Abstract
This study examined bacteria-immune interactions in a mouse model possessing microbiota-dependent immune regulatory features similar to those occurring in human atopy, colitis, and immune regulation. Associations between the abundance of several bacterial phylotypes and immunoregulatory target cell types were identified, suggesting that they may play a role in these phenotypes.
Bacteria are involved in critical aspects of immune system development and regulation (5, 23, 26, 29). Mice raised under germfree conditions exhibit a variety of abnormalities, including hypoplastic Peyer's patches, reduced numbers of IgA-producing cells, relatively unstructured spleen and lymph nodes, and hypogammaglobulinemic serum (23). Remarkably, after several weeks of exposure to standard intestinal microbiota, normal immune structure and function are restored. Mechanistic details underlying microbe-immune interactions have been recently elucidated for two common intestinal bacteria. Bacteroides thetaiotaomicron was shown to induce the angiogenin Ang4, a component of innate immunity possessing microbicidal activity against a wide range of intestinal microbes, including both bacterial and fungal pathogens (16). In addition, studies of the Bacteroides fragilis zwitterionic capsular polysaccharide A have established it as a cognate antigen of certain CD4+ T cells, which programs immune effector polarization (24) and protection of mice from infection by Helicobacter hepaticus through several immune-mediated mechanisms (25). Resident microbiota also modify the interaction of dendritic cells with regulatory T-cell populations, with resultant susceptibility to chronic inflammatory disease, like colitis (15, 28).
Recent work by Braun and colleagues has characterized a mouse model with unique immunologic features linking resident microbiota with levels of regulatory CD8+ T cells (13, 17, 39). This model is comprised of two physically isolated colonies of isogenic mice harboring distinct microbial communities: conventional floras (CF) and restricted floras (RF). CF refers to C57BL/6 mice housed in a standard specific-pathogen-free facility, while RF refers to C57BL/6 mice containing a different complement of intestinal microorganisms (13, 30), originally created by transferring several nonpathogenic anaerobic bacteria into antibiotic-treated mice (13). RF mice differ from CF mice in several immunologic phenotypes, including selective reduction of marginal zone (MZ) B cells (39), plasmacytoid dendritic cells (pDC) (13), and invariant natural killer (iNK) T cells (38a), as well as naïve CD4+ and CD8+ T cells (17). In addition, RF mice were shown to be resistant to colitis under genetic or adoptive transfer conditions that permit disease activity in CF mice (2). RF mice also cleared experimental infections by Campylobacter jejuni more slowly than did their CF counterparts (6). The resulting concept is that certain resident microbiota, which may be more abundant in RF mice than in CF mice, induce invariant Qa-1 T cells, with resultant changes in host immunoregulation and microbial surveillance (2).
An important issue raised by the foregoing observations is the identity of resident microbiota responsible for this host immunoregulatory response. The objective of this study was to develop a methodology, based on bacteria-immune interactions in the RF/CF mouse model, to identify candidate microbiota. In this study, we employed a series of experiments examining associations between the population densities of bacterial rRNA genes and several immunologic features that differ between CF and RF mice.
Mouse analyses.
These experiments utilized a mouse model comprised of two physically isolated colonies of isogenic C57BL/6 mice harboring distinct microbial communities: CF and RF. A detailed description of this model can be found in the report by Fujiwara et al. (13). A former name for RF mice was LF (limited flora) (6). A former name for CF mice was SPF (specific pathogen free) (2, 13, 17, 39). All animals were housed under specific-pathogen-free conditions and were monitored by serology or culture for the absence of a panel of viral, fungal, and bacterial pathogenic taxa, including Helicobacter spp. The animal procedures were carried out in accordance with the animal research protocols approved by the UCLA institutional animal research committee.
CD8+ T-cell populations were reduced or abolished in two types of experiments. Purified anti-NK1.1 (PK136) and anti-CD8-β (341) antibodies (from BD Biosciences, San Diego, CA) containing no preservative were administered intravenously (i.v.) into RF mice at 100 μg/mouse. Mice in control groups were injected with isotype control antibodies. Injection was repeated twice per week. The mice were sacrificed 1 week after the final injection. CD8α−/− (14), and Prf1−/− mice (37) with the C57BL/6 background were bred to contain restricted microflora by raising cesarean section-delivered pups with RF foster mothers as recently described (13).
