Abstract
Fluorescence in situ hybridization (FISH) with singly labeled rRNA-targeted oligonucleotide probes is widely applied for direct identification of microbes in the environment or in clinical specimens. Here we show that a replacement of singly labeled oligonucleotide probes with 5′-, 3′-doubly labeled probes at least doubles FISH signal intensity without causing specificity problems. Furthermore, Cy3-doubly labeled probes strongly increase in situ accessibility of rRNA target sites and thus provide more flexibility for probe design.
Since its introduction almost 20 years ago (2, 7), the identification of microorganisms by fluorescence in situ hybridization (FISH) with singly labeled rRNA-targeted probes has found widespread application in environmental and medical microbiology (1, 16). Despite being a methodologically robust technique, standard FISH suffers from several limitations (26) that may prevent successful detection of target microorganisms. One of the most frequently reported FISH problems is a low signal intensity of the detected microbes.
Dim signals can be caused by a low cellular concentration of the target molecules (16S rRNA or 23S rRNA), a feature typically found in microorganisms thriving in oligotrophic environments (19). In order to increase the sensitivity of FISH and make it suitable for the detection of microbes with a low ribosome content, several strategies have been developed (6, 13, 18, 19, 21, 25), of which catalyzed reporter deposition (CARD)-FISH has found the most widespread application. The CARD-FISH technique (18, 21) uses horseradish peroxidase (HRP)-labeled oligonucleotide probes and tyramide signal amplification and achieves a 26- to 41-fold-higher sensitivity than standard FISH (11). However, CARD-FISH is rather expensive, requires enzymatic pretreatment to allow the large horseradish peroxidase-labeled probes to penetrate the target cells (17), and causes a dramatically altered melting behavior of the probes (11).
Another frequently encountered FISH problem is the low in situ accessibility of many regions of the 16S and 23S rRNA for singly labeled probes (9, 10). Probes targeting such regions, which comprise about one-third of the Escherichia coli 16S rRNA (10), confer signals which are very dim or even below the detection limit. In order to avoid the selection of poorly accessible target sites for FISH probe design, a consensus 16S rRNA accessibility map for prokaryotes has been established based on detailed accessibility maps of two bacterial model organisms and one archaeal model organism (3). Considering this consensus map during probe design is recommended, but it excludes many probes with useful specificities from FISH applications. Furthermore, accessibility of many 16S and 23S rRNA target sites varies between different microorganisms and thus cannot yet be reliably predicted in silico. Accessibility of target sites to probes can be improved by the following: (i) use of unlabeled helper probes (8), (ii) elongation of the hybridization time up to 96 h (30), (iii) elongation of the probes, resulting in an altered ΔG°overall (30), or (iv) use of peptide nucleic acid probes (reference 26 and references therein). However, all these strategies have specific limitations. For example, the design of helper probes is often impossible for probes with broader specificities, the extension of the hybridization time might lead to unspecific probe or dye binding in complex samples, probe elongation is often not possible without narrowing its specificity, and previously published oligonucleotide probes cannot simply be converted into the expensive peptide nucleic acid probes without a dramatically changed specificity (26).
In principle, using oligonucleotide probes labeled with multiple fluorescent dyes should provide a simple means to increase the FISH signal intensity. The first experiments with multilabeled oligonucleotides were performed briefly after the introduction of the technique in microbiology but resulted in a pronounced increase in unspecific staining of nontarget organisms and/or an unexpected decrease in the signal intensity of the target organism which was attributed to quenching effects (28). Inconsistent with these data, Spear et al. (23) reported a successful increase in the signal-to-noise ratio of FISH-detected fungal cells by application of a multilabeled 18S rRNA-targeted oligonucleotide probe. In this work, we systematically evaluated the effect of 5′- and 3′-doubly labeled oligonucleotide probes (which were not included in the previously published study of multilabeled probes [28]) on the FISH signal intensity of Gram-negative and Gram-positive cells and studied the influence of double labeling on the in situ accessibility of rRNA target sites.
