Abstract
A leaching experiment, where liquid manure spiked with Salmonella enterica serovar Typhimurium (Tet+) DSM554 was applied to soil surfaces, was conducted on intact soil monoliths (60 cm in diameter and 100 cm long). A total of 6.5 × 1010 CFU was applied to each column. We found that Salmonella serovar Typhimurium could be transported to a 1-m depth in loamy soil at concentrations reaching 1.3 × 105 CFU/ml of leachate. The test strain was found in concentrations ranging from 300 to 1.35 cells/ml in loamy soil throughout the 27 days of the experiment, while concentrations below 20 cells/ml were sporadically detected in the leachates from sandy monoliths. Real-time PCR targeting invA DNA showed a clear correspondence between the total and culturable numbers of cells in the leachate, indicating that most cells leached were viable. On day 28, distribution of Salmonella serovar Typhimurium at five depths in the four monoliths was determined. The highest recovery rate, ranging from 1.5% to 3.8% of the total applied inoculum, was found in the top 0.2 m.
The spreading of liquid manure on agricultural land is an economic and practical solution for improving soil quality. However, animal manure frequently contains zoonotic pathogenic bacteria such as certain Escherichia coli, Salmonella spp., and Campylobacter spp. (8, 9, 24). Salmonellosis is one of the most frequently reported food-borne diseases in Europe, accounting for 67.5% of reported food-borne outbreaks (32). Salmonella enterica serovar Typhimurium, which accounts for 23% of nonhuman isolates (from animal, food, feed, and environmental sources) and for 18% of human isolates, was the second-most-common serotype found worldwide from 2000 to 2004 (38). Human salmonellosis has been related to the consumption of water or foods contaminated with animal manure (9, 15), which frequently contains a variety of different enteric pathogenic microorganisms (27).
Manure disposed of on agricultural land may create a risk for microbial contamination of surface water and groundwater. Field scale studies have reported manure bacteria in drainage water (5, 25, 36, 37). In addition to contamination originating from manure, pathogens in irrigation water may contaminate soil, water, crops, and, subsequently, animals and humans (7, 15). Disease outbreaks have been associated with water and food directly or indirectly contaminated with animal manure (9). Recent data from 1990 to 2004 showed that compared to beef, poultry, seafood, and eggs, fresh produce caused the second-highest number of food-borne disease outbreaks as well as the highest number of reported illnesses per outbreak (4).
When microorganisms enter the soil environment, their survival and distribution is affected by various factors, including soil texture, pH, temperature, water saturation, and the intensity of rain events (17, 28). Preferential water movement in macropores is probably the primary route by which bacteria move down through the soil (1, 2, 23). Studies of bacterial transport have been made mostly on homogenized natural soils in laboratory soil columns (2, 18, 30, 33). The purpose of our study was to perform a large-scale experiment using intact 100-cm-deep soil monoliths to compare the leaching of Salmonella serovar Typhimurium in two structurally different soils: a loamy soil with high absorptive properties and macropores and a coarse sandy soil with less-absorptive properties and less preferential flow. The monoliths were initially exposed to normal precipitation events followed by intensive precipitation, where the leaching of Salmonella serovar Typhimurium cells was observed. After the leaching experiment, the concentrations of Salmonella serovar Typhimurium cells at five depths were determined. Furthermore, we validated bacterial enumerations based on cultures on agar plates with real-time PCR to determine if a proportion of the test strain entered a nonculturable state.
MATERIALS AND METHODS
Soil monoliths.
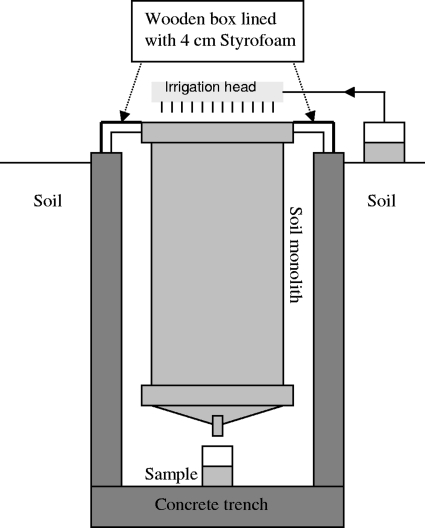
Four intact soil monoliths (60 cm in diameter and 100 cm deep) were excavated from two Danish sites. From each site, two monoliths, which throughout the manuscript will be referred to as loam A, loam B, sand A, and sand B, were excavated. The loamy soil is classified as typic argiudoll (12), and the sandy soil is classified as orthic haplohumod (12). For details on excavation and soil parameters, see Lægdsmand et al. (19). Both soils had been used for organic farming with the application of manure, the loamy soil since 1951, and the sandy soil since 1990. The loamy soil is a well-structured soil with a clay content of 21 to 39%, whereas the sandy soil is less structured and contains 5% clay. The monoliths were placed in an outdoor cement trench covered with a 40-mm Styrofoam-insulated wooden box to simulate insulation from the surrounding soil (Fig. 1). The internal soil temperature was approximately 4°C during the experiment, whereas the average air temperature was 2.9°C and −5.7°C during the day and night, respectively; temperature data are available at the DMI database (http://www.dmi.dk/dmi/vejrarkiv?region=2&year=2006&month=3).
