Abstract
Terminal restriction fragment length polymorphism (T-RFLP) is used to monitor the structural diversity of complex microbial communities in terms of richness, relative abundance, and distribution of the major subpopulations and individual members. However, discrepancies of several nucleotides between expected and experimentally observed lengths of terminal restriction fragments (T-RFs), together with the difficulty of obtaining DNA sequence information from T-RFLP profiling, often prevent accurate phylogenetic characterization of the microbial community of interest. In this study, T-RFLP analysis of DNA from an artificial assembly of five bacterial strains was carried out with a combination of two size markers with different fluorescent tags. Precise sizing of T-RFs in the 50- to 500-nucleotide range was achieved by using the same dye for both samples and size markers. Phylogenetic assignment of the component microbial strains was facilitated by coupling T-RFLP to denaturing high-performance liquid chromatography (D-HPLC) of 16S RNA gene fragments followed by direct sequencing. The proposed coupling of D-HPLC and T-RFLP provides unambiguous characterization of microbial communities containing less than 15 microbial strains.
Over the last 2 decades, the development of molecular biology tools has led to the emergence of a new discipline, molecular microbial ecology. The overall structural diversity of microbial communities can be examined easily using PCR-based strategies (6), usually targeting the 16S rRNA gene as a universal genetic marker of prokaryotes. Genotyping approaches avoid current limitations of cultivation methods, which only poorly reflect the phylogenetic diversity of microbial communities (12). The principles, technical aspects, and limitations of commonly employed methods were recently reviewed (10). Among these methods, terminal restriction fragment length polymorphism (T-RFLP) has proved to be invaluable for rapid characterization of the composition and dynamics of species-rich samples (13). Compared to other approaches, T-RFLP is semiquantitative and combines high levels of sensitivity, resolution, and reproducibility (see Table S1 in the supplemental material). Taxonomic diversity of microbial communities is evaluated by using the strain-dependent variability of restriction sites within a conserved PCR-amplified DNA fragment. The terminal restriction fragments (T-RFs) of digested PCR products appear as chromatographic peaks after size-dependent electrophoretic separation due to a fluorescent tag attached to one of the primers used for PCR. The relative abundance of peaks is evaluated, and fragment lengths are estimated using a fluorescent internal size standard comigrating with the sample (5). The estimated lengths corresponding to the T-RFLP peaks obtained are compared to databases of T-RF sizes generated by in silico digestion of known 16S rRNA gene sequences with commonly used restriction enzymes for phylogenetic assignment (13). However, estimation of T-RF lengths from experimental chromatograms is biased by the fact that differences in the electrophoretic properties of the two different fluorescent dyes used to distinguish sample fragments from the size marker significantly affect fragment migration (7, 11). Discrepancies greater than 6 nucleotides (nt), depending on the length of the fragment, have been reported between expected and experimentally estimated fragment lengths (7). This causes errors in phylogenetic assignments and may in turn lead to erroneous inferences regarding the functional aspects of the microbial communities under investigation. Another drawback of T-RFLP is the difficulty of retrieving sequence information directly from experimental T-RFs, since additional construction of representative 16S rRNA gene libraries is required to obtain such information.
Here we propose an experimental strategy to circumvent current limitations of T-RFLP and facilitate characterization of microbial communities. First, we propose an optimized protocol for T-RFLP that yields reliable T-RF sizes. Second, we describe use of denaturing high-performance liquid chromatography (D-HPLC) as an alternative to cloning in order to gain direct access to DNA sequence information. D-HPLC, an emerging technique for microbial community profiling (1, 4), enables collection of DNA fragments separated on the basis of differences in sequence, sequence length, and G+C content at a partially denaturing temperature. The unambiguous phylogenetic characterization of a model microbial assembly of five reference strains is described as proof of principle of the usefulness of the proposed strategy.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Methylobacterium extorquens CM4 (formerly named M. chloromethanicum [14]), Pseudomonas chlororaphis subsp. chlororaphis DSM 50083, Pseudomonas stutzeri KC (= DSM 7136), and Staphylococcus aureus subsp. aureus ATCC 25904 were cultivated on nutrient agar (Sigma, United States) under oxic conditions at 30°C. Clostridium acetobutylicum ATCC 824 was grown anaerobically at 25°C on solid C medium (3) in a glove box (Jacomex, France).
