Abstract
Natural noncoding small RNAs have been shown to be involved in a number of cellular processes as regulators. Using the mechanisms thus elucidated, artificial small interfering RNAs (siRNAs), ribozymes, and RNA aptamers are also expected to be potential candidates for RNA therapeutic agents. However, current techniques are too costly for industrial production of these RNAs for use as drugs. Here, we propose a new method for in vivo production of artificial RNAs using the marine phototrophic bacterium Rhodovulum sulfidophilum. Using engineered plasmids and this bacterium, which produces extracellular nucleic acids in nature, we developed a method for extracellular production of a streptavidin RNA aptamer. As the bacterium does not produce any RNases in the culture medium, at least within the cultivation period tested, the designed RNA itself is produced and retained in the culture medium of the bacterium without any specific mechanism for protection against degradation by nucleases. Here, we report that the streptavidin RNA aptamer is produced in the culture medium and retains its specific function. This is the first demonstration of extracellular production of a functional artificial RNA in vivo, which will pave the way for inexpensive production of RNA drugs.
Recent studies have indicated that many small RNAs play key roles in the regulation of gene expression and that higher-order structures in RNA sequences, such as riboswitches or ribozymes, act as regulators of mRNA expression (3, 4, 17). In addition to these natural RNA functions, artificial small interfering RNAs (siRNAs), ribozymes, and RNA aptamers are also expected to be potential candidates for RNA therapeutics (10, 20). An RNA aptamer has already been developed as an RNA drug for the inhibition of macular degradation by specifically targeting the vascular endothelial growth factor (18). In both basic studies of RNA and RNA drug production, efficient methods for preparation of homogeneous RNA molecules are very important. In the case of RNA drug production, economically efficient methods for large-scale production are required.
At present, the most reliable methods for preparation of homogeneous RNAs are in vitro transcription using T7 RNA polymerase (15) and chemical synthesis (12). These methods, however, are not suitable for preparation in large quantities because they are both costly and labor intensive. For industrial production of RNAs, in vivo production using microorganisms is thought to be the most suitable method. Recently, Ponchon and Dardel reported in vivo production of recombinant RNAs using Escherichia coli (19). They proposed a system called a “tRNA scaffold” in which the product RNA is designed to be included in a tRNA structure to obtain a homogeneous RNA product. Their product RNA is produced efficiently in homogeneous form, because the in vivo transcript containing the product sequence is processed as a tRNA by the cellular processing system, although the product contains the flanking sequences of tRNA on both sides (19). We have reported the in vivo production of a circular RNA aptamer (26). Circularization of the RNA product was accomplished in E. coli by self-splicing permuted intron-exon sequences derived from the T4 bacteriophage gene td. The circular RNA was stably produced in vivo because of its resistance to exonucleases (26). In both cases, however, modifications of RNA products, such as the tRNA scaffold or circularization, are necessary to obtain the product efficiently. In particular, the tRNA scaffold method requires RNase H treatment to remove the flanking sequences (19), which is unsuitable for industrial RNA drug production. With the use of E. coli as the host, it is always necessary to devise a method for avoiding degradation by cellular nucleases. Here, we propose an alternative host, the marine phototrophic bacterium Rhodovulum sulfidophilum, which produces nucleic acids in the culture medium (1, 24, 25). R. sulfidophilum is a purple phototrophic marine alphaproteobacterium that can grow by either anoxygenic photosynthesis or respiration with a wide variety of organic compounds as electron donors and carbon sources (8). Previously, we reported that R. sulfidophilum releases extracellular nucleic acids into the culture medium in soluble form and that the extracellular RNAs are mainly nonaminoacylated fully mature tRNAs and fragments of 16S and 23S rRNAs (1, 25). In addition, we found that the bacterium did not produce any detectable RNase in the culture medium (25). Therefore, we examined extracellular production of an artificial RNA using the bacterium.
Here, we report the extracellular production of a streptavidin RNA aptamer as an RNA drug product model using R. sulfidophilum. As the bacterium does not produce any detectable RNases in the culture medium, a linear form of streptavidin RNA aptamer without any modification, such as tRNA scaffold or circularization, could be produced in the culture medium.
MATERIALS AND METHODS
Bacterial strain and growth conditions.
The marine phototrophic bacterium R. sulfidophilum DSM 1374T (7-9) was used throughout this study. Cultivation was performed essentially by the method of Hiraishi and Ueda (8). The strain was grown anaerobically at 25°C in screw-cap test tubes filled with PYS medium (16), including 2% (wt/vol) NaCl under incandescent illumination (about 5,000 lx). For the bacteria harboring plasmids (pHSP1 or pHSR1), the medium was supplemented with streptomycin (20 μg/ml). Cell growth was evaluated by measuring the turbidity of the culture medium at 600 nm.
RNase activity assay in culture medium.
