Abstract
During recent years, the animal-associated methicillin-resistant Staphylococcus aureus clone ST398 has extensively been studied. The DNA of these isolates turned out to be refractory to SmaI restriction, and consequently, SmaI is unsuitable for subtyping this clone by standard pulsed-field gel electrophoresis (PFGE). Very recently, ST398 DNA was shown to be digested by Cfr9I, a neoschizomer of SmaI. In the present study, we employed Cfr9I PFGE on 100 German and 5 Dutch ST398 isolates and compared their PFGE profiles, protein A gene variable repeat regions (spa types), and types of the staphylococcal cassette chromosome mec (SCCmec). The isolates (from healthy carrier pigs, clinical samples from pigs, dust from farms, milk, and meat) were assigned to 35 profiles, which were correlated to the SCCmec type. A dendrogram with the Cfr9I patterns assigned all profiles to two clusters. Cluster A grouped nearly all isolates with SCCmec type V, and cluster B comprised all SCCmec type IVa and V* (a type V variant first identified as III) carriers plus one isolate with SCCmec type V. Both clusters also grouped methicillin-susceptible S. aureus isolates. The association of the majority of isolates with SCCmec type V in one large cluster indicated the presence of a successful subclone within the clonal complex CC398 from pigs, which has diversified. In general, the combination of Cfr9I PFGE with spa and SCCmec typing demonstrated the heterogeneity of the series analyzed and can be further used for outbreak investigations and traceability studies of the MRSA ST398 emerging clone.
Methicillin-resistant Staphylococcus aureus (MRSA) strains are an important cause of hospital-acquired infections worldwide (8). However, MRSA strains are not confined to health care settings, and during the last 10 years community-acquired MRSA has increasingly been reported (8). In 2003, a clone of MRSA associated with pig farming and not related to the traditional hospital- and community-acquired MRSA emerged in the Netherlands (37), where it now amounts to >30% of human MRSA cases (16). This clone has also been detected in healthy and sick animals, in food of animal origin, and in humans from other European countries, Canada, the United States, the Dominican Republic, and China (5, 7, 31, 38, 39). This emerging MRSA clone belongs to the multilocus sequence type ST398, which includes different spa types (mainly t011, t034, and t108). The majority of the ST398 isolates reported are MRSA, although methicillin-susceptible (MSSA) strains have been described as well (15, 34). Resistance to methicillin and other β-lactam antibiotics is caused by the mecA gene, which is located on a mobile genetic element, the staphylococcal cassette chromosome mec (SCCmec). The SCCmec cassette consists of the mec gene complex, the ccr gene complex, and the junkyard regions. Based on the variability and combinations of these genetic elements, several types of SCCmec and several variants of the types have been described (9). Three SCCmec types (III, IVa, and V) were identified in ST398 isolates (25). However, recent investigations have shown that some ST398 isolates typed as SCCmec type III using the method of Zhang et al. (40) proved to be type V after further sequencing (21, 35).
For typing S. aureus, pulsed-field gel electrophoresis (PFGE) of the whole genome by macrorestriction with the SmaI endonuclease is still considered as the “gold standard” (26). However, the isolates of the ST398 clone are nontypeable (NT) by PFGE using SmaI (3, 4). Consequently, comparison between these isolates and the typeable ones from humans and animals is not possible. The nontypeability is due to the action of a novel C5-cytosine methyltransferase which modifies the consensus sequence CmCNGG at the second cytosine (3, 4). Other enzymes with a different recognition sequence from SmaI have been used for PFGE typing of the ST398 clone, including EagI and ApaI (22, 28, 31, 38), but the patterns obtained cannot be compared to S. aureus patterns generated with SmaI. XmaI, a neoschizomer of SmaI that recognizes the same sequence cutting at a different position, only generates partial digestions (3, 4). Recently, the use of Cfr9I, another neoschizomer of SmaI whose activity is not reduced on ST398 methylated DNA, has been recommended. This enzyme had been successfully used for typing SmaI NT macrolide-resistant Streptococcus pyogenes isolates (6, 30), and now it is being applied for typing ST398 isolates, i.e., from human origin (5, 11, 36) and, to a lesser extent, from animals (3, 36). The aim of this study was to characterize a large collection of recent ST398 isolates by Cfr9I PFGE as well as other methods (spa typing, multilocus sequence typing [MLST], and SCCmec typing). Most of them were recovered in Germany from different sources, including animals and foods.
MATERIALS AND METHODS
Bacterial strains.
