Abstract
Enteric viruses are important pathogens found in contaminated surface waters and have previously been detected in waters of the Great Lakes. Human adenoviruses were monitored because of their high prevalence and persistence in aquatic environments. In this study, we quantified adenoviruses in wastewater, surface water, and combined sewer overflows (CSOs) by real-time PCR. Between August 2005 and August 2006, adenovirus concentrations in raw sewage, primary-treated effluent, secondary-treated effluent, and chlorinated effluent from a wastewater treatment plant in Michigan were examined. CSO samples (n = 6) were collected from a CSO retention basin in Grand Rapids, MI. Adenoviruses were detected in 100% of wastewater and CSO discharge samples. Average adenovirus DNA concentrations in sewage and CSOs were 1.15 × 106 viruses/liter and 5.35 × 105 viruses/liter, respectively. Adenovirus removal was <2 log10 (99%) at the wastewater treatment plant. Adenovirus type 41 (60% of clones), type 12 (29%), type 40 (3%), type 2 (3%), and type 3 (3%) were isolated from raw sewage and primary effluents (n = 28). Six of 20 surface water samples from recreational parks at the lower Grand River showed virus concentrations above the real-time PCR detection limit (average, 7.8 × 103 viruses/liter). This research demonstrates that wastewater effluents and wastewater-impacted surface waters in the lower Grand River in Michigan contain high levels of viruses and may not be suitable for full-body recreational activities. High concentrations of adenovirus in these waters may be due to inefficient removal during wastewater treatment and to the high persistence of these viruses in the environment.
Enteric viruses are important waterborne pathogens. They are frequently isolated from feces-contaminated water and have been linked to numerous waterborne outbreaks (9, 34, 42, 61). This group of pathogens includes adenoviruses, enteroviruses, hepatitis A virus, noroviruses, and rotavirus. In the Great Lakes region, enteric viruses were isolated from recreational beaches and groundwater for municipal usage, indicating an elevated public health risk in consuming or coming into contact with these waters (15, 69). Although recent developments in molecular detection assays substantially increase the detection of viruses from waters, from a management standpoint it is impractical to test all viruses when determining the microbial quality of water. Here we propose that adenovirus monitoring can be used to examine wastewater impacts on surface water quality.
Adenoviruses, which have a high prevalence in water, have been suggested as preferred candidates as index organisms for viral pathogens because they fit most criteria for an ideal indicator (19, 33, 38, 54). It is estimated that more than 90% of the human population is seropositive for one or more serotypes of adenoviruses (11, 68). Human adenoviruses (HAdVs) are present at a higher frequency in sewage than are other enteric viruses (54) and are excreted in high concentrations from infected patients (up to 1011 viral particles per gram of feces) (68).
Adenoviruses were first isolated from humans and identified as the causative agent of epidemic febrile respiratory disease among military recruits in the 1950s (30, 55). Human adenoviruses are the second most important viral pathogen of infantile gastroenteritis, after rotavirus (3, 10, 44, 51, 58, 62, 65). Serotypes of adenoviruses have been found to cause symptomatic infections in several organ systems, including the respiratory system (pharyngitis, acute respiratory disease, and pneumonia), eye (conjunctivitis), gastrointestinal tract (gastroenteritis), central nervous system (meningoencephalitis), and genitalia (urethritis and cervicitis) (8, 37). Human adenovirus types 40 and 41 have been associated with gastroenteritis in children, while human adenovirus type 4 is linked to persistent epidemics of acute respiratory disease in the United States (10, 49). It was estimated that 2 to 7% of all lower respiratory tract illnesses in children may be caused by adenoviruses (5, 17).
Transmission routes of adenoviruses include the fecal-oral route and inhalation of aerosols. Adenoviruses have been associated with outbreaks in different settings, including military camps (7, 36, 40), hospitals (6, 28, 32), day care centers (1, 38), and schools (27). Waterborne outbreaks due to adenoviruses have also involved swimming pools (53, 64).