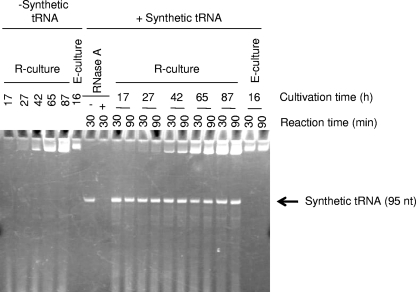
To examine whether the culture medium of R. sulfidophilum DSM 1374T contained RNase activity, an RNase assay was performed using PAGE. Cells were removed by centrifugation at various times in culture. Aliquots of 50 μl of supernatant were transferred to fresh tubes, and 1 μl of synthetic RNA (0.25 μg) was added to the tube, resulting in a total volume of 51 μl. The synthetic RNA used was a precursor of E. coli glycine tRNA 95 bases in length prepared by in vitro transcription from the appropriate template. The samples were incubated at 37°C for 30 or 90 min and then precipitated with ethanol and subjected to 10% denaturing PAGE. For comparison, the RNase activities of E. coli cultures were analyzed by the same method (Fig. 1).
FIG. 1.
RNase-free culture medium of R. sulfidophilum. The supernatant of R. sulfidophilum DSM 1374T or E. coli JM 109 culture medium was incubated with synthetic tRNA substrate (+ Synthetic tRNA) for 30 min or 90 min at 37°C as described in Materials and Methods. The reaction mixtures were analyzed by 10% denaturing PAGE. − Synthetic tRNA, no addition of the substrate to the reaction mixture; R-culture and E-culture, R. sulfidophilum culture medium and E. coli culture medium, respectively; nt, nucleotides. The cultivation time (17, 16, 27, 42, 65, or 87 h) and the reaction time (30 or 90 min) are indicated. RNase A represents the reaction mixture with RNase A instead of culture supernatant as a positive control.
Construction of plasmids.
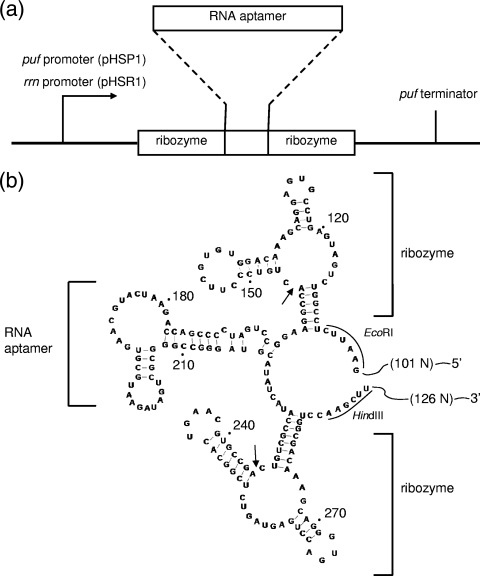
To test the self-cleavage activities of the designed hammerhead ribozyme sequences (6), we first constructed a plasmid for in vitro transcription, pGEM-5A3. The double-stranded DNA bearing the designed sequence shown in Fig. 2b was constructed by assembling synthetic oligodeoxynucleotides (purchased from Takara Bio Inc.) and DNA ligase. This DNA, containing a HindIII site at one end and an EcoRI site at the other (Fig. 2b), was digested with these two restriction enzymes and ligated into the corresponding sites of the plasmid vector pGEM-3Z (Promega) to give pGEM-5A3. The hammerhead ribozyme activity was confirmed by in vitro transcription using T7 RNA polymerase and 10% denaturing PAGE (data not shown). For in vivo production of the RNA aptamer, we constructed pHSP1 and pHSR1, which were derivatives of pCF1010, kindly provided by K. Matsuura (Tokyo Metropolitan University) (13). First, the puf promoter sequence (13, 14) was amplified from the genome of R. sulfidophilum DSM 1374T by PCR, using the primers puf-L (5′-AAGGAAAAAAGCGGCCGCCGAGGTCAACATGGTGAT-3′) and puf-R (5′-ATGAATTCCCTCGATGGTTTCAAGCACCACT-3′). The NotI and EcoRI sites in the primers are underlined. Similarly, the puf terminator sequence (14) was amplified by PCR, using PCter-L2 primer (5′-ACAAGCTTGCAAGAGCCGCCCTTTCC-3′) and PCter-R (5′-ACTCTAGAGCTCATGCCCTTGAGATCG-3′). The HindIII and XbaI sites in the primers are underlined. Then, the EcoRI-HindIII fragment containing the RNA aptamer sequence with flanker ribozyme sequences was cut out from pGEM-5A3. The EcoRI-treated fragment of the puf promoter sequence was then ligated to the EcoRI terminus of this fragment. Similarly, the HindIII-treated fragment of the puf terminator sequence was ligated to the HindIII terminus of the fragment. The resulting DNA containing the puf promoter, the RNA aptamer flanked by ribozymes, and the puf terminator was digested with NotI and XbaI. This DNA was inserted into the corresponding sites of the plasmid pCF1010 to give pHSP1. For construction of pHSR1, we first amplified the rrn promoter region from the genome of R. sulfidophilum DSM 1374T (DDBJ accession number AB513346) by PCR using the primers rrn-2L (5′-TTGGTTGCGGCCGCTCTGGCGCTTCGATTCC-3′) and rrn-R (5′-TTAATGCATGAATTCATTTCTACTTGGCGCCG-3′). The NotI and EcoRI sites in the primers are underlined. The puf promoter region of pHSP1 was replaced by this rrn promoter to give pHSR1.