A series of 109 S. aureus isolates were included in the present study. One hundred four isolates were selected from the strain collection of the National Reference Laboratory for Coagulase-Positive Staphylococci (NRL-Staph) at the Federal Institute for Risk Assessment (BfR). They had been isolated between 2004 and 2008 at different German regional laboratories or at NRL-Staph as part of various surveys and investigations. Biochemical characterization, susceptibility testing, and typing of the bacteria were conducted at NRL-Staph and the NRL for Antimicrobial Resistance (NRL-AR). MRSA isolates were confirmed by multiplex PCR (27) detecting 16S rRNA genes, the S. aureus-specific nuclease gene nuc, and the resistance gene mecA in the case of methicillin-resistant isolates (all except four isolates). The isolates had been collected from healthy carrier pigs at slaughter (42 isolates), clinical samples fromf pigs (31 isolates), dust from pig farms (10 isolates), cow's milk (five isolates), and meat from food-producing animals (16 isolates, including eight from pigs, five from turkeys, two from chickens, and one from a cow). The other five isolates came from the Netherlands and had been collected from fecal samples from pigs. The S. aureus strains NCTC 8325 and ATCC 29213 were used as controls for PFGE.
spa typing, MLST, and SCCmec typing.
DNA of the isolates was extracted using the RTP-bacteria DNA minikit (Invitek, Berlin, Germany). All isolates were analyzed by spa typing (29). Representative isolates with different spa types (14 German and 5 Dutch isolates) were additionally subjected to MLST (10). PCR products were sequenced by Qiagen (Hilden, Germany). The software Ridom Staphtype (Ridom GmbH, Würzburg, Germany) and the S. aureus MLST database (http://www.saureus.mlst.net) were used to assign spa types and sequence types (ST), respectively. The software Ridom Staphtype was also used to investigate the clonal relatedness of the different spa types, using the Based Upon Repeat Pattern (BURP) algorithm (24). The group founder within BURP clusters of at least three different spa types is defined as the spa type with the higher founder score (assigned to the spa type to which the relevant spa types and strains are more closely related) (24). MRSA isolates were typed by SCCmec using a modified protocol for the multiplex PCR described by Zhang et al. (40). This multiplex PCR identifies SCCmec types I to V. SCCmec type III isolates were further characterized by PCR amplification and sequencing as proposed by Jansen et al. (21), using primers for ccrC of SCCmec type V (20) and different regions of the SCCmec type III: ccrA/B3 (21), mN1-mN2 (which comprises ccrA3 and ccrB3), mN3-Tn554 (positions 171-146; a region spanning from transposase B to ψTn554), TnpA1016-TnpA636 (tranposase A), and merA2-merN (marker of the mercury operon) (19).
Macrorestriction-PFGE analysis.
Whole DNA from each S. aureus isolate was analyzed by SmaI and Cfr9I PFGE, as previously described (3), using a CHEF-DRIII SYS220/240 (Bio-Rad Laboratories GmbH, Munich, Germany) system, applying the Harmony protocol guidelines (26). The resulting profiles were analyzed recording the presence or absence of fragments larger than 20 kb. Those patterns showing one or more mismatching bands were considered as different and labeled “S” (SmaI profiles) or “C” (Cfr9I profiles) followed by a number. Genetic similarity between profiles was determined by the unweighted-pair method with arithmetic averages and the Jaccard's coefficient, using the software program MVSP version 3.1 (Multivariate Statistics Package for PCs; RockWare, Inc.).
Statistical methods.
To asses the discriminatory power of the typing methods applied, the discrimination indices (DIs) and their confidence intervals (CIs) were calculated using Simpson's index of diversity as previously described (13, 14, 17, 32). This index of diversity defines the probability that two unrelated strains chosen from the test population will be placed into different typing groups (17). The acceptance level of discrimination of a typing method depends on several factors, with a DI greater than 0.9 being desirable.
To compare type assignments and to measure the clustering concordance between typing methods, the Rand's and adjusted Rand's indices and the Wallace coefficients (W) were calculated as reported by Carriço et al. (6) with the BioNumerics 5.1 software (Applied-Maths, Ghent, Belgium) using the script available at http://biomath.itqb.unl.pt/ClusterComp. The Rand index is a symmetric coefficient which represented the proportion of agreement for both matches and mismatches in two partitions. The adjusted Rand index allows a better quantitative evaluation of the global congruence between the two partitions. Wallace′s coefficients provide an estimate, given a typing method, of how much new information is obtained from another typing method. A high value of Wallace's coefficient indicates that partitions defined by a given method could have been predicted from the results of another method (6).
RESULTS
In order to ensure that the isolates belonged to ST398, all of them were subjected to spa typing. Four of the 109 isolates presented spa types (t318, t337, t002, and t1430) not related to the ST398 clone. They originated from German clinical samples from pigs (2) or from meat (2) and were considered as out-group isolates for further analyses. Only two of them were MRSA: one was SCCmec type IVa, while the other was nontypeable.