DNA preparation.
Total DNA of a synthetic mixture of the five bacterial strains described above was purified and isolated from bacteria trapped in agarose plugs. An average of 108 to 109 cells were washed and resuspended in 0.5 ml TE buffer (1 mM EDTA [pH 8.0], 10 mM Tris-HCl [pH 8.0]). The suspension was mixed with an equal volume of 1% low-melting-point agarose (SeaPlaque GTG agarose; FMC Bioproducts, United States) at 50°C. Aliquots (80 μl) were dispensed into plug molds (5 by 1.5 by 10 mm; Bio-Rad, United States). After cooling on ice, the solidified agarose plugs were incubated for 20 h at 37°C in 10 ml lysis solution (0.25 M EDTA [pH 8.0], 0.1 M NaCl, 0.1 M Tris-HCl [pH 8.0], 1% N-lauryl-sarcosine) containing lysozyme (2 mg ml−1). Cell lysis was continued for another 20 h at 50°C in the same lysis solution in the presence of proteinase K (2 mg ml−1). Inactivation of proteinase K by incubation of the plugs for 30 min at 55°C in TE buffer containing phenylmethylsulfonyl fluoride (20 μg ml−1; Sigma) was followed by three 20-min washes in TE buffer at room temperature. Total DNA was recovered by digestion of the agarose with beta-agarase I (NE Biolabs, United States), using the manufacturer's recommendations, and was purified by a final dialysis step using 0.025-μm filters (VSWP type; Millipore, United States). An alternative DNA isolation protocol was also used for individual strains. A single colony corresponding to approximately 108 cells was suspended in 40 μl 0.05 M NaOH and heated at 95°C for 10 min, and the resulting solution was cooled on ice for 5 min, diluted (1/15) in sterile ultrapure water, and used as the template for PCR amplification.
PCR amplification of the 16S rRNA gene.
Primer 27f (2) that was 5′ end labeled with 6-carboxyfluorescein (6-FAM) and primer 926r (8) were used for T-RFLP analysis. For D-HPLC analysis, unlabeled primers 27f and 534r (9) were used. PCR mixtures (total volume, 50 μl) contained 1× high-fidelity PCR buffer (Bio-Rad, United States), 200 μM each of the four deoxynucleoside triphosphates, primers (0.4 μM each), 1 U iProof high-fidelity polymerase (Bio-Rad), and 1.0 ng DNA or 2 μl of a diluted lysed colony solution. Reaction tubes were placed in a PCR block (Mastercycler personal thermocycler; Eppendorf, Germany) that was preheated at 95°C before amplification. PCR amplification involved an initial denaturation at 95°C for 2 min, followed by 30 cycles of denaturation at 94°C for 20 s, annealing at 52°C for 30 s, and extension at 72°C for 30 s (T-RFLP) or 20 s (D-HPLC) and then a final 1-min extension at 72°C. PCRs were carried out in triplicate for each DNA sample and primer pair, and replicates were pooled and stored at 4°C until they were used.
T-RFLP analysis.
