Abstract
Cyanobactins are small, cyclic peptides recently found in cyanobacteria. They are formed through proteolytic cleavage and posttranslational modification of short precursor proteins and exhibit antitumor, cytotoxic, or multi-drug-reversing activities. Using genome project data, bioinformatics, stable isotope labeling, and mass spectrometry, we discovered novel cyclic peptides, anacyclamides, in 27 Anabaena strains. The lengths of the anacylamides varied greatly, from 7 to 20 amino acids. Pronounced sequence variation was also detected, and only one amino acid, proline, was present in all anacyclamides. The anacyclamides identified included unmodified proteinogenic or prenylated amino acids. We identified an 11-kb gene cluster in the genome of Anabaena sp. 90, and heterologous expression in Escherichia coli confirmed that this cluster was responsible for anacyclamide production. The discovery of anacyclamides greatly increases the structural diversity of cyanobactins.
Cyanobacteria are a prolific source of secondary metabolites with potential as drug leads or useful probes for cell biology studies (23). They include biomedically interesting compounds, such as the anticancer drug lead cryptophycin (15), and environmentally problematic hepatotoxic peptides, such as microcystins and nodularins produced by bloom-forming cyanobacteria (23). Many of these compounds contain nonproteinogenic amino acids and modified peptides and are produced by nonribosomal peptide synthesis (23, 26).
The cyanobactins are a new group of cyclic peptides recently found in cyanobacteria (4). They are assembled through posttranslational proteolytic cleavage and head-to-tail macrocyclization of short precursor proteins. The cyanobactins are low-molecular-weight cyclic peptides that contain heterocyclized amino acids and can be prenylated or contain d-amino acids (3, 4). The cyanobactins that contain heterocyclized amino acids include patellamides, ulithiacyclamides, trichamide, tenuecyclamides, trunkamides, patellins, and microcyclamides and are synthesized in this manner (3, 4, 20, 24, 28). They possess antitumor, cytotoxic, and multi-drug-reversing activities and have potential as drug leads (4, 18, 20).
Cyanobactins containing heterocyclized amino acids are found in a variety of cyanobacteria (4). A recent study demonstrated that the cyanobactin biosynthetic pathway is prevalent in planktonic bloom-forming cyanobacteria (14). However, the products of these gene clusters encoding new cyanobactins are unknown. Here we report discovery of a novel family of low-molecular-weight cyanobactins and show that these compounds are common in strains of the genus Anabaena. These anacyclamides exhibit pronounced length and sequence variation and contain unmodified or prenylated amino acids.
MATERIALS AND METHODS
Cyanobacterial strains and cultivation.
The Anabaena strains were grown in Z8 (12) medium lacking a source of combined nitrogen, except for a few cultures for which nitrogen was included in the medium (Table 1). Strains were grown in 40-ml cultures under continuous light with a photon irradiance of 5 to 12 μmol m−2 s−1 at 20 to 25°C. Z8 medium containing 34S was used to detect sulfur-containing peptides. The MgSO4·7 H2O of Z8 medium was replaced by a stable isotope of MgSO4 (catalog no. IS7080; 90 atom% 34S; Icon).
TABLE 1.
Anacyclamides and their detection by LC-MS in Anabaena strainsa
| Anabaena strain | Origin | Amino acid sequence | Length (amino acids) | Unmodified peptide |
Modified peptide (one group) |
Modified peptide (two groups) |
Accession no. | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Designation | Retention time (min) | [M+H]+ | Designation | Retention time (min) | [M+H]+ | Designation | Retention time (min) | [M+H]+ | |||||
| 90b | Lake Vesijärvi, Finland | TSQIWGSPVP | 10 | A10 | 26.48 | 1053.5 | NDf | FJ461733 | |||||
| 0TU33S16b | Lake Tuusulanjärvi, Finland | TSQIWGSPVP | 10 | A10 | 26.44 | 1053.5 | ND | GU183825 | |||||
| 0TU39S15B | Lake Tuusulanjärvi, Finland | TSQIWGSPVP | 10 | A10 | 26.46 | 1053.5 | NAg | GU183826 | |||||
| SYKE971/6c | Lake Kotojärvi, Finland | TSQIWGSPVP | 10 | A10 | 26.44 | 1053.5 | ND | GU183827 | |||||
| 299Bb,c | Lake Vesijärvi, Finland | TSQIWGSPVP | 10 | A10 | 26.45 | 1053.5 | NA | GU183828 | |||||
| 1TU31S9 | Lake Tuusulanjärvi, Finland | SSVIWGSPVP | 10 | B10 | 30.20 | 1010.5 | NA | GU183829 | |||||
| SYKE763A | Lake Tuusulanjärvi, Finland | SSVIWGSPVP | 10 | B10 | 29.08 | 1010.5 | ND | GU183830 | |||||
| 299Ab | Lake Vesijärvi, Finland | SAQWQNFGVP | 10 | C10 | 27.51 | 1115.5 | ND | GU183831 | |||||
| SYKE748A | Lake Tuusulanjärvi, Finland | SAQWQNFGVP | 10 | C10 | 27.50 | 1115.5 | ND | GU183832 | |||||
| 1TU44S9 | Lake Tuusulanjärvi, Finland | NAHWQNFGVP | 10 | D10 | 23.17 | 1151.5 | ND | GU183833 | |||||
| PH256 | Lake Knud, Denmark | YAPLQNFGVPd | 10 | E10 | 33.05 | 1087.6 | E10P | 40.98 | 1155.6 | GU183834 | |||
| BIR260 | Baltic Sea, Gulf of Finland | DNWLGEWIGIP | 11 | A11 | 40.47 | 1281.6 | ND | GU183835 | |||||
| BIR250B | Baltic Sea, Gulf of Finland | DNWLGEWIGIP | 11 | A11 | 40.49 | 1281.6 | ND | GU183836 | |||||
| BIR257 | Baltic Sea, Gulf of Finland | DNWLGEWIGIP | 11 | A11 | 40.37 | 1281.6 | ND | GU183837 | |||||
| 37b | Lake Sääskjärvi, Finland | HAFIGYDQDPTGKYP | 15 | A15 | 19.48 | 1690.8 | A15G | 35.12 | 1826.9e | GU183838 | |||
| 141b | Lake Vesijärvi, Finland | HAFIGYDQDPTGKYP | 15 | ND | 1690.8 | A15G | 35.08 | 1826.9e | GU183839 | ||||
| 1TU39S8 | Lake Tuusulanjärvi, Finland | RERFVYP | 7 | A7 | 17.12 | 948.5 | ND | GU183840 | |||||
| 1TU32S11c | Lake Tuusulanjärvi, Finland | YSNKPSDFSP | 10 | ND | 1123.5 | F10P | 30.84 | 1191.6 | GU183841 | ||||