The 105 isolates assigned to ST398 comprised 19 different spa types. The most frequent types were t011 (39 isolates; 37%), t034 (31 isolates; 29.5%), and t108 (11 isolates; 10.5%). Relationships between SCCmec and spa types are shown in Table 1. Nineteen representative isolates belonging to t011 (5 isolates), t108 (3 isolates), t1451, (2 isolates), and t034, t1250, t1255, t1457, t1928, t1985, t2346, t2510, and t2970 (one isolate each) were further characterized by MLST and ascribed to ST398. The clonal relatedness of the different ST398 spa types found was then investigated using the BURP algorithm. One large cluster included 93.3% of the isolates and 16 spa types, with t011 as the founder. Seven isolates with spa types t2510, t2576, t2970, and t5210 had no alignment in the BURP algorithm.
TABLE 1.
Relationships between SCCmec and spa typesa
| SCCmec type (no.) |
spa type (no.) |
|
|---|---|---|
| Frequent types | Others | |
| V* (11) | t034 (7), t108 | t571 (2), t1928 |
| IVa (5) | t011 (4) | t779 |
| V (80) | t011 (33), t034 (20), t108 (9) | t1255, t1451 (4), t1457, t1580, t1985, t2011, t2346 (2), t2510, t2576 (4), t2970, t5210 |
| Nontypeable (7) | t011 (2), t034 (3) | t1250, t1753 |
| Negative (2) | t034, t108 | |
Numbers of isolates are shown in parentheses if there are more than one.
Among the 105 ST398 isolates, all except two were MRSA (98%). The isolates carried typical SCCmec types associated with the clone: III (11 isolates; 10.5%), IVa (5 isolates; 4.8%), and V (80 isolates; 76.2%), identified by the Zhang et al. (40) protocol, while seven isolates were nontypeable. The 11 SCCmec type III isolates were further characterized by amplification and sequencing. This revealed the presence of ccrC of SCCmec type V, but although no amplification was found using primers for ccrA/B3 of SCCmec type III, all of these isolates harbored the transposase A gene of Tn554, which is integrated into J2 regions of SCCmec types II and III. This new type of SCCmec, related to type V, was termed “V*” in the present work.
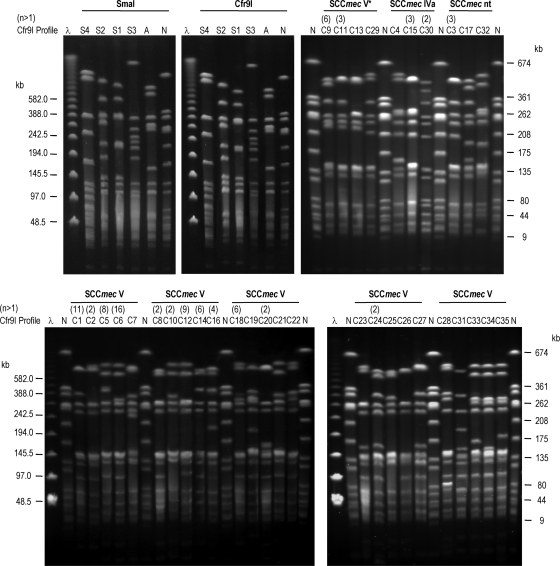
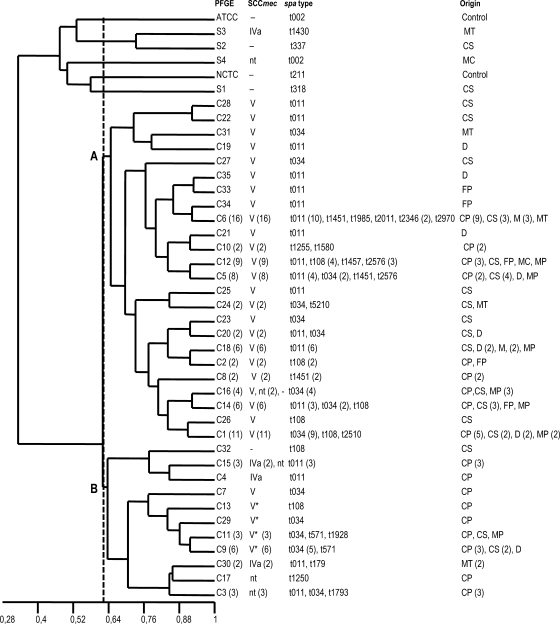
Genomic DNA of the four out-group isolates and the control strains NCTC 8325 and ATCC 29213 was digested with both SmaI and Cfr9I (Fig. 1). The two enzymes yielded identical restriction patterns. In contrast, DNA of the 105 ST398 isolates was resistant to SmaI (all of them were NT) but susceptible to Cfr9I, which generated similar patterns, differing by one to seven fragments (Fig. 1). For all ST398 isolates, 35 different Cfr9I profiles were found, many of them (19 out of 35) represented by only one isolate. However, the majority of isolates (84 out of 105) showed 16 patterns, with C6, C1, C12, and C5 being the most frequent (shown by 16, 11, 9, and 8 isolates, respectively). The relationships between Cfr9I profiles and spa and SCCmec types are compiled in Tables 2 and 3. On the basis of the Jaccard′s similarity coefficient, a dendrogram with the Cfr9I patterns was constructed (Fig. 2). At a cutoff point of 0.64, two major clusters (A and B) including all ST398 isolates were identified. Out-group and control strains were clearly separated in the dendrogram. The largest cluster, A, included 78% of the isolates, distributed into 24 profiles. This cluster comprised all except one of the SCCmec type V isolates, two of the SCCmec nontypeable isolates, and one MSSA isolate. Cluster B included 22% of the isolates distributed in three subclusters. One of them grouped the four SCCmec V* isolates and one SCCmec V isolate, while the other two contained the SCCmec IVa isolates, SCCmec nontypeable and MSSA.