FIG. 1.
Experimental setup showing a soil monolith installed in the cement trench and indicating irrigation and sample collection equipment. Irrigation water was applied with a watering can from days 1 to 3 and with a rain simulator from days 19 to 27.
Inoculum.
The manure (8.8% dry matter) was taken from a stirred-manure storage tank (from a facility for fattening pigs) and kept at 4°C for 2 days. Salmonella enterica serovar Typhimurium (Tet+) DSM554 was obtained from Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH, Braunschweig, Germany, and propagated in Luria broth (Becton Dickinson, Franklin Lakes, NJ) supplemented with 2.5 μg/ml tetracycline and incubated at 37°C for 48 h at 100 rpm. Without further washing of the culture, four batches, each containing 75 ml of culture and approximately 815 g of manure, were mixed, yielding a concentration of 8 × 107 CFU/g of manure. The total amount of the test strain added to each monolith was 6.5 ×1010 CFU. Within 1 h, each batch was applied evenly on the soil surface of one of the four monoliths, corresponding to a normal agricultural practice of 3 kg manure/m2.
Irrigation experiment.
Prior to the experimental onset, 30 mm of tap water was surface applied with a watering can. Prior to manure application, percolated water tested negative for tetracycline-resistant bacteria. During the first 3 days, 8.8, 8.8, and 5.3 mm, respectively, of tap water was added in irrigation events of 1 h each. Water was applied with a watering can. One water sample from each of the four monoliths was collected at 3, 6, 18, 21, 25, 29, 41, 45, and 49 h after manure application.
The monoliths were not monitored from day 3 to 18. On day 19, an intensive 10-day irrigation period was started. Water was applied to the soil surface using a rain simulator that ensured a uniform drop-wise application of water over the entire monolith surface. Each monolith was preirrigated with 30 mm tap water (10 mm h−1) and allowed to leach for 36 h to obtain a water content approximately corresponding to field capacity. The first liter of drainage water on days with 50 mm of precipitation was analyzed for tetracycline-resistant bacteria. The irrigation schedule was as follows: loam A received 30 mm on days 19 and 23 and 50 mm on days 21 and 25, loam B received 30 mm on days 20 and 24 and 50 mm on days 22 and 27, sand A received 30 mm on days 17 and 21 and 50 mm on days 19 and 23, and sand B received 30 mm on days 18 and 22 and 50 mm on days 21 and 25. The total volume of water added to each monolith during the 27 days was 183 mm. Water was collected in sterilized blue-cap bottles at the outlets of the monoliths (Fig. 1). The conversion from millimeters of precipitation to volume of water applied was calculated by multiplying the surface area by the millimeters of precipitation.
Salmonella serovar Typhimurium cells were enumerated in all water samples on Mueller-Hinton broth (Oxoid Limited, Hampshire, United Kingdom) (17) mixed with 1.5 (wt/wt) of Bacto agar (Becton Dickinson, Franklin Lakes, NJ) with a tetracycline concentration of 2.5 μg/ml. A series of 10-fold-dilutions was prepared, and Salmonella serovar Typhimurium cells were enumerated in the water sample from each monolith on Mueller-Hinton agar plates following incubation at 37°C for 18 to 24 h. Enumeration was done in triplicate for each water sample.
PCR assay.