The double-labeling-of-oligonucleotide-probes (DOPE)-FISH approach was initially tested with four bacterial pure cultures. These included the gamma- and betaproteobacterial Gram-negative species Escherichia coli (DSM 498) and Burkholderia cepacia (DSM 7288), respectively. In addition, the two Gram-positive bacteria Bacillus subtilis (DSM 10) and Listeria monocytogenes (strain LO28) were used. E. coli, B. cepacia, and B. subtilis were grown according to the DSMZ instructions until they reached their stationary growth phase and were fixed with paraformaldehyde (E. coli and B. cepacia) or ethanol (B. subtilis) as described elsewhere (5). L. monocytogenes strain LO28 was grown on brain heart infusion for 5 h and fixed with ethanol as outlined previously (27). The oligonucleotide probes EUB338 (targeting the 16S rRNA of most but not all bacteria), NonEUB338 (a nonsense probe), GAM42a (targeting the 23S rRNA of many members of the Gammaproteobacteria), and five E. coli-targeted probes with a low 16S rRNA accessibility (10) were obtained as singly and doubly labeled derivatives from Thermo Hybaid (Interactiva Division, Ulm, Germany). More information about the applied probes can be found at probeBase (14) and in the publication by Fuchs et al. (10). FISH was performed by following the standard protocol (5) under the conditions recommended for each probe (14). If not stated otherwise, all hybridizations were carried out with identical hybridization (4 h) and washing times (10 min), respectively. Probe-conferred signal intensities were quantified using a confocal laser scanning microscope (CLSM) (LSM 510 Meta; Zeiss, Oberkochen, Germany) and the software program daime (4) by analyzing at least 1,000 single cells per experiment. For these measurements, individual cells were detected by image segmentation via edge detection.
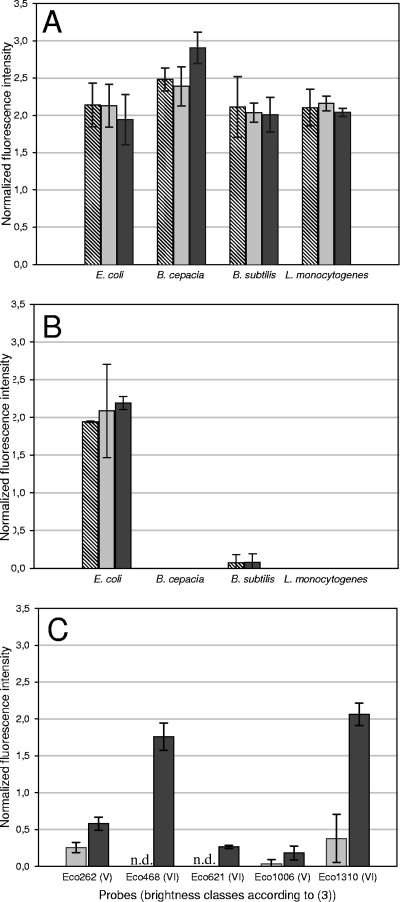
Regardless of the dye used (Cy3, Cy5, or FLUOS) and the respective target organism analyzed, hybridization with the doubly labeled probe EUB338 resulted in an increase in the FISH signal compared to the use of the singly labeled probe EUB338. For three of the four reference organisms, this increase was about 2-fold, while an even stronger signal amplification was observed for B. cepacia (Fig. 1A). Consequently, the distance between the two dye molecules in 18-nucleotide probes labeled at both ends is sufficient to avoid quenching. This is in contrast to oligonucleotides which are multiply labeled at one end or within the probe (28). Hybridization of the four reference organisms with the singly and doubly labeled derivatives of probe GAM42a confirmed these results and demonstrated that double labeling does not increase the background fluorescence of nontarget microorganisms (Fig. 1B). Consistent with these findings, hybridization of all reference organisms with a doubly labeled nonsense probe (with FLUOS, Cy3, and Cy5) resulted in signals below the detection limit of the CLSM if standard FISH settings were applied (data not shown). We also evaluated the influence of the hybridization time on the signal intensity achievable by DOPE-FISH by varying the hybridization time between 1 and 6 h. In these experiments, no significant difference in DOPE-FISH signal intensities of the Gram-positive and Gram-negative reference organisms were observed, indicating that the additional label of the DOPE-FISH probes does not dramatically influence the hybridization kinetics (data not shown).