The 0.9-kb 16S rRNA gene fragments obtained by PCR were purified from 1% agarose gels with a QIAquick gel extraction kit (Qiagen, Germany) used according to the manufacturer's recommendations. DNA samples (200 to 300 ng) were digested at 37°C for 20 h with 20 U AluI (AGCT), HhaI (GCGC), MspI (CCGG) (Fermentas, Lithuania), or RsaI (GTAC) (New England Biolabs, United States), purified with a QIAquick nucleotide removal kit (Qiagen), and resuspended in 50 μl sterile ultrapure water. Digested and purified samples of each strain were analyzed separately or pooled prior to T-RFLP analysis. DNA (10 to 50 ng) in 5 μl ultrapure water was mixed with 10 μl of a solution of Hi-Di formamide (Applied Biosystems, United Kingdom) containing 1:20 (vol/vol) 6-FAM- or carboxy-X-rhodamine (ROX)-labeled MapMarker 1000 (Bioventures, United States) (Table 1), denatured at 95°C for 5 min, and snap-cooled on ice. Denatured restriction fragments were separated by size by capillary electrophoresis using an ABI Prism 3130 XL genetic analyzer (POP 7 matrix; capillary size, 50 cm; Applied Biosystems) and the following parameters: injection voltage, 1.6 kV; injection duration, 15 s; electrophoretic voltage, 15 kV; and run time at 60°C, 60 min. T-RFLP electropherograms were analyzed with GeneScan V3.7 software (Applied Biosystems). At least two independent runs with each 6-FAM- and ROX-labeled internal size standard were performed for each DNA preparation.
TABLE 1.
Accuracy of terminal restriction fragment length determination
| Restriction enzyme | T-RF length (nt)a |
||
|---|---|---|---|
| Calculated from sequence data | With ROX-labeled markerb | With 6-FAM-labeled markerb | |
| AluI | 72 | 66.6 ± 0.2 | 72.0 ± 0.1 |
| HhaI | 207 | 205.0 ± 0.2 | 207.3 ± 0.1 |
| MspI | 490 | 489.4 ± 0.1 | 490.9 ± 0.2 |
| RsaI | 878 | 880.7 ± 0.8 | 883.3 ± 0.8 |
The fragment used was a P. stutzeri strain KC 6-FAM-labeled 16S rRNA gene fragment. The data are the averages of eight independent replicates.
The marker was the MapMarker 1000 internal size standard (50, 75, 100, 125, 150, 200, 250, 300, 350, 400, 450, 475, 500, 550, 600, 650, 700, 750, 800, 850, 900, 950, and 1,000 nt).
D-HPLC and sequencing.
D-HPLC was performed with the WAVE 3500 microbial analysis system (Transgenomic, United States), using a DNASep cartridge column and a two-buffer eluent system. Buffer A consisted of 0.1 M triethylammonium acetate (TEAA) (pH 7.0) in water, and buffer B consisted of 0.1 M TEAA (pH 7.0) in 25% (vol/vol) aqueous acetonitrile. 16S rRNA gene fragments of the five-strain assembly and separate or pooled amplicons from each of the strains were analyzed. PCR products (50 to 500 ng) were applied to the column equilibrated with 54% buffer B and separated with an oven temperature of 62°C and a flow rate of 0.45 ml min−1 by using a gradient of 60 to 69% buffer B in 18 min after initial equilibration with 60% buffer B for 30 s. Data were collected by UV detection at 260 nm and analyzed with Navigator V2.0 software (Transgenomic). Separated 16S rRNA gene fragments were recovered in 0.1- to 0.2-ml fractions using a fragment collector. D-HPLC fractions (2 μl) were used as templates in PCR amplifications with primers 27f and 534r. The PCR products obtained were purified with ExoSAP-IT (USB Corporation, United States) by following the manufacturer's instructions and were sequenced with either primer 27f or primer 534r using an ABI Prism 3130 XL sequencer (Applied Biosystems).
RESULTS AND DISCUSSION
Accuracy of T-RF size determination.
