Abstract
The cationic lytic peptide cecropin B (CB), isolated from the giant silk moth (Hyalophora cecropia), has been shown to effectively eliminate Gram-negative and some Gram-positive bacteria. In this study, the effects of chemically synthesized CB on plant pathogens were investigated. The S50s (the peptide concentrations causing 50% survival of a pathogenic bacterium) of CB against two major pathogens of the tomato, Ralstonia solanacearum and Xanthomonas campestris pv. vesicatoria, were 529.6 μg/ml and 0.29 μg/ml, respectively. The CB gene was then fused to the secretory signal peptide (sp) sequence from the barley α-amylase gene, and the new construct, pBI121-spCB, was used for the transformation of tomato plants. Integration of the CB gene into the tomato genome was confirmed by PCR, and its expression was confirmed by Western blot analyses. In vivo studies of the transgenic tomato plant demonstrated significant resistance to bacterial wilt and bacterial spot. The levels of CB expressed in transgenic tomato plants (∼0.05 μg in 50 mg of leaves) were far lower than the S50 determined in vitro. CB transgenic tomatoes could therefore be a new mode of bioprotection against these two plant diseases with significant agricultural applications.
Bacterial plant diseases are a source of great losses in the annual yields of most crops (5). The agrochemical methods and conventional breeding commonly used to control these bacterially induced diseases have many drawbacks. Indiscriminate use of agrochemicals has a negative impact on human, as well as animal, health and contributes to environmental pollution. Conventional plant-breeding strategies have limited scope due to the paucity of genes with these traits in the usable gene pools and their time-consuming nature. Consequently, genetic engineering and transformation technology offer better tools to test the efficacies of genes for crop improvement and to provide a better understanding of their mechanisms. One advance is the possibility of creating transgenic plants that overexpress recombinant DNA or novel genes with resistance to pathogens (36). In particular, strengthening the biological defenses of a crop by the production of antibacterial proteins with other origins (not from plants) offers a novel strategy to increase the resistance of crops to diseases (35, 39, 41). These antimicrobial peptides (AMPs) include such peptides as cecropins (2, 15, 20, 23-24, 27, 31, 42, 50), magainins (1, 9, 14, 29, 47), sarcotoxin IA (35, 40), and tachyplesin I (3). The genes encoding these small AMPs in plants have been used in practice to enhance their resistance to bacterial and fungal pathogens (8, 22, 40). The expression of AMPs in vivo (mostly cecropins and a synthetic analog of cecropin and magainin) with either specific or broad-spectrum disease resistance in tobacco (14, 24, 27), potato (17, 42), rice (46), banana (9), and hybrid poplar (32) have been reported. The transgenic plants showed considerably greater resistance to certain pathogens than the wild types (4, 13, 24, 27, 42, 46, 50). However, detailed studies of transgenic tomatoes expressing natural cecropin have not yet been reported.
The tomato (Solanum lycopersicum) is one of the most commonly consumed vegetables worldwide. The annual yield of tomatoes, however, is severely affected by two common bacterial diseases, bacterial wilt and bacterial spot, which are caused by infection with the Gram-negative bacteria Ralstonia solanacearum and Xanthomonas campestris pv. vesicatoria, respectively. Currently available pesticides are ineffective against R. solanacearum, and thus bacterial wilt is a serious problem.
Cecropins, one of the natural lytic peptides found in the giant silk moth, Hyalophora cecropia (25), are synthesized in lipid bodies as proteins consisting of 31 to 39 amino acid residues. They adopt an α-helical structure on interaction with bacterial membranes, resulting in the formation of ion channels (12). At low concentrations (0.1 μM to 5 μM), cecropins exhibit lytic antibacterial activity against a number of Gram-negative and some Gram-positive bacteria, but not against eukaryotic cells (11, 26, 33), thus making them potentially powerful tools for engineering bacterial resistance in crops. Moreover, cecropin B (CB) shows the strongest activity against Gram-negative bacteria within the cecropin family and therefore has been considered an excellent candidate for transformation into plants to improve their resistance against bacterial diseases.
The introduction of genes encoding cecropins and their analogs into tobacco has been reported to have contradictory results regarding resistance against pathogens (20). However, subsequent investigations of these tobacco plants showed that the expression of CB in the plants did not result in accumulation of detectable levels of CB, presumably due to degradation of the peptide by host peptidases (20, 34). Therefore, protection of CB from cellular degradation is considered to be vital for the exploitation of its antibacterial activity in transgenic plants. The secretory sequences of several genes are helpful, because they cooperate with the desired genes to enhance extracellular secretion (24, 40, 46). In the present study, a natural CB gene was successfully transferred into tomatoes. The transgenic plants showed significant resistance to the tomato diseases bacterial wilt and bacterial spot, as well as with a chemically synthesized CB peptide.
MATERIALS AND METHODS
Bacterial strains.
Plasmid cloning and amplification of the CB gene were performed in Escherichia coli strain DH5α. Plant pathogens, such as R. solanacearum PPS4, X. campestris pv. vesicatoria XVT28 (both provided by C. P. Cheng, Academia Sinica, Taiwan), and Erwinia carotovora subsp. carotovora (ATCC 13137; from the Bioresource Collection and Research Center [BCRC], Hsinchu, Taiwan), were cultured and maintained in 523 medium (0.3 g MgSO4·7H2O, 2.0 g K2HPO4, 4.0 g yeast extract, 8.0 g casein hydrolysate, 10.0 g sucrose, 15.0 g agar per liter) at 28°C. For plant transformation, Agrobacterium tumefaciens strain LBA4404 (provided by the Transgenic Plant Laboratory, Academia Sinica, Taiwan) was maintained in YEP medium (1% [wt/vol] yeast extract, 0.5% [wt/vol] NaCl, 1% [wt/vol] peptone). Antibiotics were used for plant tissue culture at the following concentrations: carbenicillin at 500 mg/liter and kanamycin at 50 mg/liter (Sigma, St. Louis, MO). Other pathogens used to test antibacterial activity in vitro were E. coli (ATCC 25922) and Salmonella enteritidis (BCRC 10744) (both from the BCRC). These bacteria were maintained in Luria-Bertani broth or nutrient broth (NB) medium.
Synthesis and construction of the CB gene.