| 1TU33S10b | Lake Tuusulanjärvi, Finland | YDDKLNLSPd | 9 | ND | 1046.5 | A9P | 33.96 | 1114.6 | GU183842 | ||||
| 202A1/35b | Lake Vesijärvi, Finland | WGNGTGLDWKLLTGGISASP | 20 | ND | 2012.0 | A20P | 35.80 | 2081 | A20PP | 41.32 | 2149 | GU183843 | |
| 202A1b | Lake Vesijärvi, Finland | WGNGTGLDWKLLTGGISASP | 20 | ND | 2012.0 | A20P | 35.75 | 2081 | A20PP | 41.21 | 2149 | GU183844 | |
| SYKE844Cc | Lake Lohjanjärvi, Finland | WGNGTGLDWKLLTGGISASP | 20 | ND | 2012.0 | A20P | 35.79 | 2081 | A20PP | 41.24 | 2149 | GU183845 | |
| 1TU28S13b,c | Lake Tuusulanjärvi, Finland | WGNGTGLDWKLLTGGISASP | 20 | ND | 2012.0 | A20P | 35.76 | 2081 | A20PP | 41.27 | 2149 | GU183846 | |
| TR232 | Baltic Sea | HQPWHAAPd | 8 | ND | 925.4 | A8P | 23.57 | 993.5 | GU183847 | ||||
| SYKE816 | Baltic Sea, Gulf of Finland | FSPDWRAP | 8 | B8 | 26.93 | 957.5 | ND | GU183848 | |||||
| PH262 | Lake Knud, Denmark | VIQHYLFP | 8 | C8 | 32.35 | 998.5 | ND | GU183849 | |||||
| SYKE844B | Lake Lohjanjärvi, Finland | LIGIMHP | 7 | B7 | 30.56 | 762.4 | ND | GU183850 | |||||
The strains, their origins, and the predicted amino acid sequences and masses, as well as retention times, for the anacyclamides detected by LC-MS are shown. We detected prenylated (68-mass-unit increase) and geranylated (136-mass-unit increase) anacyclamides in 10 strains. The numbers of amino acids detected and derivatizations are indicated in the designations of the peptides (P, prenyl; G, geranyl). The five underlined peptides were validated by comparison to a synthetic peptide (retention time for A10, 26.46 min; retention time for C10, 27.54 min; retention time for A15, 19.48 min; retention time for A7, 17.04 min; retention time for B7, 30.53 min).
Axenic.
The Z8 medium included added nitrogen.
Bold type indicates a prenylated amino acid.
[M+H]+ indicates geranylation.
ND, not detected.
NA, not analyzed.
Annotation of the acy gene cluster.
We obtained the complete genome sequence of the toxic bloom-forming freshwater cyanobacterium Anabaena sp. 90. Open reading frame (ORF) predictions were made with Glimmer, and the acy gene cluster was annotated using Artemis (Sanger Institute). Functional analysis of proteins was performed using BLASTp, and the predictions for start sites of the genes were checked manually. A putative peptide precursor gene was identified using BLASTp searches and homology of the N-terminal sequence of the 49-amino-acid AcyE protein and the precursor proteins of patellamide, tenuecyclamide, trichamide, and microcyclamide (4, 20, 24, 28).
DNA extraction, amplification, and sequencing.
Genomic DNA was extracted from the strains of Anabaena using an E.Z.N.A. plant DNA mini kit (Omega Bio-Tek, Doraville, GA). We prescreened 74 Anabaena strains for the presence of cyanobactin biosynthetic genes using a PCR approach based on the conserved subtilisin-like protease gene, as previously described (14). In order to identify an Anabaena strain producing a cyanobactin containing a sulfur amino acid, the acyE peptide precursor gene was amplified using primers preRNAF (5′GAAGAACATCCGCCCCCAACAAGTTG3′) and preRNAR (5′CTCCGCGTCGTCGCCTGCAAAAGG-3′) and primers PreF (5′-GCCTTCACCAAACCAGTCTTCTTCAT-3′) and PreR (5′-CATCGAGGCGAACCGTGCGCCAAGGGAT-3′) from the genomic DNA of Anabaena strains that were selected based on the presence of a cyanobactin biosynthetic gene fragment. PCRs were performed as previously described (14). Sequencing was carried out with an ABI 3730xl automated sequencer (Applied Biosystems) by Macrogen (Seoul, Korea). Sequences were checked and edited by using the Chromas 2.2 program (Technelysium Pty. Ltd.) and were aligned by using the Bioedit sequence alignment editor.
Cloning the acy biosynthetic gene cluster.
The entire 11-kb acy gene cluster was amplified from genomic DNA of Anabaena sp. 90 by PCR using primers patex2f (5′-ATGGATCCTGATGGACTGTAGTGTGAG-3′) and patex5r (5′-TACTCGAGAGGTTTTGGGACTCTTTAG-3′) in three 60-μl reaction mixtures containing 1× PCR buffer for Super Taq Plus (HT Biotechnology Ltd.), 200 μmol of each nucleotide (Finnzymes), 0.75 μM of each primer, 0.8 U Super Taq Plus proofreading polymerase (HT Biotechnology Ltd.), and 100 ng of Anabaena sp. 90 template DNA. The thermocycling conditions were 94°C for 2 min, followed by 30 cycles of 94°C for 30 s, 56.4°C for 30 s, and 68°C for 9 min and then a final extension at 68°C for 20 min. The PCR products were separated on a 0.6% agarose gel containing 0.5× Tris-acetate-EDTA and run for 30 min at 100 V. The gel was stained using SYBR Safe DNA gel stain (Invitrogen) and was visualized using a Dark reader (Clare Chemical Research Inc.) to avoid DNA damage from UV light during gel extraction. The 11-kb PCR product was gel extracted with a MinElute gel extraction kit (Qiagen) and cloned into the pCR 2.1-TOPO vector using a TOPO TA cloning kit (Invitrogen) with an insert-to-vector molar ratio of 3:1. The vector was used to transform chemically competent Escherichia coli One Shot TOP10 cells according to the manufacturer's instructions. The resultant pNL1901 and pNL1902 plasmids were analyzed by PCR and restriction digestion to ensure that the integrity of the insert was maintained during the cloning and amplification in E. coli. The transformants containing the 11-kb insert in the plasmid were grown overnight with shaking at 120 rpm at 28°C in 50 ml of LB medium containing 100 μg ml−1 of ampicillin (sodium salt; Sigma-Aldrich) for liquid chromatography (LC)-mass spectrometry (MS) analysis. The cells were harvested by centrifugation at 10,000 × g for 10 min (Eppendorf 5804R centrifuge) and dried with a Heto vacuum centrifuge (Heto-Holten A/S), which yielded ca. 43 mg (dry weight).