FIG. 1.
Cfr9I PFGE analysis of Staphylococcus aureus ST398 isolates. Lane λ, lambda ladder PFGE marker (New England Biolabs, Frankfurt am Main, Germany); lane A, Cfr9I profile of strain ATCC 29213; lane N, Cfr9I profile of strain NCTC 8325 included as quality control; lanes S1 to S4, SmaI profiles of out-group isolates; lanes C1 to C35, Cfr9I profiles generated from ST398 isolates.
TABLE 2.
Number of types and Simpson's index of diversity for each typing method
| Method | Typeability (%) | No. of types | Simpson's index of diversity (95% CI) |
|---|---|---|---|
| PFGE | 100 | 35 | 0.94 (0.92-0.96) |
| spa typing | 100 | 19 | 0.77 (0.71-0.83) |
| SCCmec typing | 91.4a | 5a | 0.41 (0.29-0.52) |
Seven strains were nontypeable, and two were MSSA. For the purpose of calculating Simpson's diversity, they were considered in two groups.
TABLE 3.
Comparison between typing methods used in 105 ST398 isolates
| Method/feature | Relative result by: |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Rand index |
Adjusted Rand index |
Wallace's coefficients |
||||||||||
| PFGE | spa typing | SCCmec typing | Origin | PFGE | spa typing | SCCmec typing | Origin | PFGE | spa typing | SCCmec typing | Origin | |
| PFGE | 1 | 0.76 | 0.45 | 0.71 | 1 | 0.09 | 0.07 | 0.01 | 1 | 0.45 | 0.97 | 0.30 |
| spa typing | 0.76 | 1 | 0.46 | 0.61 | 0.09 | 1 | 0.02 | 0.04 | 0.10 | 1 | 0.62 | 0.22 |
| SCCmec typing | 0.45 | 0.46 | 1 | 0.42 | 0.07 | 0.02 | 1 | 0.05 | 0.09 | 0.24 | 1 | 0.23 |
| Origin | 0.71 | 0.61 | 0.42 | 1 | 0.01 | 0.04 | 0.05 | 1 | 0.06 | 0.20 | 0.53 | 1 |
FIG. 2.
Dendrogram showing the relatedness between Cfr9I macrorestriction fragment profiles generated from the S. aureus ST398 isolates, the out-group isolates, and the control (NCTC 8325 and ATCC 29213) strains. At a Jaccard's coefficient of similarity (J) of 0.63, two clusters (labeled A and B, the latter with three subclusters) were detected grouping the ST398 isolates. The SCCmec and spa types and origin associated with each Cfr9I profile are also indicated. −, no SCCmec (MSSA isolates); CP, carrier pig at abattoir; CS, clinical sample from pig; D, dust; FP, fecal sample from pig; M, milk; MC, meat from chicken; MP, meat from pig; MT, meat from turkey.
The index of diversity and the interval coefficients of the different typing methods used are shown in Table 2. As has been shown, the most discriminative method was Cfr9I PFGE, followed by spa typing. The concordance among different typing methods and the origin of the isolates is shown in Table 3. For PFGE, the highest concordances were with spa typing and SCCmec typing. However, those methods were poor predictors of the PFGE as expressed by the Wallace coefficient (W = 0.06 to 0.10). The SCCmec was almost completely predicted by the PFGE (W = 0.97). The use of all methods revealed a high heterogeneity within the ST398 clone.