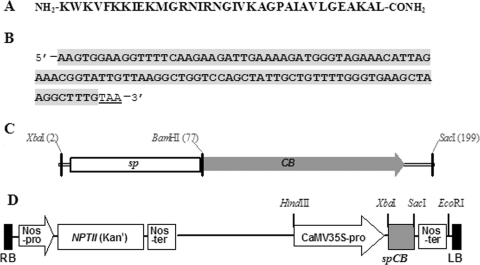
Based on the amino acid sequence of CB (Fig. 1A), the DNA sequence containing the coding region of the CB gene (Fig. 1B) was chemically synthesized (GenScript Corp., Piscataway, NJ). For improved plant expression, the codon choice was made according to the codon usage in the tomato. The primers 5′-GCTGGATCCAAGTGGAAGGTTTTC-3′ (the BamHI site is underlined) and 5′-GAGAGCTCACTATTACAAAGCCTTAG-3′ (the SacI site is underlined) were used to obtain BamHI and SacI restriction sites for subcloning. The CB gene was amplified by PCR and inserted into a pUC18 vector at the 3′ end of the secretory sequence (synthesized by GenScript Corp.) from the barley α-amylase gene in a pUC18 vector. A 199-bp fragment consisting of secretory signal peptide (sp) and CB sequences (Fig. 1C) was subcloned into the binary plant expression vector pBI121 under the control of a constitutive promoter, Cauliflower mosaic virus 35S (CaMV35S) and a terminator, NOS (nopaline synthase gene). This new construct was named pBI121-spCB (Fig. 1D). The sequence including the signal peptide and CB was confirmed by DNA sequencing.
FIG. 1.
Sequence and expression construct of CB. (A) Amino acid sequence of CB. (B) CB's nucleotide sequence containing the coding region and stop codon TAA (underlined). For improved plant expression, codon choice was made according to the codon usage in the tomato. (C) Gene construct of CB with the secretory signal peptide (sp). (D) Schematic diagram of the expression construct pBI121-spCB used for the current tomato expression. Expression of the CB gene was driven by the Cauliflower mosaic virus (CaMV) 35S promoter and the Nos (nopaline synthase gene) terminator. The NPTII gene encoding neomycin phosphotransferase II served as a selectable marker for plant transformation. RB and LB are the right and left border regions of the Agrobacterium tumefaciens Ti plasmid, respectively.
Transformation of the plasmid into A. tumefaciens.
The pBI121-spCB plasmid was introduced into A. tumefaciens LBA4404 by the freeze-thaw method (Hermann Schmidt, DNA-Cloning Service, Hamburg, Germany [http://www.DNA-Cloning-Service.de]) for plant transformation.
Plant transformation (tomato).
Tomato seeds (S. lycopersicum cv. Microtom; seeds provided by Kin-Ying To, Academia Sinica, Taiwan) were treated with a 2% (wt/vol) NaOCl, 0.1% (wt/vol) Tween solution for 30 min and rinsed five times with sterile deionized water in order to prevent any growth of microorganisms while in culture. The sterilized seeds were germinated and grown on Murashige and Skoogs (MS) medium and maintained in a growth chamber under a 16-h/8-h (light/dark) photoperiod at 26°C with a relative humidity of 55%. Cotyledon leaves from 7-day-old seedlings were excised. To stimulate the initial growth, the explants were preconditioned overnight with preculture medium (MS salt, 2% [wt/vol] sucrose, Gamborg B5 basal salt mixture with vitamins [16], 2 mg/liter benzylaminopurine, pH 5.7, 0.3% [wt/vol] phytagel, 0.25 mg/liter indole-3-acetic acid) at 26°C. The explants were then cocultivated with the overnight culture of A. tumefaciens LBA4404 containing the pBI121-spCB plasmid for 48 h at 26°C in the dark. After being washed 3 times with sterile deionized water containing 500 mg/liter carbenicillin, the explants were incubated in regeneration medium (MS salt, Nitsch vitamin [37], 3% [wt/vol] sucrose, pH 5.7, 2 mg/liter zeatin, 2 mg/liter kinetin, 0.3% [wt/vol] phytagel, 500 mg/liter carbenicillin) under a 16-h/8-h (light/dark) photoperiod at 26°C and 55% humidity for the purpose of shoot induction.
Regeneration and selection of transgenic plants (tomato).
To select transformants, the explants were subcultured once a week in regeneration medium supplemented with 50 mg/liter of kanamycin for a period of several weeks. The initial callus was observed at the site of wounding on the explants. When shoots appeared from the calli, they were separated and transferred into shoot formation medium (MS salt, Nitsch vitamin, 3% [wt/vol] sucrose, pH 5.7, 0.3% [wt/vol] phytagel, 500 mg/liter carbenicillin, supplemented with 50 mg/liter kanamycin). The regenerated shoots were then transferred and grown in Megenta boxes for root induction (1/2 MS salt, LS vitamin [30], 3% [wt/vol] sucrose, 0.7 g/liter 2-n-morpholino-ethanesulfonic acid, pH 5.7, 0.3% [wt/vol] phytagel, 500 mg/liter carbenicillin, supplemented with 50 mg/liter kanamycin). The rooted plants were then transplanted into boxes containing soil. The tomato plants were grown in a growth cabinet (16 h at 28°C and 8 h in the dark at 23°C) in a mixture of peat-vermiculite-perlite (10:1:2 [vol/vol/vol]).
DNA isolation and PCR analysis.
Genomic DNA was isolated from leaf tissue of tomato plants by using a plant genomic DNA purification kit (Genemark, Taiwan). Integration of the CB gene into the plant genome was confirmed by PCR amplification of the CB transgene using the forward primer 5′-GCTGGATCCAAGTGGAAGGTTTTC-3′ and the reverse primer 5′-GAGAGCTCACTATTACAAAGCCTTAG-3′. PCR products were analyzed on 1% (wt/vol) agarose gels, stained with ethidium bromide, and visualized under UV light.
Western blot analysis.