Luminal compartment samples were collected by obtaining 5- to 10-cm segments of the small intestine or colon, moving the luminal contents to one end of the intestinal segment with a forceps, and placing 2- to 3-cm segments of the tissues containing the condensed luminal contents in a FastDNA lysis tube containing 1 ml of cell lysis solution for yeast (CLS-Y) buffer from a FastDNA Spin kit (MP Biomedicals, Solon, OH) and immediately frozen at −70°C for future DNA extraction. Mucosal compartment samples were collected from small intestine and colon samples by using a previously described procedure to harvest intraepithelial lymphocytes (IEL) (38), except that 200 μl of the IEL preparations were removed prior to Percoll fractionation and placed in FastDNA lysis tubes containing 1 ml CLS-Y buffer from a FastDNA Spin kit (MP Biomedicals) and immediately frozen at −70°C for future DNA extraction.
Fluorochrome-antibody conjugates reactive to mouse B220, CD3ɛ, CD21, IgD, and T-cell receptor β (TCRβ) were purchased from BD Biosciences (San Diego, CA). Mouse CD1d tetramers loaded with PBS-57 were obtained from the NIH Tetramer Core Facility at the Emory Vaccine Center at Yerkes (Atlanta, GA). Enumeration of iNK T cells from hepatic lymphocytes was performed as previously described (19), using multiparameter staining for CD1d tetramers, anti-CD3ɛ, and anti-TCRβ. Enumeration of MZ B cells from splenic lymphocytes was performed as previously described (39), based on a B220+ CD21+ IgD− phenotype. Data were collected on a FACSCalibur flow cytometer (BD Biosciences) and analyzed using CellQuest software.
Bacterial analyses.
DNA was extracted from luminal and mucosal compartment samples by using the FastDNA Spin kit (MP Biomedicals) as described by the manufacturer, using the CLS-Y buffer and a 40-s bead-beading step with a FastPrep instrument setting of 6. DNA was further purified and size fractionated by electrophoresis in 1% agarose gels. DNA larger than 3 kb was excised without exposure to UV or ethidium bromide and recovered using the QIAquick gel extraction kit (Qiagen, Valencia, CA) following the manufacturer's instructions, except that the gel pieces were not exposed to heat.
Bacterial 16S-23S rRNA intergenic fragments were amplified in 25-μl PCR mixtures with the following final concentrations or amounts: 1 μl of luminal or mucosal DNA, 0.04 μM forward primer (1406F, TGYACACACCGCCCGT) (20), 0.4 μM reverse primer (23SR, GGGTTBCCCCATTCRG), and 12.5 μl Taq 2× master mix (New England Biolabs, Ipswich, MA) (3). All reagents were combined and heated at 94°C for 5 min. Thirty-five cycles of PCR were then performed at 94°C for 20 s, 52°C for 30 s, and 72°C for 45 s, and 72°C for 5 min. PCRs were performed in an MJ Research PTC-200 thermal cycler. The PCR products were resolved by electrophoresis on 2% agarose gels, stained with ethidium bromide, and photographed under UV light.
Nucleotide sequences of selected bacterial 16S-23S rRNA intergenic fragments were determined using the ABI BigDye Terminator v3.1 cycle sequencing kit and an ABI 3730xl DNA analyzer (Applied Biosystems, Foster City, CA). Sequence identities were determined using BLAST (NCBI) (1). Some of the ribosomal intergenic spacer analysis (RISA) bands contained several distinct nucleotide sequences and were not examined further in this study.