Loam A water samples from day 1 to day 25 were tested by real-time PCR, where volumes of 45-ml water samples were centrifuged (3,900 × g, 15 min, 5°C). The pellet was dissolved in 1 ml 0.010 M phosphate buffer (5.7 ml 1 M NaH2PO4·H2O plus 4.2 ml 1 M Na2HPO4·2H2O in 1,000 ml water [pH 7.4]) and frozen at −80°C. DNA extraction was done with a FastDNA Spin kit for soil (BIO 101, Vista, CA). We followed the manufacturer's recommendations except for the beating step and an additional freeze-thaw step. The beating step was changed from 30 s at a speed of 5.5 in a FastPrep FP120 to four times at 30 s at a speed of 4.0. The freeze-thaw step was 1 h at −80°C followed by 30 min at 37°C. Real-time PCRs were carried out in an iCycler iQ (Bio-Rad, Hercules, CA), where quantification was based on an internal standard 10-fold dilution of the test strain DNA extract. The samples were made in triplicate, including three negative controls. The primers used were invA forward, 5′-ACAGTGCTCGTTTACGACC-3′, and invA reverse, 5′-ACTGGTACTGATCGATAAT-3′ (16). The 20-μl reaction mixtures contained 0.08 μl 0.4 μM forward primer, 0.08 μl 0.4 μM reverse primer, 10 μl master mix (DyNAmo HS SYBR green qPCR kit; Finnzymes, Finland), 2 μl bovine serum albumin (New England Biolabs, Inc., Ipswich, MA), 6.34 μl distilled H2O, and 1.5 μl DNA sample. The following adapted protocol was used (16): 12 min at 95°C for enzyme activation, 40 cycles of 30 s each at 95°C for denaturation, 30 s at 55°C for primer annealing, 30 s at 72°C for elongation, and 15 s at 77°C for quantification of the invA product. The protocol was finished with one 6-min cycle at 72°C for elongation and a melting-curve analysis generated by analyzing the amount of double-stranded DNA after each 0.5°C increase in the temperature, up to 95°C.
Soil distribution.
At day 28, soil from five depths was recovered, and 20 liters of soil from each depth was carefully aseptically mixed in a separate plastic container in an end-over-end mixer for 20 min. Soil samples were collected from the following depths: 0 to 20 cm, 20 to 30 cm, 30 to 40 cm, 40 to 50 cm, and 50 to100 cm. One gram of soil was taken from the homogenized sample and mixed with 9.5 ml 0.010 M phosphate buffer on a vortex mixer for 10 s and plated on Mueller-Hinton agar with 25 μg/ml tetracycline.
Statistics.
Results were analyzed by analysis of variance (ANOVA) by using SigmaStat, version 3.11 (Systat Software, Inc., San Jose, CA).
RESULTS AND DISCUSSION
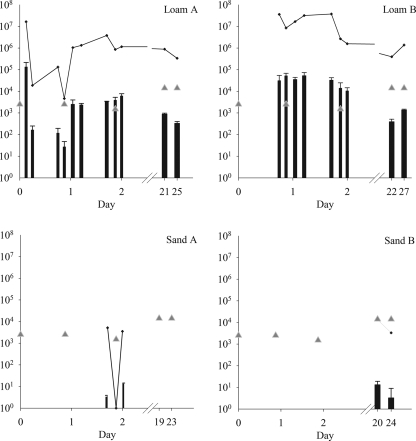
The loamy monoliths had significantly (P < 0.001) higher concentrations of Salmonella serovar Typhimurium cells in drainage water than the sandy monoliths (Fig. 2). Salmonella serovar Typhimurium was detected in all water samples from loam A, with a breakthrough concentration of 1.3 × 105 ± 8.2 × 104 CFU/ml in the first water sample. Loam B had a breakthrough concentration of 3.1 × 104 ± 2.3 × 104 CFU/ml after 18 h.
FIG. 2.
The concentrations (CFU/ml) of viable Salmonella serovar Typhimurium DSM554 in drainage water during the 27 days after liquid manure application are shown by black bars. The detection limit was 3.3 CFU/ml. Values below 200 CFU/ml should be read as positive samples rather than precise quantifications. Each bar represents the mean of triplicate plate counts per water sample; error bars indicate standard deviations. Gray triangles show irrigation times and amounts of water applied in milliliters. Black lines show the total numbers of CFUs of the test strain collected (determined by multiplying the concentration by the volume of water).