FIG. 1.
(A) Effect of double labeling of the EUB338 probe on the FISH signal intensity of four reference organisms. For each organism, the signal intensity conferred by a doubly labeled EUB338 probe was normalized to the signal intensity obtained with the same probe as a singly labeled derivative. Hatched, light-gray, and dark-gray bars depict results with the Cy3-, Cy5- and FLUOS-labeled probe EUB338, respectively. (B) Effect of double labeling of the probe Gam42a (in the presence of the unlabeled competitor probe Bet42a, specific for most members of the Betaproteobacteria [14]) on the FISH signal intensities of four reference organisms. For each organism, the signal intensity conferred by the doubly labeled probe Gam42a was normalized to the signal intensity obtained for E. coli with the same probe as a singly labeled derivative. Hatched, light-gray, and dark-gray bars depict results with the Cy3-, Cy5-, and FLUOS-labeled probe, respectively. The weak unspecific signals observed with some DOPE-FISH probes for B. subtilis are also detectable at comparable intensities with singly labeled probes (data not shown). (C) Cy3-doubly labeled but not FLUOS-doubly labeled probes improve in situ accessibility of E. coli 16S rRNA target sites. E. coli was hybridized with five probes representing brightness classes V and VI (3). FISH signals were recorded for Cy3-singly and -doubly labeled probes and normalized to the FISH signal obtained for E. coli with the singly labeled probe EUB338. Light-gray and dark-gray bars depict results with Cy3-singly and doubly labeled probes, respectively. FLUOS-singly and -doubly labeled probes showed no signal. For all panels, all experiments were performed in triplicate. Error bars indicate the standard deviation. ND, not detectable.
Although DOPE-FISH worked well with all tested reference organisms, it should be noted that the Gram-positive species L. monocytogenes showed a signal amplification only if it was treated with lysozyme (according to Wagner et al. [27]) prior to the application of the doubly labeled probe. Without this enzymatic pretreatment, application of the singly labeled probe EUB338 resulted in a stronger signal than was seen with the doubly labeled probe EUB338, indicating that for some Gram-positive microorganisms with dense cell walls, double labeling impairs probe penetration. The observation that lysozyme treatment of L. monocytogenes also enhanced the probe-conferred signal of the singly labeled EUB338 probe confirmed that the cell wall of this organism after ethanol fixation is also not freely permeable to singly labeled probes (27; also data not shown). Enzymatic pretreatment of fixed microbial cells is also routinely applied for successful application of CARD-FISH, but since this method uses peroxidase-labeled probes which are much larger than DOPE-FISH probes, such treatments are also used for Gram-negative bacteria (11). Although many microorganisms are detectable by CARD-FISH with enhanced signal intensities, appropriate pretreatment protocols are not yet available for all microbes. For example, the sheathed filamentous methane oxidizer Crenothrix polyspora can easily be detected by standard FISH (24), but only very few cells within the filaments show a signal after CARD-FISH even if harsh permeabilization pretreatments are applied (see Fig. S1A in the supplemental material), a phenomenon known for sheathed microorganisms (12). In contrast, detection of all Crenothrix cells with more than 2-fold-increased signal intensity is readily possible by DOPE-FISH (see Fig. S1B).