Seven bacterial phylotypes were enumerated using sequence-selective quantitative PCR (qPCR). Real-time qPCR assays were performed using a Bio-Rad iCycler MyiQ real-time detection system (Bio-Rad Laboratories Inc., Hercules, CA) and SYBR green detection with iCycler iQ PCR plates and optical flat eight-cap strips (Bio-Rad Laboratories Inc). Twenty-five-microliter reaction mixtures contained the following reagents: 50 mM Tris (pH 8.3), 500 μg/ml bovine serum albumin (BSA), 2.5 mM MgCl2, 250 μM each deoxynucleoside triphosphate (dNTP), 400 nM each primer, 1 μl of template DNA, 2 μl of 10× SYBR green I (Invitrogen, Carlsbad, CA), and 1.25 U Taq DNA polymerase. The primers, amplicon sizes, and other information on the phylotypes can be found in Table S1 in the supplemental material. Sequence-selective primers were designed either by (i) locating DNA sequences that were conserved among the rRNA gene sequences within each phylotype and which had few, if any, identical matches to rRNA gene sequences from unrelated taxonomic groups or by (ii) using the recently developed PRISE software (12). The thermal cycling conditions were 94°C for 5 min; 36 to 38 cycles of 94°C for 20 s, X°C for 30 s, and 72°C for 30 s; and 72°C for 10 min, where X = 65.3°C for Lactobacillus 456, 63.7°C for Clostridium 2041 and Barnesiella 116, 65.9°C for Akkermansia 671, 63.3°C for Enterobacter 144, 60.7°C for Turicibacter 458, and 61.9°C for Bacteroides 110. At each cycle, accumulation of PCR product was measured by monitoring the increase in fluorescence of the double-stranded DNA-binding SYBR green dye. rRNA gene levels in the sample DNA were quantified by interpolation from a standard curve comprised of a dilution series of cloned rRNA genes. To increase the likelihood that the real-time signals were produced by amplification of the target sequences, for each sequence-selective PCR assay, fragments were cloned into pGEM-T (Promega, Madison, WI) and the nucleotide sequences of two clones were determined; these experiments confirmed that the target sequences were being amplified (data not shown).
Associations between the bacterial rRNA gene quantities and lymphocyte levels were examined using correlation analyses (Minitab 15; State College, PA). Comparisons of the bacterial rRNA gene quantities from the CD8+ T-cell experiments were performed using two-tailed Student's t tests (Excel; Microsoft, Redmond, WA).
Experimental strategy.
The long-term objective of this research is to understand the roles that microorganisms play in the differing immunologic phenotypes expressed by CF and RF mice. Toward this end, we (i) identified bacterial phylotypes whose population densities were different in CF and RF mice, (ii) examined the relationships between the population densities of these phylotypes and the abundance of target immune cell types (MZ B and iNK T cells) in CF and RF mice, and (iii) investigated the relationships between the phylotype population densities in RF mice and RF mice with reduced or abolished CD8+ T-cell populations.
Comparison of intestinal bacteria in CF and RF mice.
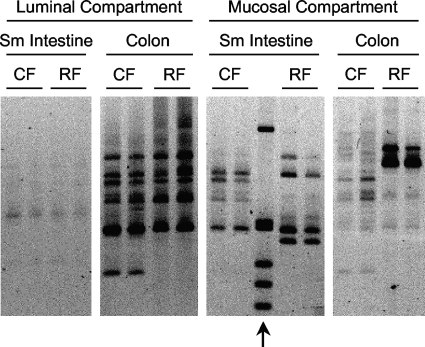
Ribosomal intergenic spacer analyses were used to examine the bacterial communities in CF and RF mice (Fig. 1). These analyses showed that there were distinct differences in bacterial composition between CF and RF mice. Most of these differences were found in the mucosal compartments, which were examined by collecting and analyzing the intestinal cells that were released by dithiothreitol treatments of small intestine and colonic tissues. These analyses also showed that within each mouse type, there were differences in bacterial composition between the small intestine and colon as well as between the luminal and mucosal compartments.
FIG. 1.
Bacterial community profiles of various intestinal regions and compartments from CF and RF mice. Profiles were obtained by RISA. For each mouse type-intestinal region combination, bacterial communities from two different mice are shown. Arrow indicates a 1-kb DNA ladder (Invitrogen) (top band = 1,018 bp; bottom band = 298 bp). Sm, small.
Gel bands exhibiting different intensities between CF and RF mice in these rRNA gene analyses, as well as others, were excised, cloned, and sequenced. Phylotypes were given designations based on their closest cultured relative (see Table S1 in the supplemental material). For each of the phylotypes, sequence-selective qPCR assays were developed to enable their enumeration in subsequent experiments.
Associations between bacterial phylotypes and MZ B or iNK T cells.