Fresh leaves of wild-type or transgenic tomato plants were ground in liquid nitrogen and then homogenized in sterile deionized water. After incubation at 70°C for 60 min, the debris was removed by centrifugation at 10,000 × g for 5 min. For Western blot analysis, the supernatant containing soluble proteins was precipitated with 5% (wt/vol) trichloroacetic acid (TCA) at 4°C for 10 min. The protein precipitates were washed three times in cold acetone and resuspended in 0.1 N NaOH. Protein samples were separated by 10% (wt/vol) SDS-PAGE and electrotransferred onto a polyvinylidene difluoride (PVDF) nylon membrane (Millipore Corp., Burlington, MA). Immunodetection was performed using polyclonal rabbit anti-CB serum (Cashmere Biotech, Taipei, Taiwan) as the primary antibody at 1:3,000 dilution and a goat anti-rabbit alkaline phosphatase-conjugated secondary antibody (Chemicon International, Inc., Temecula, CA) at a dilution of 1:8,000. Antigen-antibody complexes were detected using Chemiluminescence Reagent Plus (Perkin Elmer, Shelton, CT), and the images were recorded on X-ray film.
In vitro bioassays of antibacterial activity: liquid growth inhibition assay. (i) Antibacterial efficacy of synthetic CB peptide.
Liquid cultures of test bacteria were grown overnight prior to the assay and adjusted to 1 × 106 CFU/ml with 2 ml MH medium (Mueller-Hinton broth: 2.0 g of beef extract, 17.5 g acid digest of casein, and 1.5 g of soluble starch per liter). CB peptide was dissolved in sterile deionized water to a concentration of 128 μg/ml (33.4 μM) as a stock solution. The stock solution was then serially diluted to the different concentrations used for the experiments. A known amount of CB (SynPep Corp., Dublin, CA) was added to a bacterial suspension and incubated in a rotary shaker at 125 rpm at 28°C. After incubation for 17 h, the optical density at 600 nm (OD600) was recorded. The antibacterial activity was determined by measuring the percentage survival of bacteria. The survival rate (percent) of bacteria was calculated by changes in optical density: the test OD600 (bacterial growth in the presence of peptide) divided by the reference OD600 (bacterial growth in the absence of peptide). The peptide concentration causing 50% survival of each pathogen derived from dose-response curves (constructed by using 11 different concentrations of synthetic CB) for growth inhibition was designated the S50. The antibiotics ampicillin and kanamycin were used as controls.
(ii) Antibacterial activities of CB transgenic-plant extracts.
The tested bacteria were grown overnight and adjusted to 1 × 104 CFU with 800 μl NB medium. Protein extracts (700 μl) isolated from 300 mg of leaves of transgenic and wild-type tomato plants were added to the bacterial suspension and incubated in a rotary shaker at 125 rpm and at a temperature appropriate for the specific bacteria. After incubation for 17 h, the OD600 was recorded. Three independent experiments with separate preparation and three replicates for each experiment were performed.
In vivo plant bioassay for resistance to bacterial wilt.
R. solanacearum PPS4 was grown for 72 h at 30°C on NB medium. The bacterial suspension for inoculation was prepared and adjusted until the OD600 = 0.3 (108 CFU/ml). Thirty milliliters of the bacterial suspension was poured onto each 6-week-old tomato plant (wild-type and transgenic [second generation, or T2] plants) grown in a greenhouse, and they were kept in a growth chamber at 30°C. Disease development and the symptoms of the plants were then recorded on the 14th day after infection. Disease readings were made according to the following numerical grades: 0, no symptoms; 1, one leaf partially wilted; 2, two or three leaves wilted; 3, all except the top 2 or 3 leaves wilted; 4, all leaves wilted; 5, death (49). The experiment was repeated twice.
In vivo plant bioassay for resistance to bacterial spot.
X. campestris pv. vesicatoria XVT28 was grown for 72 h at 30°C on NB agar medium. The bacterial suspension for inoculation was prepared in distilled water containing 0.1% (wt/vol) carboxymethyl cellulose (CMC) and adjusted until the OD600 = 0.3 (108 CFU/ml). More than eight fully expanded leaves of 6-week-old greenhouse-grown tomato plants (wild-type and transgenic [T2] plants) were soaked in the bacterial suspension for 30 s. The inoculated plants were then incubated in a growth chamber at 30°C. Disease development and symptoms were recorded on the 14th day after infection. The numbers of spots on the 5th and 6th leaves were calculated. For the quantification of X. campestris pv. vesicatoria in the infected regions, mutant strains of X. campestris pv. vesicatoria resistant to rifampin were selected on NB agar medium supplemented with 100 μg/ml rifampin and used for inoculation. After 14 days of infection, the extracts from the leaves were dispersed on NB agar medium supplemented with 100 μg/ml rifampin. After incubation for 48 to 72 h, bacterial growth was monitored by counting viable CFU. The experiment was repeated twice.
RESULTS
Effectiveness of chemically synthesized cecropin B against three plant pathogens.
CB has long been reported to possess lytic activity against several Gram-negative bacteria, such as E. coli, Salmonella enteritidis, Klebsiella pneumoniae, and Pseudomonas aeruginosa (10, 11). To confirm that CB would provide protection against major bacterial diseases of the tomato, the effectiveness of a synthetic CB peptide against two common tomato pathogens, R. solanacearum and X. campestris pv. vesicatoria, and the plant pathogen E. carotovora were investigated by liquid growth inhibition assay (Table 1). The S50 of CB for the plant pathogen E. carotovora (1.32 μg/ml) was lower than those of control samples, like ampicillin and kanamycin. Furthermore, CB was extremely potent against the pathogen X. campestris pv. vesicatoria XVT28, with an S50 of only 0.29 μg/ml. The S50 of CB (529.6 μg/ml) against R. solanacearum PPS4 was higher than the S50s of antibiotic controls.
TABLE 1.
S50s of synthetic CB and the antibiotics ampicillin and kanamycin for the tomato bacterial pathogens Ralstonia solanacearum, Xanthomonas campestris pv. vesicatoria, and the Gram-negative bacterium Erwinia carotovora (18, 28)a
| Pathogen | S50 [μg/ml (μM)] |
||
|---|---|---|---|
| Ampicillin | Kanamycin | CB | |
| R. solanacearum | 1.03 (2.8) | 0.44 (0.76) | 529.6 (138) |
| X. campestris pv. vesicatoria | 6.56 (17.7) | 0.46 (0.78) | 0.29 (0.08) |
| Erwinia carotovora | 3.73 (10.0) | 1.96 (3.36) | 1.32 (0.34) |
Liquid cultures of test bacteria were grown overnight prior to the assay and adjusted to 1 × 106 CFU/ml. A known amount of CB was added to a bacterial suspension and incubated in a rotary shaker at 125 rpm at 28°C. After incubation for 17 h, the OD600 was recorded. The S50s of pathogens were represented by single measurements.