Fluorochromes routinely used in T-RFLP include 6-carboxyfluorescein (6-FAM) (molecular weight, 389) and carboxy-X-rhodamine (ROX) (molecular weight, 548). Internal size standards labeled with either fluorochrome comigrated with the fragments obtained from a 6-FAM-labeled 0.9-kb PCR product derived from the P. stutzeri KC 16S rRNA gene and digested with endonucleases AluI, HhaI, MspI, and RsaI, which are commonly used in T-RFLP. The expected lengths of the T-RFs generated spanned a broad size range (72, 207, 490, and 878 nt, respectively) (Table 1). The fragment lengths deduced from the results of the experiments clearly highlight the poor match between the expected and experimentally deduced T-RF lengths when the ROX-labeled size marker was used for calibration of 6-FAM-labeled T-RFs (Table 1). Conversely, size determination with the matched 6-FAM-labeled internal standard gave accurate results for fragments less than 500 nt long. Size estimates were wrong for longer T-RFs (>500 nt) irrespective of the fluorochrome used, which was due to the incorrect assumption by the analysis software that migration is linearly proportional to fragment size (13).
T-RFLP was then performed with an artificial assembly of five known bacterial strains with restriction enzymes AluI and HhaI. The strains tested included Gram-positive bacteria (C. acetobutylicum ATCC 824, a spore-forming strict anaerobe, and S. aureus ATCC 25904, a non-spore-forming facultative anaerobe), as well as Gram-negative proteobacteria (M. extorquens CM4 and members of two species of the genus Pseudomonas, P. chlororaphis DSM 50083 and P. stutzeri KC) (Table 2 and Fig. 1A). Accurate T-RF lengths ranging from 70 to 350 nt were again obtained for the peaks of all five strains when the identically labeled internal size standard was used as a reference.
TABLE 2.
16S rRNA gene data for the strains studied
| Strain | 16S rRNA gene characteristics |
T-RF size (nt) |
GenBank accession no. | |||||
|---|---|---|---|---|---|---|---|---|
| Gene copy no. | WAVE-PCR fragmenta |
Sequence data |
Exptl datab |
|||||
| Size (bp) | G+C content (%) | AluI | HhaI | AluI | HhaI | |||
| Clostridium acetobutylicum ATCC 824 | 11 | 491 | 53.8 | 229 | 223 | 229.2 ± 0.1 | 223.1 ± 0.1 | NC003030 |
| Methylobacterium extorquens CM4 | 5 | 472 | 56.1 | 145 | 343 | 145.6 ± 0.1 | 344.0 ± 0.1 | NC011757 |
| Pseudomonas chlororaphis subsp. chlororaphis DSM 50083 | 4-7c | 521 | 53.6 | 72 | 207 | 72.0 ± 0.1 | 207.3 ± 0.1 | EF620458 |
| Pseudomonas stutzeri KC | 4-7c | 521 | 53.2 | 72 | 207 | 72.0 ± 0.1 | 207.3 ± 0.1 | AF063219 |
| Staphylococcus aureus subsp. aureus ATCC 25904 | 5 | 534 | 51.1 | 74 | 238 | 73.6 ± 0.2 | 238.7 ± 0.1 | NC009641 |
Fragment obtained with PCR primers 27f and 534r.
Data obtained with 6-FAM-labeled PCR primer 27f, primer 926r, and 6-FAM-labeled size standard.
Estimated from data for other sequenced Pseudomonas strains (accession no. NC009646, NC004129, NC002947, NC009434, NC007005, NC008027, and NC009439).
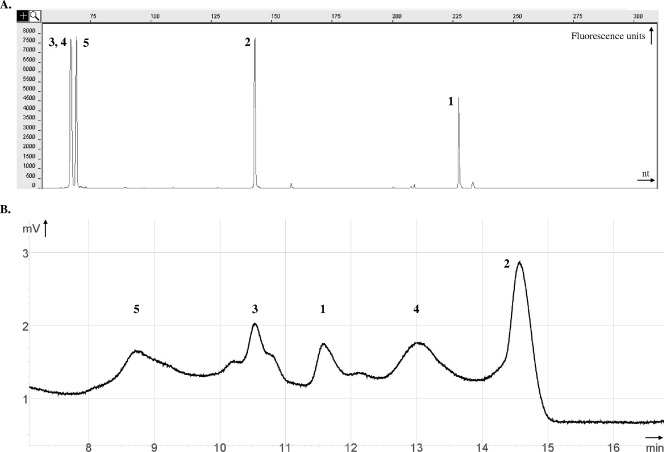
FIG. 1.