It is hypothesized that combined sewer overflows (CSOs; where storm water and untreated sewage are combined) may contribute high concentrations of waterborne pathogens, especially viruses, which in turn may pose an adverse risk to human health. In older cities of Michigan, such as Detroit, East Lansing, and Grand Rapids, major contributors to microbial contamination of surface water during high-rainfall events include discharges from sanitary sewer systems and combined sewer systems. The federal government's effort to control CSOs started in 1994, when the U.S. EPA published the CSO Control Policy as the national framework. In Michigan, the first CSO policy was drafted by the Department of Environmental Quality in 1983. However, the first noncontested permit requiring a long-term CSO correction program was issued to the Grand Rapids wastewater treatment plant (WWTP) only in Fall 1988, following a large CSO event in the city that affected water quality downstream, in Grand Haven (50). To date, Michigan communities have eliminated 75% of the 613 untreated CSO outfalls that existed in the year 1988, and the remaining 25% are scheduled for correction/elimination through implementation of long-term control plans. However, water quality after CSO or any sewage spill remains a public health concern to individuals via recreational water exposure at recreational parks and beaches downstream of discharge sites.
The aim of this study was to evaluate the presence and concentration of adenoviruses in sewage and in the Grand River in the state of Michigan. Raw sewage, wastewater effluent, CSO discharges, and surface water in the lower Grand River were surveyed for the occurrence and concentration of human adenoviruses. Real-time PCR was used for quantification. Predominant adenovirus genotypes in sewage were determined, and the efficiency of virus removal during wastewater treatment was evaluated.
MATERIALS AND METHODS
Sample collection and concentration.
Grab water samples were collected from the East Lansing WWTP approximately once a month between August 2005 and August 2006. Wastewater treatment processes in the East Lansing WWTP consist of primary treatment (sedimentation), secondary treatment (aeration, activated sludge and secondary sedimentation), and tertiary treatment (chlorination, rapid gravity sand filters, dechlorination, and postfiltration aeration).
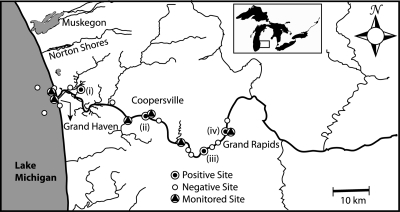
Surface water samples were collected from the lower Grand River in Kent and Ottawa counties during a 1-week intensive study in June 2005 and intermittently between June 2005 and August 2007. A total of 16 surface water samples were collected from different sites at the lower Grand River during the intensive study (Fig. 1). Following the intensive study, several sites were chosen for continued monitoring because of high bacterial indicator concentrations (data not shown) and detection of HAdV DNA from two of these sites (Deer Creek and Sixth Street Park). Our sample collection locations included three Lake Michigan beach sites (Rosy Mound, North Shore, and North Beach Park) and four park sites (Riverside Park, Deer Creek Park, Grand River Park, and Sixth Street Park).
FIG. 1.
Sampling sites along the lower Grand River during the intensive study in June 2005. The map shows sites that were positive for adenoviruses (•), sites that were negative for adenoviruses (○), and sites selected for continued monitoring between 2005 and 2007 (▴).
Between February and June 2008, six combined sewer samples were collected from the Market Avenue Retention Basin in Grand Rapids, MI, within 24 h of overflowing.
The procedures of Haramoto et al. (25) were used for water filtration and concentration of viruses. Briefly, between 500 ml and 2 liters of wastewater (for raw sewage and primary effluent, 100 to 200 ml was filtered) or surface water was filtered through a 90-mm, type HA, negatively charged membrane (Millipore, Billerica, MA) with a 0.45-μm pore size. A volume of 100 ml of 0.5 mM H2SO4 was then passed through the membrane, and viral particles were eluted with 20 ml of 1 mM NaOH. Eluates were stored in a tube containing 0.1 ml of 50 mM H2SO4 and 0.1 ml of 100× Tris-EDTA (TE) buffer for neutralization before further concentration.
For further purification and concentration, eluates were centrifuged in Amicon Ultra 100K concentrator columns (Millipore). The final volume of concentrated eluate recovered for each sample was between 200 μl and 700 μl. Concentrates were stored at −80°C. Concentrated samples were used for extraction of viral nucleic acids and purification with a DNeasy tissue kit (Qiagen, Valencia, CA) following the manufacturer's protocol. Purified viral DNA was eluted in 60 μl of RNase-free water.