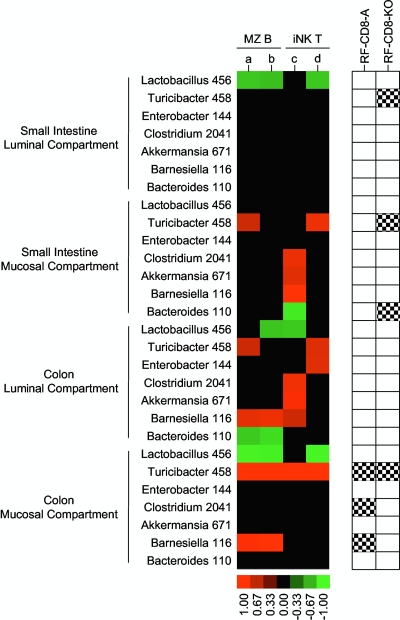
As a first step in examining the interactions between bacteria and the immunologic features in these mice, we measured the population densities of the bacterial phylotypes and the amounts of MZ B and iNK T cells in CF and RF mice. Phylotypes were enumerated in the luminal and mucosal compartments of the small intestine and colon. Immune cells were enumerated in spleen and liver tissues. Associations between the amounts of the phylotypes and immune cells in CF and RF mice are depicted in a heat map (colored portions of Fig. 2). Such associations could indicate bacterial populations that are driving, or being altered by, the immunologic features of these mice.
FIG. 2.
Associations between the amounts of the bacterial phylotypes and MZ B and iNK T cells in CF and RF mice. Phylotypes were measured in several intestinal regions and compartments by using sequence-selective qPCR assays (see Table S1 in the supplemental material for more information on the phylotypes). Lymphocytes were measured by flow cytometry (a = %MZ B cells [CD21 + IgMhi] in spleen, b = %MZ B cells [CD21 + IgD] in spleen, c = NK1.1 + % in liver, and d = CD1d tetramer + % in liver). Phylotypes whose abundance correlated (P < 0.05) with lymphocyte levels are shown as colored blocks. The strengths of the trends (Pearson's correlation coefficients) are depicted by the brightness of the colors (see scale bar). n = 8 for the luminal and mucosal compartments of the small intestine and the luminal compartment of the colon, and n = 4 for the mucosal compartment of the colon. The right side of the figure shows phylotypes that increased (black and white checkered boxes) in RF mice treated with CD8 and NK1.1 antibodies (RF-CD8-A) or RF mice with a CD8+ T-cell knockout (RF-CD8-KO) compared to RF mice; these results are from the experiments depicted in Fig. 3 and are presented here to facilitate interpretation of all three sets of experiments.
In a very simple scenario, where one type of bacteria from a single location is causing the immunologic differences between RF and CF mice, one would expect to find significant associations between this organism and all of the immune cell types that are divergent in these mice. Intriguingly, an examination of the heat map (Fig. 2) shows only one phylotype-location combination that fits this criterion: Turicibacter 458 from the mucosal compartment of the colon (four immune cell types consistently divergent in RF and CF mice were measured in the experiments represented in Fig. 2). Two other phylotype-location combinations come close to fitting this model, as they exhibited significant associations (P < 0.05) for three immune cell types and relatively high probability values for the fourth—Barnesiella 116 from the luminal compartment of the colon (P = 0.172) and Lactobacillus 456 from the mucosal compartment of the colon (P = 0.115). If the interactions between the bacteria and the immunologic features involve more than one bacterium, then this heat map could be interpreted to show several bacteria putatively involved in these immunologic features. Note that these association experiments are only the first in a series of investigations endeavoring to understand the bacteria-immune interactions in this mouse model and that subsequent experimentation will be needed to assess causality and elucidate mechanistic interactions. In the following, we describe how bacteria could be influencing the immunologic differences between CF and RF mice.
How bacteria could influence MZ B and iNK T cells in CF and RF mice.
The principal immunologic feature that differs between the two mouse types involves CD8+ T cells. RF mice have reduced populations of iNK T and MZ B cells and pDC, all of which appear to be caused by cytolytic CD8+ T cells (13, 39; Wei et al., submitted). Conversely, in CF mice, relatively low levels of CD8+ T cells are associated with higher levels of MZ B and iNK T cells.
In a very nonspecific model, bacteria could influence these immunologic features in four general different ways. In RF mice, high levels (or high activity) of a specific bacterium could facilitate the killing of MZ B and iNK T cells through some positive regulating mechanism. Conversely, low levels (or low activity) of a specific bacterium could facilitate the killing through some negative regulating mechanism. For the CF mice, one could envision similar but opposite scenarios.
In a more specific model, which is consistent with the our current knowledge of RF mice (13, 39; Wei et al., submitted), the immunologic changes observed in RF mice could be caused by bacteria infecting MZ B and iNK T cells (see Figure S1A in the supplemental material). Here, CD8+ T cells would be reducing MZ B- and iNK T-cell populations via a cytolytic mode of action that involves targeting the infected bacterial cells in a major histocompatibility complex class I (MHC-I) antigen-specific manner.