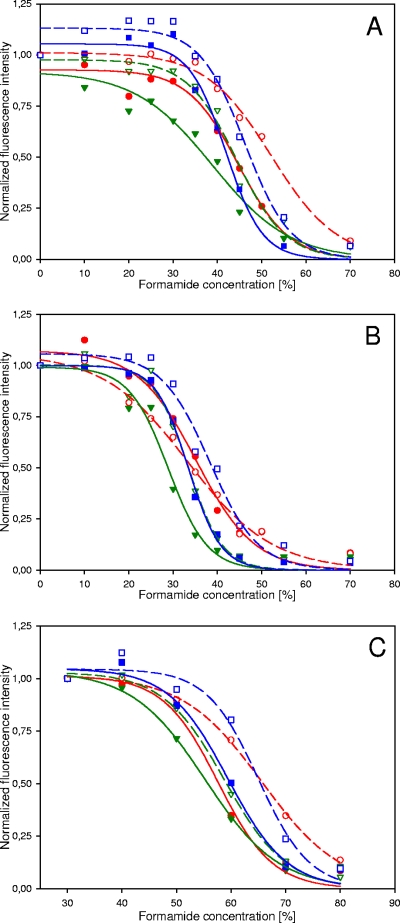
Prior research has demonstrated that probe labeling with horseradish peroxidase for CARD-FISH dramatically alters the melting behavior of oligonucleotide probes (11). Therefore, we recorded melting curves for Cy3-, Cy5-, and FLUOS-singly and -doubly labeled probes EUB338 and Gam42a by applying increasingly stringent conditions in the hybridization and wash steps (5). Interestingly, these experiments showed that the FLUOS singly labeled probes formed less-stable duplexes with their target sequences than the respective Cy3- and Cy5-labeled probes (Fig. 2). This effect, which is consistent with recent data on the stabilizing effect of various fluorophores on model probe-target duplexes (15), indicates that FLUOS-labeled FISH probes are generally applied under more-stringent conditions than Cy3- or Cy5-labeled probes. Cy3- and Cy5-doubly labeled probes displayed with their target organisms probe dissociation profiles similar to those of the respective FLUOS singly labeled probes, demonstrating that Cy3 or Cy5 double labeling does not further stabilize but rather moderately weakens the probe-target hybrid. Consistent with these findings, doubly FLUOS-labeled probes showed the lowest Tm (Fig. 2). Importantly, double labeling of probe GAM42a did not adversely affect mismatch discrimination, as shown by its dissociation profiles if in situ hybridizations were performed at various stringencies with B. cepacia containing a single mismatch in the probe target site of its 23S rRNA. (Fig. 2B). These results indicate that the specificities of DOPE-FISH probes can be regarded as identical to those of standard singly labeled FISH probes.
FIG. 2.
Comparison of probe dissociation profiles of singly and doubly labeled probes. For each profile, the microscopic settings were adjusted for the lowest formamide concentration and subsequently kept constant. Dashed and solid lines represent sigmoid fittings for singly and doubly labeled probes, respectively. (A) Dissociation profiles of the singly and doubly labeled probe Gam42a with E. coli as the target organism. Empty circles, squares, and triangles represent data obtained with the Cy3-, Cy5-, and FLUOS-singly labeled probe GAM42a, respectively. Filled circles, squares, and triangles depict the data measured for the Cy3-, Cy5-, and FLUOS-doubly labeled probe GAM42a, respectively. (B) Dissociation profiles of the singly and doubly labeled probe Gam42a with B. cepacia as a nontarget organism having a single mismatch to probe GAM42a. Empty circles, squares, and triangles represent data obtained with the Cy3-, Cy5-, and FLUOS-singly labeled probe GAM42a, respectively. Filled circles, squares, and triangles depict the data measured for the Cy3-, Cy5-, and FLUOS-doubly labeled probe GAM42a, respectively. The melting curves for FLUOS-singly labeled and Cy5-doubly labeled probes are almost identical and thus overlap. In the presence of the unlabeled probe Bet42a as a competitor, no probe-conferred signal was recordable for both singly and doubly labeled GAM42a probes. (C) Dissociation profiles of the singly and doubly labeled probe EUB338 with E. coli as the target organism. Empty circles, squares, and triangles represent data obtained with the Cy3-, Cy5-, and FLUOS-singly labeled probe EUB338, respectively. Filled circles, squares, and triangles depict the data measured for the Cy3-, Cy5-, and FLUOS-doubly labeled probe EUB338, respectively. For all panels, error bars are not shown since they were always smaller than the symbols.