FIG. 2.
Construction of plasmids pHSP1 and pHSR1 and nucleotide sequence and secondary-structure model of the RNA expressed. (a) Organization of RNA aptamer (streptavidin RNA aptamer) expression sites designed in the plasmids. The plasmids pHSP1 and pHSR1 contained the puf promoter and the rrn promoter, respectively. (b) Predicted secondary structure of the expressed RNA. The streptavidin RNA aptamer with flanking hammerhead ribozyme sequences is shown. The self-cleavage sites expected in hammerhead ribozyme reactions are indicated by small arrows. 101N and 126N indicate the predicted additional lengths expressed from the plasmid pHSR1 (rrn promoter). The EcoRI and HindIII sites used for cloning (see Materials and Methods) are indicated.
Conjugation techniques.
The plasmid pHSP1 or pHSR1 was introduced into R. sulfidophilum cells by conjugation with the mobilizing strain E. coli S17-1, as described previously (22).
Preparation of intracellular and extracellular RNAs.
Cultivated cells were collected by centrifugation, and the supernatant was transferred to a fresh tube. The nucleic acid fraction of the supernatant was precipitated with ethanol. The nucleic acid fraction thus collected was incubated with 0.5% (wt/vol) SDS and 0.05 μg/μl proteinase K (Sigma) at 37°C for 60 min to remove proteins. The preparation was then extracted with phenol-chloroform, and nucleic acids were collected by ethanol precipitation (21). This sample was treated with DNase I (RNase-free grade; Promega), and the extracellular RNAs were phenolized to remove DNase I and precipitated with ethanol. In some cases, proteinase K treatment was omitted, and instead, guanidine-HCl treatment and extraction with phenol-chloroform were performed. There was essentially no difference between these two methods to remove proteins from the RNA preparations. For the intracellular RNAs, the collected cells were resuspended in 0.3 M sodium acetate (pH 5.0) and extracted with equal volumes of sterilized water-saturated phenol. After centrifugation, the aqueous phase was transferred to new tubes, and the nucleic acids were precipitated with ethanol. The preparation was treated with DNase I, phenolized, and precipitated with ethanol as described above.
Quantitative analyses of streptavidin RNA aptamer production.
The amount of streptavidin RNA aptamer produced was measured by reverse transcription (RT)-PCR. We performed reverse transcription with a ThermoScript RT-PCR System (Invitrogen) with the primer Apt-R2-Et (5′-AATATGCATCCCGGCCCGCGACTATCTTAC-3′). Apt-R2-Et is hybridizable to positions 194 to 222 of the aptamer sequence (Fig. 2b). PCR was performed using the streptavidin RNA aptamer-specific primers Apt-L-A (5′-ATAGGCCTGATCCCCGACCAGAATCATGCAAGTG-3′) and Apt-R2-Et. The sequence of Apt-L-A corresponds to positions 161 to 192 of the aptamer sequence (Fig. 2b). The DNAs thus amplified were subjected to 6% nondenaturing PAGE. The amplified DNA is expected to have a size of 65 bp. The gels were stained with ethidium bromide, band intensities were evaluated using Image Gauge (Fuji Film Co.), and the amounts of the streptavidin RNA aptamer produced were determined by comparison with the intensities of the standard samples. The authentic-standard bands were prepared from the precisely quantified in vitro transcript of the streptavidin RNA aptamer. The results are shown as the averages of at least three independent assays.
Northern blotting analysis.
To determine the sizes of the RNA products, Northern blotting analysis was performed. Extracellular RNAs were prepared from the cultures (200 ml) in early log phase as described above. The RNAs were separated by 10% denaturing PAGE and transferred onto positively charged membranes. A portion of the RNA preparation was incubated with refolding buffer prior to electrophoresis to enhance self-cleavage by hammerhead ribozyme activity. This self-cleavage reaction mixture (refolding mixture) contained 50 mM HEPES-KOH (pH 7.4), 10 mM MgCl2, 100 mM NaCl, and 1.4 mg of extracellular RNA from the culture in a total volume of 5 ml. The mixture was first incubated at 75°C for 5 min and then at 37°C for 30 min. The probe Apt-R2-Et (see above) was labeled at the 5′ end with [γ-32P]ATP and T4 polynucleotide kinase. Hybridization was carried out overnight at 50°C in DIG Easy Hyb buffer (Roche). Washing was performed in 2× SSC buffer (17.53 g/liter sodium chloride, 8.82 g/liter sodium citrate, 0.1% [wt/vol] SDS) at room temperature and 0.1× SSC buffer (0.89 g/liter sodium chloride, 0.44 g/liter sodium citrate, 0.1% [wt/vol] SDS) at 50°C. Bands were detected by autoradiography using a BAS-1800 Bioimaging Analyzer System (Fuji Film Co.).