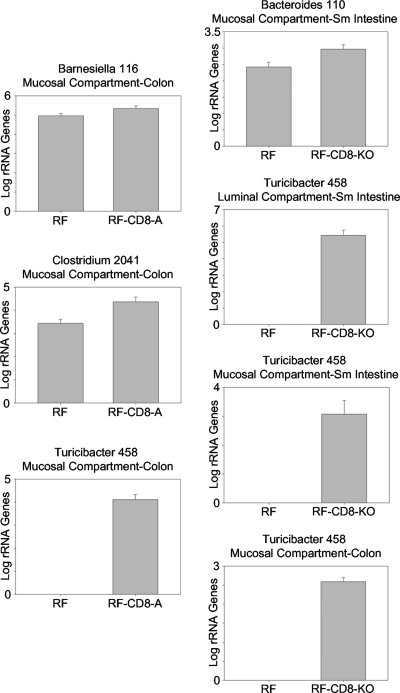
To further investigate the bacteria-immune system interactions in this mouse model, we performed two types of experiments that measured the population densities of the bacterial phylotypes in both standard RF mice and RF mice with reduced or abolished CD8+ T-cell populations. The purpose of these experiments was to narrow down the list of bacteria that may be involved in these interactions—the organisms consistently exhibiting associations with the phenotypes being the ones most likely to be involved in the interactions. The first experiment utilized antibodies to deplete CD8+ T cells in RF mice, while the second employed CD8+ knockout mice. In both experiments, when the number of CD8+ T cells was reduced or abolished, significant increases in bacterial population densities were detected in six phylotype-location combinations (Fig. 3; see also the checkered black and white boxes on the right side of Fig. 2). In the antibody experiment, Barnesiella 116, Clostridium 2041, and Turicibacter 458 exhibited increases in their population densities in the treated animals. In the knockout mice, the population of Turicibacter 458 increased in three of the four intestinal compartments while that of Bacteroides 110 increased in the mucosal compartment of the small intestine. When these results are combined with those presented in the heat map (Fig. 2), the number of phylotype-location combinations exhibiting an association with both an immune cell type and one of the CD8 treatments is reduced to only four—Bacteroides 110 and Turicibacter 458 in the small intestine and Barnesiella 116 and Turicibacter 458 in the colon—all of which were located in the mucosal compartments of these tissues.
FIG. 3.
Bacterial phylotypes whose population densities changed after reducing or abolishing CD8+ T-cell populations in RF mice. Left column, CD8-NK1.1 antibody-treated RF mice (RF-CD8-A) compared to RF mice (P < 0.05). Right column, RF mice containing a CD8 knockout (RF-CD8-KO) compared to RF mice (P < 0.05). Phylotypes were measured by sequence-selective qPCR. n = 4 to 6 for RF-CD8-A mice and 2 to 4 for RF mice and RF-CD8-KO mice. Columns show means and standard errors.
In the context of the MHC-I antigenic targeting model, if a specific bacterium was infecting MZ B and iNK T cells and its presence was leading to antigenic targeting by CD8+ T cells, then reducing or removing the population of CD8+ T cells should lead to increases in the population densities of the MZ B cells and iNK T cells as well as the infecting bacterium (see Figure S1B in the supplemental material). As described above, several phylotype-location combinations are consistent with these criteria and therefore the antigenic targeting model (Fig. 3; see also the checkered black and white boxes on the right side of Fig. 2). However, one unexplained factor remains, which is that all of the phylotypes examined in this study inhabit both CF and RF mice. Thus, if the infected immune cells are being antigenically targeted by CD8+ T cells, why is this occurring to a much greater extent in RF mice?
There are several ways to explain this apparent inconsistency. Though the immunological differences in the CF and RF mice likely involve differences in microbial populations, it is possible that none of the bacteria examined in this study are the ones causing the differences. It is also possible that multiple organisms and mechanisms are contributing to this process. For example, one organism may be involved in triggering the antigenic targeting of MZ B and iNK T cells, while another may be involved in either activating CD8+ T cells or causing a selective uptake of the antigenically targeted bacteria into MZ B and iNK T cells. A third explanation is that bacteria possessing identical rRNA genes but different physiological properties (18) are present in our mice and are confounding the results. For example, a relative of Turicibacter 458, Erysipelothrix rhusiopathiae, can undergo morphological changes resulting in variations in its virulence and antigenic properties (8).