Chemical analysis.
Cells of Anabaena strains were collected from the 40-ml cultures by centrifugation at 7,000 × g for 7 min. The collected cells were freeze-dried with Supermodulyo (Edwards High Vacuum International) or dried with a Heto vacuum centrifuge, which yielded 5 to 12 mg (dry weight). The dried Anabaena cells, as well as E. coli transformants, were extracted with 1 ml of methanol (HiperSolv for high-performance liquid chromatography [HPLC]; BDH Laboratory Supplies) in 2-ml plastic tubes containing glass beads (cell disruption medium; 0.5-mm-diameter glass beads; Scientific Industries Inc.). Each mixture was homogenized by shaking it with a FastPrep cell disrupter (Bio 101, Thermo Electron Corporation, Qbiogene, Inc.) for 30 s at a speed of 5 m s−1. The mixture was centrifuged at 10,000 × g for 5 min, and the supernatant was used for LC-MS analysis.
The methanol extracts were analyzed with a high-performance liquid chromatograph combined with a mass spectrometer (Agilent 1100 series LC/MSD with Ion Trap XCT Plus and an electrospray ion source) in order to detect low-molecular-weight peptides. Peptides were separated from the extracts by HPLC using a Phenomenex C18 (2) column (2.0 mm by 150 mm; particle size, 5 μm). The mobile phase gradient consisted of 0.1% aqueous (water purified with Milli-Q Plus; Millipore) formic acid (50% solution in water; Fluka, Sigma-Aldrich) (solvent A) and 0.1% formic acid in isopropyl alcohol (Sigma-Aldrich) (solvent B). Two different settings were used; one setting was used for screening methanolic extracts of Anabaena cells, and the other setting (values in parentheses below) was used for further structural characterization of natural and synthetic peptides. The percentage of solvent B was increased from 5 to 50% in 60 min. A flow rate of 0.15 ml min−1 was used, and the columns were heated to 40°C during separation. The positive-ion mode of electrospray ionization was used. The pressure of the nebulizer gas (N2) was 30 lb/in2 (35 lb/in2), the drying gas flow rate was 8 liters min−1, and the temperature was 350°C. The capillary voltage was set at 5,000 V (4,500 V), and the capillary offset value was 300 V. A skimmer potential of 85 V (100 V) and a trap drive value of 144 (111) were used. Spectra were recorded using a scan range from m/z 100 to m/z 2,200. Identification of the anacyclamides was based on the molecular weights calculated from the predicted peptide AcyE precursor amino acid sequences and the assigned fragment ion patterns of MSn (n = 1 to 3) spectra. A comparison with the retention time and MS data for a synthetic reference was used in five cases (Beijing SBS Genetech Co., Ltd., China; anacyclamide B7 from JPT Peptide Technologies GmbH, Germany). The level of biosynthetic anacyclamide A10 in Anabaena sp. 90 was determined by LC-MS using synthetic peptide that was >98% pure and the m/z 1053.5 (MH+) signal.
Nucleotide sequence accession numbers.
The sequence of the acy gene cluster has been deposited in the GenBank database under accession number FJ461733. All acyE gene sequences have been deposited in the GenBank database under accession numbers GU183825 to GU183850.
RESULTS
Characterization of acy genes in Anabaena sp. 90.
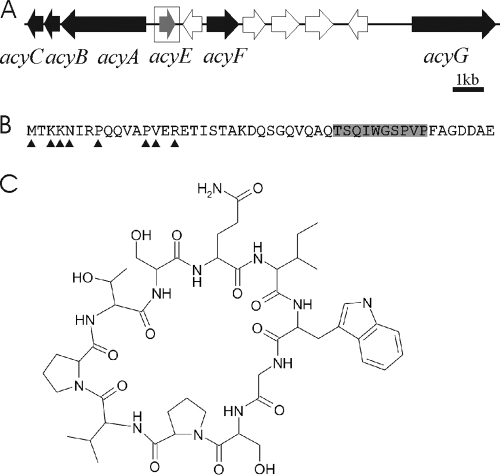
We identified an 11-kb gene cluster encoding two subtilisin-like proteases during preliminary assembly and annotation of the Anabaena sp. 90 genome. The 11 genes in the acy gene cluster are organized in two operons with a putative bidirectional promoter (Fig. 1A and Table 2). The acy gene cluster is bordered by genes encoding a signal recognition particle GTPase and ribosomal protein S16. AcyA and AcyG have homology to the proteases involved in the biosynthesis of patellamides, trunkamide, trichamide, tenuecyclamide, patellin, and microcyclamides (Table 2). AcyE is a 49-amino-acid protein with N-terminal homology to the peptide precursor proteins in the other cyanobactin pathways (Fig. 2). Eight amino acids are conserved in the N-terminal leader sequence in all eight precursor proteins (Fig. 1B and 2). In addition, AcyB, AcyC, and AcyF are homologous to proteins that are encoded by almost all cyanobactin gene clusters but have unknown functions (Table 2). There are five ORFs between acyE and acyG that encode hypothetical and conserved proteins. It is not clear if these ORFs encode proteins that participate in the biosynthesis of an anacyclamide. Interestingly, the acy gene cluster encoded no homologs of the PatD, McaD, TriA, and TenD proteins, which are postulated to catalyze the heterocyclization of serine, threonine, and cysteine to thiazolines and oxazolines. Furthermore, the AcyG protein lacks the oxidase domains of PatG homologs, which are hypothesized to catalyze the oxidation of thiazolines and oxazolines to thiazoles and oxazoles in cyanobactin biosynthesis.