Nevertheless, during the first 3 days of the experiment, the highest proportion of Salmonella serovar Typhimurium cells leached through loam B, where a total of 1.35 × 108 CFU of Salmonella serovar Typhimurium was leached within 49 h. This number was calculated by multiplying drainage volumes and test strain concentrations. Despite the high number of leached test strain cells in loam B, this number correlates to only 0.2% of the total number of Salmonella serovar Typhimurium cells applied to the soil surface, provided that no regrowth occurred.
The low recovery rate may be partly explained by the use of a culture collection strain, as previous studies have shown a rapid die-off upon inoculation of a pure culture into manure (6, 11). A decimal reduction time (D-value) of 15 days was found for Salmonella in liquid manure from pigs (13), whereas a D-value (in days) of 2 was found for Salmonella in unincorporated liquid pig manure on an arable sandy loam (14). Malik et al. (20) found that Salmonella enterica serotype Anatum did not leach to the subsurface drainage and explained this finding by low rates of survival and filtration. Adhesion was found to be an important factor controlling the extent of bacterial movement (10).
Concentrations up to 1.3 × 105 CFU/ml were measured in the leachates from the loamy monoliths. Concentrations ranging from 3 × 102 to 1.3 × 103 CFU/ml were still recovered 3 weeks after the initial inoculation. In sand A, it was possible to detect Salmonella serovar Typhimurium in the water sample after 49 h, and for sand B, on days 18 and 22. Concentrations of the test strain were below 13 CFU/ml in leachates from sandy monoliths. The difference in the results for the two soil types can be explained by a higher degree of preferential flow in the loamy soil than in the sandy soil. A previous study with the same soil monoliths found a greater preferential flow in the loamy soil. This was shown with a bromide tracer, which was detected in the drainage water within the first 10 mm of precipitation for the loamy soils and after 60 mm for the sandy soils (19). Other studies underline that preferential flow is important in the vertical transport of fecal bacteria (22, 31, 35). The test strain was also found in the leachates of the sandy monoliths, but later and in lower concentrations. Based on this work, it can be concluded that in soils where preferential flow occurs, bacterial pathogens like the Salmonella serovar Typhimurium strain can be transported to a depth of at least 1 m. This fast transport of the test stain was found after a normal precipitation event of only 8.8 mm. Liquid manure application onto wet soil followed by a single precipitation event therefore poses a risk for pathogen contamination of the groundwater or surface water.
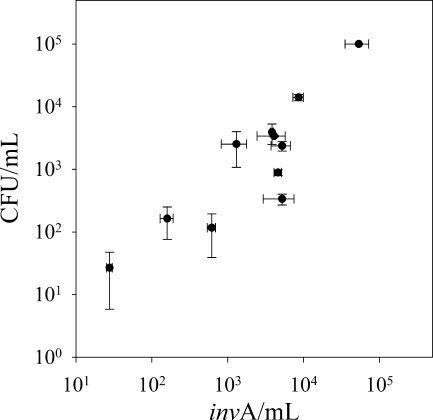
The CFU data based on plating of the tetracycline-resistant Salmonella test strain was verified by direct real-time PCR quantification of the invA gene (16) in order to verify whether the test strain lost cultivability. Real-time PCR results for loam A soil were plotted against CFU enumerations obtained by selective plating and showed a significant relation between the results obtained by direct DNA quantification and plating-based enumeration (P = 0.249 in a paired t test) (Fig. 3). These findings indicate a high level of cultivability of the test strain after it leached through the soil monolith. The soil monoliths were kept outside at ambient temperature and moisture conditions, likely increasing survival of bacterial pathogens (3). It has previously been shown that DNA quantification resulted in numbers comparable to CFUs in wet soils, while a clear drop in CFUs was found in dry soil (26). The high level of cultivability contradicts other findings for Salmonella spp. entering a natural environment, where the viable but nonculturable (VBNC) state has been induced in soil (21, 34) and water (29).
FIG. 3.
Comparison between CFU counts and real-time PCR detection of Salmonella serovar Typhimurium DSM554 in water samples from loam A. CFU/ml was determined by plating on a selective agar, and the amount of invA DNA/ml was determined from DNA extracts of a dilution series of a Salmonella strain. Each point represents the mean of triplicate quantifications from one sample; error bars indicate ± the standard deviations.