In order to analyze whether the in situ accessibility of rRNA target sites to doubly labeled probes differs from that to those labeled with only one dye, we tested five probes targeting E. coli 16S rRNA. These probes were described as yielding only very dim signals with standard FISH as a consequence of limited target site accessibility and were thus assigned to the lowest brightness classes, V or VI (3, 10). Consistently, standard FISH with these five singly labeled probes (Cy3 and FLUOS) gave no or very weak signals (Fig. 1C). Unexpectedly, however, Cy3-doubly labeled derivatives of these probes produced much brighter signals (Fig. 1C), and for some of the probes (Eco468 and Eco1310), the DOPE-FISH signal intensity was higher than that measured for the Cy3-singly labeled probe EUB338 (Fig. 1C). One could speculate that a Cy3 label at the 3′ end and not the double labeling might be responsible for the improved accessibility of rRNA target sites for Cy3 DOPE-FISH probes. However, since selected probes (Eco262, Eco468, and Eco1310) labeled with a single Cy3 molecule at the 3′ end did not result in increased fluorescence, this hypothesis can be rejected (data not shown). Interestingly, FLUOS-labeled DOPE-FISH probes did not show increased fluorescence, strongly indicating that the improved accessibility of Cy3 DOPE-FISH probes depends on the chemical structure of the fluorophore. This is consistent with the observation that Cy5 double labeling of the five E. coli probes also resulted in improved probe accessibilities (data not shown). While Cy3 and Cy5 double labeling decreases the probe-target duplex stability (Fig. 2), it apparently helps to resolve secondary or tertiary structures responsible for poor in situ accessibility of rRNA target sites. It is tempting to speculate that binding of Cy3 or Cy5 to double-stranded rRNA regions, analogous to the previously reported intercalation of certain cyanine class dyes in DNA (29) or other modes of nucleic acid binding by cyanine dyes (15), contributes to this phenomenon.
The improved accessibility of rRNA target sites for Cy3 DOPE-FISH probes offers more flexibility for probe design because it enables the use of probes with excellent specificity but low standard FISH signal intensity for the successful in situ detection of microbes. This advantage of DOPE-FISH is nicely demonstrated by the probe Ntspa175 (5′-GAC CAG GAG CCG TAT GCG-3′), which targets the 16S rRNA (GenBank accession no. GU229885) of an uncultured nitrite oxidizer of the genus Nitrospira thriving in activated sludge. At 25% formamide in the standard FISH hybridization buffer (5), this probe is highly specific as demonstrated by Clone-FISH (22) using another activated sludge-derived 16S rRNA Nitrospira-like sequence with a single mismatch to probe Ntspa175 as a nontarget control (data not shown). Standard FISH of activated sludge with the Cy3-labeled probe Ntsp175, which targets the 175-to-193 region in the 16S rRNA, resulted in the detection of Nitrospira microcolonies with very variable FISH signal intensities. A considerable number of stained microcolonies had extremely dim FISH signals, indicating that these cells had a ribosome content too low to be reliably detectable by a standard FISH probe of a low brightness class. DOPE-FISH of the same sample with the Cy3-doubly labeled probe Ntspa175 led to a pronounced increase in signal intensity of the target cells (see Fig. S2 in the supplemental material) without causing increased background fluorescence if standard confocal-microscope settings were applied. In accordance with this observation, the relative biovolume-abundance of the detectable Ntspa175-stained population compared to the biovolume of those cells labeled by the Nitrospira genus-specific probe Ntspa662 (14) in the activated sludge increased by a factor of 1.81 ± 0.1 if a doubly labeled Ntspa175 probe was used (measurements made by the software package daime using confocal-microscope images as described previously [4]).
In summary, DOPE-FISH with commercially available doubly labeled oligonucleotide probes is a straightforward modification of the standard FISH procedure which increases the signal intensity of standard FISH probes by at least a factor of 2 without causing specificity problems. Importantly, the influence of DOPE-FISH on the dissociation profile of probes is not larger than that caused by a dye switch from Cy3 to FLUOS if singly labeled probes are used for FISH. Thus, previously optimized hybridization and washing conditions for published probes can be applied for DOPE-FISH. Since DOPE-FISH unlocks previously inaccessible target sites on the rRNA, this new FISH approach offers more options for the design of specific probes, a task which becomes increasingly difficult with the rapid growth of rRNA databases (20).
Supplementary Material
Acknowledgments
This work was funded by Austrian Science Fund FWF grant P20775 to K.S.
Stephanie Füreder is gratefully acknowledged for culturing and fixing L. monocytogenes, Christian Baranyi for help with statistical analyses, and Doris Steger for helpful discussions.
