at the beach studied frequently exceed the regulatory guidelines for indicator bacteria during high tide, particularly in the summer months (21, 41). It is therefore important to determine whether pathogens are also present at levels that pose a significant health risk to bathers.
Supplementary Material
Acknowledgments
This research was supported by the Florida Department of Health (contract CORAM), by the NSF-NIEHS Oceans and Human Health Program (grants NIEHS P50 ES12736 and NSF OCE 0432368/0911373), by the NSF REU Program (grant OCE 0432368), and by the NSF SGER Program (grant OCE 0554402). We thank IDEXX Corporation for their support of this project through provision of supplies needed for the chromogenic substrate analysis of enterococci.
We acknowledge Roger Fujioka for his advice on S. aureus analysis and Moataz Eltoukhy for his assistance with the statistical analysis of the results. We also acknowledge Sabrina Garcia, Julie Armstrong, Yang Deng, and Hasan Abdel Fattah for their assistance in the University of Miami laboratory and during field sample collection.
Footnotes
Published ahead of print on 4 December 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Abdelzaher, A., H. M. Solo-Gabriele, C. Palmer, and M. Wright. 2009. Simultaneous concentration of enterococci and coliphage from marine waters using a dual layer filtration system. J. Environ. Qual. 38:2468-2473. [DOI] [PubMed] [Google Scholar]
- 2.Abdelzaher, A., H. M. Solo-Gabriele, M. E. Wright, and C. J. Palmer. 2008. Sequential concentration of bacteria and viruses from marine waters using a dual membrane system. J. Environ. Qual. 37:1648-1655. [DOI] [PubMed] [Google Scholar]
- 3.Abdelzaher, A., M. Wright, T. Scott, G. Lucasik, H. M. Solo-Gabriele, A. Bonilla, T. Bonilla, and C. Palmer. 2008. Dual layer filtration system for concentrating fecal indicators and pathogens from marine waters, poster 390. Proc. ASLO (Am. Soc. Limnol. Oceanogr.) Spec. Session OHH Oceans Hum. Health.
- 4.Alm, E. W., J. Burke, and E. Hagan. 2006. Persistence and potential growth of the fecal indicator bacteria, Escherichia, in shoreline sand at Lake Huron. J. Great Lakes Res. 32:401-405. [Google Scholar]
- 5.American Public Health Association. 1998. Standard methods for the examination of water and wastewater, 20th ed. American Public Health Association, Washington, DC.
- 6.Betancourt, W. Q., and J. B. Rose. 2004. Drinking water treatment processed for removal of Cryptosporidium and Giardia. J. Vet. Parasitol. 126:219-234. [DOI] [PubMed] [Google Scholar]
- 7.Beversdorf, L. J., S. M. Bornstein-Forst, and S. L. McLellan. 2007. The potential for beach sand to serve as a reservoir for Escherichia coli and the physical influences on cell die-off. Appl. Environ. Microbiol. 102:1372-1381. [DOI] [PubMed] [Google Scholar]
- 8.Boehm, A. B. 2007. Enterococci concentrations in diverse coastal environments exhibit extreme variability. Environ. Sci. Technol. 41:8227-8232. [DOI] [PubMed] [Google Scholar]
- 9.Boehm, A. B., N. J. Ashbolt, J. M. Colford, L. E. Dunbar, L. E. Fleming, M. A. Gold, J. A. Hansel, P. R. Hunter, A. M. Ichida, C. D. McGee, J. A. Soller, and S. B. Weisberg. 2008. A sea change ahead for recreational water quality criteria. J. Water Health 7:9-20. [DOI] [PubMed] [Google Scholar]
- 10.Bonilla, T. D., K. Nowosielski, M. Cuvelier, A. Hartza, M. Greenb, N. Esiobub, D. S. McCorquodalea, J. M. Fleisher, and A. Rogerson. 2007. Prevalence and distribution of fecal indicator organisms in South Florida beach sand and preliminary assessment of health effects associated with beach sand exposure. Mar. Pollut. Bull. 54:1472-1482. [DOI] [PubMed] [Google Scholar]
- 11.Bonilla, T. D., K. Nowosielski, N. Esiobu, D. S. McCorquodale, and A. Rogerson. 2006. Species assemblages of Enterococcus indicate potential sources of fecal bacteria at a south Florida recreational beach. Mar. Pollut. Bull. 52:807-810. [DOI] [PubMed] [Google Scholar]
- 12.Brownell, M. B., V. J. Harwood, R. C. Kurz, S. M. McQuaig, J. Lukasik, and T. M. Scott. 2007. Confirmation of putative stormwater impact on water quality at a Florida beach by microbial source tracking methods and structure of indicator organism populations. Water Res. 41:3747-3757. [DOI] [PubMed] [Google Scholar]
- 13.Byappanahalli, M., and R. Fujioka. 2004. Indigenous soil bacteria and low moisture may limit but allow faecal bacteria to multiply and become a minor population in tropical soils. Water Sci. Technol. 50:27-32. [PubMed] [Google Scholar]
- 14.Cabelli, V. 1983. Health effects criteria for marine recreational waters. U.S. EPA report EPA-600/1-80-031. U.S. Environmental Protection Agency, Cincinnati, OH.
