Abstract
Ratiometric pre-rRNA analysis (RPA) detects the replenishment of rRNA precursors that occurs rapidly upon nutritional stimulation of bacterial cells. In contrast to DNA detection by PCR, RPA distinguishes viable from inactivated bacteria. It exhibits promise as a molecular viability test for pathogens in water and other environmental samples.
As a tool for detecting bacteria in environmental samples, the PCR is limited in part by the false-positive detection of nonviable bacteria and free DNA. One solution to this problem involves treating bacteria with DNA intercalators that penetrate inactivated cells and inhibit PCR amplification (15). These methods require careful titration, and performance varies with sample and disinfection conditions (8, 16). An alternative is to detect microbial RNA, which is less stable than DNA in the environment (10-12, 14, 17-19, 23). However, mRNA can be difficult to detect, while mature rRNA is stable within inactivated cells.
Microbial rRNA precursors (pre-rRNA) comprise an alternative RNA target (2-4, 11, 17, 23). Pre-rRNAs have leader and tail sequences that are enzymatically removed during rRNA maturation. Pre-rRNA sequences are phylogenetically specific, which facilitates their detection in complex samples. In growing bacterial cells, pre-rRNAs constitute a significant fraction of the total rRNA and are much easier to detect than mRNA. Upon cessation of growth, pre-rRNA synthesis ceases but maturation continues, draining pre-rRNA pools. Pre-rRNA has been used as a steady-state indicator of bacterial physiology (2, 11); however, this strategy is compromised by the complex interplay of pre-rRNA synthesis and processing (2).
The present study exploited the replenishment of pre-rRNA that occurs immediately upon the nutritional stimulation of nutrient-limited bacterial cells. Species-specific pre-rRNA was measured in samples after brief exposure to culture medium. Values that exceeded those seen in nonstimulated control samples indicated the presence of viable cells. This ratiometric pre-rRNA analysis (RPA) approach was tested on the rapidly growing opportunistic pathogen Aeromonas hydrophila (generation time [g] = 1 h) and the slowly growing actinomycete Mycobacterium avium (g = 20 h).
For both organisms, real-time quantitative PCRs (RT-qPCRs) targeted the 5′ pre-rRNA leader region. Primers were designed to straddle the 5′ mature rRNA terminus. Primers for cDNA synthesis and reverse PCR primers recognized semiconserved sequences within the mature rRNA. Forward primers recognized predicted species-specific sequences within the 5′ leader. Therefore, amplification required intact specific pre-rRNA as templates (see Table S1 in the supplemental material).
Primers targeted to the M. avium complex (MAC) consistently yielded the expected amplification products when applied to 19 genotypically diverse isolates of M. avium subsp. hominissuis and M. intracellulare (1, 9), the two most significant human pathogens within the MAC. Nucleic acids from M. terrae, M. gastri, M. smegmatis, M. nonchromogenicum, M. phlei, and M. vaccae did not yield amplification products. Specificity of pre-rRNA primers for A. hydrophila, with the exclusion of the closely related fish pathogen A. salmonicida subsp. salmonicida, was predicted by BLAST analysis of the NCBI nonredundant database.
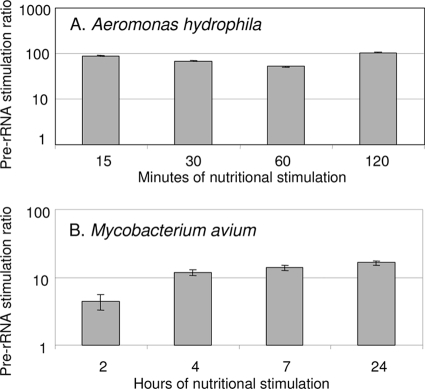
To assess the time course of pre-rRNA replenishment upon nutritional stimulation, early-stationary-phase A. hydrophila ATCC 7966 cells were washed, resuspended in autoclaved tap water (ATW), and incubated for 7 days with aeration at 28°C. Early-stationary-phase cells of M. avium subsp. hominissuis strain HMC02 were washed, resuspended in ATW, and then incubated for 14 days with aeration at 37°C. These conditions were designed to drain pre-rRNA pools in simulated water supply environments. To conduct RPA, bacteria in water were divided into two aliquots and centrifuged. One pellet was resuspended in culture medium (nutritional stimulation) and the other in ATW (control). Final cell densities were approximately 106 CFU/ml. Nutrient broth was used for nutritional stimulation of A. hydrophila, and Middlebrook 7H9 medium with 10% ADC supplement was used for M. avium subsp. hominissuis. After incubation for various periods of time, cells were lysed by high-energy bead beating, RNA was isolated by acidified phenol-chloroform (5, 13, 22), and pre-rRNA was measured by RT-qPCR. Pre-rRNA copy numbers were not calculated due to the lack of authentic pre-rRNA standards. However, ratios of RT-qPCR values in stimulated and control samples were calculated following normalization to genomic DNA standard curves. Pre-rRNA stimulation was very rapid in both organisms. Fifteen minutes and 4 h were adequate for near-maximum stimulation in A. hydrophila and M. avium, respectively (Fig. 1).