DISCUSSION
Several studies carried out during the last 2 years about the prevalence of MRSA belonging to the MLST ST398 demonstrated that this clone is widely spread in farm animals, animal-derived foods, pets, and humans (7, 8, 23, 35, 39). These findings triggered EU-wide investigations into the prevalence of MRSA in pigs during 2008 (1, 2). For the present study, more than one hundred isolates mainly obtained between 2004 and 2008 from animals, foods, and environmental samples in different parts of Germany were investigated. One hundred five of them were assigned to the clonal complex 398, on the basis of resistance to SmaI restriction, spa typing, and, in selected isolates, MLST (24). The t011, t034, and t108 types were the most frequent in the series, as has been described for the clone (7, 8, 25, 35, 38). However, a total of 19 spa types were found, suggesting that this clonal complex is continually evolving. Applying the protocol of Zhang et al. (4), most of the MRSA isolates proved to contain SCCmec type V, as reported in other studies performed in both Germany and the Netherlands (8, 33, 38), but SCCmec type IVa and SCCmec type III isolates were also found. However, recent studies, in which sequencing was applied, have demonstrated that most of the isolates previously identified as positive for SCCmec type III by the Zhang et al. (40) method belonged to type V (21). In our study, all of the SCCmec type III carriers harbored ccrC of type V together with the tnpA gene from Tn554 (associated with types II and III) but no other regions of SCCmec type III (18), hence representing a new variant of type V.
One of the aims of the study was to apply the Cfr9I PFGE method for typing a large collection of ST398 isolates. In contrast to XmaI, Cfr9I is not affected by modification of the CCCGGG recognition sequence through the action of the cytosine methyltransferase produced by the ST398 clone. The partial sequence of this DNA methylase (motifs IV to VII) shows high similarity (74 to 71%) to different C5-cytosine methyltransferases with the same recognition sequence, including M1.ScrFI from Lactococcus lactis and M.SpyI and M.SpyM6 from S. pyogenes (3).
The Cfr9I PFGE method allowed direct comparison with SmaI profiles of S. aureus and clearly separated the out-group and control strains from the ST398 isolates. This method was more discriminative than spa typing (DI of 0.94 versus 0.77), and distributed the 105 isolates into 35 genetically closely related profiles that grouped into two clusters (A and B). As expressed by the Wallace coefficient, SCCmec could be accurately predicted by Cfr9I PFGE, since the probability of isolates having the same Cfr9I profile also sharing the same SCCmec type was 97% (W = 0.97). Most of the SCCmec type V isolates clustered in A, while the remaining types clustered in B. Within clusters A and B, a limited number of isolates were nontypeable with the Zhang et al. protocol (40), sharing in some cases the Cfr9I profile of MRSA isolates. These strains could have once carried the SCCmec cassette of their neighbors in the dendrogram and then become nontypeable through further evolution. Moreover, MSSA isolates coexisted with MRSA isolates in clusters A and B, and one MRSA and one MSSA isolate showed the same Cfr9I profile, demonstrating the common background between ST398 MRSA and MSSA. Previous studies using Cfr9I typing focused mainly on human clinical isolates (5, 11, 36). Bhat et al. (5) used the Cfr9I PFGE method to type 28 human isolates from the United States and the Dominican Republic and found 14 profiles closely related to each other. Fanoy et al. (11) applied the same method to seven isolates from a residential care facility in the Netherlands with SCCmec type IVa and showed quite similar patterns to those included in cluster B. van Wamel et al. (36) analyzed by this method isolates from humans and animals (7 from horses, 10 from pigs, and one from a cow with mastitis) and found a successful ST398 clade constituted mostly by SCCmec type V isolates. Other authors also found within the clone a strong correlation between SCCmec and PFGE using other endonucleases (28). With regard to the spa types, some authors have proposed that the spa type of ST398 isolates is related to SCCmec; thus, the t011 strains have been primarily associated with SCCmec type IVa and the spa type t108 strains with SCCmec type V (25). In our study, most of the t011 (84.6%), t034 (71%), and t108 (81.8%) isolates clustered in A and carried SCCmec type V. On the other hand, there was no t011 among the SCCmec type V* isolates. This is in line with results from a study on MRSA in pigs in five German abattoirs. Only three of 212 isolates of spa type t011 in that study carried this new SCCmec type, while 195 carried SCCmec type V (33).
With regard to the origin of the isolates (Fig. 2), although some PFGE profiles were shown by isolates with the same origin (i.e., carrier pigs from Germany), others were shown by isolates from Germany with different origins (carrier pigs, clinical samples, meat, or dust) or by isolates from Germany and the Netherlands. The isolates originating from pigs and pig products could be found in both clusters, and the isolates from foods of other origins (turkeys, chickens, and cattle) shared profiles with isolates from pig samples. Accordingly, there is no clear separation in pig-related and non-pig-related clones, suggesting that the ST398 strains are probably transferred from pigs to cattle and/or poultry and vice versa.
The application of the macrorestriction-PFGE performed with Cfr9I in nontypeable SmaI ST398 isolates revealed a relatively high heterogeneity within the MLST ST398 clone, although a close genetic relationship among these isolates is still observed. As suggested in other studies (36), MSSA ST398 strains could have acquired the different SCCmec cassettes at different times, as happens in other S. aureus lineages (9, 12), and they could have evolved later due to loss and/or acquisition of recognition sites for Cfr9I, changes in the repeats within the spa region, and also by altering or losing the SCCmec cassette.