- 15.Chang, J. C., S. F. Ossoff, D. C. Lobe, M. H. Dorfman, C. M. Dumais, R. G. Qualls, and J. D. Johnson. 1985. UV inactivation of pathogenic and indicator microorganisms. Appl. Environ. Microbiol. 49:1361-1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Charoenca, N., and R. Fujioka. 1993. Assessment of Staphylococcus bacteria in Hawaii's marine recreational waters. Water Sci. Technol. 27:283. [Google Scholar]
- 17.Colford, J. M., Jr., T. J. Wade, K. C. Schiff, C. C. Wright, J. F. Griffith, S. K. Sandhu, S. Burns, J. Hayes, M. Sobsey, G. Lovelace, and S. Weisberg. 2007. Water quality indicators and the risk of illness at non-point source beaches in Mission Bay, California. Epidemiology 18:27-35. [DOI] [PubMed] [Google Scholar]
- 18.DeLeon, R., Y. S. Shieh, R. S. Baric, and M. D. Sobsey. 1990. Detection of enteroviruses and hepatitis A virus in environmental samples by gene probes and polymerase chain reaction, p. 833-853. In Proceedings of Water Quality Conference: 1990, San Diego. American Water Works Association, Washington, DC.
- 19.Desmarais, T. R., H. M. Solo-Gabriele, and C. J. Palmer. 2002. Influence of soil on fecal indicator organisms in a tidally influenced subtropical environment. Appl. Environ. Microbiol. 68:1165-1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dufour, A. P., O. Evans, T. D. Behymer, and R. Cantu. 2006. Water ingestion during swimming activities in a pool: a pilot study. J. Water Health 4:425-430. [PubMed] [Google Scholar]
- 21.Durbin, M. E., A. M. Zaher, N. F. Heybeck, H. M. Solo-Gabriele, S. Elmir, K. D. Goodwin, and C. Sinigalliano. 2005. The inter-tidal zone is the source of enterococci to a subtropical recreational beach, abstr. Q-322. Abstr. 105th Gen. Meet. Am. Soc. Microbiol. American Society for Microbiology, Washington, DC.
- 22.Elmir, S., M. Wright, A. Abdelzaher, H. Solo-Gabriele. L. Fleming, G. Miller, M. Rybolowik, P. Shih, S. Pillai, J. Cooper, and E. Quaye. 2007. Quantitative evaluation of bacteria released by bathers in a marine water. Water Res. 41:3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fowler, T. L., R. S. Fujioka, A. D. Tice, A. K. Ishikawa, and B. S. Yoneyanna. 2004. Modification of CHROMagar Staphylococcus aureus to enumerate Staphylococcus aureus from marine recreational waters in Hawaii, abstr. Q-516. Abstr. 104th Gen. Meet. Am. Soc. Microbiol. American Society for Microbiology, Washington, DC.