Another issue that remains unresolved is which, if any, of the bacteria have the ability to infect MZ B and iNK T cells—a feature that is important for the MHC-I antigenic targeting model. The phylotype that best fits this criterion is Turicibacter 458, as this bacterium is in the same family as E. rhusiopathiae (22), which can infect chondrocytes and hepatocytes (11, 35) as well as survive in macrophages (31, 33) and polymorphonuclear leukocytes (34). To date, there are very few reports describing Turicibacter species and their potential interactions with hosts. Turicibacter sanguinis was isolated from blood from a man experiencing acute appendicitis (4). Turicibacter rRNA gene sequences have also been PCR amplified from human feces (21) and ileal pelvic pouch contents from a subject with ulcerative colitis (10). More work is clearly needed to determine if Turicibacter 458, or any of the other phylotypes examined in this study, are causative agents in the immunologic phenotypes expressed by RF mice.
It is also notable that we identified associations between bacteria in the intestine and MZ B- and iNK T-cell populations in the liver and spleen. These results suggest that bacteria inhabiting the intestine may influence the immune system in a systemic manner. This is consistent with other investigations (27, 32), which, for example, have shown that an expansion of anaerobic bacteria in the small intestine was associated with mucosal and nonmucosal immune activation (9).
Experimental design consideration.
These studies, along with other reports (7, 36), highlight an important experimental design consideration. Namely, a better understanding of host-microbe interactions will require investigations that examine greater numbers of smaller compartments. In our studies, distinct differences in bacterial composition were detected in various intestinal compartments. More importantly, associations between these bacteria and the immunologic features were compartment specific and primarily occurred in the mucosal compartment—a niche that is rarely examined. Given both the small size of microorganisms and our limited knowledge of how and where such organisms interact with the host, a critical experimental design element will certainly include examining niches at a scale more commensurate with the organisms under investigation.
Nucleotide sequence accession numbers.
Nucleotide sequences of the bacterial 16S-23S rRNA intergenic fragments analyzed in this study were deposited in GenBank (NCBI) as follows: Akkermansia 671, accession no. FJ538174; Barnesiella 116, FJ538173; Bacteroides 110, accession no. FJ538175; Clostridium 2041, accession no. FJ538172; Enterobacter 144, accession no. FJ538171; Lactobacillus 456, accession no. EU375461; and Turicibacter 458, accession no. EU375462.
Supplementary Material
Acknowledgments
This work was funded in part by grants from the Edythe and Eli Broad Medical Research Program and by NIH grants DK046763, DK069434, and AI078885. Flow cytometry was performed at the Jonsson Comprehensive Cancer Center core facility (NIH grant CA016042-34).
Footnotes
Published ahead of print on 11 December 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aranda, R., B. C. Sydora, P. L. McAllister, S. W. Binder, H. Y. Yang, S. R. Targan, and M. Kronenberg. 1997. Analysis of intestinal lymphocytes in mouse colitis mediated by transfer of CD4+, CD45RBhigh T cells to SCID recipients. J. Immunol. 158:3464-3473. [PubMed] [Google Scholar]
- 3.Borneman, J., and E. W. Triplett. 1997. Molecular microbial diversity in soils from eastern Amazonia: evidence for unusual microorganisms and microbial population shifts associated with deforestation. Appl. Environ. Microbiol. 63:2647-2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bosshard, P. P., R. Zbinden, and M. Altwegg. 2002. Turicibacter sanguinis gen. nov., sp. nov., a novel anaerobic, Gram-positive bacterium. Int. J. Syst. Evol. Microbiol. 52:1263-1266. [DOI] [PubMed] [Google Scholar]
- 5.Bouskra, D., C. Brézillon, M. Bérard, C. Werts, R. Varona, I. G. Boneca, and G. Eberl. 2008. Lymphoid tissue genesis induced by commensals through NOD1 regulates intestinal homeostasis. Nature 456:507-510. [DOI] [PubMed] [Google Scholar]
- 6.Chang, C., and J. F. Miller. 2006. Campylobacter jejuni colonization of mice with limited enteric flora. Infect. Immun. 74:5261-5271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dethlefsen, L., and D. A. Relman. 2007. The importance of individuals and scale: moving towards single cell microbiology. Environ. Microbiol. 9:8-10. [DOI] [PubMed] [Google Scholar]
- 8.Ewald, F. W. 1981. The genus Erysipelothrix, p. 1688-1700. In M. P. Starr, H. Stolp, H. G. Truper, A. Balows, and H. G. Schlegel (ed.), The prokaryotes. Springer-Verlag, New York, NY.