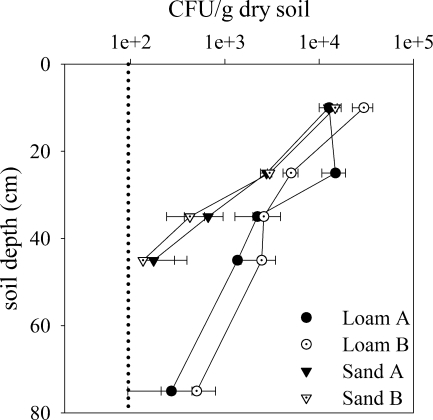
When the numbers of Salmonella serovar Typhimurium cells found in the five depths were compared, there was a statistically significant difference (P < 0.001, as determined by two-way ANOVA) between the two soil types. The total percentages of cells recovered from the five depths based on the applied inoculum were as follows: loam A, 3.0%; loam B, 4.7%; sand A, 1.8%; and loam B, 2.0%. The concentrations of Salmonella serovar Typhimurium cells varied among the five different depths in all four monoliths, with a decrease in concentration with depth (P < 0.001) (Fig. 4). For both soil types, the recovery rate was highest in the top 0.2 m, ranging between 10,000 and 20,000 CFU/g dry soil. The recovery rates in the top 0.2 m were as follows: loam A, 1.6%; loam B, 3.8%; sand A, 1.5%; and loam B, 1.8%. This high degree of filtration in the upper soil layers in both soils poses a risk for contamination of vegetable crops. Islam et al. (15) found that Salmonella enterica serovar Typhimurium survived in manure-amended soils for more than 200 days and was detected on radish and carrots for 84 and 203 days, respectively, after manure application.
FIG. 4.
The number of S. Typhimurium DSM554 (CFU/g dry soil) at depths of 0 to 20, 20 to 30, 30 to 40, 40 to 50 and 50 to 100 cm 28 days after liquid manure application. Depths plotted are averages. The detection limit was 95 CFU/g. Each point represents the mean of triplicate quantifications from one sample; error bars indicate ± the standard deviations.
The structural differences of the loamy and sandy soils clearly influence the transport of Salmonella serovar Typhimurium cells through the two soil types. The test strain was not found in the lower 0.5 m of both of the sandy monoliths, which may be explained by the lack of preferential flow systems in this soil type. The sandy monoliths contain 92% sand, which may explain why the test strain was found at 0.5 m depth. Detection of the test strain in the top 50 cm contradicts other studies with homogenized soils, where the lack of structure led to a higher degree of filtration. The test strain was found at all five depths in the loamy soils due to the macropores of the soil and thereby bypassed the finely textured matrix soil. Despite the high numbers of cells found in the lower parts of the loamy monoliths and the high concentrations found in leachates from these soils, we cannot conclude that leaching of Salmonella serovar Typhimurium is a problem only in structured soils because the total water volume was not replaced in the experiment (183 mm was added, while theoretically about 330 mm was required).
However, the amounts of water applied for the duration of the experiment (27 days) to both loamy and sandy monoliths were the same and correspond to approximately one-quarter of the average annual precipitation in Denmark, making the study a worst-case study. The survival time of Salmonella serovar Typhimurium in soils will also be of paramount importance in modeling the potential leaching of this bacterium through sandy soils. Several studies have found that Salmonella spp. can survive between 56 and 180 days in manure-applied soil (11, 14). From the present study, it can be concluded that Salmonella serovar Typhimurium leached through large loamy soil monoliths in concentrations up to 105 CFU/ml and that the pathogen remained culturable for at least a month under moist and cold conditions. Between 1.5% and 3.8% of the applied test strain was still viable in the top 0.2 m after 28 days and may pose a risk for crop contamination.
Acknowledgments
This study was supported by the Pathos Project funded by the Strategic Research Council of Denmark (ENV 2104-07-0015).
We thank Pia Jacobsen, Stig Rasmussen, and Mette Munch for skilled technical assistance.
Footnotes
Published ahead of print on 18 December 2009.
REFERENCES
- 1.Artz, R. R. E., J. Townend, K. Brown, W. Towers, and K. Killham. 2005. Soil macropores and compaction control the leaching potential of Escherichia coli O157:H7. Environ. Microbiol. 7:241-248. [DOI] [PubMed] [Google Scholar]
- 2.bu-Ashour, J., D. M. Joy, H. Lee, H. R. Whiteley, and S. Zelin. 1998. Movement of bacteria in unsaturated soil columns with macropores. Trans. ASAE 41:1043-1050. [Google Scholar]
- 3.Cools, D., R. Merckx, K. Vlassak, and J. Verhaegen. 2001. Survival of E. coli and Enterococcus spp. derived from pig slurry in soils of different texture. Appl. Soil Ecol. 17:53-62. [Google Scholar]
- 4.DeWaal, C. S., and Bhuiya, F. 2007. Outbreak Alert! Center for Science in the Public Interest, Washington, DC. http://www.cspinet.org/foodsafety/outbreak_alert.pdf.