Acknowledgments
We thank U. Kämpe and J. Beutlich for their support. We also are grateful to the different German Regional Laboratories, as well as to D. Mevius and K. Veldman (Central Veterinary Institute of Waningen, Lelystad, the Netherlands), for providing the isolates or samples that were included in the study.
The project was supported by the Federal Institute for Risk Assessment (BfR-2008/45-004 and 41-001), the German Ministry for Food Agriculture and Consumer Protection (2808HS032), and the Fondo de Investigaciones Sanitarias (PI052489 and PI080656) from the Spanish Ministry of Science and Innovation. M. A. Argudín, a Ph.D. student, was the recipient of grant FPU AP-2004-3641 from the Ministry of Science and Innovation, Spain, cofunded by the European Social Fund. She performed a short stay at the Department of Biological Safety of the Federal Institute for Risk Assessment (BfR), Berlin, Germany, supported by the same grant.
Footnotes
Published ahead of print on 18 December 2009.
REFERENCES
- 1.Anonymous. 2008. Commission Decision 2008/55/EC concerning a financial contribution of the Community towards a survey on the prevalence of Salmonella spp. and methicillin-resistant Staphylococcus aureus in herds of breeding pigs to be carried out in the Member States. Off. J. Eur. Union. http://eurlex.europa.eu/LexUriServ/LexUriServ.do?uri=OJ:L:2008:014:0010:0025:EN:PDF.
- 2.Anonymous. 2009. Joint scientific report of ECDC, EFSA and EMEA on methicillin-resistant Staphylococcus aureus (MRSA) in livestock, companion animals and food. www.health.gov.mt/fsc/fschome_files/efsa_biohaz_report_301_joint_mrsa_en.pdf.
- 3.Argudín, M. A., M. R. Rodicio, and B. Guerra. 2009. The emerging methicillin-resistant Staphylococcus aureus ST398 clone can easily be typed using Cfr9I SmaI neoschizomer. Lett. Appl. Microbiol. 50:127-130. [DOI] [PubMed] [Google Scholar]
- 4.Bens, C. C. P. M., A. Voss, and C. H. W. Klaassen. 2006. Presence of a novel DNA methylation enzyme in methicillin-resistant Staphylococcus aureus isolates associated with pig farming leads to uninterpretable results in standard pulsed-field gel electrophoresis analysis. J. Clin. Microbiol. 44:1875-1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhat, M., C. Dumortier, B. S. Taylor, M. Miller, G. Vasquez, J. Yunen, K. Brudney, E. J. Sánchez, C. Rodriguez-Taveras, R. Rojas, P. Leon, and F. D. Lowy. 2009. Staphylococcus aureus ST398, New York City and Dominican Republic. Emerg. Infect. Dis. 15:285-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carriço, J. A., C. Silva-Costa, J. Melo-Cristino, F. R. Pinto, H. de Lencastre, J. S. Almeida, and M. Ramirez. 2006. Illustration of a common framework for relating multiple typing methods by application to macrolide-resistant Streptococcus pyogenes. J. Clin. Microbiol. 44:2524-2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Boer, E., J. T. Zwarkrui-Nahauis, B. Wit, X. W. Huijsdens, A. J. de Neeling, T. Bosch, R. A. van Oosterom, A. Vila, and A. E. Heuvelink. 2009. Prevalence of methicillin-resistant Staphylococcus aureus in meat. Int. J. Food Microbiol. 134:52-56. [DOI] [PubMed] [Google Scholar]
- 8.de Neeling, A. J., M. J. M. van den Broek, E. C. Spalburg, M. G. van Santen-Verheuvel, W. D. C. Dam-Deisz, H. C. Boshuizen, A. W. van de Giessen, E. van Duijkeren, and X. W. Huijsdens. 2007. High prevalence of methicillin-resistant Staphylococcus aureus in pigs. Vet. Microbiol. 122:366-372. [DOI] [PubMed] [Google Scholar]
- 9.Deurenberg, R. H., and E. E. Stobbering. 2008. The evolution of Staphylococcus aureus. Infect. Genet. Evol. 8:747-763. [DOI] [PubMed] [Google Scholar]
- 10.Enright, M. C., N. P. Day, C. E. Davies, S. J. Peacock, and B. G. Spratt. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fanoy, E., L. C. Helmhout, W. L. van der Vaart, K. Weijdema, M. G. van Santen-Verheuvel, S. F. Thijsen, A. J. de Neeling, W. J. van Wamel, S. H. Maňásková, and J. L. Kingma-Thijssen. 2009. An outbreak of non-typeable MRSA within a residential care facility. Euro Surveill. 14:pii=19080. [PubMed] [Google Scholar]