- 9.Fagarasan, S., M. Muramatsu, K. Suzuki, H. Nagaoka, H. Hiai, and T. Honjo. 2002. Critical roles of activation-induced cytidine deaminase in the homeostasis of gut flora. Science 298:1424-1427. [DOI] [PubMed] [Google Scholar]
- 10.Falk, A., C. Olsson, S. Ahrné, G. Molin, D. Adawi, and B. Jeppsson. 2007. Ileal pelvic pouch microbiota from two former ulcerative colitis patients, analysed by DNA-based methods, were unstable over time and showed the presence of Clostridium perfringens. Scand. J. Gastroenterol. 42:973-985. [DOI] [PubMed] [Google Scholar]
- 11.Franz, B., M. E. Davies, and A. Horner. 1995. Localization of viable bacteria and bacterial antigens in arthritic joints of Erysipelothrix rhusiopathiae-infected pigs. FEMS Immunol. Med. Microbiol. 12:137-142. [DOI] [PubMed] [Google Scholar]
- 12.Fu, Q., P. Ruegger, E. Bent, M. Chrobak, and J. Borneman. 2008. PRISE (PRImer SElector): software for designing sequence-selective PCR primers. J. Microbiol. Methods 72:263-267. [DOI] [PubMed] [Google Scholar]
- 13.Fujiwara, D., B. Wei, L. L. Presley, S. Brewer, M. McPherson, M. A. Lewinski, J. Borneman, and J. Braun. 2008. Systemic control of plasmacytoid dendritic cells by CD8+ T cells and commensal microbiota. J. Immunol. 180:5843-5852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fung-Leung, W. P., M. W. Schilham, A. Rahemtulla, T. M. Kundig, M. Vollenweider, J. Potter, W. van Ewijk, and T. W. Mak. 1991. CD8 is needed for development of cytotoxic T cells but not helper T cells. Cell 65:443-449. [DOI] [PubMed] [Google Scholar]
- 15.Garrett, W. S., G. M. Lord, S. Punit, G. Lugo-Villarino, S. K. Mazmanian, S. Ito, J. N. Glickman, and L. H. Glimcher. 2007. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell 131:33-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hooper, L. V., T. S. Stappenbeck, C. V. Hong, and J. I. Gordon. 2003. Angiogenins: a new class of microbiocidal proteins involved in innate immunity. Nat. Immunol. 4:269-273. [DOI] [PubMed] [Google Scholar]
- 17.Huang, T., B. Wei, P. Velazquez, J. Borneman, and J. Braun. 2005. Commensal microbiota alter the abundance and TCR responsiveness of splenic naïve CD4+ T lymphocytes. Clin. Immunol. 117:221-230. [DOI] [PubMed] [Google Scholar]
- 18.Jaspers, E., and J. Overmann. 2004. Ecological significance of microdiversity: identical 16S rRNA gene sequences can be found in bacteria with highly divergent genomes and ecophysiologies. Appl. Environ. Microbiol. 70:4831-4839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kronenberg, M. 2005. Toward an understanding of NKT cell biology: progress and paradoxes. Annu. Rev. Immunol. 23:877-900. [DOI] [PubMed] [Google Scholar]
- 20.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. Wiley, New York, NY.
- 21.Licht, T. R., B. Madsen, and A. Wilcks. 2007. Selection of bacteria originating from a human intestinal microbiota in the gut of previously germ-free rats. FEMS Microbiol. Lett. 277:205-209. [DOI] [PubMed] [Google Scholar]
- 22.Ludwig, W., K. H. Schleifer, and W. B. Whitman. 2009. Revised road map to the phylum Firmicutes. In P. De Vos, G. Garrity, D. Jones, N. R. Krieg, W. Ludwig, F. A. Rainey, K. H. Schleifer, and W. B. Whitman (ed.), Bergey's manual of systematic bacteriology, 2nd ed., vol. 3. Springer-Verlag, New York, NY.