Electrophoretic mobility shift assay.
To confirm whether the function of streptavidin RNA aptamer produced by R. sulfidophilum was maintained, we performed an electrophoretic mobility shift assay. We used a culture (300 ml) in early log phase of the producing strain harboring pHSR1. Extracellular RNA was prepared as described above. The RNAs were subjected to 10% denaturing PAGE. The bands corresponding to 50 to 100 nucleotides in length were cut, and the RNAs in the gel pieces were eluted and purified. This RNA preparation was further purified using streptavidin agarose. The RNA preparation was labeled at the 5′ end with [γ-32P]ATP and T4 polynucleotide kinase and then purified by 10% denaturing PAGE. The radiolabeled RNA (500 cpm) was incubated with 1 μM streptavidin in binding buffer at room temperature for 1 h in a total volume of 2 μl in the presence or absence of 3.3 mM biotin. After incubation, the samples were subjected to 6% nondenaturing PAGE. The radioactive bands were detected by autoradiography using a BAS-1800 Bioimaging Analyzer System (Fuji Film Co.).
RESULTS
Rhodovulum sulfidophilum does not release RNases into the culture medium.
Previously, we reported that R. sulfidophilum strain DSM 2351 produced nucleic acids extracellularly and that most of the tRNAs released into the culture medium retained the mature 3′-terminal CCA sequence (1, 25), although these sequences are usually very susceptible to ubiquitous exonucleases. These observations suggested that the bacterium may be useful for fermentative extracellular production of artificial RNA drugs. However, methods for gene manipulation in R. sulfidophilum strain DSM 2351 have not yet been developed. Therefore, we used another strain, R. sulfidophilum DSM 1374T, which has been used in photosynthesis studies and for which gene manipulation methods have been established (13, 14). The amount of natural extracellular tRNA produced by this strain is about 0.2 μg/ml of culture medium at 100 h of cultivation. This is only slightly less than that of the strain DSM 2351. The other properties, such as the cell growth rate and the extracellular RNA species produced, are almost the same in the two strains (1). We also confirmed that the strain DSM 1374T does not produce RNases in the culture medium. RNase activities in the culture media from R. sulfidophilum DSM 1374T and E. coli JM 109 were assayed using a synthetic tRNA precursor as a substrate. As shown in Fig. 1, almost no degradation of the synthetic tRNA occurred in the incubation mixtures containing R. sulfidophilum DSM 1374T cultures at any cultivation time tested, whereas complete degradation was observed in the reaction mixture containing E. coli culture. Therefore, we used the strain R. sulfidophilum DSM 1374T throughout this study.
Construction of plasmids for production of streptavidin RNA aptamer.
To examine whether artificial RNAs are also produced extracellularly by this organism, we chose the streptavidin RNA aptamer (23) as a model product. As there was no detectable RNase activity in the culture medium of R. sulfidophilum DSM 1374T, we designed a plasmid to produce a mature form of streptavidin RNA aptamer directly in the culture medium. As shown in Fig. 2, we constructed the streptavidin RNA aptamer expression plasmids pHSP1 and pHSR1 containing the streptavidin RNA aptamer sequence as derivatives of pCF1010 (11). The designed aptamer sequence was flanked on both sides by self-cleaving hammerhead ribozyme sequences (6). The hammerhead ribozyme is a self-cleaving sequence originating from plant virusoids. This self-cleavage is an indispensable step for virusoid maturation and occurs at a specific site on the RNA sequence without the help of any protein enzyme (6). Here, we expected to produce a self-processed mature form of the aptamer using this self-cleaving hammerhead ribozyme (Fig. 2). The artificial gene was designed to be transcribed by the puf promoter and the puf terminator (13, 14) in pHSP1, whereas the sequence in pHSR1 was designed to be transcribed by the rrn promoter and the puf terminator (Fig. 2). We isolated and cloned the rrn promoter from the genome of R. sulfidophilum DSM 1374T (unpublished data) (see Materials and Methods). R. sulfidophilum DSM 1374 containing either plasmid is expected to produce the streptavidin RNA aptamer as the mature form (the form processed by hammerhead ribozyme self-cleavage activity). The expected self-cleavage sites are indicated by small arrows in Fig. 2b. To confirm these self-cleaving ribozyme activities, we constructed another plasmid, designated pGEM-5A3, for in vitro transcription (see Materials and Methods) and analyzed the products transcribed from this plasmid. Electrophoretic analyses of the in vitro transcript from pGEM-5A3 showed perfect self-cleavage at the expected sites during transcription (data not shown).
Detection of the streptavidin RNA aptamer in the culture medium.
