Abstract
Recently, we have demonstrated that the 26-47 segment of Salmonella enterica serovar Typhimurium flagellin is capable of mediating flagellar export. In order to reveal whether other parts of the N-terminal region have any significant influence on secretion, a series of plasmids were constructed containing the lac promoter followed by the 26-47, 2-65, or 2-192 portion of Salmonella flagellin, to which various heterologous proteins of different size were fused (18 constructs overall). Essentially, all three segments could drive protein export; however, the nature of the attached polypeptide also had a significant effect on secretion efficiency. When low export efficiency was observed, it was mainly caused by inclusion body formation. Our data provide strong support for the idea that a short segment within the disordered N-terminal region of axial proteins is recognized by the flagellar type III export machinery. The 26-47 segment of flagellin contains all of the necessary information to direct translocation of attached polypeptide chains. This short (positions 26 to 47) flagellin segment attached to recombinant proteins can be used for secreted protein expression. Certain fusion proteins that are easily degraded within the cells were found to be intact in the medium, implying a potential application of this expression system for proteins with high proteolytic susceptibility.
The bacterial flagellum is a biological nanomachine for locomotion. A membrane embedded molecular motor rotates a long helical filament that works as a propeller driving the bacterium through the liquid environment. The filamentous portion of flagellum extends from the cytoplasm to the cell exterior and involves several substructures: the rod, the hook, the hook-filament junction, the long helical filament, and a cap at the filament tip.
The flagellar proteins forming the structures lying beyond the cytoplasmic membrane are synthesized in the cell and exported sequentially by the flagellum-specific protein export apparatus from the cytoplasm to the site of assembly at the distal end of the growing filament (23). Thousands of subunits must be translocated through the narrow (20 to 25 Å wide) central channel of the flagellum (42) in a mostly unfolded conformation. It is puzzling how subunits can be efficiently transported through the hollow core of filaments over large (10- to 15-μm) distances. The hydrophilic inner surface of the channel is supposed to be essential for rapid and efficient transport. It is an interesting question whether the export channel is specialized for the delivery of flagellar proteins, or whether it is capable of passing a wide variety of polypeptide chains.
The flagellar protein export system is thought to exist at the cytoplasmic face of the basal body to distinguish flagellar proteins from other cytoplasmic proteins and to facilitate their transportation (28). Since the identification and enzymatic characterization of FliI ATPase as a component of the flagellar protein export system, it had been thought that the flagellar protein export is driven by the energy of ATP hydrolysis (37). Recent studies, however, have clearly shown that the proton motive force across the cytoplasmic membrane is responsible for driving the export process that involves unfolding of export substrate proteins and translocation of the unfolded chains with the help of the FliI hexamer ring complex (27, 31).
The flagellar protein export system belongs to the family of the type III secretion systems (T3SSs) (4), which also include those for secretion of virulence factors by a wide variety of pathogenic bacteria (15). The nature of the signal directing flagellar protein secretion is still debated because the protein substrates have no cleavable signal sequences or do not share any obvious consensus sequence (12). It has been suggested that the recognition of flagellar export substrates may involve mRNA signals (25), but a growing amount of evidence indicates that the signal is located in the disordered N-terminal region of the secreted proteins (6, 11, 12, 19, 33, 36, 40). For example, residues 38 to 58 of Caulobacter crescentus flagellar hook protein were found to be essential for secretion (19). Similarly, it has been demonstrated that the export signal of the hook scaffolding protein FlgD of Escherichia coli is located exclusively within the N-terminal 71 amino acids (40). Single amino acid substitutions within the N-terminal region of the anti-sigma factor FlgM (6), which is also exported by the flagellum-specific export apparatus, severely impair its export.
Our previous experiments have suggested that the 26-47 disordered segment of Salmonella flagellin contains the recognition signal for the flagellar export machinery (36). When this segment was attached to the small CCP2 domain of the human C1r complement protein, the fusion construct was secreted into the culture medium. The objective of the present study was to explore whether other N-terminal portions of flagellin have any significant influence on secretion. We also aimed to reveal whether the recognition signal can facilitate translocation of a wide variety attached foreign proteins through the flagellum-specific export pathway, which may open up the possibility to use the flagellar export system to secrete heterologous proteins overexpressed in bacteria.
MATERIALS AND METHODS
Strains, plasmids, and gene synthesis.
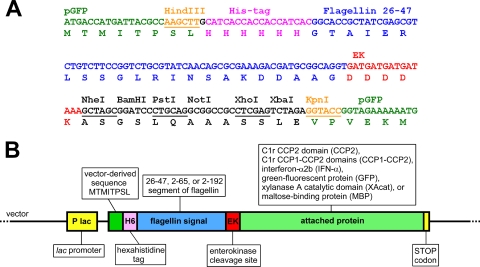
The Salmonella enterica serovar Typhimurium ΔfliC strain, SJW2536 (30), was kindly provided by Kenji Oosawa (Nagoya University, Nagoya, Japan). The pGFP plasmid was purchased from Clontech, and the pTWIN-MBP1 plasmid was from New England Biolabs. Genes for human C1r (9), human IFN-α2b (unpublished data), and Thermotoga maritima xylanase A (16) have been available in our laboratory. The gene cassette containing a polyhistidine tag, the 26-47 serovar Typhimurium flagellin segment, and the enterokinase (EK) site was made by gene synthesis (GeneScript). The cassette was placed between the HindIII and KpnI sites of pGFP, resulting in the pVJGFPa plasmid (Fig. 1A). Synthetic genes of similar cassettes (GeneScript) containing the 2-65 and the 2-192 segments were placed between the HindIII and NheI sites into pVJGFPa, replacing the short segment, and resulting in plasmids pVJGFPb and pVJGFPc. Five genes, CCP2, CCP1-CCP2, alpha interferon (IFN-α), XAcat, and maltose-binding protein (MBP), were amplified by PCR using primers listed in Table 1. The PCR products were digested with NheI and BamHI enzymes, except for MBP, which was digested with NheI and XhoI. The products then were cloned into each of the pVJGFPa, pVJGFPb, and pVJGFPc vectors resulting in the plasmids listed in Table 2 . E. coli Top10 (from Invitrogen) was used for general cloning and plasmid production.
FIG. 1.