Genotyping analyses of five reference strains by (A) T-RFLP (AluI restriction profile) and (B) D-HPLC. Peak 1, C. acetobutylicum ATCC 824; peak 2, M. extorquens CM4; peak 3, P. chlororaphis subsp. chlororaphis DSM 50083; peak 4, P. stutzeri KC; peak 5, S. aureus subsp. aureus ATCC 25904. See Materials and Methods for details concerning chromatographic conditions.
Complementing T-RFLP with D-HPLC.
As sequence data necessary for taxon identification cannot be directly obtained by T-RFLP, D-HPLC was performed in a complementary approach to analyze the model consortium described above. A 470- to 530-bp fragment of the 16S rRNA gene (Table 2) was amplified with primers 27f and 534r, which were previously used as universal eubacterial oligonucleotides in T-RFLP and denaturing gradient gel electrophoresis (DGGE) (5, 9) but to our knowledge have not been used previously for D-HPLC analysis. A DNA fragment size of approximately 500 bp is well suited for D-HPLC separation, whereas longer PCR amplicons are recommended for T-RFLP in order to include a larger number of restriction sites. In the present study, the same forward primer was used for T-RFLP and D-HPLC, which allowed straightforward correlation of the results obtained by the two approaches. Five major distinct D-HPLC peaks were obtained with DNA from the five-strain model mixture (Fig. 1B). Each peak corresponded to a single genotype, as verified by separate analysis of individual strains (data not shown). The results thus validate the choice of the 27f/534r primer pair for analysis of mixed microbial communities by D-HPLC. Fractions containing the different fragments were collected and sequenced with both primers used for amplification. The sequences obtained matched expected GenBank entries (Table 2). Notably, the D-HPLC approach allowed discrimination of the two Pseudomonas strains present in the artificial community, P. stutzeri KC and P. chlororaphis subsp. chlororaphis DSM 50083, which cannot be distinguished by T-RFLP (Fig. 1A and 1B). These two strains exhibit 93% sequence identity in the amplified fragment, but it is known that even single-nucleotide differences can be detected by D-HPLC (1). Thus, D-HPLC may often be a powerful alternative to the more time-consuming construction of gene banks to obtain sequence data. However, the resolution of D-HPLC is usually poorer than that of T-RFLP, and migration parameters have to be optimized for each sample. Indeed, the reported maximum number of separated DNA fragments when D-HPLC is used is about 20 (4), whereas up to 450 peaks can theoretically be discriminated by T-RFLP, with single-nucleotide resolution in the 50- to 500-nt range (see Table S1 in the supplemental material). Thus, in the approach proposed here, T-RFLP yields a detailed fingerprint of a microbial consortium, while D-HPLC provides direct access to sequence information about its composition.
Conclusions.
The results presented here suggest that each T-RFLP sample should be run twice with an internal size standard labeled with two different fluorescent dyes for accurate microbial community fingerprinting using this method. The first run, with different fluorochromes for the internal marker and sample fragments, facilitates determination of the total number of fragment lengths and their relative distribution. Performing a second run using the same fluorescent label for both the sample and internal sizing standard then enables accurate determination of length in the 50- to 500-nucleotide range. The optimized T-RFLP protocol has been used for microbial diversity analysis of groundwater samples containing more than 250 operational taxonomic units (OTUs) (C. Penny, T. Nadalig, C. Gruffaz, S. Vuilleumier, and F. Bringel, unpublished data). Complementary D-HPLC analysis allows workers to distinguish differences between T-RFs that are the same length and to obtain sequence information directly after profiling. Here, this two-pronged genotyping approach proved to be effective for an assembly of five taxa, and it should be applicable for up to 15 different strains. A similar strategy is currently being used for characterization of microbial consortia in enrichment cultures obtained from polluted sites.