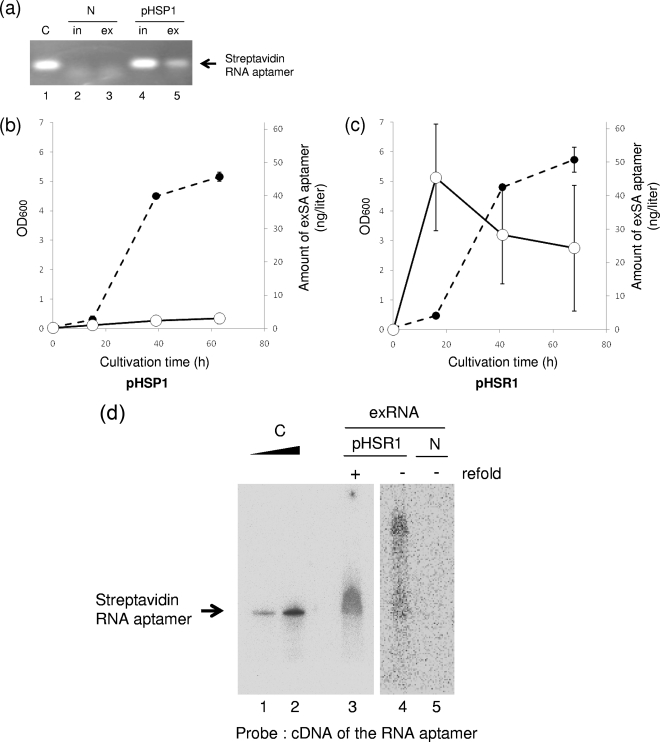
R. sulfidophilum DSM 1374T was transformed with the plasmid pHSP1 by conjugation with E. coli S17-1 (22) as described in Materials and Methods. The transformed cells were cultivated under anaerobic conditions in the light. First, we examined whether the streptavidin RNA sequence was released from the cell into the culture medium. After 42 h of cultivation (stationary phase), the cells were removed by centrifugation and the nucleic acids in the supernatant (culture medium) were collected by ethanol precipitation. The RNA fraction was purified by DNase I and proteinase K treatment and phenolization as described in Materials and Methods. The RNA fraction thus prepared was subjected to RT-PCR analysis to detect the streptavidin RNA aptamer sequence. The sequence could be amplified from the extracellular RNA preparation by RT-PCR (Fig. 3a, lane 5), indicating that the RNA aptamer sequence was indeed produced in the culture medium. Figure 3a also shows that the intracellular RNA preparation from this transformant contained the RNA aptamer sequence (lane 4), whereas none of the nucleic acid preparations from nontransformed cells contained the sequence (lanes 2 and 3). When the reverse transcription reaction prior to PCR was omitted, the aptamer sequence could not be amplified from any of the RNA preparations (data not shown). These observations indicated that the streptavidin RNA aptamer sequence was expressed in this transformant and released into the culture medium. We also confirmed that R. sulfidophilum DSM 1374T transformed with another plasmid containing the rrn promoter, pHSR1, released the streptavidin RNA aptamer sequence into the culture medium (data not shown).
FIG. 3.
Production and release of the streptavidin RNA aptamer into the culture medium. (a) Detection of the streptavidin RNA aptamer sequence. Using streptavidin RNA aptamer-specific primers, the aptamer sequences were amplified by RT-PCR from intracellular (in) and extracellular (ex) RNA preparations from R. sulfidophilum harboring pHSP1 in the early stationary phase of growth. N and pHSP1 represent the RNA preparations from R. sulfidophilum DSM 1374T only (no plasmid) and the bacterium transformed with pHSP1, respectively. C indicates a positive control using the in vitro transcript of the streptavidin RNA aptamer as a template for RT-PCR. (b and c) Extracellular production of the streptavidin RNA aptamer. Time courses of extracellular production of the RNA aptamer from the bacteria harboring pHSP1 (b) and pHSR1 (c) are shown. The amounts of RNA aptamers were measured by quantitative RT-PCR as described in Materials and Methods. Closed circles, cell growth (optical densities at 600 nm [OD600]); open circles, amounts of extracellular streptavidin RNA (exSA) aptamer (ng/liter culture). (d) Northern blotting analysis of the streptavidin RNA aptamer produced. To confirm the size of the RNA, Northern analysis of extracellular-RNA preparations was performed using a cDNA probe for the streptavidin RNA aptamer. C indicates a positive control using the in vitro transcript of the streptavidin RNA aptamer. Clear, sharp bands are shown. This aptamer has a size of 80 nucleotides and can be used as an accurate size marker. N and pHSR1 represent the extracellular RNA preparations (exRNA) from R. sulfidophilum DSM 1374T only (no plasmid) and the bacterium transformed with pHSR1, respectively; refold indicates additional incubation to enhance self-cleavage (see the text).
Extracellular production of the streptavidin RNA aptamer.