- 12.Gomes, A. R., H. Westh, and H. de Lencastre. 2006. Origins and evolution of methicillin-resistant Staphylococcus aureus clonal lineages. Antimicrob. Agents Chemother. 50:3237-3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grundmann, H., S. Hori, and G. Tanner. 2001. Determining confidence intervals when measuring genetic diversity and the discriminatory abilities of typing methods for microorganisms. J. Clin. Microbiol. 39:4190-4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grundmann, H., S. Hori, M. C. Enright, C. Webster, A. Tami, E. J. Feil, and T. Pitt. 2002. Determining the genetic structure of the natural population of Staphylococcus aureus: a comparison of multilocus sequence typing with pulsed-field gel electrophoresis, randomly amplified polymorphic DNA analysis, and phage typing. J. Clin. Microbiol. 40:4544-4546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guardabassi, L., M. Stegger, and R. Skov. 2007. Retrospective detection of methicillin-resistant and susceptible Staphylococcus aureus ST398 in Danish slaughter pigs. Vet. Microbiol. 122:384-386. [DOI] [PubMed] [Google Scholar]
- 16.Huijsdens, X. W., T. Bosch, M. G. van Santen-Verheuvel, E. Spalburg, G. N. Pluister, M. van Luit, M. E. Heck, A. Haenen, and A. J. de Neeling. 2009. Molecular characterisation of PFGE non-typeable methicillin-resistant Staphylococcus aureus in The Netherlands, 2007. Euro Surveill. 14:pii=19335. [DOI] [PubMed] [Google Scholar]
- 17.Hunter, P. R., and M. A. Gaston. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson′s index of diversity. J. Clin. Microbiol. 26:2464-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.International Working Group on the Classification of Staphylococcal Cassette Chromosome Elements (IWG-SCC). 2009. Classification of staphylococcal cassette chromosome mec (SCCmec): guidelines for reporting novel SCCmec elements. Antimicrob. Agents Chemother. 53:4961-4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ito, T., Y. Katayama, K. Asada, N. Mori, K. Tsutsumimoto, C. Tiensasitorn, and K. Hiramatsu. 2001. Structural comparison of three types of staphylococcal cassette chromosome mec integrated in the chromosome in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 45:1323-1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ito, T., X. Xue Ma, F. Takeuchi, K. Okuma, H. Yuzawa, and K. Hiramatsu. 2004. Novel type V staphylococcal cassette chromosome mec driven by a novel cassette chromosome recombinase, ccrC. Antimicrob. Agents Chemother. 48:2637-2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jansen, M. D., A. T. A. Box, and A. C. Fluit. 2009. SCCmec-typing in methicillin-resistant Staphylococcus aureus strains of animal origin. Emerg. Infect. Dis. 15:136-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kadlec, K., R. Ehricht, S. Monecke, U. Steinacker, H. Kaspar, J. Mankertz, and S. Schwarz. 2009. Diversity of antimicrobial resistance pheno- and genotypes of methicillin-resistant Staphylococcus aureus ST398 from diseased swine. J. Antimicrob. Chemother. 64:1156-1164. [DOI] [PubMed] [Google Scholar]
- 23.Meemken, D., C. Cuny, W. Witte, U. Eichler, R. Staudt, and T. Blaha. 2008. Occurrence of MRSA in pigs and in humans involved in pig production—preliminary results of a study in the northwest of Germany. Dtsch. Tierarztl. Wochenschr. 115:132-139. [PubMed] [Google Scholar]
- 24.Mellmann, A., T. Weniger, C. Berssenbrügge, J. Rothgänger, M. Sammeth, J. Stoye, and D. Harmsen. 2007. Based Upon Repeat Pattern (BURP): an algorithm to characterize the long-term evolution of Staphylococcus aureus populations based on spa polymorphisms. BMC Microbiol. 7:98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morgan, M. 2008. Methicillin-resistant Staphylococcus aureus and animals: zoonosis or humanosis? J. Antimicrob. Chemother. 62:1181-1187. [DOI] [PubMed] [Google Scholar]
- 26.Murchan, S., M. E. Kaufmann, A. Deplano, R. de Ryck, M. Struelens, C. E. Zinn, V. Fussing, S. Salmenlinna, J. Vuopio-Varkila, N. El Solh, C. Cuny, W. Witte, P. T. Tassios, N. Legakis, W. van Leeuwen, A. van Belkum, A. Vindel, I. Laconcha, J. Garaizar, S. Haeggman, B. Olsson-Liljequist, U. Ransjo, G. Coombes, and B. Cookson. 2003. Harmonization of pulsed-field gel electrophoresis protocols for epidemiological typing of strains of methicillin-resistant Staphylococcus aureus: a single approach developed by consensus in 10 European laboratories and its application for tracing the spread of related strains. J. Clin. Microbiol. 41:1574-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poulsen, A. B., R. Skov, and L. V. Pallesen. 2003. Detection of methicillin-resistance in coagulase-negative staphylococci and in staphylococci directly from simulated blood cultures using the EVIGENE MRSA Detection Kit. J. Antimicrob. Chemother. 51:419-421. [DOI] [PubMed] [Google Scholar]