Constructs. (A) Part of the sequence of pVJGFPa shown as an example. A synthetic DNA fragment was placed between the HindIII and KpnI sites of pGFP, resulting in the pVJGFPa plasmid. The pVJGFPb and pVJGFPc plasmids differ only in the flagellin segment. (B) Schematic representation of all constructs. The initial pVJGFP series plasmids already contain the GFP gene. Other genes, all ending with a stop codon, were placed between the NheI and BamHI or XhoI sites as described in Materials and Methods.
TABLE 1.
Primers used for the amplification of genes prior to subcloning into pVJGFP series plasmids
| Proteina | Direction | Sequence (5′-3′)b |
|---|---|---|
| CCP2 | Forward | GCGCGCTAGCTGTGGGCAGCCCCGAAAC |
| CCP2 | Reverse | GCGCGGATCCTTAGCACCGAGGAATCTTCTC |
| CCP1-CCP2 | Forward | GCGCGCTAGCTGCCCCCAGCCCAAGAC |
| CCP1-CCP2 | Reverse | Same as the CCP2 reverse primer |
| IFN-α | Forward | CGCGGCTAGCATGGCCTTGACCTTTGC |
| IFN-α | Reverse | CGCGGGATCCTCATTCCTTACTTCTTAAACTTTC |
| XAcat | Forward | CGCGGCTAGCATGATACCTGCTCTGAAAGAAG |
| XAcat | Reverse | GCGCGGATCCCTACTCAGGTGCCACTATCG |
| MBP | Forward | GCGCGCTAGCGAAGAAGGTAAACTGGTAATCTG |
| MBP | Reverse | GCGCCTCGAGTCAAGTCTGCGCGTCTTTCAGGG |
CCP2, human C1r CCP2 domain; CCP1-CCP2, human C1r CCP1-CCP2 domain pair; IFN-α, human IFN-α2b; GFP; A. victoria GFP; XAcat, T. maritima xylanase A catalytic domain; MBP, E. coli MBP.
The parts of the sequences corresponding to coding regions are indicated in boldface, and restriction sites are underlined. The NheI and BamHI restriction sites were used for all genes, except for MBP, where NheI and XhoI sites were used.
TABLE 2.
Levels of expressed (undegraded) fusion proteins within the cells and in the medium of serovar Typhimurium SJW2536 harboring the indicated plasmids at an OD600 of ∼1a
| Signal | Plasmid | Attached protein | Calculated mol mass (kDa) | Level in the cells (mg/liter) |
Level in the medium (mg/liter) | Export efficiency (%) |
||
|---|---|---|---|---|---|---|---|---|
| Total | Soluble fraction | Total | Soluble fraction | |||||
| 26-47 | pVJCCPa | CCP2 | 14 | 1 (W) | 0 (ND) | 4.0 | 80 | 100 |
| pVJdubCCPa | CCP1-CCP2 | 21 | 19 | 0 (ND) | 1.2 | 5.9 | 100 | |
| pVJIFNa | IFN-α | 26 | 4.3 | 0 (ND) | 0 (ND) | 0.0 | NC | |
| pVJGFPa | GFP | 33 | 33 | 4 | 0.8 | 2.4 | 17 | |
| pVJXAa | XAcat | 43 | 12 | 12 | 0.8 | 6.2 | 6.2 | |
| pVJMBPa | MBP | 45 | 57 | 55 | 4.6 | 7.5 | 7.7 | |
| 2-65 | pVJCCPa | CCP2 | 18 | 1 (W) | 0 (ND) | 2.0 | 67 | 100 |
| pVJdubCCPa | CCP1-CCP2 | 25 | 24 | 0 (ND) | 0.6 | 2.4 | 100 | |
| pVJIFNa | IFN-α | 31 | 1 (W) | 0.5 (W) | 0.2 (W) | 17 | 29 | |
| pVJGFPa | GFP | 38 | 43 | 2 | 0.6 | 1.4 | 23 | |
| pVJXAa | XAcat | 47 | 0 (ND) | 0 (ND) | 0 (D) | NCb | NC | |
| pVJMBPa | MBP | 49 | 7 (MD) | 5 (MD) | 0.4 (MD) | 5.4 | 7.4 | |
| 2-192 | pVJCCPa | CCP2 | 32 | 15 | 3 | 4.0 | 21 | 57 |
| pVJdubCCPa | CCP1-CCP2 | 39 | 30 | 1 | 1.0 | 3.2 | 50 | |
| pVJIFNa | IFN-α | 44 | 16 | 1 | 0.6 | 3.6 | 37 | |
| pVJGFPa | GFP | 52 | 44 | 3 | 1.0 | 2.2 | 25 | |
| pVJXAa | XAcat | 61 | 0 (D) | 0 (D) | 0 (D) | NC | NC | |
| pVJMBPa | MBP | 63 | 12 (MD) | 3 (MD) | 0.1 (W, MD) | 0.8 | 3 | |
The full protein names are listed in Table 1. W, detected by Western blotting only. These concentrations were estimated based on the Western blot band intensities. These values are just rough estimates. ND, not detectable on Coomassie blue-stained gels or by Western blotting. D, only degradation products can be detected either on the Coomassie blue-stained gels and/or by Western blotting. MD, major degradation products were also observed but are not included here.
NC, not calculated (division by zero).
Transformation and growth of serovar Typhimurium SJW2536.
To make electrocompetent serovar Typhimurium SJW2536, cells were grown in LB until the optical density at 600 nm (OD600) reached about 0.6, and then they were washed twice with ice-cold sterile water and suspended in the same containing 10% glycerol. For electroporation, 50 to 200 ng of plasmid DNA was used, and a 2.5-kV voltage was applied with a 5-ms time constant. Transformed cells were plated onto LB agar containing 100 μg of ampicillin/ml.
For protein expression, a 1% (vol/vol) overnight Salmonella culture was inoculated into LB medium containing 100 μg of ampicillin/ml, and the cells were grown for 3 to 4 h (OD600 ≈ 0.6) when 0.4 mM IPTG (isopropyl-β-d-thiogalactopyranoside) was added. Samples were taken after an additional 3 to 4 h of incubation, when the OD600 reached about 1.
SDS-PAGE and Western blotting.