- 5.Faust, M. A. 1982. Relationship between land-use practices and fecal bacteria in soils. J. Environ. Qual. 11:141-146. [Google Scholar]
- 6.Franz, E., A. D. van Diepeningen, O. J. de Vos, and A. H. C. van Bruggen. 2005. Effects of cattle feeding regimen and soil management type on the fate of Escherichia coli O157:H7 and Salmonella enterica serovar typhimurium in manure, manure-amended soil, and lettuce. Appl. Environ. Microbiol. 71:6165-6174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gagliardi, J. V., and J. S. Karns. 2000. Leaching of Escherichia coli O157:H7 in diverse soils under various agricultural management practices. Appl. Environ. Microbiol. 66:877-883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gerba, C. P., and J. E. Smith. 2005. Sources of pathogenic microorganisms and their fate during land application of wastes. J. Environ. Qual. 34:42-48. [PubMed] [Google Scholar]
- 9.Guan, T. Y., and R. A. Holley. 2003. Pathogen survival in swine manure environments and transmission of human enteric illness: a review. J. Environ. Qual. 32:383-392. [DOI] [PubMed] [Google Scholar]
- 10.Harvey, R. W., and S. P. Garabedian. 1991. Use of colloid filtration theory in modeling movement of bacteria through a contaminated sandy aquifer. Environ. Sci. Technol. 25:178-185. [Google Scholar]
- 11.Holley, R. A., K. M. Arrus, K. H. Ominski, M. Tenuta, and G. Blank. 2006. Salmonella survival in manure-treated soils during simulated seasonal temperature exposure. J. Environ. Qual. 35:1170-1180. [DOI] [PubMed] [Google Scholar]
- 12.Holliman, M., and A. Bridenhall (ed.). 1999. Keys to soil taxonomy. Natural Resources Conservation Service, U.S. Department of Agriculture, Washington, DC.
- 13.Hutchison, M. L., L. D. Walters, A. Moore, and S. M. Avery. 2005. Declines of zoonotic agents in liquid livestock wastes stored in batches on-farm. J. Appl. Microbiol. 99:58-65. [DOI] [PubMed] [Google Scholar]
- 14.Hutchison, M. L., L. D. Walters, A. Moore, K. M. Crookes, and S. M. Avery. 2004. Effect of length of time before incorporation on survival of pathogenic bacteria present in livestock wastes applied to agricultural soil. Appl. Environ. Microbiol. 70:5111-5118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Islam, M., J. Morgan, M. P. Doyle, S. C. Phatak, P. Millner, and X. P. Jiang. 2004. Fate of Salmonella enterica serovar Typhimurium on carrots and radishes grown in fields treated with contaminated manure composts or irrigation water. Appl. Environ. Microbiol. 70:2497-2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jacobsen, C. S., and W. E. Holben. 2007. Quantification of mRNA in Salmonella sp. seeded soil and chicken manure using magnetic capture hybridization RT-PCR. J. Microbiol. Methods 69:315-321. [DOI] [PubMed] [Google Scholar]
- 17.Joy, D. M., H. Lee, C. M. Reaume, H. R. Whiteley, and S. Zelin. 1998. Microbial contamination of subsurface tile drainage water from field applications of liquid manure. Can. Agric. Eng. 40:153-160. [Google Scholar]
- 18.Kim, S. B. 2006. Numerical analysis of bacterial transport in saturated porous media. Hydrol. Process. 20:1177-1186. [Google Scholar]
- 19.Laegdsmand, M., A. L. Gimsing, B. W. Strobel, J. C. Sorensen, O. H. Jacobsen, H. Christian, and B. Hansen. 2007. Leaching of isothiocyanates through intact soil following simulated biofumigation. Plant Soil 291:81-92. [Google Scholar]
- 20.Malik, Y. S., G. W. Randall, and S. M. Goyal. 2004. Fate of Salmonella following application of swine manure to tile-drained clay loam soil. J. Water Health 2:97-101. [PubMed] [Google Scholar]
- 21.Marsh, P., N. Z. Morris, and E. M. H. Wellington. 1998. Quantitative molecular detection of Salmonella typhimurium in soil and demonstration of persistence of an active but non-culturable population. Fems. Microbiol. Ecol. 27:351-363. [Google Scholar]