- 28.Rasschaert, G., W. Vanderhaeghen, I. Dewaele, N. Janež, X. Huijsdens, P. Butaye, and M. Heyndrickx. 2009. Comparison of fingerprinting methods for typing methicillin-resistant Staphylococcus aureus sequence type 398. J. Clin. Microbiol. 47:3313-3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shopsin, B., M. Gomez, S. O. Montgomery, D. H. Smith, M. Waddington, D. E. Dodge, D. A. Bost, M. Riehman, S. Naidich, and B. N. Kreiswirth. 1999. Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. J. Clin. Microbiol. 37:3556-3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Silva-Costa, C., M. Ramirez, and J. Melo-Cristino. 2006. Identification of macrolide-resistant clones of Streptococcus pyogenes in Portugal. Clin. Microbiol. Infect. 12:513-518. [DOI] [PubMed] [Google Scholar]
- 31.Smith, T. C., M. J. Male, A. L. Harper, J. S. Kroeger, G. P. Tinkler, E. D. Moritz, A. W. Capuano, L. A. Herwaldt, and D. J. Diekema. 2009. Methicillin-resistant Staphylococcus aureus (MRSA) strain ST398 is present in Midwestern U.S. swine and swine workers. PLoS One 4:e4258. doi: 10.1371/journal.pone.0004258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Struelens, M. J., A. Bauernfeind, A. Van Belkum, D. Blanc, B. D. Cookson, L. Dijkshoorn, N. El Solh, J. Etienne, J. Garaizar, P. Gerner-Smidh, N. Legakis, H. de Lencastre, M. H. Nicolas, T. L. Pitt, U. Römling, V. Rosdahl, and W. Witte. 1996. Consensus guidelines for appropriate use and evaluation of microbial epidemiologic typing systems. Clin. Microbiol. Infect. 2:2-11. [DOI] [PubMed] [Google Scholar]
- 33.Tenhagen, B. A., A. Fetsch, B. Stührenberg, G. Schleuter, B. Ackermann, G. Schleuter, B. Guerra, J. A. Hammerl, S. Hertwig, J. Kowall, U. Kämpe, J. Bräunig, A. Schroeter, A. Käsbohrer, and B. Appel. 2009. Prevalence of MRSA types in slaughter pigs in different German abattoirs. Vet. Rec. 165:589-593. [DOI] [PubMed] [Google Scholar]
- 34.van Belkum, A., D. C. Melles, J. K. Peeters, W. B. van Leeuwen, E. van Duijkeren, X. W. Huijsdens, E. Spalburg, A. J. de Neeling, H. A. Verbrugh, and Dutch Working Party on Surveillance and Research of MRSA-SOM. 2008. Methicillin-resistant and -susceptible Staphylococcus aureus sequence type 398 in pigs and humans. Emerg. Infect. Dis. 14:479-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Loo, I., X. Huijsdens, E. Tiemersma, A. de Neeling, N. van de Sande-Bruinsma, D. Beaujean, A. Voss, and J. Kluytmans. 2007. Emergence of methicillin-resistant Staphylococcus aureus of animal origin in humans. Emerg. Infect. Dis. 13:1834-1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Wamel, W. J. B., S. Hansenová Maňásková, A. C. Fluit, H. Verbrugh, A. J. de Neeling, E. van Duijkeren, and A. van Belkum. 2009. Short term micro-evolution and PCR-detection of methicillin-resistant and -susceptible Staphylococcus aureus sequence type 398. Eur. J. Clin. Microbiol. Infect. Dis. [Epub ahead of print.] doi: 10.1007/s10096-009-0816-3. [DOI] [PMC free article] [PubMed]
- 37.Voss, A., F. Loeffen, J. Bakker, C. Klaassen, and M. Wulf. 2005. Methicillin-resistant Staphylococcus aureus in pig farming. Emerg. Infect. Dis. 11:1965-1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Witte, W., B. Strommenger, C. Stanek, and C. Cuny. 2007. Methicillin-resistant Staphylococcus aureus ST398 in humans and animals, central Europe. Emerg. Infect. Dis. 13:255-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wulf, M., and A. Voss. 2008. MRSA in livestock animals—an epidemic waiting to happen? Clin. Microbiol. Infect. 14:519-521. [DOI] [PubMed] [Google Scholar]
- 40.Zhang, K., J. A. McClure, E. Sameer, T. Louie, and J. M. Conly. 2005. Novel multiple PCR assay for characterization and concomitant subtyping of staphylococcal cassette chromosome mec types I to IV in methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 43:5026-5033. [DOI] [PMC free article] [PubMed] [Google Scholar]