The cells were harvested by centrifugation, resuspended in 1/10 volume of water, and then lysed in an equal volume of standard SDS-PAGE sample buffer by heating and vortexing. The cell culture medium was concentrated 50-fold in spin concentrators (Millipore) before an equal volume of standard SDS-PAGE reducing sample buffer was added. The samples were analyzed on 12% gels by using the standard Tris-glycine buffer system. Then, 20-μl portions of the samples above were used for Coomassie blue-stained gels, and 10-μl portions were used for the Western blots. For the Western blots, after transfer to nitrocellulose membranes, the fusion proteins were detected by using a monoclonal anti-polyhistidine-alkaline phosphatase conjugate antibody (catalog no. A-5588; Sigma). For the analysis of inclusion bodies and soluble intracellular proteins, the cells were disrupted by ultrasonic treatment. Cells were harvested by centrifugation and resuspended in a 1/5 volume of sonication buffer (50 mM NaCl, 50 mM Tris-HCl [pH 8.0], 1 mM EDTA, 1% Triton X-100), and then 10% (wt/vol) of glass beads (catalog no. G8893; Sigma) were added, and the cells were sonicated extensively (six times for 15 s at maximum power on ice in a final volume of 1 ml). The soluble and insoluble fractions were separated by centrifugation, the pellet was washed once with sonication buffer, and finally the pellet was resuspended in the same volume as the soluble fraction. To portions of the lysates and the inclusion body suspensions, equal volumes of standard SDS-PAGE reducing sample buffer were added; the samples were then heated, and 20-μl aliquots were analyzed by SDS-PAGE and Western blotting.
Basal lysis of serovar Typhimurium SJW2536.
In order to determine the spontaneous lysis of the host cells, the relative levels of Salmonella's own MBPs were determined from the cells and from the medium. The endogenous intracellular MBP of Salmonella lacks any secretion signal; therefore, it can leak into the medium only by cell lysis. Cells and medium of 400 ml of SJW2536 culture in LB (OD600 ≈ 1) were separated by centrifugation. The cells were lysed by extensive ultrasonic treatment, and the clarified lysate was made up to a 400-ml final volume with 200 mM NaCl, 20 mM Tris-HCl (pH 7.4), and 1 mM EDTA (column buffer). The medium (400 ml) was composed of NaCl, Tris-HCl (pH 7.4), and EDTA to the same concentrations as in the column buffer. Both were loaded to 1-ml amylose resin columns (New England Biolabs catalog no. E8022), washed with column buffer, and eluted in 3 ml of the same buffer containing 10 mM maltose. Purified MBP from the cells (15 μl plus sample buffer per lane) was analyzed by SDS-PAGE and Western blotting. To obtain a detectable amount of protein, a 750-μl aliquot of MBP purified from the medium was precipitated with 20% trichloroacetic acid before SDS-PAGE and Western blot analysis. MBP was detected by using mouse monoclonal anti-MBP (Sigma catalog no. M1321) and anti-mouse-IgG alkaline phosphatase conjugate (Sigma catalog no. A2429) antibodies. Based on the observed band intensities and the amounts loaded onto the gels, the spontaneous cell lysis was estimated to be ca. 0.4%.
Protein levels and export efficiencies.
Secretion of the various fusion proteins by serovar Typhimurium, which varied in size from 9 to 40 kDa, was examined by estimating the concentration of the fusion proteins within the cells and in the cell culture supernatant applying densitometric analysis of the Coomassie blue-stained SDS-polyacrylamide gels. The band intensities were corrected with the intensity of the host protein band with the same apparent molecular weight in the neighboring lane. The bands of the molecular weight marker, which contain known amounts of the respective proteins, were used as a reference to determine the amount of the fusion protein in each lane. The concentrations were calculated from the loaded volume and the determined amount. Since the amount of the Coomassie blue stain bound to the same amount of different proteins might be somewhat different, this method does not provide an accurate measure of the levels of the expressed fusion proteins; nevertheless, it allows determination of the relative levels of the same fusion protein in the cells and in the medium. In some cases the fusion products could only be detected by Western blotting that was used to identify the His-tagged polypeptide chains. In these cases (e.g., 2-65-IFN-α and 2-192-MBP) the export efficiencies and at least one of the concentration values were estimated based on the Western blot intensities, because the corresponding band on the Coomassie blue-stained gel could not be identified. At least 1 mg of fusion protein/liter in the cells and 0.2 mg/liter in the medium is required for a clear detection on Coomassie blue-stained gels because of the host protein background. These upper limits were taken into account when the concentration values were estimated, since bands detected only by Western blot are probably equal or lower than these values. Export percent efficiencies were calculated from the levels of the fusion proteins in the cells and in the medium as follows: [(amount of intact protein in the medium)/(amount of intact protein in the medium + amount of intact protein in the cells)] × 100. This simple equation does not include degradation products; hence, it may underestimate the “true” export efficiency if the protein is degraded in the medium after secretion. Also, degradation in the cells may result in the loss of the export signal; therefore, the protein may get trapped inside the cells.
EK cleavage and purifications.
Serovar Typhimurium SJW2536 harboring pVJMBPa was grown as described above. The cells were pelleted at 5,000 × g for 15 min at 4°C, and the supernatant was used for the subsequent steps. A total of 500 ml of the medium containing 26-47-MBP was buffer exchanged by diafiltration (using a 10-kDa cutoff Sartorius Sartocon slice cassette) to 400 mM NaCl, 10 mM imidazole, 50 mM sodium phosphate [pH 7.5], 0.01% NaN3). At the same time, the volume was reduced to about 200 ml. The sample was loaded onto a 5-ml Ni-NTA Superflow column (Qiagen), washed, and then eluted with the same buffer but containing 400 mM imidazole. The two 5-ml fractions containing 26-47-MBP were combined, and the buffer was exchanged by dialysis with 50 mM Tris-HCl, 0.2 mM CaCl2 [pH 7.5], and 0.1% Tween 20. Recombinant EK (Sigma catalog no. E-4906) was added at a 26-47-MBP/EK ratio of ca. 20:1 (wt/wt) to a solution of 0.1 to 0.15 mg of 26-47-MBP/ml, followed by incubation for 30 h at room temperature. Then, 5 ml of the digested protein was loaded onto a 1-ml amylose resin column (New England Biolabs catalog no. E8022), washed with 200 mM NaCl-20 mM Tris-HCl [pH 7.4]-1 mM EDTA, and eluted with the same buffer containing 10 mM maltose. The majority of the eluted MBP was found in three of the 1-ml fractions.
RESULTS AND DISCUSSION
Constructs.