Selection of transgenic tomato plants and their progeny (PCR analysis).
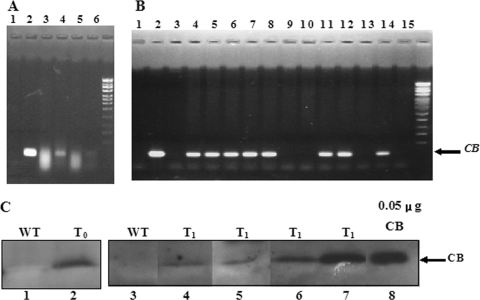
Transgenic tomato plants carrying a pBI121-spCB construct were generated and grown on medium containing 50 mg/liter kanamycin, and five independent kanamycin-resistant T0 (original) transgenic lines were selected for further analysis. Integration of the CB gene into the tomato genome was confirmed by PCR. The following control experiments were carried out: PCR amplification in the absence of a template (Fig. 2A and B, lanes 1) as a negative control, genomic DNA isolated from wild-type tomatoes as a template (Fig. 2A, lane 3, and B, lane 15), and plasmid pBI121-spCB DNA as a template (Fig. 2A and B, lanes 2, as positive controls). Three individual kanamycin-resistant T0 transgenic lines were selected for the analysis of total genomic DNA, as shown in Fig. 2A. Among them, lines 1 and 3 gave positive PCR results (Fig. 2A, lanes 4 and 6). The T0 transgenic line 1 was therefore selected for further testing for the presence of the CB gene in the next generation (T1, or first generation). As shown in Fig. 2B, among 11 T1 transgenic plants (lanes 4 to 14), only three plants (lanes 9, 10, and 13) gave negative PCR results. The outcome of segregation (the ratio of positive to negative responses was 8:3) was close to the theoretical value of 3:1, which indicated that the T0 transgenic plant probably included a single copy of CB.
FIG. 2.
Analysis of the CB gene and peptide in the transgenic tomato. (A and B) PCR analysis of the CB gene in wild-type and transgenic tomatoes. Genomic DNA isolated from 10 mg of fresh leaves from transgenic tomatoes (T0 plants, panel A, lanes 4 to 6; T1 plants, panel B, lanes 4 to 14) and nontransgenic control plants (A, lane 3, and B, lane 15) were used as templates for PCRs. The PCR products were analyzed on 1% (wt/vol) agarose gels. (A and B) Lanes 1, no template (negative control); lanes 2, PCR product amplified from plasmid pBI121-spCB (positive control); lane 3 in panel B is the PCR product amplified from plasmid pBI121 without the CB gene (negative control). (C) Analysis of peptide CB expressed in transgenic tomatoes. The levels of CB peptide in transgenic tomato plants were determined by Western hybridization analysis. Soluble protein extracts from 50 mg of fresh leaves of transgenic tomatoes (T0 plant, lane 2; T1 plants, lanes 4 through 7) and nontransgenic control plants (wild type [WT], lanes 1 and 3) were Western hybridized using a polyclonal antibody raised against CB. Lane 8 is a positive control with 0.05 μg of synthetic CB peptide.
Expression of CB in transgenic tomato plants (Western blot analysis).
Transgenic tomato plants, including T0 and T1 generations, carrying the CB gene were analyzed for peptide expression in plants by Western hybridization assay (Fig. 2C). An antibody-reactive signal at 4.0 kDa comigrated with ∼50 ng synthetic CB (Fig. 2C, lane 8). The positive signals were detected in the transgenic T0 (Fig. 2C, lane 2) and T1 (Fig. 2C, lanes 4 to 7) tomatoes, but not in nontransgenic plants (Fig. 2C, lanes 1 and 3).
Plant pathogen resistance of CB-expressing tomato.
To evaluate the resistance of transgenic plants to plant pathogens, control (wild-type) and transgenic T2 tomato plants were challenged with R. solanacearum and X. campestris pv. vesicatoria. At an inoculum concentration of 108 CFU/ml of R. solanacearum, the symptoms of the disease bacterial wilt were observed in wild-type plants from the 10th day after infection (Fig. 3A). By the 14th day after infection, all leaves of wild-type plants had wilted (Fig. 3B), leading to plant death. T2 transgenic tomato plants expressing CB were healthy and showed no symptoms of bacterial wilt on the 10th day (Fig. 3C) and the 14th day (Fig. 3D). The rating scale of disease (49) for the wild type can be set at 2 (two or three leaves wilted after 10th day of infection), 3 (all except the top 2 or 3 leaves wilted after the 12th day of infection), 4 (all leaves wilted after the 14th day of infection), or 5 (death after the 21st day of infection). However, the rating scale of disease for transgenic (T2) tomatoes was zero (no symptoms even after the 21st day of infection).
FIG. 3.