Time course and quantitative analyses of streptavidin RNA aptamer production by these transformants were performed. The amounts of product in the culture medium at various time points in culture were determined by quantitative RT-PCR. Time course curves are shown in Fig. 3b and c. Both transformants produced the streptavidin RNA aptamer, and one containing the rrn promoter (pHSR1) showed efficient production (Fig. 3c). In the case of the pHSP1-containing transformant, the maximum level of production (3 ng/liter of culture) was observed at 63 h of cultivation (stationary phase) (Fig. 3b). However, the level produced by the pHSR1-containing transformant was 45 ng/liter of culture (Fig. 3c), and the maximum level of production was observed at 16 h of cultivation (early log phase) (Fig. 3c). This is reasonable, because the rrn promoter is activated by the FIS protein, which is expressed only in early log phase (2, 5). These observations indicated that the rrn promoter functions much more efficiently than the puf promoter.
The size of the streptavidin RNA aptamer released into the extracellular environment was measured by Northern blot analysis. For this experiment, the extracellular RNA preparation from the strain harboring pHSR1 was used. As shown in lane 4 of Fig. 3d, a main band of 80 nucleotides (the expected size of the streptavidin RNA aptamer) and longer smear bands were observed. These smears were thought to be unprocessed bands. When the extracellular-RNA preparation was incubated with buffer for enhancement of self-cleavage, only a single band of 80 nucleotides was observed (Fig. 3d, lane 3). As a large amount of RNA preparation (1.4 mg) was loaded in each lane, a broad band was observed. However, it is obvious that the RNA product underwent self-processing with additional incubation. If efficient purification or a high yield of the RNA aptamer is established, a clearer single band may be shown. This self-cleaving reaction mixture contained 50 mM HEPES-KOH (pH 7.4), 10 mM MgCl2, 100 mM NaCl, and 1.4 mg of extracellular RNA from the culture in a total volume of 5 ml. The mixture was first incubated at 75°C for 5 min and then at 37°C for 30 min. Although it is at present difficult to determine the precise structure of the aptamer by direct RNA sequencing because of the low yield of the product, the observations indicated that the designed RNA sequence was expressed and produced extracellularly by the organism.
The streptavidin RNA aptamer produced retained its function.
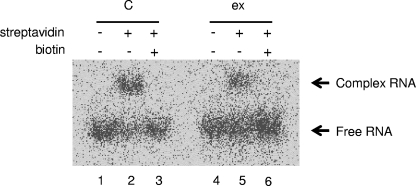
We confirmed the binding ability of the extracellularly produced streptavidin RNA aptamer by electrophoretic mobility shift assay. For this experiment, the extracellular RNA preparation from the strain harboring pHSR1 was used. As described in Materials and Methods, partially purified streptavidin RNA aptamer was labeled with [γ-32P]ATP and T4 polynucleotide kinase, mixed with streptavidin, and subjected to native PAGE. Figure 4 shows that part of the purified preparation bound to streptavidin as an up-shifted band (lane 5), and its binding was completely inhibited by the addition of 3.3 mM biotin (lane 6). These observations indicated that the streptavidin RNA aptamer extracellularly produced by R. sulfidophilum retains its binding ability and that this binding is specific to streptavidin, because it was inhibited by biotin.
FIG. 4.
Electrophoretic mobility shift assay of the product. To confirm the aptamer function of the extracellularly produced RNA, an electrophoretic mobility shift assay was performed. Partially purified and radiolabeled extracellular RNA preparations (ex) were incubated with 1 μM streptavidin in the presence (lanes 3 and 6) or absence (lanes 1, 2, 4, and 5) of 3.3 mM biotin and analyzed by native PAGE. An autoradiogram of the gel is shown. C indicates a positive control using the in vitro transcript of the streptavidin RNA aptamer. The positions of free and complex RNAs are indicated.
DISCUSSION
We have described the extracellular production of an artificial RNA using the bacterium R. sulfidophilum harboring a genetically engineered plasmid. This is the first demonstration of extracellular production of an artificial RNA. To date, such RNAs have been synthesized by in vitro transcription (15) or chemical synthesis (12). Compared to these methods, our in vivo extracellular production has a number of advantages. First, the in vivo method is much more economical, as it does not require the use of expensive enzymes, synthetic DNA templates, and chemicals, which are indispensable for in vitro transcription or chemical synthesis of RNAs. Second, the time-consuming and labor-intensive handling is not necessary, as this method requires only cultivation of the bacterium for synthesis. Moreover, the culture volume can be scaled up easily, which is necessary for large-scale industrial preparation. In vivo expression and preparation of artificial RNAs have been reported previously (19, 26). We developed a method for in vivo production of circular RNAs using E. coli (26). As the RNAs produced by this method are circular molecules, the products are stably maintained in the cells, because such circular molecules are resistant to cellular exonucleases. Ponchon and Dardel also reported the production of recombinant RNAs in vivo using E. coli (19). In this case, the RNA products contained tRNA sequences on both sides to avoid degradation by endogenous RNases. In contrast, recombinant RNA products can be produced without any modifications in R. sulfidophilum, as the bacterium expresses almost no RNases. In addition, the present method using R. sulfidophilum is distinguished from the methods using E. coli by the release of the product into the extracellular environment. Using this property, it is possible to develop a continuous production system in one vessel because collection of the cells is not necessary to obtain the products.