DNA fragments encoding the 26-47, 2-65, or 2-192 segment of serovar Typhimurium flagellin were prepared by gene synthesis. The synthetic constructs contained portions encoding a His6 tag before, and an EK cleavage site after the flagellar segment. The His6 tag facilitates easy purification and aids detection by Western blotting, whereas the EK cleavage site allows removal of the tag if necessary. The synthetic genes were placed into the lac promoter based pGFP plasmid in frame with the green fluorescent protein (GFP) gene. The resulting plasmids were named pVJGFPa, pVJGFPb, and pVJGFPc, respectively (Fig. 1A). These initial plasmids are already capable of expressing fusion proteins made of the flagellar signals and jellyfish (Aequorea victoria) GFP. Genes of various heterologous proteins, all ending with a stop codon, were placed into each of the pVJGFP plasmids in frame with the coding sequence the flagellin segment right after the EK cleavage site. The inserted genes encoded the following proteins or protein domains: human C1r CCP2 domain (CCP2), human C1r CCP1-CCP2 domain pair (CCP1-CCP2), human alpha2b interferon (IFN-α), T. maritima xylanase A catalytic domain (XAcat), and E. coli MBP. Proteins 9 to 40 kDa in size (14 to 63 kDa with the attached segments) were selected in order to test the size limit for the flagellar export apparatus to unfold and export proteins. Of the chosen proteins, four were eukaryotic, three contained disulfide bridges, two were comparable in size with flagellin (51 kDa), and none of them was export substrate of a T3SS or contained any secretion elements besides the attached segment. Overall, 18 constructs were made, including the initial plasmids (Fig. 1B). The names of the different plasmids are listed in the first column of Table 2. From here on fusion proteins are referred to by the signal and the short name of the attached protein, e.g., 2-192-CCP2 refers to the human C1r CCP2 domain attached to the 2-192 flagellin segment supplemented with the extra tags. It is important to note that except for gene segments encoding the 26-47, 2-62, or 2-192 portions of flagellin, none of the constructs contained any other flagellar nucleotide sequences from the coding or noncoding regions.
Factors influencing the export efficiency.
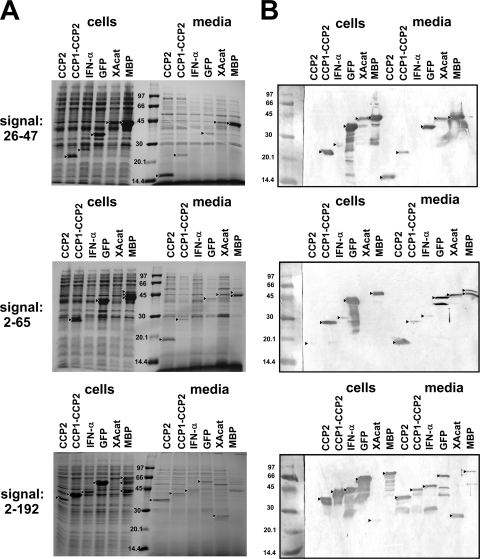
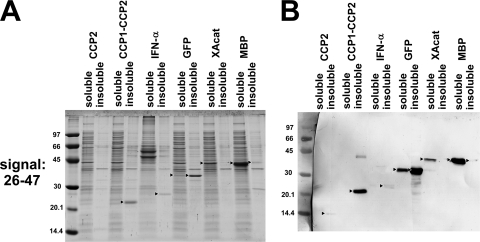
The secretion yield of the fusion proteins was determined by comparing the relative levels of a given protein in the medium and in the cells obtained from the densitometric analysis of Coomassie stained gels (Fig. 2A and Table 2). The appropriate bands were identified by Western blotting detecting the His tag present on all fusion products (Fig. 2B). In eight cases the export yield of the total intact protein was rather low (below 5%) or nothing at all, and in eleven cases the concentration of the exported fusion proteins was <1 mg/liter or 0. In contrast, the flagellar system exports flagellin very efficiently: 6 to 10 mg of monomeric flagellin/liter was secreted at a similar cell density in mutant strains (13, 14, 18), and flagellin was the major protein in the medium. In our experiments, we observed comparable levels in four cases (26-47-CCP2, 2-65-CCP2, 2-192-CCP2, and 26-47-MBP), where the produced fusion protein was the major protein in the medium, while in two other cases (2-65-MBP and 2-192-XAcat) a degradation product of the produced fusion protein appeared to be the most abundant component. Low export and high intracellular expression is best exemplified with GFP. It is expressed at a high level (33 to 44 mg/liter) with all three flagellar segments intracellularly, whereas only 0.6 to 1 mg/liter is exported to the medium. GFP is known to form intracellular aggregates (35); therefore, we separated the cell lysates to soluble and insoluble fractions and analyzed them by SDS-PAGE. GFP was found to be expressed >85% in the form of inclusion bodies. Taking this into account, soluble GFP is exported with >17% efficiency to the medium by any of the signals used as opposed to approximately a mere 2% of the total expressed protein. The analysis was extended to all constructs, and the results were added to Table 2. As an example, Fig. 3 shows the results of SDS-PAGE and Western blot analyses of the soluble and insoluble cellular fractions obtained with constructs containing the 26-47 signal.
FIG. 2.
SDS-PAGE and Western blot analysis of cell lysates and cell culture supernatants of serovar Typhimurium SJW2536 transformed with plasmids expressing fusion proteins with different flagellin signal segments. Small triangles point to the intact expressed fusion proteins and to major degradation products found in amounts that exceed those of the corresponding intact proteins. In the case of 2-192-XAcat, only a degradation product can be detected. (A) Coomassie blue-stained SDS-12% polyacrylamide gels. Each lane represents cells from 100 μl and medium from 500 μl of cell culture. Before analysis, the cells were centrifuged and resuspended in a small volume of water, while the cell culture media were concentrated on spin concentrators. (B) Western blots. Each lane represents cells from 50 μl and medium from 250 μl cell culture that were treated as described above. After transfer to nitrocellulose membranes, the fusion proteins were detected with an anti-polyhistidine monoclonal antibody conjugated to alkaline phosphatase. The contrast of the pictures was increased for better visibility. The molecular weight marker was stained with Ponceau S.
FIG. 3.