Footnotes
Published ahead of print on 4 December 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Amann, R., and B. M. Fuchs. 2008. Single-cell identification in microbial communities by improved fluorescence in situ hybridization techniques. Nat. Rev. Microbiol. 6:339-348. [DOI] [PubMed] [Google Scholar]
- 2.Amann, R. I., L. Krumholz, and D. A. Stahl. 1990. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J. Bacteriol. 172:762-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Behrens, S., C. Ruhland, J. Inacio, H. Huber, A. Fonseca, I. Spencer-Martins, B. M. Fuchs, and R. Amann. 2003. In situ accessibility of small-subunit rRNA of members of the domains Bacteria, Archaea, and Eucarya to Cy3-labeled oligonucleotide probes. Appl. Environ. Microbiol. 69:1748-1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Daims, H., S. Lücker, and M. Wagner. 2006. daime, a novel image analysis program for microbial ecology and biofilm research. Environ. Microbiol. 8:200-213. [DOI] [PubMed] [Google Scholar]
- 5.Daims, H., K. Stoecker, and M. Wagner. 2005. Fluorescence in situ hybridization for the detection of prokaryotes, p. 213-239. In A. M. Osborn and C. J. Smith (ed.), Advanced methods in molecular microbial ecology. Bios-Garland, Abingdon, United Kingdom.
- 6.DeLong, E. F., L. T. Taylor, T. L. Marsh, and C. M. Preston. 1999. Visualization and enumeration of marine planktonic archaea and bacteria by using polyribonucleotide probes and fluorescent in situ hybridization. Appl. Environ. Microbiol. 65:5554-5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DeLong, E. F., G. S. Wickham, and N. R. Pace. 1989. Phylogenetic stains: ribosomal RNA-based probes for the identification of single cells. Science 243:1360-1363. [DOI] [PubMed] [Google Scholar]
- 8.Fuchs, B. M., F. O. Glöckner, J. Wulf, and R. Amann. 2000. Unlabeled helper oligonucleotides increase the in situ accessibility to 16S rRNA of fluorescently labeled oligonucleotide probes. Appl. Environ. Microbiol. 66:3603-3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fuchs, B. M., K. Syutsubo, W. Ludwig, and R. Amann. 2001. In situ accessibility of Escherichia coli 23S rRNA to fluorescently labeled oligonucleotide probes. Appl. Environ. Microbiol. 67:961-968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fuchs, B. M., G. Wallner, W. Beisker, I. Schwippl, W. Ludwig, and R. Amann. 1998. Flow cytometric analysis of the in situ accessibility of Escherichia coli 16S rRNA for fluorescently labeled oligonucleotide probes. Appl. Environ. Microbiol. 64:4973-4982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoshino, T., L. S. Yilmaz, D. R. Noguera, H. Daims, and M. Wagner. 2008. Quantification of target molecules needed to detect microorganisms by fluorescence in situ hybridization (FISH) and catalyzed reporter deposition-FISH. Appl. Environ. Microbiol. 74:5068-5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kubota, K., H. Imachi, S. Kawakami, K. Nakamura, H. Harada, and A. Ohashi. 2008. Evaluation of enzymatic cell treatments for application of CARD-FISH to methanogens. J. Microbiol. Methods 72:54-59. [DOI] [PubMed] [Google Scholar]
- 13.Lee, S., C. Malone, and P. F. Kemp. 1993. Use of multiple 16S rRNA-targeted fluorescent probes to increase signal strength and measure cellular RNA from natural planktonic bacteria. Mar. Ecol. Prog. Ser. 101:193-201. [Google Scholar]
- 14.Loy, A., F. Maixner, M. Wagner, and M. Horn. 2007. probeBase—an online resource for rRNA-targeted oligonucleotide probes: new features 2007. Nucleic Acids Res. 35:D800-D804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moreira, B. G., Y. You, M. A. Behlke, and R. Owczarzy. 2005. Effects of fluorescent dyes, quenchers, and dangling ends on DNA duplex stability. Biochem. Biophys. Res. Commun. 327:473-484. [DOI] [PubMed] [Google Scholar]