- 23.Macpherson, A. J., and N. L. Harris. 2004. Interactions between commensal intestinal bacteria and the immune system. Nat. Rev. Immunol. 4:478-485. [DOI] [PubMed] [Google Scholar]
- 24.Mazmanian, S. K., C. H. Liu, A. O. Tzianabos, and D. L. Kasper. 2005. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 122:107-118. [DOI] [PubMed] [Google Scholar]
- 25.Mazmanian, S. K., J. L. Round, and D. L. Kasper. 2008. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature 453:620-625. [DOI] [PubMed] [Google Scholar]
- 26.Noverr, M. C., and G. B. Huffnagle. 2004. Does the microbiota regulate immune responses outside the gut? Trends Microbiol. 12:562-568. [DOI] [PubMed] [Google Scholar]
- 27.O'Mahony, C., P. Scully, D. O'Mahony, S. Murphy, F. O'Brien, A. Lyons, G. Sherlock, J. MacSharry, B. Kiely, F. Shanahan, and L. O'Mahony. 2008. Commensal-induced regulatory T cells mediate protection against pathogen-stimulated NF-kappaB activation. PLoS Pathog. 4:e1000112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rakoff-Nahoum, S., J. Paglino, F. Eslami-Varzaneh, S. Edberg, and R. Medzhitov. 2004. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell 118:229-241. [DOI] [PubMed] [Google Scholar]
- 29.Round, J. L., and S. K. Mazmanian. 2009. The gut microbiota shapes intestinal immune responses during health and disease. Nat. Rev. Immunol. 9:313-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scupham, A. J., L. L. Presley, B. Wei, E. Bent, N. Griffith, M. McPherson, F. Zhu, O. Oluwadara, N. Rao, J. Braun, and J. Borneman. 2006. Abundant and diverse fungal microbiota in the murine intestine. Appl. Environ. Microbiol. 72:793-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shimoji, Y., Y. Yokomizo, and Y. Mori. 1996. Intracellular survival and replication of Erysipelothrix rhusiopathiae within murine macrophages: failure of induction of the oxidative burst of macrophages. Infect. Immun. 64:1789-1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shreiner, A., G. B. Huffnagle, and M. C. Noverr. 2008. The “microflora hypothesis” of allergic disease. Adv. Exp. Med. Biol. 635:113-134. [DOI] [PubMed] [Google Scholar]
- 33.Timoney, J. 1969. The inactivation of Erysipelothrix rhusiopathiae in macrophages from normal and immune mice. Res. Vet. Sci. 10:301-302. [PubMed] [Google Scholar]
- 34.Timoney, J. 1970. The inactivation of Erysipelothrix rhusiopathiae in pig buffy-coat leucocytes. Res. Vet. Sci. 11:189-190. [PubMed] [Google Scholar]
- 35.Todorov, T., A. Toshkov, M. Anastassova-Kristeva, Y. Martinova, and L. Kancheva. 1980. Histoautoradiographic studies of 3H-thymidine labelled Erysipelothrix rhusiopathiae introduced into albino mice. Zentralbl. Bakteriol. A 246:499-505. [PubMed] [Google Scholar]
- 36.Turnbaugh, P. J., R. E. Ley, M. Hamady, C. M. Fraser-Liggett, R. Knight, and J. I. Gordon. 2007. The human microbiome project. Nature 449:804-810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van den Broek, M. E., D. Kagi, F. Ossendorp, R. Toes, S. Vamvakas, W. K. Lutz, C. J. Melief, R. M. Zinkernagel, and H. Hengartner. 1996. Decreased tumor surveillance in perforin-deficient mice. J. Exp. Med. 184:1781-1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Van der Heijden, P. J., and W. Stok. 1987. Improved procedure for the isolation of functionally active lymphoid cells from the murine intestine. J. Immunol. Methods 103:161-167. [DOI] [PubMed] [Google Scholar]
- 38a.Wei, B., G. Wingender, D. Fujiwara, D.Y. Chen, M. McPherson, S. Brewer, J. Borneman, M. Kronenberg, and J. Braun. 2010. Commensal microbiota and CD8+ T cells shape the formation of invariant NKT cells. J. Immunol. doi: 10.4049/jimmunol.0902620. [DOI] [PMC free article] [PubMed]
- 39.Wei, B., T. T. Su, H. Dalwadi, R. P. Stephan, D. Fujiwara, T. T. Huang, S. Brewer, L. Chen, M. Arditi, J. Borneman, D. J. Rawlings, and J. Braun. 2008. Resident enteric microbiota and CD8+ T cells shape the abundance of marginal zone B cells. Eur. J. Immunol. 38:3411-3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.