Soluble cellular expression versus inclusion body formation exemplified with samples from cells expressing fusion proteins containing the 26-47 flagellin segment. Cells were disrupted in the presence of Triton X-100 by ultrasonic treatment, and the soluble and insoluble protein fractions were separated by centrifugation. (A) Coomassie blue-stained SDS-12% polyacrylamide gel. Each lane represents the supernatant or the pellet from cell lysates equivalent to 50 μl of the original cell culture. (B) Western blot. Each lane represents the supernatant or the pellet from cell lysates obtained from 50 μl of the original cell culture. After transfer to nitrocellulose, the fusion proteins were detected with an anti-polyhistidine monoclonal antibody conjugated to alkaline phosphatase. The contrast of the pictures was increased for better visibility. The molecular weight marker was stained with Ponceau S.
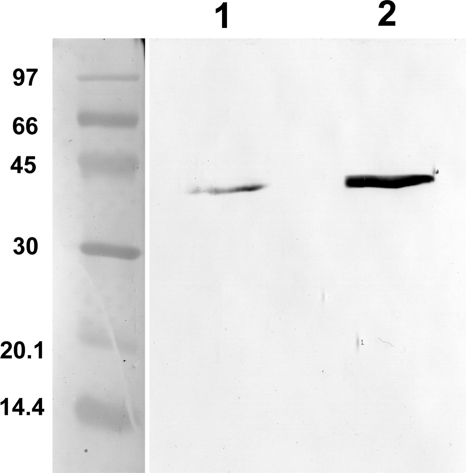
The basal lysis of Salmonella SJW2536 host cells was estimated from the relative levels of Salmonella's endogenous MBP within the cells and in the medium. The endogenous intracellular MBP of Salmonella lacks any secretion signal; therefore, it can leak into the medium only by cell lysis. The highly soluble MBP was purified from the cells and from the medium on amylose resin in an identical setup for both the medium and the cells; therefore, the loss during purification is similar in both cases. The purified and at the same time concentrated MBP was analyzed by SDS-PAGE and Western blotting (Fig. 4). Based on the observed band intensities and the amounts loaded onto the gels, spontaneous cell lysis is estimated to be ca. 0.4%. Based on this observation, export yields of greater than ca. 1% can be considered significant. Of the 18 fusion proteins examined, 14 fell into this category. In the remaining four cases, degradation or inclusion body formation prevented significant export of the intact protein.
FIG. 4.
Basal lysis of the host serovar Typhimurium SJW2536. Endogenous MBP of Salmonella lacking any signal sequence was purified from the cells and from the medium as described in Materials and Methods. Purified samples were analyzed by Western blotting. After transfer to nitrocellulose, MBP was detected with mouse monoclonal anti-MBP and anti-mouse-IgG alkaline phosphatase conjugate antibodies. The molecular weight marker was stained with Ponceau S. Lane 1, MBP purified from 100 ml of cell culture medium; lane 2, MBP purified from cells obtained from 2 ml of culture. Based on the band intensities and the amounts loaded onto the gels, spontaneous cell lysis was estimated to be ca. 0.4%.
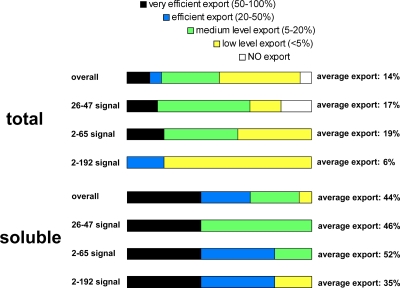
It is important to note that protein degradation, inclusion body formation, and various expression levels complicate interpretation of the data. Nevertheless, the general trend seems to be that the short (26-47 and 2-65) segments resulted in somewhat better export yield, and the shortest (26-47) signal gave the highest protein levels in the medium. Overall, 14 of 18 fusion proteins were exported into the medium of serovar Typhimurium with significant efficiency. In one case (26-47-IFN-α) all of the expressed recombinant protein was found in the form of inclusion bodies, and no export was observed, whereas in the cases of 2-65-XAcat and 2-192-XAcat only degradation products were detected mainly in the medium. In the case of 2-192-MBP, intact protein export is detectable but is not significant, probably due to degradation. In our study we focused on the secretion of intact fusion proteins, since degradation products may be exported with quite different efficiencies or not at all if the flagellar segment is removed. In most cases, the intact fusion proteins could be detected in the medium, indicating that each attached signal, regardless of the length, can mediate protein export, provided that at least a fraction of the protein in question is soluble. In four cases the export was so efficient that 100% of the soluble fusion protein was found in the medium. In all four of these cases the short (26-47 or 2-65) signals were used. On the other hand, the longest signal (the 2-192 signal) gave somewhat higher protein levels in the medium than the 2-65 one, and for IFN-α only the longest signal produced significant levels of the fusion protein in the medium. The results are summarized on Fig. 5 comparing the export efficiencies of the total expressed and the soluble fusion proteins.
FIG. 5.
Summary of the results. The export efficiencies of the total expressed and the soluble fraction of the fusion proteins showing their dependence on the attached flagellin segment length were determined. The bar length represents the fraction of cases with a given range of efficiency.
Export signal.
The structure of the bacterial flagellum and the components of the export system are fairly well characterized (5, 24, 26); however, the signal governing the export of flagellar proteins is still somewhat mysterious. The flagellar secretion system (flagellar T3SS) is closely related to the T3SSs of virulence factors of pathogenic bacteria. Salmonella has two such systems encoded by the salmonella pathogenicity islands 1 and 2 (SPI-1 T3SS and SPI-2 T3SS). Likely, the mechanisms governing export by the different pathways shares some common characteristics. Proteins secreted by any of the type III secretion pathways do not possess a classical signal sequence. Certain studies suggested that the signal sequence is found at the mRNA level (1, 25) or that the combination of an N-terminal signal and a structural element in the mRNA is required (3). On the other hand, a growing number of data support that the signal is contained at the protein level, and it is located at or near the N terminus (6, 12, 17, 19, 22, 36, 39, 40). The identified signal sequences of different flagellar proteins, however, do not show any significant sequence similarity, which indicates that determinants should be at higher than the primary structural level.