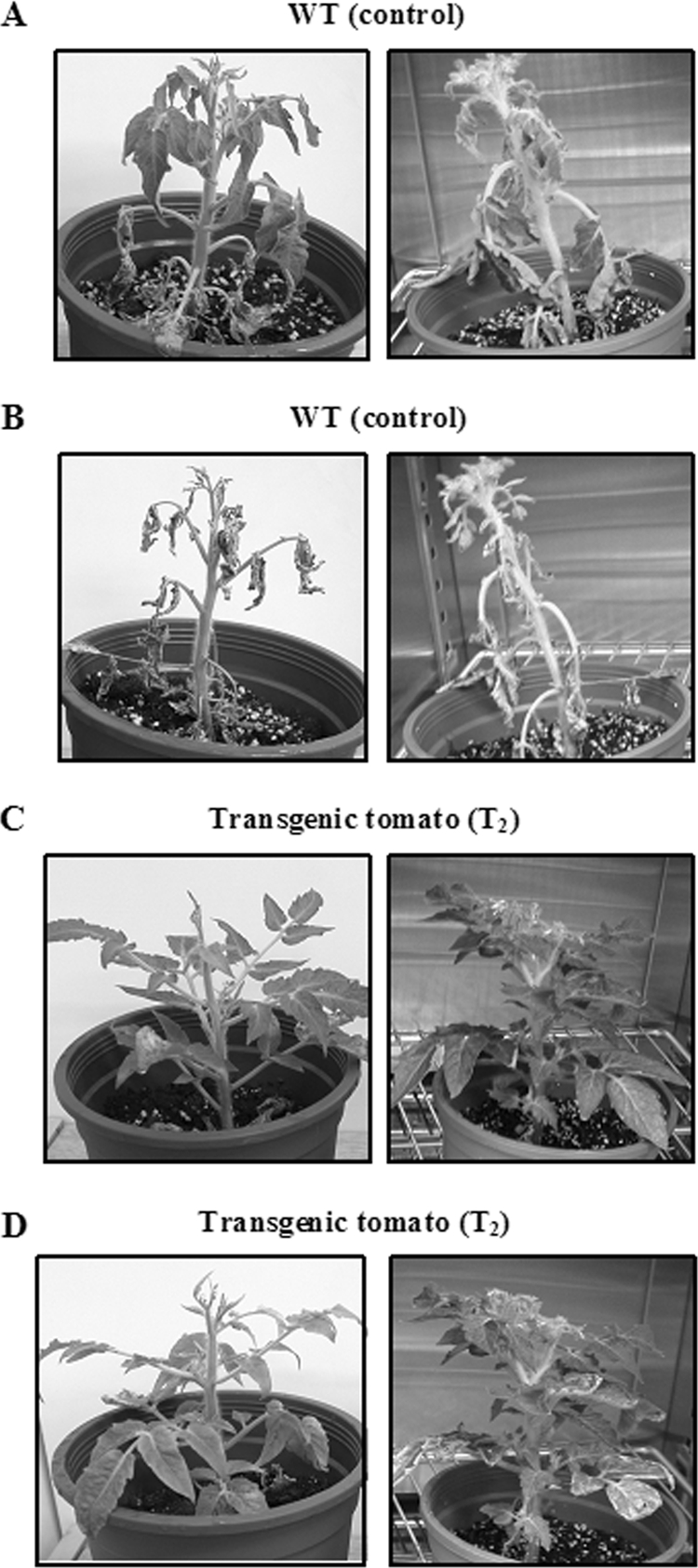
Enhanced resistance to the disease bacterial wilt in CB transgenic tomatoes. Six-week-old tomato plants, including wild-type (WT) and transgenic (T2) plants, were challenged with 30-ml suspensions of the pathogen Ralstonia solanacearum (108 CFU/ml). Disease development and symptoms in wild-type (A and B) and transgenic (C and D) tomatoes were recorded on different days. The photographs were taken on the 10th (A and C) and 14th (B and D) days after infection and are representative of duplicate analyses.
The symptoms of bacterial spot, caused by X. campestris pv. vesicatoria, were observed clearly in wild-type plants on the 14th day after infection. The numbers of spots on the 5th and 6th leaves for wild-type and transgenic plants were 42.7 and 11, respectively. Although spots appeared on the leaves of CB transgenic plants, the number of spots was 74% less than for the wild type. After 14 days of infection, the infected leaves were ground with distilled water in order to calculate the quantity of pathogens. The bacterial population in the spotted regions of transgenic plants (the number of pathogens on the 5th and 6th leaves was 189,800/g of leaves) was 49% less than those on the wild-type plants (the number of pathogens on the 5th and 6th leaves was 97,100/g of leaves). These results imply a strong correlation between disease symptoms (the number of spots) and pathogen quantity (the number of X. campestris pv. vesicatoria cells in the spotted region).
In vitro antibacterial activities of transgenic tomato plant extracts.
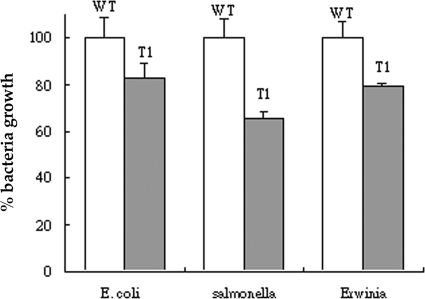
The ability of protein extracts isolated from the leaves of transgenic tomato plants to inhibit the growth of bacteria was determined by liquid growth inhibition assay. The growth of three different indicator bacteria, namely, E. coli, S. enteritidis, and E. carotovora, was examined. Incubation with total protein extracts isolated from 300 mg of leaves of transgenic tomatoes (Fig. 4, T 1) showed an inhibition range from 16.8% to 34.5% on the growth of bacteria (E. coli, S. enteritidis, and E. carotovora). Protein extracts from wild-type tomato plants (Fig. 4) did not display any antimicrobial activity.
FIG. 4.
In vitro antimicrobial assay of the effects of tomato leaf extracts on bacteria. Total protein extracts prepared from 300 mg of tomato leaves were used to investigate growth inhibition in the bacteria E. coli, Salmonella, and Erwinia. After incubation for 17 h, the bacterial growth in the presence of protein extracts from transgenic tomatoes (T1) was recorded by the OD600 and normalized to the OD600 of bacteria grown in the presence of protein extracts from wild-type (WT) tomatoes. The data are the means and the standard deviations of three replicates.
DISCUSSION
For the application of peptide antibiotics in agriculture, one of the effective methods is through the transgenic expression of these simple peptides by standard molecular-cloning techniques. Although a number of peptide antibiotics have extensive modifications (e.g., lantibiotics) (45), most of these peptides have functions based on their primary sequences. Thus, these simple peptides have become popular targets for engineering plant resistance to microbial pathogens, and recently, there has been a growing interest in developing antimicrobial peptides for disease resistance in plants (7, 19, 48). The characteristic features of the cationic lytic peptides cecropins are their net positive charge, an affinity for negatively charged prokaryotic membrane phospholipids over neutrally charged eukaryotic membranes, and the ability to form aggregates that disrupt the bacterial membrane (6, 21). It is difficult for the pathogen to develop resistance against the cecropin-like peptides in planta. Several previous studies have indicated that CB-derived peptides possess in vitro activity against several bacterial and cancer cells (10, 11). Furthermore, the antibacterial activities of synthetic CB against some well-known Gram-negative bacteria, such as E. coli, S. enteritidis, K. pneumoniae, and P. aeruginosa, were investigated in the current experiments. The results showed that CB has an S50 between 1.29 μg/ml and 1.51 μg/ml (data not shown). Moreover, CB was very effective at killing the tomato pathogen X. campestris pv. vesicatoria, with an S50 as low as 0.29 μg/ml (∼0.08 μM) (Table 1). The CB gene is therefore an ideal candidate for genetic improvement to engineer resistance to pathogens in transgenic tomato plants.