FIG. 1.
Anacyclamide biosynthetic pathway in Anabaena sp. 90. (A) acy gene cluster from Anabaena sp. 90. The genes indicated by black and gray arrows have homology to genes present in other cyanobactin gene clusters. The acyE gene encoding the precursor peptide is indicated by a gray arrow and is enclosed in a box. The genes indicated by open arrows encode conserved and hypothetical proteins without known or predicted functions. (B) AcyE peptide precursor from Anabaena sp. 90 with the hypervariable region of the 49-amino-acid protein encoding the mature anacyclamide shaded, indicating the position of cleavage and macrocyclization. The eight conserved amino acids in the AcyE leader peptide and other known cyanobactin precursor leader peptides are indicated arrowheads. (C) Structure of the decapeptide anacyclamide A10 from Anabaena sp. 90.
TABLE 2.
Functional assignment of the predicted proteins encoded by the acy gene cluster of Anabaena sp. 90 (accession no. FJ461733) and homology with proteins encoded by the microcyclamide (accession no. AM774406 and AM931579), trichamide (accession no. CP000393), tenuecyclamide (accession no. EU290741), patellamide (accession no. AY986476), and patellin (accession no. EU290742) biosynthetic gene clusters
|
Anabaena sp. 90 |
Proteins encoded by cyanobactin biosynthetic gene clusters in other cyanobacteria |
||||||
|---|---|---|---|---|---|---|---|
| Protein | Length (amino acids) | Predicted function | Microcyclamide | Trichamide | Tenuecyclamide | Patellamide | Patellin |
| AcyC | 62 | Unknown | McaC | TenC | PatC | TruC | |
| AcyB | 67 | Unknown | McaB | TriJ | TenB | PatB | TruB |
| AcyA | 658 | Subtilisin-like protease | McaA | TriH | TenA | PatA | TruA |
| AcyE | 49 | Precursor protein | McaE | TriG | TenE | PatE | TruE |
| ORF5 | 277 | Transposase-like protein | TriE | ||||
| AcyF | 296 | Unknown | McaF | TenF | PatF | TruF1/TruF2 | |
| ORF7 | 131 | Unknown | |||||
| ORF8 | 227 | Unknown | |||||
| ORF9 | 47 | Unknown | |||||
| ORF10 | 201 | Unknown | |||||
| AcyG | 710 | Subtilisin-like protease | McaG | TriK | TenG | PatG | TruG |
FIG. 2.
Peptide precursor proteins from Nostoc spongiaeforme var. tenue (TenE), Lyngbya aesturarii PCC 8106 (LynE) (4), Prochloron (PatE) (20), Microcystis aeruginosa PCC7806 (McaE1), Microcystis aeruginosa NIES 298 (McaE2) (28), Prochloron (TruE) (4), Trichodesmium erythraeum IMS 101 (TriG) (24), and Anabaena sp. 90 (AcyE). The homologous amino acids are indicated by asterisks. The areas of the precursor protein which form the mature cyanobactin are shaded.
AcyD peptide precursor diversity in strains of Anabaena.
The absence of the conserved cleavage sites in the AcyE precursor protein in Anabaena sp. 90 made direct prediction of the peptide product of the acy gene cluster impossible (Fig. 2). We used a novel approach to identify the product of the acy gene cluster. Anabaena strains were screened using PCR primers specific for the acyA protease gene in order to identify which Anabaena strains might produce cyanobactins. Subsequently, the acyE gene, encoding the precursor protein, was amplified from these Anabaena strains and sequenced (Table 1). The entire acyE gene was successfully amplified from 26 strains and sequenced. The N-terminal leader sequences and the 8 amino acids at the C terminus were very similar in all peptide precursors (data not shown). We identified a 7- to 20-amino-acid region that exhibited pronounced sequence variation between the conserved N- and C-terminal regions of the AcyE precursor protein. We postulated that the mature cyanobactin is formed from this region of the precursor peptide. A single strain (Anabaena SYKE 844B) was identified in which methionine, a sulfur-containing amino acid, was encoded in this hypervariable region of the acyE gene. We hypothesized that this strain would produce a cyanobactin that could be detected using 34S stable isotope labeling without knowing the exact amino acid composition of the peptide.
LC-MS identification of anacyclamides.