Interestingly, monomeric flagellin has been shown to be secreted by the SPI-1 T3SS and even translocated into the host cell cytosol (32, 34). Normally, flagellin is exported primarily by the flagellar T3SS; it polymerizes to form the filament, and only a small fraction of monomeric flagellin is leaked into the medium. Further research is required to clarify to what extent is the SPI-1 T3SS is involved in the secretion of the fusion proteins presented here. Involvement of the SPI-2 T3SS is unlikely because it is not active in Salmonella cultured in LB medium (7). Flagellin is not the only protein that can be exported by more than just one pathway. For example, SopE, which is secreted specifically by the SPI-1 T3SS, can be exported by the flagellar T3SS as well when its interaction with its cognate chaperone is abolished by mutations (10). It seems likely that the basic mechanism by which the different type III systems recognize their substrates is closely related, and the signal recognized by one system can be recognized by the other under certain circumstances (21). Determinants other than the N-terminal polypeptide signal may be required for targeting the specific protein to the appropriate export system (10).
We have previously identified a short segment spanning from the 26 to 47 amino acids of S. enterica serovar Typhimurium flagellin as a minimal export signal (36). Other partially or fully overlapping segments (1 to 35 and 1 to 48) have also been shown to facilitate export of an attached reporter protein (34). The objective of the present study was to explore whether there is any benefit, in terms of secretion efficiency, to using larger N-terminal portions of flagellin than the identified minimal signal. One reporter gene is, however, not sufficient to characterize the effect of signal length on protein export, since every construct is different in terms of expression level, inclusion body formation, and degradation. To average out the influence of these factors on the export yield, we used six different proteins to adequately answer this question. Three different segments of flagellin (26 to 47, 2 to 65, and 2 to 192) were tested, fused to the N terminus of each selected protein (18 constructs overall). The first ∼65 residues of flagellin, which represent the entire N-terminal unstructured region, were chosen because terminal disorder is thought to be the common characteristics of export substrates (29, 38), whereas approximately the first 190 residues encompass the highly conserved N-terminal part of the molecule (2), and the first report used about the same fragment that demonstrated that the N-terminal part of flagellin contains the signal (20). It was found that any of the attached segments resulted in export of nearly all proteins with various yields. The major reason hindering secretion was inclusion body formation of certain fusion proteins. When only the soluble fraction of the expressed proteins was considered, the export efficiency was fairly high with all signals. The average efficiency was 44% overall, and 46, 52, and 35% with the 26-47, 2-65, and 2-192 segments, respectively (Fig. 5). A quantitative comparison (Table 2) suggested that the specific characteristics of a given fusion protein rather than the attached signal caused differences in export efficiency. An important feature of the DNA constructs is that they do not contain any flagellar nucleotide sequences from the untranslated or coding region other than the ones encoding the 26-47, 2-62, or 2-192 segment. Our data provide strong support for the idea that a short segment within the N-terminal region of axial proteins is recognized by the flagellar type III export machinery and no mRNA signal is required.
Flagellar axial proteins have to be delivered to their assembly destination at the tip of the growing structure. Since the diameter of the central channel of the flagellum is only ∼2 nm, proteins to be exported must be largely unfolded for entry into and translocation through the channel. The successive process of unfolding and translocation of export substrates is driven by the proton motive force across the cell membrane (27, 31), i.e., the flagellar secretion apparatus functions as a proton-driven protein exporter. Our experiments show that the 22-residue long export signal contains all of the necessary information to direct translocation of export substrates or attached heterologous polypeptide chains. It is interesting that an additional sequence element such as the His6 tag present in all constructs is tolerated N-terminal to the signal sequence.
Prospects for biotechnological applications.
Secretion of recombinant proteins overexpressed in bacteria is still not solved satisfactorily. T3SSs have the potential to be used as effective tools for the secretion of recombinant proteins (8, 11, 25, 36, 41). A major advantage of secreted protein expression is that cell disruption is no longer required before purification, and the starting material is less contaminated with host proteins, lipopolysaccharides, and nucleic acids. It gives a chance that certain proteins which are degraded or form intracellular aggregates can be expressed in intact and soluble form. Solubility, however, does not necessarily mean that the protein of interest is correctly folded. Axial flagellar proteins are translocated in a largely unfolded state through the narrow export channel by the flagellar T3SS. It is plausible to assume that fusion proteins containing the export signal are also transported to the medium in an unfolded form, where they may or may not adopt their native conformation. Anyway, the lack of competing aggregation facilitates the folding process. The flagellin-deficient serovar Typhimurium strain SJW2536 was used in our experiments (30). As a result, the export apparatus was not overloaded with the most abundant flagellar protein, flagellin. Since SJW2536 can assemble only the basal body and the hook part of the flagellum, the expressed fusion proteins do not need to travel through a very long filament.
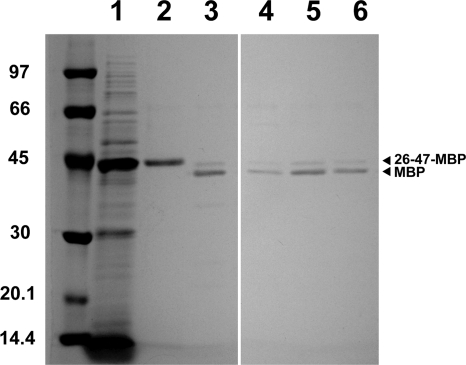
Our constructs were designed to contain a His6 tag at the N terminus. This modification enables easy detection and purification in one step on a Ni2+ affinity matrix. Another feature of the constructs is that they contain an EK cleavage site right after the flagellin segment. The EK site enables selective removal of the preceding flagellar and other sequence elements from the expressed protein. As an example, purification of 26-47-MBP was characterized in detail mainly because it was secreted to the medium at the highest concentration (although not with the highest efficiency), and its binding to amylose resin, followed by elution with maltose, makes it easy to check whether it is correctly folded. 26-47-MBP was purified with an ∼60% yield on a Ni-NTA Superflow column from the medium of SJW2536 harboring pVJMBPa (Fig. 6). After purification, the secretion tag was removed by digestion with EK. The EK-treated protein was loaded onto an amylose-agarose resin. Approximately 60% of MBP was bound and could be eluted from the column (Fig. 6), indicating that at least this portion was correctly folded. Another examined protein, 26-47-CCP2, however, remained unfolded or at least did not adopt a native-like fold after secretion (data not shown). This protein is curiously hardly detectable within the cells but found at a high concentration in the medium. A plausible explanation for this observation is that the unfolded 26-47-CCP2 that was not exported is rapidly degraded in the cells.
FIG. 6.