In this study, transgenic tomato plants constitutively expressing the CB gene from H. cecropia were generated by Agrobacterium-mediated transformation. In vitro, extracts of the CB peptide expressed in tomatoes showed broad-spectrum antimicrobial activity. The CB produced by the transgenic tomato plants was biologically active and inhibited the growth of bacteria (E. coli, S. enteritidis, and E. carotovora) (Fig. 4). In in vivo challenge studies, transgenic tomatoes showed improved resistance to disease caused by bacterial pathogens, viz., R. solanacearum (Fig. 3) and X. campestris, thus showing the potential of CB as a promising tool for the development of multiple-disease resistance in plants.
Transgenic tomatoes expressing the CB gene can very effectively resist infection by the pathogen X. campestris pv. vesicatoria. This result confirmed the in vitro outcome of a low S50 (as low as 0.29 μg/ml) (Table 1) of CB against the pathogen. A higher concentration of CB was needed to effectively inhibit the growth of the pathogen R. solanacearum in vitro (S50, 529.6 μg/ml) (Table 1). The levels of CB expressed in transgenic plants (∼0.05 μg in 50 mg of leaf material) are far smaller than the S50 determined in vitro; therefore, even a low-level constitutive expression of CB may help the plant's innate immune defenses to generate the increased resistance seen in vivo (Fig. 3). More detailed resistance studies remain to be conducted in the future.
Owens (43) reported that transgenic proteins could be degraded by plant endogenous peptidase, and expressed peptides were therefore assumed to be ruined in plants (44). Many previous reports supported this argument and reported that cecropin expression was not detected in transgenic plants, blaming the failure of cecropin to increase its abundance in transgenic plants on cellular degradation by plant peptidase, thus limiting the quantity of peptide available to kill the pathogen (2, 15, 20). However, other reports showed an alternative method in which making certain modifications to peptides (such as in their sequences or combining them with other peptides as chimeras) has made it possible to avoid such degradation and to allow sufficient levels of peptides to accumulate to resist pathogens in plants (24, 27, 42, 44). Another alternative way to prevent peptide degradation in plants is to secrete the peptide from the cell. Signal peptides of several genes, such as the PR1a gene in tobacco, the α-amylase gene in barley, and the chitinase gene in rice, can cooperate with the desired genes to allow extracellular secretion (24, 40, 46). A CB modified by the simple addition of the secretory signal peptide of the α-amylase gene to the N terminus of the CB sequence was used in this experiment. The CB peptide proved to have excellent antimicrobial activity.
Western hybridization analysis of total protein extracts from CB expression lines provided strong evidence that CB accumulated in tomato plants. The level of accumulation of CB peptide detected in total protein extracts prepared from leaves of transgenic lines was moderate, estimated at about 1 μg (= 261 pmol) per gram of fresh leaves (Fig. 2C) (the estimation was made by comparing the signal density of a transgenic plant expressing CB with the signal density of a known weight of synthetic CB). This result is comparable with the expression (250 pmol/g) of the antibacterial peptide sarcotoxin IA in transgenic tobacco plants (40). Furthermore, one of the most serious concerns regarding the use of lytic peptides in enhancing plant defense against invading pathogens is the possible toxicity to the plant. The present investigation showed that the CB gene expressed in tomato plants had no deleterious effects on the transgenic plants. Plant morphology, plant growth, and yields of fruits and seeds (data not shown) were normal. In most cases, the minimum lethal concentration of cecropin derivatives required for toxicity to plant protoplast, intact cells, and tissues is much higher than that required to kill the bacterial cells (34, 38). For example, the lethal concentration of a modified cecropin SB37 on the protoplast of tomatoes (S. lycopersicum cv. “Red Cherry” [38]) was reported to be about 4.5 μM, which is far beyond the amount of CB expressed in the transgenic tomato (<1 ng/mg of leaf material) (Fig. 2C) or much higher than those of CB analogs transferred into other transgenic plants (24, 27, 42, 46). The expression of CB in tomato by the method described here therefore is considered safe for the plant.
In summary, the expression of CB in tomato may effectively protect the plant against two common bacterial diseases: bacterial wilt and bacterial spot. The process by which the CB-containing transgenic tomato plant was developed in the present study, therefore, may be extended to other transgenic crop plants exhibiting resistance to plant pathogens.
Acknowledgments
This work was partially supported by a grant from the National Science and Technology Program for Agricultural Biotechnology in Taiwan and the National Science Council in Taiwan (NSC96-2320-B-001-022-MY3).
We thank C. P. Cheng at the Agricultural Biotechnology Research Center, Academia Sinica, for providing tomato pathogens.
Footnotes
Published ahead of print on 4 December 2009.