FIG. 1.
Time course of nutritional stimulation of pre-rRNA in A. hydrophila (A) and M. avium strain 104 (B) cells. Pre-rRNA stimulation ratios are the ratios of pre-rRNA in stimulated samples relative to that in control samples, measured by RT-qPCR. Values are means and SDs of two or more experiments per time point. To conduct RT-qPCR on extracted RNA, cDNA was first generated using the Superscript III system (Invitrogen) and cleaned using a Qiagen PCR purification kit (catalog no. 28104). Amplification of cDNA was performed using the Applied Biosystems Power SYBR green mix. Reactions were conducted in triplicate at two different dilutions to ensure quantitative readouts. Amplifications were run in 96-well plates on an ABI Prism RT-7500 as follows: 10 min at 95°C and 40 cycles of 15 s at 95°C, 30 s at 60°C, and 30 s at 72°C using 9600 emulation. ABI SDS software was used to set cycle threshold values.
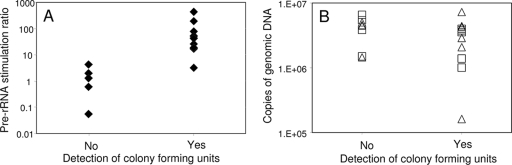
To assess the specificity of RPA for viable cells, sodium hypochlorite was used to generate suspensions with various ratios of viable and inactivated A. hydrophila cells. Briefly, A. hydrophila cells in water were diluted in ATW to an optical density at 600 nm of 0.1 and a 5% sodium hypochlorite reagent was added in various amounts to yield calculated initial chlorine concentrations ranging from 0 to 4 mg/liter. Suspensions were incubated at 28°C for 14 to 18 h with aeration. Percent viability (ratios of viable and inactivated cells) was determined posttreatment by plating on nutrient agar, and RPA was conducted as described earlier. In addition, DNA was quantified by standard qPCR with the same primers used for RPA. In a typical experiment (Table 1), samples with percent viabilities of 96.3%, 26.9%, and 0.02% exhibited pre-rRNA stimulation ratios of ≥2 ± 1 standard deviation (SD). Samples with no detectable viable cells (0% viability) exhibited pre-rRNA stimulation ratios of 1 or less. In contrast, qPCR detection of A. hydrophila genomic DNA was strongly positive in all treated and untreated samples. In four RPA experiments conducted as described for Table 1 (n = 18 chlorine-treated and untreated samples with various percent viabilities), pre-rRNA stimulation ratios seen in samples with no detectable CFU were significantly lower (P = 0.0026 by the Mann-Whitney U test) than those seen in samples with detectable CFU. No such correlation was seen when genomic DNA was quantified (Fig. 2).
TABLE 1.
Pre-rRNA stimulation ratio and genomic DNA in suspensions of A. hydrophila with various ratios of viable and inactivated cells
| Initial hypochlorite concn (mg/liter) | Final no. of CFU/ml | % Viabilitya | Pre-rRNA stimulation ratiob | No. of genomic DNA copiesc (106) |
|---|---|---|---|---|
| 0 | 963,000 | 96.3 | 3.0 ± 0.2 | 1.4 ± 0.4 |
| 1 | 279,000 | 27.9 | 17.2 ± 3.6 | 3.8 ± 0.5 |
| 2 | 190 | 0.02 | 73.0 ± 54.2 | 4.0 ± 0 |
| 3 | 0 | 0 | 0.6 ± 1.0 | 3.8 ± 0.5 |
| 4 | 0 | 0 | 0.04 ± 0.1 | 5.4 ± 0.6 |
Normalized to estimated 1 × 106 input bacteria.
Mean ± SD of three replicate samples.
Extracted by phenol-chloroform method; mean ± SD of replicate two samples.
FIG. 2.
Correlation between the presence of viable A. hydrophila cells and the pre-rRNA stimulation ratio (A) or genomic DNA quantified by qPCR (B) in chlorine-treated laboratory suspensions. Pre-rRNA stimulation ratios (A) are the ratios of pre-rRNA in stimulated samples relative to that in control samples measured by RT-qPCR. Values are means of three measurements per sample. Numbers of genomic DNA copies (B) were quantified by qPCR normalized to a genomic DNA standard curve. DNA was measured in stimulated (open squares) and nonstimulated (open triangles) samples.