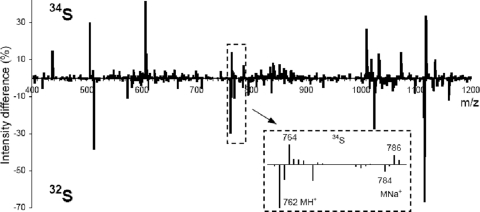
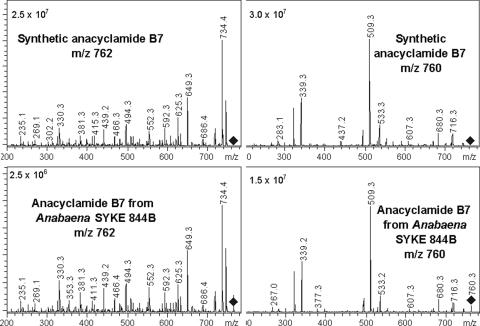
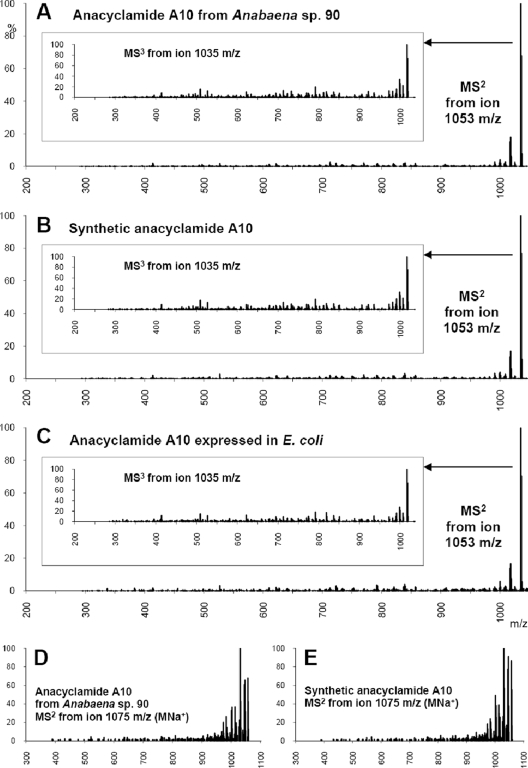
In order to test this hypothesis, Anabaena sp. SYKE 844B was cultivated in medium containing either inorganic 34S or 32S as the principal source of sulfur. Cell extracts from both cultures were analyzed by LC-MS, and the sum mass spectra, containing all of the ions of the chromatogram, were compared (Fig. 3). We detected shifts of 2 mass units in mass spectra of the labeled (34S) and unlabeled (32S) extracts. A 2-mass-unit shift corresponding to one sulfur atom and one methionine was found. The m/z 762 (MH+) and 784 (MNa+) ions in the unlabeled extract were shifted to the m/z 764 and 786 ions in the 34S-labeled extract. The MS2 spectra of the m/z 762 and 764 ions were identical, with the exception of a shift originating from the 34S atom. An ion mass of m/z 762 corresponds to a protonated anacyclamide B7 peptide (Fig. 3 and Table 1). This was confirmed by comparison to synthetic anacyclamide B7. The synthetic and biosynthetic peptides had the same retention time (Fig. 4), and the product ion spectra for positive (m/z 762) and negative (m/z 760) precursor ions of the peptides were identical (Fig. 5). Identification of the product of the acy gene cluster from Anabaena sp. SYKE 844B made it possible to predict the sites of proteolytic cleavage in all AcyE peptide precursors and calculate a predicted mass based on assumptions concerning cyclization and the amino acid content. We predicted that the product of the Anabaena sp. 90 acy gene cluster is a decapeptide with a monoisotopic molecular mass of 1,052 mass units, anacyclamide A10 (Fig. 1C). The corresponding m/z 1053 (MH+) and m/z 1075 (MNa+) ions were detected in Anabaena sp. 90 cell extracts by LC-MS. The biosynthetic and synthetic protonated anacyclamide A10 (MH+ and MH+-H2O) produced very similar spectra when tandem mass spectrometry was used (Fig. 6). The biosynthetic and chemically synthesized anacyclamides had the same retention time, indicating that all amino acids were in the l configuration.
FIG. 3.
Shift in mass detected from comparison of mass spectra for extracts of Anabaena sp. strain SYKE 844B cultivated on normal medium (32S) (spectrum below the line) and on medium containing 34S (spectrum above the line), presented as mirror images. The m/z 764 peak is the protonated ion from 34S-labeled anacyclamide B7, and the m/z 786 peak is its Na adduct; the corresponding 32S ions are the m/z 762 and 784 ions, respectively.
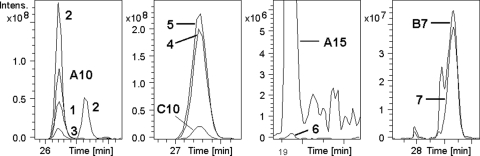
FIG. 4.
Ion chromatograms for the protonated biosynthetic and synthetic reference anacyclamides A10, C10, A15, and B7. Ion masses and the anacyclamides are shown in Table 1. Chromatogram 1, Anabaena strain 90; chromatogram 2, Anabaena strain 0TU33S16; chromatogram 3, Anabaena strain SYKE971/6; chromatogram 4, Anabaena strain 299A; chromatogram 5, Anabaena strain SYKE748A; chromatogram 6, Anabaena strain 37; chromatogram 7, Anabaena strain SYKE 844B. The peak eluting later in the ion chromatogram of strain 0TU33S16 originated from a cyclic peptide belonging to another peptide family.
FIG. 5.
Comparison of the product ion spectra for the m/z 762.4 (MH+) and m/z 760.4 (M-H−) molecular ions of synthetic and biosynthetic (Anabaena SYKE 844B) anacyclamide B7.
FIG. 6.
Comparison of biosynthetic anacyclamide A10 of Anabaena sp. 90 to a synthetic standard and anacyclamide A10 produced in E. coli. (A to C) Mass spectra of (A) protonated biosynthetic anacyclamide produced by Anabaena sp. 90, (B) synthetic anacyclamide, and (C) biosynthetic anacyclamide produced by E. coli(pNLA90). (D and E) Mass spectra of (D) sodiated anacyclamide A10 from Anabaena sp. 90 and (E) a synthetic standard.
The predicted anacyclamides were detected in cell extracts of the Anabaena strains studied (Table 1). Altogether, 18 anacyclamides were unique (Table 1). Three more synthetic anacyclamides (anacyclamides C10, A15, and A7) were analyzed to ensure that the structural interpretation was correct. The retention times of the biosynthetic and synthetic anacyclamides were the same (Fig. 4), and the product ion spectra of MH+ were practically identical (data not shown). Two arginine residues in anacyclamide A7 induced the formation of an intense doubly charged M+2H2+ ion and a highly characteristic product ion spectrum, which unequivocally identified the structure.