In the present study, the maximum yield of the streptavidin RNA aptamer was 45 ng/liter of culture (Fig. 3c). In a preliminary experiment, however, a yield of 100 ng/liter of culture was obtained under aerobic conditions in the dark (data not shown). This value is not yet practical even for laboratory use, but several approaches are expected to yield improvements. In fact, the yield has been greatly improved by replacement of the puf promoter with the rrn promoter (compare Fig. 3b and c). It is possible that the yield can be improved by constitutive expression of the FIS protein in the cell, because this protein is known to activate the rrn promoter (5). In addition, increasing the copy number of the plasmid vector may be effective, although at present the copy number of pHSR1 is rather low (approximately 1 to 10 per cell).
We showed that R. sulfidophilum harboring pHSR1 also produces the streptavidin RNA aptamer inside the cells, and the recovered RNA has full aptamer activity (data not shown). The total amount of the RNA aptamer produced was calculated to be 7 μg from 1 liter of culture. In E. coli, using a high-copy-number plasmid and an efficient transcription system, we obtained 24 μg of circular RNA aptamer from 1 liter of culture (26). The value of 7 μg from 1 liter of culture gives hope for future improvements, because this result for R. sulfidophilum is from an experiment using a low-copy-number plasmid. The streptavidin RNA aptamer seems to be stably maintained in R. sulfidophilum cells even if the product is a linear RNA molecule without any modification, such as circularization. This indicates that the level of intracellular RNase activity of R. sulfidophilum may also be extremely low, as in the culture medium (Fig. 1).
At the beginning of this project, we also confirmed that R. sulfidophilum had the ability to produce and retain lacZ mRNA of E. coli in the culture medium using a plasmid containing the puf promoter and lacZ mRNA sequence (data not shown). These observations indicated that there is almost no RNase activity in the culture medium of R. sulfidophilum, as mRNA is thought to be the most susceptible of the RNAs to RNases. In addition, this is evidence that R. sulfidophilum can produce not only the streptavidin RNA aptamer, but also other artificial RNAs.
Although the RNA aptamer is also produced inside the cells, as described above, extracellular production is thought to be very beneficial for the future development of an efficient process-engineering system for industrial production of RNA drugs, because harvest and disruption of cells are not necessary. Previously, we reported that extracellular nucleic acids of R. sulfidophilum are produced by partial cell lysis of a subpopulation of bacteria and that the lysis may be controlled by a physiological mechanism (24). If we can control this mechanism and construct a system for product collection from the culture media, a continuous RNA drug production system can be created. Another strain of the genus Rhodovulum (strain PS88) has been reported to produce extracellular natural RNAs in a yield of 62.5 mg/g dry cells (27). This genus has great potential for extracellular-RNA production in large quantities. Further physiological investigations and optimization of the conditions will lead to the development of more efficient recombinant-RNA production systems. Thus, R. sulfidophilum is a highly promising organism for RNA drug production, because the organism produces almost no RNases.
Acknowledgments
This work was supported in part by a grant for scientific research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (to Y.K.). H.S. was supported by a Research Fellowship from the Japan Society for the Promotion of Science (JSPS) for Young Scientists.
We thank A. Hiraishi (Toyohashi University of Technology) for discussion, K. Matsuura (Tokyo Metropolitan University) for the kind gift of the strain of R. sulfidophilum and plasmids, and E. Sakai for technical assistance.
Footnotes
Published ahead of print on 4 December 2009.