REFERENCES
- 1.Alan, A. R., and E. D. Earle. 2002. Sensitivity of bacterial and fungal plant pathogens to the lytic peptides, MSI-99, magainin II, and cecropin B. Mol. Plant Microbe Interact. 15:701-708. [DOI] [PubMed] [Google Scholar]
- 2.Allefs, S., D. Florack, C. Hoogendoorn, and W. Stiekema. 1995. Erwinia soft rot resistance of potato cultivars transformed with a gene construct coding for antimicrobial peptide cecropin B is not altered. Am. Potato J. 72:437-445. [Google Scholar]
- 3.Allefs, S., E. R. Jong, D. Florack, C. Hoogendoorn, and W. Stiekema. 1996. Erwinia soft rot resistance of potato cultivars expressing antimicrobial peptide tachyplesin I. Mol. Breed. 2:97-105. [Google Scholar]
- 4.Arce, P., M. Moreno, M. Gutierrez, M. Gebauer, P. Dell'Orto, H. Torres, I. Acuna, P. Oliger, A. Venegas, X. Jordana, J. Kalazich, and L. Holuigue. 1999. Enhanced resistance to bacterial infection by Erwinia carotovora subsp. atroseptica in transgenic potato plants expressing the attacin or the cecropin SB-37 genes. Am. J. Potato Res. 76:169-177. [Google Scholar]
- 5.Baker, B., P. Zambryski, B. Staskawicz, and S. P. Dinesh-Kumar. 1997. Signaling in plant-microbe interactions. Science 276:726-733. [DOI] [PubMed] [Google Scholar]
- 6.Biggin, P. C., and M. S. Sansom. 1999. Interactions of alpha-helices with lipid bilayers: a review of simulation studies. Biophys. Chem. 76:161-183. [DOI] [PubMed] [Google Scholar]
- 7.Broekaert, W. F., B. P. A. Cammue, M. F. C. De Bolle, K. Thevissen, G. W. De Samblanx, and R. W. Osborn. 1997. Antimicrobial peptides from plants. Crit. Rev. Plant Sci. 16:297-323. [Google Scholar]
- 8.Carmona, M. J., A. Molina, J. A. Fernandez, J. J. Lopez-Fando, and F. Garcia-Olmedo. 1993. Expression of the alpha-thionin gene from barley in tobacco confers enhanced resistance to bacterial pathogens. Plant J. 3:457-462. [DOI] [PubMed] [Google Scholar]
- 9.Chakrabarti, A., T. R. Ganapathi, P. K. Mukherjee, and V. A. Bapat. 2003. MSI-99, a magainin analogue, imparts enhanced disease resistance in transgenic tobacco and banana. Planta 216:587-596. [DOI] [PubMed] [Google Scholar]
- 10.Chen, H. M., S. C. Chan, J. C. Lee, C. C. Chang, M. Murugan, and R. W. Jack. 2003. Transmission electron microscopic observations of membrane effects of antibiotic cecropin B on Escherichia coli. Microsc. Res. Tech. 62:423-430. [DOI] [PubMed] [Google Scholar]
- 11.Chen, H. M., W. Wang, D. Smith, and S. C. Chan. 1997. Effects of the anti-bacterial peptide cecropin B and its analogs, cecropins B-1 and B-2, on liposomes, bacteria, and cancer cells. Biochim. Biophys. Acta 1336:171-179. [DOI] [PubMed] [Google Scholar]
- 12.Christensen, B., J. Fink, R. B. Merrifield, and D. Mauzerall. 1988. Channel-forming properties of cecropins and related model compounds incorporated into planar lipid membranes. Proc. Natl. Acad. Sci. U. S. A. 85:5072-5076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coca, M., G. Penas, J. Gomez, S. Campo, C. Bortolotti, J. Messeguer, and B. S. Segundo. 2006. Enhanced resistance to the rice blast fungus Magnaporthe grisea conferred by expression of a cecropin A gene in transgenic rice. Planta 223:392-406. [DOI] [PubMed] [Google Scholar]
- 14.DeGray, G., K. Rajasekaran, F. Smith, J. Sanford, and H. Daniell. 2001. Expression of an antimicrobial peptide via the chloroplast genome to control phytopathogenic bacteria and fungi. Plant Physiol. 127:852-862. [PMC free article] [PubMed] [Google Scholar]
- 15.Florack, D., S. Allefs, R. Bollen, D. Bosch, B. Visser, and W. Stiekema. 1995. Expression of giant silkmoth cecropin B genes in tobacco. Transgenic Res. 4:132-141. [DOI] [PubMed] [Google Scholar]
- 16.Gamborg, O. L., R. A. Miller, and K. Ojima. 1968. Nutrient requirements of suspension cultures of soybean root cells. Exp. Cell Res. 50:151-158. [DOI] [PubMed] [Google Scholar]
- 17.Gao, A. G., S. M. Hakimi, C. A. Mittanck, Y. Wu, B. M. Woerner, D. M. Stark, D. M. Shah, J. Liang, and C. M. Rommens. 2000. Fungal pathogen protection in potato by expression of a plant defensin peptide. Nat. Biotechnol. 18:1307-1310. [DOI] [PubMed] [Google Scholar]
- 18.Hancock, R. E. 2007. The complexities of antibiotic action. Mol. Syst. Biol. 3:142-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hancock, R. E., and R. Lehrer. 1998. Cationic peptides: a new source of antibiotics. Trends Biotechnol. 16:82-88. [DOI] [PubMed] [Google Scholar]
- 20.Hightower, R., C. Baden, E. Penzes, and P. Dunsmuir. 1994. The expression of cecropin peptide in transgenic tobacco does not confer resistance to Pseudomonas syringae pv tabaci. Plant Cell Rep. 13:295-299. [DOI] [PubMed] [Google Scholar]
- 21.Houston, M. E., Jr., L. H. Kondejewski, D. N. Karunaratne, M. Gough, S. Fidai, R. S. Hodges, and R. E. Hancock. 1998. Influence of preformed alpha-helix and alpha-helix induction on the activity of cationic antimicrobial peptides. J. Pept. Res. 52:81-88. [DOI] [PubMed] [Google Scholar]
- 22.Huang, Y., and J. H. McBeath. 1994. Differential activation of the bean chalcone synthase gene in transgenic tobacco by compatible and incompatible strains of Pseudomonas solanacearum. Plant Sci. 103:41-49. [Google Scholar]
- 23.Huang, Y., J. H. McBeath, H. Lockwood, and L. D. Owens. 1992. Expression of cecropin B controlled by plant inducible promoters in transgenic tobacco plants confers enhanced resistance to Pseudomonas solanacearum. Phytopathology 82:1101. [Google Scholar]
- 24.Huang, Y., R. O. Nordeen, M. Di, L. D. Owens, and J. H. McBeath. 1997. Expression of an engineered cecropin gene cassette in transgenic tobacco plants confers disease resistance to Pseudomonas syringae pv. tabaci. Phytopathology 87:494-499. [DOI] [PubMed] [Google Scholar]