Secreted expression: a potential biotechnological application. The 26-47-MBP fusion protein was purified from the medium by Ni2+-affinity chromatography, cleaved by EK, and finally purified on amylose-agarose. Lane 1, concentrated medium (equivalent to 500 μl of original medium) of SJW2536 harboring pVJMBPa; lane 2, ∼0.7 μg of 26-47-MBP purified on Ni-NTA Superflow; lane 3, ∼0.5 μg of 26-47-MBP treated with EK (at a 26-47-MBP/EK ratio of 20:1 [wt/wt]) for 30 h at room temperature; lanes 4 to 6, fractions containing the EK-treated MBP eluted from amylose-agarose beads with a maltose-containing buffer.
These preliminary experiments indicate that fusion proteins carrying N-terminal flagellin segments can be efficiently secreted into the medium of serovar Typhimurium SJW2536 and purified with ease. As demonstrated by 26-47-MBP, they may acquire the correct fold after secretion. This expression system also has a potential for unfolded or unstructured proteins that are easily degraded intracellularly but may escape proteolysis if secreted efficiently into the medium.
Conclusions.
In summary, all of the examined N-terminal flagellin segments were recognized by the export apparatus and could mediate protein export. Our data reinforce the hypothesis that the export signal for the flagellar T3SS is located at the protein level near the N terminus of export substrates. The identified minimal, 22-residue-long signal in flagellin contains all of the essential information to direct export of attached heterologous proteins by the flagellar export system. Our results demonstrate that the export channel is not specialized for the delivery of flagellar proteins but capable of passing a wide variety of polypeptide chains. N-terminal flagellin segments containing the export signal combined with a His6 tag can be attached to heterologous proteins, facilitating their secreted expression and easy purification from the medium.
Acknowledgments
We thank Kenji Oosawa for the SJW2536 strain, Márton Megyeri for help with cloning of the IFN-α2b gene, Katalin Szilágyi for the refolded C1r CCP2, Balázs Major for technical help, and Szilárd Kamondi for the cloned XAcat gene.
The support of Hungarian Scientific Research Fund (OTKA) grants NI61915 and NK77978, National Development Agency grant KMOP-1.1.2-07/1-2008-0003, and Hungarian National Office for Research and Technology (NKTH-KPI) grant GVOP-3.1.1-0386/3.0 is acknowledged.
Footnotes
Published ahead of print on 11 December 2009.
REFERENCES
- 1.Anderson, D. M., and O. Schneewind. 1997. A mRNA signal for the type III secretion of Yop proteins by Yersinia enterocolitica. Science 278:1140-1143. [DOI] [PubMed] [Google Scholar]
- 2.Beatson, S. A., T. Minamino, and M. J. Pallen. 2006. Variation in bacterial flagellins: from sequence to structure. Trends Microbiol. 14:151-155. [DOI] [PubMed] [Google Scholar]
- 3.Blaylock, B., J. A. Sorg, and O. Schneewind. 2008. Yersinia enterocolitica type III secretion of YopR requires a structure in its mRNA. Mol. Microbiol. 70:1210-1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blocker, A., K. Komoriya, and S.-I. Aizawa. 2003. Type III secretion systems and bacterial flagella: insights into their function from structural similarities. Proc. Natl. Acad. Sci. U. S. A. 100:3027-3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chevance, F. F. V., and K. T. Hughes. 2008. Coordinating assembly of a bacterial macromolecular machine. Nat. Rev. Microbiol. 6:455-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chilcott, G. S., and K. T. Hughes. 1998. The type III secretion determinants of the flagellar anti-transcription factor, FlgM, extend from the amino terminus into the anti-σ28 domain. Mol. Microbiol. 30:1029-1040. [DOI] [PubMed] [Google Scholar]
- 7.Deiwick, J., T. Nikolaus, S. Erdogan, and M. Hensel. 1999. Environmental regulation of Salmonella pathogenicity island 2 gene expression. Mol. Microbiol. 31:1759-1773. [DOI] [PubMed] [Google Scholar]
- 8.Derouazi, M., B. Toussaint, L. Quénée, O. Epaulard, M. Guillaume, R. Marlu, and B. Polack. 2008. High-yield production of secreted active proteins by the Pseudomonas aeruginosa type III secretion system. Appl. Environ. Microbiol. 74:3601-3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dobó, J., P. Gál, K. Szilágyi, S. Cseh, Z. Lörincz, V. N. Schumaker, and P. Závodszky. 1999. One active C1r subunit is sufficient for the activity of the complement C1 complex: stabilization of C1r in the zymogen form by point mutations. J. Immunol. 162:1108-1112. [PubMed] [Google Scholar]
- 10.Ehrbar, K., B. Winnen, and W.-D. Hardt. 2006. The chaperone binding domain of SopE inhibits transport via flagellar and SPI-1 TTSS in the absence of InvB. Mol. Microbiol. 59:248-264. [DOI] [PubMed] [Google Scholar]
- 11.Gál, P., B. Végh, P. Závodszky, and F. Vonderviszt. 2006. Export signals. Nat. Biotechnol. 24:900-901. [DOI] [PubMed] [Google Scholar]
- 12.Hirano, T., T. Minamino, K. Namba, and R. M. Macnab. 2003. Substrate specificity classes and the recognition signal for Salmonella type III flagellar export. J. Bacteriol. 185:2485-2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Homma, M., H. Fujita, S. Yamaguchi, and T. Iino. 1984. Excretion of unassembled flagellin by Salmonella typhimurium mutants deficient in hook-associated proteins. J. Bacteriol. 159:1056-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ikeda, T., R. Kamiya, and S. Yamaguchi. 1983. Excretion of flagellin by a short-flagella mutant of Salmonella typhimurium. J. Bacteriol. 153:506-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Journet, L., K. T. Hughes, and G. R. Cornelis. 2006. Type III secretion: a secretory pathway serving both motility and virulence. Mol. Membr. Biol. 22:41-50. [DOI] [PubMed] [Google Scholar]