- 16.Moter, A., and U. B. Goebel. 2000. Fluorescence in situ hybridization (FISH) for direct visualization of microorganisms. J. Microbiol. Methods 41:85-112. [DOI] [PubMed] [Google Scholar]
- 17.Pernthaler, A., and J. Pernthaler. 2007. Fluorescence in situ hybridization for the identification of environmental microbes. Methods Mol. Biol. 353:153-164. [DOI] [PubMed] [Google Scholar]
- 18.Pernthaler, A., J. Pernthaler, and R. Amann. 2002. Fluorescence in situ hybridization and catalyzed reporter deposition for the identification of marine bacteria. Appl. Environ. Microbiol. 68:3094-3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pernthaler, A., C. M. Preston, J. Pernthaler, E. F. DeLong, and R. Amann. 2002. Comparison of fluorescently labeled oligonucleotide and polynucleotide probes for the detection of pelagic marine bacteria and archaea. Appl. Environ. Microbiol. 68:661-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pruesse, E., C. Quast, K. Knittel, B. M. Fuchs, W. Ludwig, J. Peplies, and F. O. Glöckner. 2007. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35:7188-7196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schönhuber, W., B. Zarda, S. Eix, R. Rippka, M. Herdman, W. Ludwig, and R. Amann. 1999. In situ identification of cyanobacteria with horseradish peroxidase-labeled, rRNA-targeted oligonucleotide probes. Appl. Environ. Microbiol. 65:1259-1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schramm, A., B. M. Fuchs, J. L. Nielsen, M. Tonolla, and D. A. Stahl. 2002. Fluorescence in situ hybridization of 16S rRNA gene clones (Clone-FISH) for probe validation and screening of clone libraries. Environ. Microbiol. 4:713-720. [DOI] [PubMed] [Google Scholar]
- 23.Spear, R. N., S. Li, E. V. Nordheim, and J. H. Andrews. 1999. Quantitative imaging and statistical analysis of fluorescence in situ hybridization (FISH) of Aureobasidium pullulans. J. Microbiol. Methods 35:101-110. [DOI] [PubMed] [Google Scholar]
- 24.Stoecker, K., B. Bendinger, B. Schöning, P. H. Nielsen, J. L. Nielsen, C. Baranyi, E. R. Toenshoff, H. Daims, and M. Wagner. 2006. Cohn's Crenothrix is a filamentous methane oxidizer with an unusual methane monooxygenase. Proc. Natl. Acad. Sci. U. S. A. 103:2363-2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trebesius, K., R. Amann, W. Ludwig, K. Mühlegger, and K.-H. Schleifer. 1994. Identification of whole fixed bacterial cells with nonradioactive 23S rRNA-targeted polynucleotide probes. Appl. Environ. Microbiol. 60:3228-3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wagner, M., M. Horn, and H. Daims. 2003. Fluorescence in situ hybridisation for the identification and characterisation of prokaryotes. Curr. Opin. Microbiol. 6:302-309. [DOI] [PubMed] [Google Scholar]
- 27.Wagner, M., M. Schmid, S. Juretschko, K. H. Trebesius, A. Bubert, W. Goebel, and K. H. Schleifer. 1998. In situ detection of a virulence factor mRNA and 16S rRNA in Listeria monocytogenes. FEMS Microbiol. Lett. 160:159-168. [DOI] [PubMed] [Google Scholar]
- 28.Wallner, G., R. Amann, and W. Beisker. 1993. Optimizing fluorescent in situ hybridization with rRNA-targeted oligonucleotide probes for flow cytometric identification of microorganisms. Cytometry 14:136-143. [DOI] [PubMed] [Google Scholar]
- 29.Yarmoluk, S. M., S. S. Lukashov, T. Y. Ogul'Chansky, M. Y. Losytskyy, and O. S. Kornyushyna. 2001. Interaction of cyanine dyes with nucleic acids. XXI. Arguments for half-intercalation model of interaction. Biopolymers 62:219-227. [DOI] [PubMed] [Google Scholar]
- 30.Yilmaz, L. S., H. E. Okten, and D. R. Noguera. 2006. Making all parts of the 16S rRNA of Escherichia coli accessible in situ to single DNA oligonucleotides. Appl. Environ. Microbiol. 72:733-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.