- 25.Hultmark, D., A. Engstrom, H. Bennich, R. Kapur, and H. G. Boman. 1982. Insect immunity: isolation and structure of cecropin D and four minor antibacterial components from Cecropia pupae. Eur. J. Biochem. 127:207-217. [DOI] [PubMed] [Google Scholar]
- 26.Jaynes, J. M., G. R. Julian, G. W. Jeffers, K. L. White, and F. M. Enright. 1989. In vitro cytocidal effect of lytic peptides on several transformed mammalian cell lines. Pept. Res. 2:157-160. [PubMed] [Google Scholar]
- 27.Jaynes, J. M., P. Nagpala, L. Destéfano-Beltrán, J. H. Huang, J. Kim, T. Denny, and S. Cetiner. 1993. Expression of a cecropin B lytic peptide analog in transgenic tobacco confers enhanced resistance to bacterial wilt caused by Pseudomonas solanacearum. Plant Sci. 89:43-53. [Google Scholar]
- 28.Kohanski, M. A., D. J. Dwyer, B. Hayete, C. A. Lawrence, and J. J. Collins. 2007. A common mechanism of cellular death induced by bactericidal antibiotics. Cell 130:797-810. [DOI] [PubMed] [Google Scholar]
- 29.Liang, H., C. M. Catranis, C. E. Maynard, and W. A. Powell. 2002. Enhanced resistance to the poplar fungal pathogen, Septoria musiva, in 395 hybrid poplar clones transformed with genes encoding antimicrobial peptides. Biotechnol. Lett. 24:383-389. [Google Scholar]
- 30.Linsmaier, E. M., and F. Skoog. 1965. Organic growth factor requirements of tobacco tissue cultures. Physiol. Plant. 18:100-127. [Google Scholar]
- 31.Liu, Q., J. Ingersoll, L. Owens, S. Salih, R. Meng, and F. A. Hammerschlag. 2001. Response of transgenic Royal Gala apple (Malus-domestica Borkh.) shoots carrying a modified cecropin MB39 gene, to Erwinia amylovora. Plant Cell Rep. 20:306-312. [Google Scholar]
- 32.Mentag, R., M. Luckevich, M. J. Morency, and A. Seguin. 2003. Bacterial disease resistance of transgenic hybrid poplar expressing the synthetic antimicrobial peptide D4E1. Tree Physiol. 23:405-411. [DOI] [PubMed] [Google Scholar]
- 33.Mills, D., and F. A. Hammerschlag. 1993. Effect of cecropin B on peach pathogens, protoplasts, and cells. Plant Sci. 93:143-150. [Google Scholar]
- 34.Mills, D., F. A. Hammerschlag, R. O. Nordeen, and L. D. Owens. 1994. Evidence for the breakdown of cecropin B by proteinases in the intercellular fluid of peach leaves. Plant Sci. 104:17-22. [Google Scholar]
- 35.Mitsuhara, I., H. Matsufuru, M. Ohshima, H. Kaku, Y. Nakajima, N. Murai, S. Natori, and Y. Ohashi. 2000. Induced expression of sarcotoxin IA enhanced host resistance against both bacterial and fungal pathogens in transgenic tobacco. Mol. Plant Microbe Interact. 13:860-868. [DOI] [PubMed] [Google Scholar]
- 36.Mourgues, F., M. N. Brisset, and E. Chevreau. 1998. Strategies to improve plant resistance to bacterial diseases through genetic engineering. Trends Biotechnol. 16:203-210. [DOI] [PubMed] [Google Scholar]
- 37.Nitsch, J. P., and C. Nitsch. 1969. Haploid plants from pollen grains. Science 163:85-87. [DOI] [PubMed] [Google Scholar]
- 38.Nordeen, R. O., S. L. Sinden, J. M. Jaynes, and L. D. Owens. 1992. Activity of cecropin SB37 against protoplasts from several plant species and their bacterial pathogens. Plant Sci. 82:101-107. [Google Scholar]
- 39.Oard, S. V., and F. M. Enright. 2006. Expression of the antimicrobial peptides in plants to control phytopathogenic bacteria and fungi. Plant Cell Rep. 25:561-572. [DOI] [PubMed] [Google Scholar]
- 40.Ohshima, M., I. Mitsuhara, M. Okamoto, S. Sawano, K. Nishiyama, H. Kaku, S. Natori, and Y. Ohashi. 1999. Enhanced resistance to bacterial diseases of transgenic tobacco plants overexpressing sarcotoxin IA, a bactericidal peptide of insect. J. Biochem. 125:431-435. [DOI] [PubMed] [Google Scholar]
- 41.Osusky, M., L. Osuska, R. E. Hancock, W. W. Kay, and S. Misra. 2004. Transgenic potatoes expressing a novel cationic peptide are resistant to late blight and pink rot. Transgenic Res. 13:181-190. [DOI] [PubMed] [Google Scholar]
- 42.Osusky, M., G. Zhou, L. Osuska, R. E. Hancock, W. W. Kay, and S. Misra. 2000. Transgenic plants expressing cationic peptide chimeras exhibit broad-spectrum resistance to phytopathogens. Nat. Biotechnol. 18:1162-1166. [DOI] [PubMed] [Google Scholar]
- 43.Owens, L. D. 1995. Overview of gene availability, identification and regulation. Hort. Sci. 30:957-961. [Google Scholar]
- 44.Owens, L. D., and T. M. Heutte. 1997. A single amino acid substitution in the antimicrobial defense protein cecropin B is associated with diminished degradation by leaf intercellular fluid. Mol. Plant Microbe Interact. 10:525-528. [DOI] [PubMed] [Google Scholar]
- 45.Sahl, H. G., and G. Bierbaum. 1998. Lantibiotics: biosynthesis and biological activities of uniquely modified peptides from gram-positive bacteria. Annu. Rev. Microbiol. 52:41-79. [DOI] [PubMed] [Google Scholar]
- 46.Sharma, A., R. Sharma, M. Imamura, M. Yamakawa, and H. Machii. 2000. Transgenic expression of cecropin B, an antibacterial peptide from Bombyx mori, confers enhanced resistance to bacterial leaf blight in rice. FEBS Lett. 484:7-11. [DOI] [PubMed] [Google Scholar]
- 47.Smith, F. D., D. M. Gadoury, J. M. Van Eck, A. Blowers, J. C. Sanford, J. Van der Meij, and R. Eisenreich. 1998. Enhanced resistance to powdery mildew in transgenic poinsettia conferred by antimicrobial peptides. Phytopathology 88:S83. [Google Scholar]
- 48.Tossi, A., L. Sandri, and A. Giangaspero. 2000. Amphipathic, alpha-helical antimicrobial peptides. Biopolymers 55:4-30. [DOI] [PubMed] [Google Scholar]
- 49.Winstead, N. N., and A. Kelman. 1952. Inoculation techniques for evaluating resistance to Pseudomonas solanacearum. Phytopathology 42:628-634. [Google Scholar]
- 50.Yevtushenko, D. P., R. Romero, B. S. Forward, R. E. Hancock, W. W. Kay, and S. Misra. 2005. Pathogen-induced expression of a cecropin A-melittin antimicrobial peptide gene confers antifungal resistance in transgenic tobacco. J. Exp. Bot. 56:1685-1695. [DOI] [PubMed] [Google Scholar]