- 16.Kamondi, S., A. Szilágyi, L. Barna, and P. Závodszky. 2008. Engineering the thermostability of a TIM-barrel enzyme by rational family shuffling. Biochem. Biophys. Res. Commun. 374:725-730. [DOI] [PubMed] [Google Scholar]
- 17.Karavolos, M. H., A. J. Roe, M. Wilson, J. Henderson, J. J. Lee, D. L. Gally, and C. M. A. Khan. 2005. Type III secretion of the Salmonella effector protein SopE is mediated via an N-terminal amino acid signal and not an mRNA sequence. J. Bacteriol. 187:1559-1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Komoriya, K., N. Shibano, T. Higano, N. Azuma, S. Yamaguchi, and S.-I. Aizawa. 1999. Flagellar proteins and type III-exported virulence factors are the predominant proteins secreted into the culture media of Salmonella typhimurium. Mol. Microbiol. 34:767-779. [DOI] [PubMed] [Google Scholar]
- 19.Kornacker, M. G., and A. Newton. 1994. Information essential for cell-cycle-dependent secretion of the 591-residue Caulobacter hook protein is confined to a 21-amino-acid sequence near the N terminus. Mol. Microbiol. 14:73-85. [DOI] [PubMed] [Google Scholar]
- 20.Kuwajima, G., I. Kawagishi, M. Homma, J.-I. Asaka, E. Kondo, and R. M. Macnab. 1989. Export of an N-terminal fragment of Escherichia coli flagellin by a flagellum-specific pathway. Proc. Natl. Acad. Sci. U. S. A. 86:4953-4957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee, S. H., and J. E. Galán. 2004. Salmonella type III secretion-associated chaperones confer secretion-pathway specificity. Mol. Microbiol. 51:483-495. [DOI] [PubMed] [Google Scholar]
- 22.Lloyd, S. A., M. Norman, R. Rosqvist, and H. Wolf-Watz. 2001. Yersinia YopE is targeted for type III secretion by N-terminal, not mRNA, signals. Mol. Microbiol. 39:520-531. [DOI] [PubMed] [Google Scholar]
- 23.Macnab, R. M. 2003. How bacteria assemble flagella. Annu. Rev. Microbiol. 57:77-100. [DOI] [PubMed] [Google Scholar]
- 24.Macnab, R. M. 2004. Type III flagellar protein export and flagellar assembly. Biochim. Biophys. Acta 1694:207-217. [DOI] [PubMed] [Google Scholar]
- 25.Majander, K., L. Anton, J. Antikainen, H. Lång, M. Brummer, T. K. Korhonen, and B. Westerlund-Wikström. 2005. Extracellular secretion of polypeptides using a modified Escherichia coli flagellar secretion apparatus. Nat. Biotechnol. 23:475-481. [DOI] [PubMed] [Google Scholar]
- 26.Minamino, T., and K. Namba. 2004. Self-assembly and type III protein export of the bacterial flagellum. J. Mol. Microbiol. Biotechnol. 7:5-17. [DOI] [PubMed] [Google Scholar]
- 27.Minamino, T., and K. Namba. 2008. Distinct roles of the FliI ATPase and proton motive force in bacterial flagellar protein export. Nature 451:485-488. [DOI] [PubMed] [Google Scholar]
- 28.Minamino, T., K. Imada, and K. Namba. 2008. Mechanisms of type III protein export for bacterial flagellar assembly. Mol. Biosyst. 4:1105-1115. [DOI] [PubMed] [Google Scholar]
- 29.Namba, K. 2001. Roles of partly unfolded conformations in macromolecular self-assembly. Genes Cells 6:1-12. [DOI] [PubMed] [Google Scholar]
- 30.Ohnishi, K., Y. Ohto, S.-I. Aizawa, R. M. Macnab, and T. Iino. 1994. FlgD is a scaffolding protein needed for flagellar hook assembly in Salmonella typhimurium. J. Bacteriol. 176:2272-2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paul, K., M. Erhardt, T. Hirano, D. F. Blair, and K. T. Hughes. 2008. Energy source of flagellar type III secretion. Nature 451:489-492. [DOI] [PubMed] [Google Scholar]
- 32.Roy, C. R., and D. S. Zamboni. 2006. Cytosolic detection of flagellin: a deadly twist. Nat. Immunol. 2006 7:549-551. [DOI] [PubMed] [Google Scholar]
- 33.Stafford, G. P., L. B. D. Evans, R. Krumscheid, P. Dhillon, G. M. Fraser, and C. Hughes. 2007. Sorting of early and late flagellar subunits after docking at the membrane ATPase of the type III export pathway. J. Mol. Biol. 374:877-882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun, Y.-H., H. G. Rolán, and R. M. Tsolis. 2007. Injection of flagellin into the host cell cytosol by Salmonella enterica serotype typhimurium. J. Biol. Chem. 282:33897-33901. [DOI] [PubMed] [Google Scholar]
- 35.Tsien, R. Y. 1998. The green fluorescent protein. Annu. Rev. Biochem. 67:509-544. [DOI] [PubMed] [Google Scholar]
- 36.Végh, B. M., P. Gál, J. Dobó, P. Závodszky, and F. Vonderviszt. 2006. Localization of the flagellum-specific secretion signal in Salmonella flagellin. Biochem. Biophys. Res. Commun. 345:93-98. [DOI] [PubMed] [Google Scholar]
- 37.Vogler, A. P., M. Homma, V. M. Irikura, and R. M. Macnab. 1991. Salmonella typhimurium mutants defective in flagellar filament regrowth and sequence similarity of FliI to F0F1, vacuolar, and archaebacterial ATPase subunits. J. Bacteriol. 173:3564-3572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vonderviszt, F., R. Ishima, K. Akasaka, and S.-I. Aizawa. (1992) Terminal disorder: a common structural feature of the axial proteins of bacterial flagellum? J. Mol. Biol. 226:575-579. [DOI] [PubMed] [Google Scholar]
- 39.Warren, S. M., and G. M. Young. 2005. An amino-terminal secretion signal is required for YplA export by the Ysa, Ysc, and flagellar type III secretion systems of Yersinia enterocolitica biovar 1B. J. Bacteriol. 187:6075-6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weber-Sparenberg, C., P. Pöplau, H. Brookman, M. Rochón, C. Möckel, M. Nietschke, and H. Jung. 2006. Characterization of the type III export signal of the flagellar hook scaffolding protein FlgD of Escherichia coli. Arch. Microbiol. 186:307-316. [DOI] [PubMed] [Google Scholar]
- 41.Widmaier, D. M., D. Tullman-Ercek, E. A. Mirsky, R. Hill, S. Govindarajan, J. Minshull, and C. A. Voigt. 2009. Engineering the Salmonella type III secretion system to export spider silk monomers. Mol. Syst. Biol. 5:309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yonekura, K., S. Maki-Yonekura, and K. Namba. 2003. Complete atomic model of the bacterial flagellar filament by electron cryomicroscopy. Nature 424:643-650. [DOI] [PubMed] [Google Scholar]