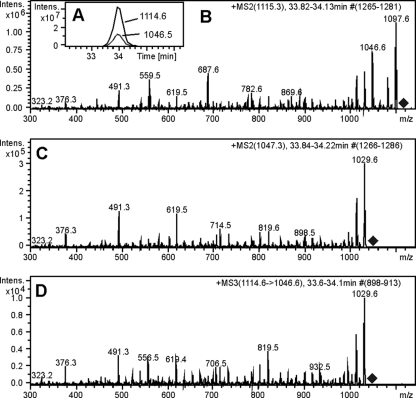
At first we were able to detect 11 of the 18 different types of predicted anacyclamides in the strains of Anabaena (Table 2). Heterocyclized variants of anacyclamides were not detected in any of the strains, suggesting that the products of the acy gene cluster are unmodified cyclic peptides. However, it became apparent that seven of the anayclamides were produced in a derivatized form. Anacyclamides F10P, A9P, A20P, and A8P with masses that were 68 mass units greater than the masses predicted from the AcyE precursor peptide sequence were detected, while anacyclamide A20PP with a mass that was 136 mass units (2 × 68 mass units) greater than the predicted mass was also detected. Unmodified anacyclamide was not produced together with the derivatized peptide. Anacyclamide E10P was 68 mass units larger than predicted, and anacyclamide A15G was 136 mass units larger than predicted. Loss of 68 mass units was not observed for anacyclamide A15G. The unmodified anacyclamides eluted earlier than the derivatized anacyclamides, which demonstrated the hydrophobic nature of the structural units (R) with masses of 68 and 136 mass units (Table 1). Another characteristic describing these groups is the weak bonds to the peptidic structure in anacyclamides E10P, A15G, F10P, A9P, and A8P as the groups were partially lost in the ion source. Coelution of the M+H+ and M-R+H+ ions showed that M-R+H+ is probably formed from M+H+ via loss of 68 mass units during ionization in the ion source (Fig. 7A). The product ion spectrum for the M+H+ ion contained an intense M-R+H+ ion peak, and this is evidence that the M-R+H+ ion present in the ion source originated from the M+H+ ion (Fig. 7B). Finally, the evidence indicating that the structure of the M-R+H+ ion present in the ion source and the structure of the M-R+H+ ion formed via fragmentation in the trap from the M+H+ ion are identical is that the two M-R+H+ ions produced nearly identical product ion spectra (Fig. 7C and D). Data obtained for other derivatized anacyclamides unambiguously proved that anacyclamides E10P, A15G, F10P, A9P, and A8P contained loosely bound groups with masses of 68 or 136 mass units (Fig. 7). Anacyclamides A20P and A20PP lost the 68-mass-unit groups only when they were fragmented in the ion trap. Detailed analysis of MS2 and MS3 fragmentation patterns suggested that the 68-mass-unit group is located in tyrosine or tryptophan in anacyclamides E10P, A9P, and A8P (Table 1).
FIG. 7.
Chromatographic and mass spectrometric data showing the presence of a structural unit with a mass of 68 mass units in anacyclamide A9P. (A) Ion chromatograms of m/z 1114.6 and m/z 1046.5 ions. (B) Product ion spectrum for protonated derivatized anacyclamide A9P m/z 1114.6 ion. (C and D) Product ion spectra for the m/z 1046.5 protonated ion formed (C) in the ion source and (D) in the ion trap.
Heterologous expression.
Cyanobacteria are difficult to manipulate genetically, and Anabaena sp. 90 is recalcitrant to genetic manipulation. In order to demonstrate that the acy gene cluster is responsible for the production of anacylamide, we amplified the 11-kb acy gene cluster by PCR, cloned the entire cluster into the pCR2.1 plasmid, and used the plasmid to transform E. coli TOP10. Clones containing the acy gene cluster in both orientations (pNLA901 and pNLA902) were identified by restriction analysis and used for heterologous expression of anacyclamide. We cultivated E. coli TOP10(pNLA901) and E. coli TOP10(pNLA902) overnight at 28°C in 50 ml of LB medium containing 100 μg ml−1 ampicillin. We detected the predicted anacyclamide A10 in cell extracts of E. coli TOP10 carrying either plasmid. The ion masses of anacyclamide A10 in Anabaena sp. 90 were m/z 1053 (MH+) and m/z 1075 (MNa+), and the corresponding ions were found in E. coli TOP10(pNLA901) and E. coli TOP10(pNLA902) but not in the negative control (Fig. 6). The retention time of anacyclamide A10 produced in E. coli was also identical (±0.04 min) to the retention time of the synthetic and biosynthetic peptides from Anabaena sp. 90. The successful heterologous expression of anacyclamide A10 in E. coli confirmed that the acy genes are responsible for anacyclamide biosynthesis. The amount of anacyclamide A10 produced by Anabaena sp. 90 was 100 ng mg−1 (dry weight) of biomass, and the amount produced by E. coli TOP10(pNLA901) 5 ng mg−1 (dry weight) of biomass.
DISCUSSION
Cyanobactins are recently described low-molecular-weight cyclic peptides that contain heterocyclized amino acids and are formed through proteolytic cleavage of short precursor proteins (4). Some cyanobactins also contain disulfide bridges, prenylated amino acids, or d-amino acids (3, 4). Here we describe the discovery of anacyclamides, a new family of cyanobactins produced by strains of the genus Anabaena. The cyclic anacyclamides are composed of 7 to 20 amino acids, and the sequence variation is so pronounced that just a single conserved amino acid is present in all anacyclamides (Table 1). However, we found no evidence that anacyclamides contain heterocyclized amino acids. In keeping with this, we did not find genes encoding the proteins predicted to catalyze heterocyclization or oxidation of the heterocycles in the acy gene cluster of Anabaena sp. 90 (Table 2).
We found anacyclamides comprised of just unmodified proteinogenic amino acids and also a number of anacyclamides comprised of amino acids derivatized with structural units having masses of 68 or 136 (2 × 68) mass units. Mass spectrometric and chromatographic elution data presented here suggest that these structural units are prenyl (C5H8) and geranyl (C10H16) groups attached to the side chain heteroatom of the amino acid. Neutral loss of 68 mass units is characteristic of O-prenyl-substituted flavonoids (8), as detected for derivatized anacyclamides E10P, A9P, F10P, A20P, A20PP, and A8P. O-Prenyl-, N-prenyl-, and N-geranyl-substituted peptides have been reported for cyanobacteria and other marine organisms. Myriastramides and prenylagaramides contain O-prenylated tyrosine (6, 16), mollamides contain O-prenylated serine or threonine (5), cyclomarins contain N-prenylated tryptophan (21), microguanidine contains N-geranylated arginine (9), and aeruginoguanidines contain N-prenylated and geranylated arginine (10). Our findings broaden the known structural variation of cyanobactins to include unmodified and prenylated homodetic cyclic peptides lacking heterocyclized amino acids and exhibiting pronounced length and sequence variation.