REFERENCES
- 1.Ando, T., H. Suzuki, S. Nishimura, T. Tanaka, A. Hiraishi, and Y. Kikuchi. 2006. Characterization of extracellular RNAs produced by the marine photosynthetic bacterium Rhodovulum sulfidophilum. J. Biochem. 139:805-811. [DOI] [PubMed] [Google Scholar]
- 2.Azam, T. A., A. Iwata, A. Nishimura, S. Ueda, and A. Ishihama. 1999. Growth phase-dependent variation in protein composition of the Escherichia coli nucleoid. J. Bacteriol. 181:6361-6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Breaker, R. R. 2004. Natural and engineered nucleic acids as tools to explore biology. Nature 432:838-845. [DOI] [PubMed] [Google Scholar]
- 4.Chu, C. Y., and T. M. Rana. 2007. Small RNAs: regulators and guardians of the genome. J. Cell Physiol. 213:412-419. [DOI] [PubMed] [Google Scholar]
- 5.Dryden, S. C., and S. Kaplan. 1993. Identification of cis-acting regulatory regions upstream of the rRNA operons of Rhodobacter sphaeroides. J. Bacteriol. 175:6392-6402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Forster, A. C., and R. H. Symons. 1987. Self-cleavage of plus and minus RNAs of a virusoid and a structural model for the active sites. Cell 49:211-220. [DOI] [PubMed] [Google Scholar]
- 7.Hansen, T. A., and H. Veldkamp. 1973. Rhodopseudomonas sulfidophila, nov. spec., a new species of the purple nonsulfur bacteria. Arch. Mikrobiol. 92:45-48. [DOI] [PubMed] [Google Scholar]
- 8.Hiraishi, A., and Y. Ueda. 1994. Intrageneric structure of the genus Rhodobacter: transfer of Rhodobacter sulfidophilus and related marine species to the genus Rhodovulum gen. nov. Int. J. Syst. Bacteriol. 44:15-23. [Google Scholar]
- 9.Hiraishi, A., and Y. Ueda. 1995. Isolation and characterization of Rhodovulum strictum sp. nov. and some other purple nonsulfur bacteria from colored blooms in tidal and seawater pools. Int. J. Syst. Bacteriol. 45:319-326. [DOI] [PubMed] [Google Scholar]
- 10.Khan, A. U. 2006. Ribozyme: a clinical tool. Clin. Chim. Acta 367:20-27. [DOI] [PubMed] [Google Scholar]
- 11.Lee, J. K., and S. Kaplan. 1995. Transcriptional regulation of puc operon expression in Rhodobacter sphaeroides. J. Biol. Chem. 270:20453-20458. [PubMed] [Google Scholar]
- 12.Marshall, W. S., and R. J. Kaiser. 2004. Recent advances in the high-speed solid phase synthesis of RNA. Curr. Opin. Chem. Biol. 8:222-229. [DOI] [PubMed] [Google Scholar]
- 13.Masuda, S., K. V. P. Nagashima, K. Shimada, and K. Matsuura. 2000. Transcriptional control of expression of genes for photosynthetic reaction center and light-harvesting proteins in the purple bacterium Rhodovulum sulfidophilum. J. Bacteriol. 182:2778-2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Masuda, S., M. Yoshida, K. V. P. Nagashima, K. Shimada, and K. Matsuura. 1999. A new cytochrome subunit bound to the photosynthetic reaction center in the purple bacterium, Rhodovulum sulfidophilum. J. Biol. Chem. 274:10795-10801. [DOI] [PubMed] [Google Scholar]
- 15.Milligan, J. F., D. R. Groebe, G. W. Witherell, and O. C. Uhlenbeck. 1987. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 15:8783-8798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nagashima, K. V. P., A. Hiraishi, K. Shimada, and K. Matsuura. 1997. Horizontal transfer of genes coding for the photosynthetic reaction centers of purple bacteria. J. Mol. Evol. 45:131-136. [DOI] [PubMed] [Google Scholar]
- 17.Nellen, W., and C. Hammann. 2005. Small RNAs: analysis and regulatory functions. Nucleic acids and molecular biology. Springer-Verlag, Heidelberg, Germany.
- 18.Ng, E. W. M., D. T. Shima, P. Calias, E. T. Cunningham, Jr., D. R. Guyer, and A. P. Adamis. 2006. Pegaptanib, a targeted anti-VEGF aptamer for ocular vascular disease. Nat. Rev. Drug Discov. 5:123-132. [DOI] [PubMed] [Google Scholar]
- 19.Ponchon, L., and F. Dardel. 2007. Recombinant RNA technology: the tRNA scaffold. Nat. Methods 4:571-576. [DOI] [PubMed] [Google Scholar]
- 20.Que-Gewirth, N. S., and B. A. Sullenger. 2007. Gene therapy progress and prospects: RNA aptamers. Gene Ther. 14:283-291. [DOI] [PubMed] [Google Scholar]
- 21.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 22.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Biotechnology 1:37-45. [Google Scholar]
- 23.Srisawat, C., and D. R. Engelke. 2001. Streptavidin aptamers: affinity tags for the study of RNAs and ribonucleoproteins. RNA 7:632-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suzuki, H., M. Daimon, T. Awano, S. Umekage, T. Tanaka, and Y. Kikuchi. 2009. Characterization of extracellular DNA production and flocculation of the marine photosynthetic bacterium Rhodovulum sulfidophilum. Appl. Microbiol. Biotechnol. 84:349-356. [DOI] [PubMed] [Google Scholar]
- 25.Suzuki, H., S. Umekage, T. Tanaka, and Y. Kikuchi. 2009. Extracellular tRNAs of the marine photosynthetic bacterium Rhodovulum sulfidophilum are not aminoacylated. Biosci. Biotechnol. Biochem. 73:425-427. [DOI] [PubMed] [Google Scholar]
- 26.Umekage, S., and Y. Kikuchi. 2009. In vitro and in vivo production and purification of circular RNA aptamer. J. Biotechnol. 139:265-272. [DOI] [PubMed] [Google Scholar]
- 27.Watanabe, M., K. Sasaki, Y. Nakashimada, T. Kakizono, N. Noparatnaraporn, and N. Nishio. 1998. Growth and flocculation of a marine photosynthetic bacterium Rhodovulum sp. Appl. Microbiol. Biotechnol. 50:682-691. [Google Scholar]