- 22.Mawdsley, J. L., R. D. Bardgett, R. J. Merry, B. F. Pain, and M. K. Theodorou. 1995. Pathogens in livestock waste, their potential for movement through soil and environmental pollution. Appl. Soil Ecol. 2:1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McMurry, S. W., M. S. Coyne, and E. Perfect. 1998. Fecal coliform transport through intact soil blocks amended with poultry manure. J. Environ. Qual. 27:86-92. [Google Scholar]
- 24.Oliver, D. M., C. D. Clegg, P. M. Haygarth, and A. L. Heathwaite. 2005. Assessing the potential for pathogen transfer from grassland soils to surface waters. Adv. Agro. 85:125-180. [Google Scholar]
- 25.Patni, N. K., H. R. Toxopeus, and P. Y. Jui. 1985. Bacterial quality of runoff from manured and non-manured cropland. Trans. ASAE 28:1871-1878. [Google Scholar]
- 26.Pedersen, J. C., and C. S. Jacobsen. 1993. Fate of Enterobacter cloacae JP120 and Alcaligenes eutrophus AEO106(pRO101) in soil during water stress: effects on culturability and viability. Appl. Environ. Microbiol. 59:1560-1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pell, A. N. 1997. Manure and microbes: public and animal health problem? J. Dairy Sci. 80:2673-2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saini, R., L. J. Halverson, and J. C. Lorimor. 2003. Rainfall timing and frequency influence on leaching of Escherichia coli RS2G through soil following manure application. J. Environ. Qual. 32:1865-1872. [DOI] [PubMed] [Google Scholar]
- 29.Santo Domingo, J. W., S. Harmon, and J. Bennett. 2000. Survival of Salmonella species in river water. Curr. Microbiol. 40:409-417. [DOI] [PubMed] [Google Scholar]
- 30.Schafer, A., P. Ustohal, H. Harms, F. Stauffer, T. Dracos, and A. J. B. Zehnder. 1998. Transport of bacteria in unsaturated porous media. J. Contam. Hydrol. 33:149-169. [Google Scholar]
- 31.Smith, M. S., G. W. Thomas, R. E. White, and D. Ritonga. 1985. Transport of Escherichia coli through intact and disturbed soil columns. J. Environ. Qual. 14:87-91. [Google Scholar]
- 32.Tirado, C., and K. Schmidt. 2001. WHO surveillance programme for control of foodborne infections and intoxications: preliminary results and trends across greater Europe. J. Infect. 43:80-84. [DOI] [PubMed] [Google Scholar]
- 33.Torkzaban, S., S. S. Tazehkand, S. L. Walker, and S. A. Bradford. 5 April 2008, posting date. Transport and fate of bacteria in porous media: coupled effects of chemical conditions and pore space geometry. Water Resour. Res. 44:W04403. doi: 10.1029/2007WR006541. [DOI]
- 34.Turpin, P. E., K. A. Maycroft, C. L. Rowlands, and E. M. H. Wellington. 1993. Viable but non-culturable salmonellas in soil. J. Appl. Bacteriol. 74:421-427. [DOI] [PubMed] [Google Scholar]
- 35.Unc, A., and M. J. Goss. 2003. Movement of faecal bacteria through the vadose zone. Water Air Soil Pollut. 149:327-337. [Google Scholar]
- 36.Unc, A., and M. J. Goss. 2004. Transport of bacteria from manure and protection of water resources. Appl. Soil Ecol. 25:1-18. [Google Scholar]
- 37.Vinten, A. J. A., D. R. Lewis, D. R. Fenlon, K. A. Leach, R. Howard, I. Svoboda, and I. Ogden. 2002. Fate of Escherichia coli and Escherichia coli O157 in soils and drainage water following cattle slurry application at 3 sites in southern Scotland. Soil Use Manag. 18:223-231. [Google Scholar]
- 38.World Health Organization. 2006. WHO global salm-surv progress report 2000-2005: building capacity for laboratory-based foodborne disease surveillance and outbreak detection and response. World Health Organization, Geneva, Switzerland. http://www.who.int/salmsurv/links/GSSProgressReport2005.pdf.