The pronounced chemical diversity of anacyclamides that we observed is achieved through hypervariation at the nucleotide level in the region of the acyE gene encoding the mature peptide. At present it is not clear how the length and sequence variation is achieved at the genetic level in nature. The hypervariation suggests that there is strong selection for rapid evolution, maintenance, and diversification of anacyclamides in Anabaena. The biosynthetic machinery for assembling cyanobactins resembles that of bacteriocin protein toxins found in a wide range of bacteria (13). It is not clear why Anabaena produces anacyclamides, but, analogous to bacteriocins, they may be produced for chemical warfare (1). The anacyclamides might participate in interstrain competition or act as antimicrobial agents against bacteria. Anabaena produces a range of toxins and enzyme inhibitors widely believed to act as grazing deterrents (23).
Cyanobacteria are a rich source of low-molecular-weight peptides with a broad range of structural variations. Strains of the cyanobacterial genus Planktothrix produce cyclic peptides containing only unmodified proteinogenic amino acids, including planktocyclin (2), oscillapeptin (7), and agardhipeptin (22). Strains of the genus Planktothrix also produce prenylated cyclic peptides called prenylagaramides (16). Microcystis aeruginosa produces unmodified and prenylated versions of the undecapeptide kawaguchipeptin that exhibit antimicrobial activity (11). Interestingly, all of these compounds contain a proline residue (2, 7, 11, 16, 22). Anacyclamides exhibit considerable sequence variation, in contrast to other cyanobactins, and the terminal proline is the only amino acid conserved in all anacyclamides (Table 1). The presence of proline may be necessary for cleavage and/or macrocyclization of the anacyclamide precursor peptide by the AcyA and AcyG proteases. In an analogous situation the presence of a conserved asparagine residue is necessary for both cleavage and macrocyclization of the precursor in plant cyclopetide biosynthesis (19). The biosynthetic origins of the unmodified and prenylated cyclic peptides produced by Planktothrix and Microcystis are unclear, but the chemical similarities suggest that such peptides may be synthesized in a fashion similar to anacyclamides.
Low-molecular-weight cyclic peptides consisting of unmodified proteinogenic amino acids have been found in a broad range of organisms, including cyanobacteria (2, 22) and plants (25). Unmodified cyclic peptides have a range of interesting pharmaceutical activities, including antimicrobial, antiplatelet, antimalarial, immunosuppressive, antileukemic, and enzyme-inhibiting activities (2, 17, 22, 25). Thus, it has been established that cyclic peptides such as anacyclamides can exhibit interesting bioactivities and warrant further investigation. Research on the chemical diversity and biosynthetic potential of anacyclamides may lead to useful biotechnological and industrial applications. Cyanobacteria are notoriously difficult to manipulate, and strains of the bloom-forming genus Anabaena are recalcitrant to genetic manipulation. To date there have been just a few examples of expression of a cyanobacterial metabolite in a heterologous host (3, 4, 20, 27). The heterologous expression of the anacyclamide gene cluster in an E. coli surrogate host opens the possibility of determining the role of each gene in the acy gene cluster in the assembly of the cyclic peptide, as well as the production of a library of recombinant cyclic anacyclamides.
In conclusion, we detected 18 anacyclamide variants in 27 strains of Anabaena originating from different habitats using mass spectrometry. The anacyclamides identified contained unmodified proteinogenic or prenylated amino acids. In contrast to previously described cyanobactins, the length of the anacyclamides varied greatly, from 7 to 20 amino acids, and there was pronounced sequence diversity with conservation of just a single amino acid, proline. Such great length variation or sequence diversity of cyanobactins has not been reported previously for cyanobacteria. In addition, heterologous expression of the gene cluster in E. coli provides further avenues for detailed characterization of the machinery, as well as its utilization in production of novel compounds belonging to this biomedically interesting class of cyclic peptides. This work also emphasizes the power of genome mining, culture collections, and labeling experiments, as well as a combination of molecular and chemical tools, for discovering new families of bioactive compounds.
Acknowledgments
We are grateful to Lyudmila Saari for her valuable help in handling the cultures. We thank Hao Wang, who kindly assisted with annotation of the acy gene cluster from Anabaena sp. 90. We also thank Jarkko Rapala for providing the SYKE strains used in this study.
This work was supported by grants from the Academy of Finland to D.P.F. (grant 1212943) and to K.S. (Research Center of Excellence grant 53305, grant 118637, and Academy Professors grant 214457). N.L. is a student at Viikki Graduate School in Biosciences.
Footnotes
Published ahead of print on 11 December 2009.
REFERENCES
- 1.Baba, T., and O. Schneewind. 1998. Instruments of microbial warfare: bacteriocin synthesis, toxicity and immunity. Trends Microbiol. 6:66-71. [DOI] [PubMed] [Google Scholar]
- 2.Baumann, H. I., S. Keller, F. E. Wolter, G. J. Nicholson, G. Jung, R. D. Susmuth, and F. Jüttner. 2007. Planktocyclin, a cyclooctapeptide protease inhibitor produced by the freshwater cyanobacterium Planktothrix rubescens. J. Nat. Prod. 70:1611-1615. [DOI] [PubMed] [Google Scholar]
- 3.Donia, M. S., B. J. Hathaway, S. Sudek, M. G. Haygood, M. J. Rosovitz, J. Ravel, and E. W. Schmidt. 2006. Natural combinatorial peptide libraries in cyanobacterial symbionts of marine ascidians. Nat. Chem. Biol. 2:729-735. [DOI] [PubMed] [Google Scholar]
- 4.Donia, M. S., J. Ravel, and E. W. Schmidt. 2008. A global assembly line for cyanobactins, Nat. Chem. Biol. 4:341-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Donia, M. S., B. Wang, D. C. Dunbar, P. V. Desai, A. Patny, M. Avery, and M. T. Hamann. 2008. Mollamides B and C, cyclic hexapeptides from the Indonesian tunicate Didemnum molle. J. Nat. Prod. 71:941-945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Erickson, K. L., K. R. Gustafson, D. J. Milanowski, L. K. Pannell, J. R. Klose, and M. R. Boyd. 2003. Myriastramides A-C, new modified cyclic peptides from the Philippines marine sponge Myriastra clavosa. Tetrahedron 59:10231-10238. [Google Scholar]
- 7.Fujii, K., K. Sivonen, E. Naganawa, and K.-I. Harada. 2000. Non-toxic peptides from toxic cyanobacteria, Oscillatoria agardhii. Tetrahedron 56:725-733. [Google Scholar]
- 8.Garo, E., J.-L. Wolfender, K. Hostettmann, W. Hiller, S. Antus, and S. Mavi. 1998. Prenylated flavanones from Monotes engleri: on-line structure elucidation by LC/UV/NMR. Helv. Chim. Acta 81:754-763. [Google Scholar]
- 9.Gesner-Apter, S., and S. Carmeli. 2008. Three novel metabolites from bloom of the cyanobacterium Microcystis sp. Tetrahedron 64:6628-6634. [Google Scholar]
- 10.Ishida, K., H. Matsuda, Y. Okita, and M. Murakami. 2002. Aeruginoguanidines 98-A-98-C: cytotoxic unusual peptides from the cyanobacterium Microcystis aeruginosa. Tetrahedron 58:7645-7652. [Google Scholar]
- 11.Ishida, K., H. Matsuda, M. Murakami, and K. Yamaguchi. 1997. Kawaguchipeptin B, an antibacterial cyclic undecapeptide from the cyanobacterium Microcystis aeruginosa. J. Nat. Prod. 60:724-726. [DOI] [PubMed] [Google Scholar]
- 12.Kotai, J. 1972. Instructions for preparation of modified nutrient solution Z8 for algae. Norwegian Institute for Water Research, Oslo, Norway.