- 24.Fujioka, R. S., H. Hashimoto, E. B. Siwak, and R. Young. 1981. Effect of sunlight on survival of indicator bacteria in seawater. Appl. Environ. Microbiol. 41:690-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gaillot, O., M. Wetsch, N. Fortineau, and P. Berche. 2000. Evaluation of CHROMagar Staph. aureus, a new chromogenic medium, for isolation and presumptive identification of Staphylococcus aureus from human clinical specimens. J. Clin. Microbiol. 38:1587-1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gordon, K. V., M. C. Vickery, A. DePaola, C. Staley, and V. J. Harwood. 2008. Real-time PCR assays for quantification and differentiation of Vibrio vulnificus strains in oysters and water. Appl. Environ. Microbiol. 74:1704-1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Griffin, D. W., K. A. Donaldson, J. H. Paul, and J. B. Rose. 2003. Pathogenic human viruses in coastal waters. Clin. Microbiol. Rev. 16:129-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guy, R. A., P. Payment, U. J. Krull, and P. A. Horgen. 2003. Real-time PCR for quantification of Giardia and Cryptosporidium in environmental water samples and sewage. Appl. Environ. Microbiol. 69:5178-5185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haile, R. W., J. S. Witte, M. Gold, R. Cressey, C. McGee, R. C. Millikan, A. Glasser, N. Harawa, C. Ervin, P. Harmon, J. Harper, J. Dermand, J. Alamillo, K. Barretr, M. Nides, and G. Wang. 1999. The health effects of swimming in ocean water contaminated by storm drain runoff. Epidemiology 10:355-363. [PubMed] [Google Scholar]
- 30.Haugland, R. A., S. C. Seifring, L. J. Wymer, K. P. Brenner, and A. P. Dufour. 2005. Comparison of Enterococcus measurements in freshwater at two recreational beaches by quantitative polymerase chain reaction and membrane filter culture analysis. Water Res. 39:559-568. [DOI] [PubMed] [Google Scholar]
- 31.Jiang, S. C., and W. Chu. 2004. PCR detection of pathogenic viruses in southern California urban rivers. J. Appl. Microbiol. 97:17-28. [DOI] [PubMed] [Google Scholar]
- 32.Jiang, S., R. Noble, and W. P. Chui. 2001. Human adenoviruses and coliphages in urban runoff-impacted coastal waters of Southern California. Appl. Environ. Microbiol. 67:179-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiang, S., R. Noble, W. Chu, and J. He. 2007. Seasonal detection of human viruses and coliphage in Newport Bay, California. Appl. Environ. Microbiol. 73:6468-6474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaysner, C. A., and A. DePaola. May 2004, posting date. Chapter 9. Vibrio. In Bacteriological analytical manual online. U.S. Food and Drug Administration, Washington, DC. http://www.fda.gov/Food/ScienceResearch/LaboratoryMethods/BacteriologicalAnalyticalManualBAM/ucm070830.htm.
- 35.Lipp, E. K., S. R. Farrah, and J. B. Rose. 2001. Assessment and impact of microbial fecal pollution and human enteric pathogens in a coastal community. Mar. Pollut. Bull. 42:286-293. [DOI] [PubMed] [Google Scholar]
- 36.McQuaig, S. M., T. M. Scott, V. J. Harwood, S. R. Farrah, and J. O. Lukasik. 2006. Detection of human-derived fecal pollution in environmental waters by use of a PCR-based human polyomavirus assay. Appl. Environ. Microbiol. 72:7567-7574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Noble, R. T., and J. A. Fuhrman. 2001. Enteroviruses detected by reverse transcriptase polymerase chain reaction from the coastal waters of Santa Monica Bay, California: low correlation to bacterial indicator levels. Hydrobiologia 460:175-184. [Google Scholar]
- 38.Prüss, A. 1998. Review of epidemiological studies on health effects from exposure to recreational water. Int. J. Epidemiol. 27:1-9. [DOI] [PubMed] [Google Scholar]
- 39.Samra, Z., O. Ofir, and J. Bahar. 2004. Optimal detection of Staphylococcus aureus from clinical specimens using a new chromogenic medium. Diagn. Microbiol. Infect. Dis. 49:243-247. [DOI] [PubMed] [Google Scholar]
- 40.Scott, T. M., T. M. Jenkins, J. Lukasik, and J. B. Rose. 2005. Potential use of a host associated molecular marker in Enterococcus faecium as an index of human fecal pollution. Environ. Sci. Technol. 39:283-287. [PubMed] [Google Scholar]
- 41.Shibata, T., H. Solo-Gabriele, L. Fleming, and S. Elmir. 2004. Monitoring marine recreational water quality using multiple microbial indicators in an urban tropical environment. Water Res. 38:3119-3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shuval, H. 2003. Estimating the global burden of thalassogenic diseases: human infectious diseases caused by wastewater pollution of the marine environment. J. Water Health 2:53-64. [PubMed] [Google Scholar]