Qualitative detection of human adenoviruses.
Samples from the intensive study in 2005 were qualitatively assayed for the presence of human adenoviruses. Nested PCR was performed by amplifying the open reading frame of the hexon gene of adenoviruses, using the primers described by Allard et al. (2), as described by Fong et al. (14). Amplicons of 300 and 142 bp were produced from the first and second rounds of PCR, respectively. HAdV type 40 (ATCC VR-931) was used as the positive control, and molecular-grade nuclease-free water was used as a no-template negative control.
Detection and quantification of human adenoviruses.
A real-time TaqMan PCR assay was performed for the quantification of HAdV DNA in water samples, following the protocol of Xagoraraki et al. (69). The forward primer (JTVXF), reverse primer (JTVXR), and TaqMan probe (JTVXP) designed by Jothikumar et al. (35) were used (Table 1) and targeted the hexon gene of HAdV. All amplification reactions were carried out in duplicate, using LightCycler TaqMan master mix (Roche Applied Sciences, Indianapolis, IN). qPCR standards were set up by cloning a section of the HAdV40 hexon gene in a plasmid vector (pCR4-TOPO), using a Topo TA cloning kit for sequencing (Invitrogen, Carlsbad, CA), as described by Xagoraraki et al. (69). PCR products were selected randomly for visualization by gel electrophoresis on a 2%-average-strength Omnipur agarose gel (EM Science, Darmstadt, Germany). The gel was stained with gelStar nucleic acid stain and viewed under UV light. Real-time PCR assays were performed in a Roche LightCycler 2.0 instrument (Roche Applied Sciences). Viral DNA extracts and standards were each run at least in triplicate. All qPCR runs included a negative control reaction (PCR-grade H2O without template) and a positive control reaction. Uracil-DNA glycosylase (Roche Applied Sciences) was added to each reaction mix to prevent carryover contamination. The crossing point (Cp) of each PCR was automatically determined by the LightCycler 4.0 software and used to calculate the hexon gene concentration. The concentrations of HAdVs in the river water samples from the studied recreational sites were converted from hexon gene copies per liter to viruses per liter, using a ratio of 1 (i.e., each HAdV particle consists of one copy of the hexon gene, as previously reported). The detection limit of this real-time PCR assay was determined to be 10 copies/PCR through serial dilution of a cloned PCR amplicon (69).
TABLE 1.
Primers and probes used for detection of adenovirus DNA in water samplesa
| Assay | Primer or probe | Name (polarity) | Sequence (5′-3′) | Tm (°C) | Amplicon size (bp) | Reference |
|---|---|---|---|---|---|---|
| HAdV nested PCR (detection) | Primer | Ad-A1 (+) | GCCGCAGTGGTCTTACATGCACATC | 60 | 300 | 2 |
| Primer | Ad-A2 (−) | CAGCACGCCGCGGATGTCAAAGT | 60 | 300 | 2 | |
| Nested primer | Ad-B1 (+) | GCCACCGAGACGTACTTCAGCCTG | 60 | 143 | 2 | |
| Nested primer | Ad-B2 (−) | TTGTACGAGTACGCGGTATCCTCGCGGTC | 60 | 143 | 2 | |
| HAdV TaqMan PCR | Primer | JTVXF (+) | GGACGCCTCGGAGTACCTGAG | 68 | 95 | 35 |
| Primer | JTVXR (−) | ACIGTGGGGTTTCTGAACTTGTT | 63 | 95 | 35 | |
| Probe | JTVXP (+) | CTGGTGCAGTTCGCCCGTGCCA | 78 | 35 | ||
| HAdV nested PCR | Primer | AdhexF1 (+) | TICTTTGACATICGIGGIGTICTIGA | 46 | 764-896 | 45 |
| (sequencing) | Primer | AdhexR1 (−) | CTGTCIACIGCCTGRTTCCACA | 53 | 764-896 | 45 |
| Nested primer | AdhexF2 (+) | GGYCCYAGYTTYAARCCCTAYTC | 45 | 688-821 | 45 | |
| Nested primer | AdhexR2 (−) | GGTTCTGTCICCCAGAGARTCIAGCA | 59 | 688-821 | 45 |
In all cases, the hexon gene was amplified.