To field test RPA for A. hydrophila, 300-ml water samples were collected from surface water sites in Seattle, WA. Three-hundred-milliliter samples were concentrated by filtration following standard protocols (6, 7). Aliquots resuspended in water were diluted twofold in 2× nutrient broth (stimulated sample) or ATW (control). After 1 h of incubation, particulates and bacteria were concentrated and RPA was conducted. Viable counts were also obtained by plating onto ampicillin-dextrin agar with vancomycin as described previously (6). Suspected A. hydrophila colonies were confirmed by PCR for the ast gene (7).
In total, three freshwater samples and one salt water sample were analyzed. The freshwater samples yielded between 280 and 798 CFU/ml A. hydrophila, and RPA results ranging from 4.8 ± 1.4 to 39.8 ± 12.8 (mean stimulated/control ratio). Autoclaved freshwater samples yielded no CFU and no A. hydrophila pre-rRNA signals. The salt water sample had 6 CFU/ml A. hydrophila, but no A. hydrophila pre-rRNA was detected. Therefore, in its current form, RPA applied to natural samples had a detection limit between 6 and 280 CFU/ml.
In practical terms, RPA can conducted by dividing a sample into two equal aliquots, one of which is nutritionally stimulated while the other is held in water or buffer. After stimulation for ≤1 generation time, species-specific pre-rRNA is quantified ratiometrically. The nutritional stimulation step is not of sufficient duration for even modest amplification of pathogen numbers. Thus, RPA is not a culture enrichment.
Ideally, a threshold pre-rRNA stimulation ratio of 1.0 would indicate the presence viable target cells. However, the laboratory samples in Fig. 2, which were densely populated with A. hydrophila cells (approximately 106/ml), included some samples with no detectable CFU of A. hydrophila and mean pre-rRNA stimulation ratios approaching 4.0. Our cultivation methods may not have been sufficiently sensitive to detect all of the viable cells in these samples. Alternatively, the samples may have contained viable but not culturable cells (20, 21) that were capable of pre-rRNA synthesis but not of colony formation. As in any detection method, threshold values for positivity are likely to be affected by sample type and pathogen load. Lower threshold values may apply to dilute natural samples.
In its present form, RPA is not quantitative. Signals that were outside of the accurate linear range of RT-qPCR, such as strongly positive stimulated samples, sometimes yielded ratios that were not numerically proportionate to viable counts. Optimization and standardization for dilute samples may enable quantitative readouts.
In summary, our results support the feasibility of RPA as a means to specifically detect viable pathogens in environmental samples. The method reduced the number of false-positive results obtained with samples containing only dead bacterial cells and DNA. It should also reduce the number of false positives caused by laboratory contamination of samples or PCR reagents. RPA is robust and built upon a physiological feature of all bacteria. It may prove broadly useful in food and water safety analysis, either by itself or as an adjunct to other tools.
Supplementary Material
Acknowledgments
This publication was developed under STAR Research Assistance agreements FP91698201-0 and R833011 awarded by the U.S. Environmental Protection Agency (EPA).
This publication has not been formally reviewed by the EPA. The views expressed herein are solely ours, and the EPA does not endorse any products or commercial services mentioned in this publication.
Footnotes
Published ahead of print on 30 November 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Cangelosi, G. A., R. Freeman, K. N. Lewis, D. Livingston-Rosanoff, S. J. Milan, and S. Goldberg. 2004. Evaluation of a high throughput rep-PCR system for DNA fingerprinting of Mycobacterium tuberculosis and Mycobacterium avium complex. J. Clin. Microbiol. 42:2685-2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cangelosi, G. A., and W. H. Brabant. 1997. Depletion of pre-16S rRNA in starved Escherichia coli cells. J. Bacteriol. 179:4457-4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cangelosi, G. A., W. H. Brabant, T. B. Britschgi, and C. K. Wallis. 1996. Detection of rifampin- and ciprofloxacin-resistant Mycobacterium tuberculosis by using species-specific assays for precursor rRNA. Antimicrob. Agents Chemother. 40:1790-1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cangelosi, G. A., A. M. Hamlin, R. Marin, and C. A. Scholin. 1997. Detection of stable pre-rRNA in toxigenic Pseudo-nitzschia species. Appl. Environ. Microbiol. 63:4859-4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cangelosi, G. A., J. S. Do, R. Freeman, J. G. Bennett, M. Semret, and M. A. Behr. 2006. The two-component regulatory system mtrAB is required for morphotypic multidrug resistance in Mycobacterium avium. Antimicrob. Agents Chemother. 50:461-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Environmental Protection Agency Office of Water. 2001. Method 1605: Aeromonas in finished water by membrane filtration using ampicillin-dextrin agar with vancomycin (ADA-V). EPA/821/R-01/034. United States Environmental Protection Agency Office of Science and Technology, Washington, DC.