Supplementary Material
Acknowledgments
This work was supported by funds from Programme Pluri-Formations and the Contrat-Plan Etat-Région to REALISE, Network of Laboratories in Engineering and Science for the Environment in the Alsace Région (France), and by the EC2CO Program of the Institut National des Sciences de l'Univers. Support from the National Research Fund of Luxembourg for Ph.D. and researcher mobility grants to C.P. (grants Ext-BFR-05-085 and FNR-08-AM2c-21) is gratefully acknowledged.
Footnotes
Published ahead of print on 30 November 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Barlaan, E. A., M. Sugimori, S. Furukawa, and K. Takeuchi. 2005. Profiling and monitoring of microbial populations by denaturing high-performance liquid chromatography. J. Microbiol. Methods 61:399-412. [DOI] [PubMed] [Google Scholar]
- 2.Edwards, U., T. Rogall, H. Blöcker, M. Emde, and E. C. Böttger. 1989. Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 17:7843-7853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gälli, R., and P. L. McCarty. 1989. Biotransformation of 1,1,1-trichloroethane, trichloromethane, and tetrachloromethane by a Clostridium sp. Appl. Environ. Microbiol. 55:837-844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goldenberg, O., S. Herrmann, G. Marjoram, M. Noyer-Weidner, G. Hong, S. Bereswill, and U. B. Göbel. 2007. Molecular monitoring of the intestinal flora by denaturing high performance liquid chromatography. J. Microbiol. Methods 68:94-105. [DOI] [PubMed] [Google Scholar]
- 5.Liu, W. T., T. L. Marsh, H. Cheng, and L. J. Forney. 1997. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl. Environ. Microbiol. 63:4516-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Malik, S., M. Beer, M. Megharaj, and R. Naidu. 2008. The use of molecular techniques to characterize the microbial communities in contaminated soil and water. Environ. Int. 34:265-276. [DOI] [PubMed] [Google Scholar]
- 7.Marsh, T. L. 2005. Culture-independent microbial community analysis with terminal restriction fragment length polymorphism. Methods Enzymol. 397:308-329. [DOI] [PubMed] [Google Scholar]
- 8.Muyzer, G., A. Teske, C. O. Wirsen, and H. W. Jannasch. 1995. Phylogenetic relationships of Thiomicrospira species and their identification in deep-sea hydrothermal vent samples by denaturing gradient gel electrophoresis of 16S rDNA fragments. Arch. Microbiol. 164:165-172. [DOI] [PubMed] [Google Scholar]
- 9.Muyzer, G., E. C. de Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nocker, A., M. Burr, and A. K. Camper. 2007. Genotypic microbial community profiling: a critical technical review. Microb. Ecol. 54:276-289. [DOI] [PubMed] [Google Scholar]
- 11.Pandey, J., K. Ganesan, and R. K. Jain. 2007. Variations in T-RFLP profiles with differing chemistries of fluorescent dyes used for labeling the PCR primers. J. Microbiol. Methods 68:633-638. [DOI] [PubMed] [Google Scholar]
- 12.Rappé, M. S., and S. J. Giovannoni. 2003. The uncultured microbial majority. Annu. Rev. Microbiol. 57:369-394. [DOI] [PubMed] [Google Scholar]
- 13.Schütte, U. M. E., Z. Abdo, S. J. Bent, C. Shyu, C. J. Williams, J. D. Pierson, and L. J. Forney. 2008. Advances in the use of terminal restriction fragment length polymorphism (T-RFLP) analysis of 16S rRNA genes to characterize microbial communities. Appl. Microbiol. Biotechnol. 80:365-380. [DOI] [PubMed] [Google Scholar]
- 14.Studer, A., C. McAnulla, R. Büchele, T. Leisinger, and S. Vuilleumier. 2002. Chloromethane-induced genes define a third C1 utilization pathway in Methylobacterium chloromethanicum CM4. J. Bacteriol. 184:3476-3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.