- 43.Sinigalliano, C. D., M. L. Gidley, T. Shibata, D. Whitman, T. H. Dixon, E. Laws, A. Hou, D. Bachoon, L. Brand, L. Amaral-Zettler, R. J. Gast, G. F. Steward, O. D. Nigro, R. Fujioka, W. Q. Betancourt, G. Vithanage, J. Mathews, L. E. Fleming, and H. M. Solo-Gabriele. 2007. Impacts of hurricanes Katrina and Rita on the microbial landscape of the New Orleans area. Proc. Natl. Acad. Sci. U. S. A. 104:9029-9034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sinton, L. W., C. H. Hall, P. A. Lynch, and R. J. Davies-Colley. 2002. Sunlight inactivation of fecal indicator bacteria and bacteriophages from waste stabilization pond effluent in fresh and saline waters. Appl. Environ. Microbiol. 68:1122-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Solo-Gabriele, H., M. Wolfert, and C. Palmer. 2000. Sources of Escherichia coli in a subtropical coastal environment. Appl. Environ. Microbiol. 66:230-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Steiner, T. S., N. M. Thielmen, and M. D. Guerrant. 1997. Protozoal agents: what are the dangers for the public water supply? Annu. Rev. Med. 48:329-340. [DOI] [PubMed] [Google Scholar]
- 47.Sunderland, D., T. K. Graczyk, L. Tamang, and P. N. Breysse. 2007. Impacts of bathers on levels of Cryptosporidium parvum oocysts and Giardia lamblia cysts in recreational beach waters. Water Res. 41:3483-3489. [DOI] [PubMed] [Google Scholar]
- 48.U.S. Environmental Protection Agency. 1995. Method for detection and enumeration of Clostridium perfringens in water and sediments by membrane filtration. Publication EPA/600-R-95/030. U.S. EPA Office of Research and Development, Washington, DC.
- 49.U.S. Environmental Protection Agency. 2002. Escherichia coli (E. coli) in water by membrane filtration using modified membrane-thermotolerant Escherichia coli agar (modified mTEC). Publication EPA 821-R-02-023. U.S. EPA Office of Research and Development, Washington, DC.
- 50.U.S. Environmental Protection Agency. 2002. Method 1600: membrane filter test method for enterococci in water. Publication EPA-821-R-02-022. U.S. Environmental Protection Agency, Washington, DC.
- 51.U.S. Environmental Protection Agency. 2004. Water quality standards for coastal and Great Lakes recreation waters: proposed rule. Fed. Regist. 69:41720-41742. [Google Scholar]
- 52.U.S. Environmental Protection Agency. 2005. Method 1623: Cryptosporidium and Giardia in water by filtration/IMS/FA. Publication EPA 815-R-05-002. U.S. Environmental Protection Agency, Washington, DC.
- 53.Van Elsas, J. D., and K. Smalla. 1997. Methods for sampling soil microbes, p. 383-390. In C. J. Hurst, G. R. Knudsen, M. J. McInernery, L. D. Stetzenbach, and M. V. Walter (ed.), Manual of environmental microbiology. ASM Press, Washington, DC.
- 54.Wade, T. J., N. Pai, J. N. Eisenberg, and J. M. Colford, Jr. 2003. Do U.S. Environmental Protection Agency water quality guidelines for recreational waters prevent gastrointestinal illness? A systematic review and meta-analysis. Environ. Health Perspect. 8:1102-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weisberg, S., A. Dufour, F. Galt, M. Gold, M. Noble, R. Noble, E. Reichard, P. Roberts, J. Rose, and D. Rosenblatt. 2000. Huntington Beach closure investigation. Technical review USCSG-TR-01-2000. Sea Grant, University of Southern California, San Diego, CA.
- 56.Wetz, J. J., E. K. Lipp, D. W. Griffin, J. Lukasik, D. Wait, M. D. Sobsey, T. M. Scott, and J. B. Rose. 2004. Presence, infectivity, and stability of enteric viruses in seawater: relationship to marine water quality in the Florida Keys. Mar. Pollut. Bull. 48:698-704. [DOI] [PubMed] [Google Scholar]
- 57.Whitman, R. L., D. A. Shively, H. Pawlik, M. B. Nevers, and M. N. Byappanahalli. 2003. Occurrence of Escherichia coli and enterococci in Cladophora (Chlorphyta) in nearshore water and beach sand of Lake Michigan. Appl. Environ. Microbiol. 69:4714-4719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Whitman, R. L., M. B. Nevers, G. C. Korinek, and M. N. Byappanahalli. 2004. Solar and temporal effects on Escherichia coli concentration at a Lake Michigan swimming beach. Appl. Environ. Microbiol. 70:4276-4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wright, M. E., H. M. Solo-Gabriele, S. Elmir, and L. E. Fleming. 2009. Microbial load from animal feces at a recreational beach. Mar. Pollut. Bull. 58:1649-1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.