- 7.Environmental Protection Agency Office of Water. 2006. Aeromonas: human health criteria document. United States Environmental Protection Agency Office of Science and Technology, Washington, DC.
- 8.Gedalanga, P., and B. Olson. 2009. Development of a quantitative PCR method to differentiate between viable and nonviable bacteria in environmental water samples. Appl. Microbiol. Biotechnol. 82:587-596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Horan, K. L., R. Freeman, K. Weigel, M. Semret, S. Pfaller, T. C. Covert, D. van Soolingen, S. C. Leao, M. A. Behr, and G. A. Cangelosi. 2006. Isolation of the genome sequence strain Mycobacterium avium 104 from multiple patients over a 17-year period. J. Clin. Microbiol. 44:783-789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Licht, T. R., T. Tolker-Nielsen, K. Holmstrom, K. A. Krogfelt, and S. Molin. 1999. Inhibition of Escherichia coli precursor-16S rRNA processing by mouse intestinal contents. Environ. Microbiol. 1:23-32. [DOI] [PubMed] [Google Scholar]
- 11.Lu, T., P. G. Stroot, and D. B. Oerther. 2009. Reverse transcription of 16S rRNA to monitor ribosome-synthesizing bacterial populations in the environment. Appl. Environ. Microbiol. 75:4589-4598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Molin, S., and M. Giskov. 1999. Application of molecular tools for in situ monitoring of bacterial growth activity. Environ. Microbiol. 1:383-391. [DOI] [PubMed] [Google Scholar]
- 13.Mostowy, S., C. Cleto, D. R. Sherman, and M. A. Behr. 2004. The Mycobacterium tuberculosis complex transcriptome of attenuation. Tuberculosis 84:197-204. [DOI] [PubMed] [Google Scholar]
- 14.Muttray, A. F., and W. W. Mohn. 1999. Quantitation of the population size and metabolic activity of a resin acid degrading bacterium in activated sludge using slot-blot hybridization to measure the rRNA:rDNA ratio. Microb. Ecol. 38:348-357. [DOI] [PubMed] [Google Scholar]
- 15.Nocker, A., and A. K. Camper. 2009. Novel approaches toward preferential detection of viable cells using nucleic acid amplification techniques. FEMS Microbiol. Lett. 291:137-142. [DOI] [PubMed] [Google Scholar]
- 16.Nocker, A., K. E. Sossa, and A. K. Camper. 2007. Molecular monitoring of disinfection efficacy using propidium monoazide in combination with quantitative PCR. J. Microbiol. Methods 70:252-260. [DOI] [PubMed] [Google Scholar]
- 17.Oerther, D. B., J. Pernthaler, A. Schramm, R. Amann, and L. Raskin. 2000. Monitoring precursor 16S rRNAs of Acinetobacter spp. in activated sludge wastewater treatment systems. Appl. Environ. Microbiol. 66:2154-2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Patel, B. K., D. K. Banerjee, and P. D. Butcher. 1993. Determination of Mycobacterium leprae viability by polymerase chain reaction amplification of 71-kDa heat-shock protein mRNA. J. Infect. Dis. 168:799-800. [DOI] [PubMed] [Google Scholar]
- 19.Poulsen, L. K., G. Ballard, and D. A. Stahl. 1993. Use of rRNA fluorescence in situ hybridization for measuring the activity of single cells in young and established biofilms. Appl. Environ. Microbiol. 59:1354-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pruzzo, C., R. Tarsi, M. Mar Lleo, C. Signoretto, M. Zampini, L. Pane, R. R. Colwell, and P. Canepari. 2003. Persistence of adhesive properties in Vibrio cholerae after long-term exposure to sea water. Environ. Microbiol. 5:850-858. [DOI] [PubMed] [Google Scholar]
- 21.Rahman, I., M. Shahamat, M. A. Chowdhury, and R. R. Colwell. 1996. Potential virulence of viable but nonculturable Shigella dysenteriae type 1. Appl. Environ. Microbiol. 62:115-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sherman, D. R., M. Voskuil, D. Schnappinger, R. Liao, M. I. Harrell, and G. K. Schoolnik. 2001. Regulation of the Mycobacterium tuberculosis hypoxic response gene encoding alpha-crystallin. Proc. Nat. Acad. Sci. U. S. A. 98:7534-7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stroot, P. G., and D. B. Oerther. 2003. Elevated precursor 16S rRNA levels suggest the presence of growth inhibitors in wastewater. Water Sci. Technol. 47:241-250. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.