- 13.Lee, S. W., D. A. Mitchell, A. L. Markley, M. E. Hensler, D. Gonzalez, A. Wohlrab, P. C. Dorrestein, V. Nizet, and J. E. Dixon. 2008. Discovery of a widely distributed toxin biosynthetic gene cluster. Proc. Natl. Acad. Sci. U. S. A. 105:5879-5884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leikoski, N., D. P. Fewer, and K. Sivonen. 2009. Widespread occurrence and lateral transfer of the cyanobactin biosynthetic gene cluster in cyanobacteria. Appl. Environ. Microbiol. 75:853-857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Magarvey, N. A., Z. Q. Beck, T. Golakoti, Y. Ding, U. Huber, T. K. Hemscheidt, D. Abelson, R. E. Moore, and D. H. Sherman. 2006. Biosynthetic characterization and chemoenzymatic assembly of the cryptophycins. Potent anticancer agents from cyanobionts. ACS Chem. Biol. 1:766-779. [DOI] [PubMed] [Google Scholar]
- 16.Murakami, M., Y. Itou, K. Ishida, and H. J. Shin. 1999. Prenylagaramides A and B, new cyclic peptides from two strains of Oscillatoria agardhii. J. Nat. Prod. 62:752-755. [DOI] [PubMed] [Google Scholar]
- 17.Pomilio, A. B., M. E. Battista, and A. A. Vitale. 2006. Naturally-occurring cyclopeptides: structures and bioactivity. Curr. Org. Chem. 10:2075-2121. [Google Scholar]
- 18.Salvatella, X., J. M. Caba, F. Albericio, and E. Giralt. 2003. Solution structure of the antitumor candidate trunkamide A by 2D NMR and restrained simulated annealing methods. J. Org. Chem. 68:211-215. [DOI] [PubMed] [Google Scholar]
- 19.Saska, I., A. D. Gillon, N. Hatsugai, R. G. Dietzgen, I. Hara-Nishimura, M. A. Anderson, and D. J. Craik. 2007. An asparaginyl endopeptidase mediates in vivo protein backbone cyclization. J. Biol. Chem. 282:29721-29728. [DOI] [PubMed] [Google Scholar]
- 20.Schmidt, E. W., J. T. Nelson, D. A. Rasko, S. Sudek, J. A. Eisen, M. G. Haygood, and J. Ravel. 2005. Patellamide A and C biosynthesis by a microcin-like pathway in Prochloron didemni, the cyanobacterial symbiont of Lissoclinum patella. Proc. Natl. Acad. Sci. U. S. A. 102:7315-7320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schultz, A. W., D.-C. Oh, J. R. Carney, R. T. Williamson, D. W. Udwary, P. R. Jensen, S. J. Gould, W. Fenical, and B. S. Moore. 2008. Biosynthesis and structures of cyclomarins and cyclomarazines, prenylated cyclic peptides of marine actinobacterial origin. J. Am. Chem. Soc. 130:4507-4516. [DOI] [PubMed] [Google Scholar]
- 22.Shin, H. J., H. Matsuda, M. Murakami, and K. Yamaguchi. 1996. Agardhipeptins A and B, two new cyclic hepta- and octapeptides, from the cyanobacterium Oscillatoria agardhii (NIES-204). Tetrahedron 52:13129-13136. [Google Scholar]
- 23.Sivonen, K., and T. Börner. 2008. Bioactive compounds produced by cyanobacteria, p. 159-197. In A. Herraro and E. Flores (ed.), The cyanobacteria: molecular biology, genomics and evolution. Caister Academic Press, Norfolk, United Kingdom.
- 24.Sudek, S., M. G. Haygood, D. T. A. Youssef, and E. W. Schmidt. 2006. Structure of trichamide, a cyclic peptide from the bloom-forming cyanobacterium Trichodesmium erythraeum, predicted from the genome sequence. Appl. Environ. Microbiol. 72:4382-4387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tan, N. H., and J. Zhou. 2006. Plant cyclopeptides. Chem. Rev. 106:840-895. [DOI] [PubMed] [Google Scholar]
- 26.Welker, M., and H. von Döhren. 2006. Cyanobacterial peptides—nature's own combinatorial biosynthesis. FEMS Microbiol. Rev. 30:530-563. [DOI] [PubMed] [Google Scholar]
- 27.Ziemert, N., K. Ishida, A. Liaimer, C. Hertweck, and E. Dittmann. 2008. Ribosomal synthesis of tricyclic depsipeptides in bloom-forming cyanobacteria. Angew. Chem. Int. Ed. Engl. 47:7756-7759. [DOI] [PubMed] [Google Scholar]
- 28.Ziemert, N., K. Ishida, P. Quillardet, C. Bouchier, C. Hertweck, N. Tandeau de Marsac, and E. Dittmann. 2008. Microcyclamide biosynthesis in two strains of Microcystis aeruginosa: from structure to genes and vice versa. Appl. Environ. Microbiol. 74:1791-1797. [DOI] [PMC free article] [PubMed] [Google Scholar]