Nested PCR and adenovirus DNA sequencing.
Adenovirus species present in water samples were identified by amplifying and sequencing the hypervariable region 1 to 6 of the adenovirus hexon gene. Samples that were positive by the real-time PCR assay were amplified using a conventional PCR assay prior to sequencing. Primers used and PCR conditions were as described by Lu and Erdman (45) (Table 1; primers AdhexF1 and AdhexR1 yield amplicons ranging in size from 764 to 896 bp). If insufficient DNA was amplified from the first PCR, a second PCR was performed using internal primers AdhexF2 and AdhexR2, which yield amplicons of between 688 and 821 bp (45). The sensitivity of this conventional nested PCR assay was determined by limiting-dilution experiments with pure adenovirus 40 stock in cell culture lysates (∼108 viral particles ml−1). Virus stocks were used to extract DNA, serially diluted to 100 virus DNA copies/reaction (as quantified by real-time PCR), and assayed by conventional PCR. The detection limit of this conventional PCR was 10 to 100 virus DNA copies/reaction.
Amplicons were visualized on an agarose gel and purified using a QiaQuick DNA purification kit (Qiagen) prior to sequencing. All HAdV-positive samples were identified by sequencing of at least two independent PCR products in both directions, i.e., each nucleotide was determined at least four times. Sequencing was performed by the Research Technology Support Facility at Michigan State University. Sequences were determined on an ABI Prism 3100 genetic analyzer (Applied Biosystems, Foster City, CA).
Sequence analysis.
The sequences obtained were aligned with hexon protein sequences of selected adenovirus prototype strains available from GenBank, using web-based ClustalW (http://www.ebi.ac.uk/clustalw/) with the following default settings: gap opening of 10, gap extension of 0.2, and hydrophilic residue gap penalties enabled. For analysis of serotypes, approximately 600-bp sequences of the PCR products were used. Amino acid alignment of the sequences was performed in ClustalW. A phylogenetic tree was then constructed using the neighbor-joining method. The branching confidence was estimated by bootstrapping with 1,000 resamplings in MEGA 4.1 (beta). The GenBank accession numbers of adenovirus prototypes used for alignment and phylogenetic analysis were as follows: species A, human adenovirus type 12 (AB330093), type 18 (DQ149610), and type 31 (AB330112); species B1, human adenovirus type 3 (EF494650) and type 7 (AC000018); species B2, adenovirus type 35 (AC000019); species C, adenovirus type 1 (AC000017) and type 2 (EU867481); species D, adenovirus type 8 (AB361058), type 19 (AB330133), and type 51 (AB330132); species E, adenovirus type 4 (AB330085); and species F, adenovirus type 40 (AB330121) and type 41 (EF429128).
Virus detection limits and recovery efficiency.
The sensitivity of the virus adsorption-elution method of Haramoto et al. (25) was evaluated in seeded studies. In the first experiment, the efficiency of the method in recovering naturally occurring adenovirus in sewage was tested. Two different volumes (700 ml and 3.8 liters) of MilliQ water were inoculated with raw sewage (collected from the East Lansing wastewater treatment plant) at sewage-to-MilliQ water ratios of 1:100,000, 1:10,000, 1:1,000, 1:100, and 1:10. Inoculated water was mixed at room temperature for at least an hour before being processed. Seeded samples were concentrated, extracted, and detected by nested PCR and real-time PCR, following previously described protocols. For each concentration, three to five replicates were tested. For the sewage-seeded experiment, viruses were eluted directly from the membrane with 1 mM NaOH and neutralized with H2SO4 and Tris-EDTA buffers.
In the second seeded experiment, known concentrations (7 × 104 to 7 × 105 PCR detection units) of adenovirus type 40 were inoculated into MilliQ and surface water. Samples were filtered through a cation-coated HA membrane, and viruses were eluted from the membrane in NaOH as described previously. The adenovirus concentration recovered was determined by quantitative PCR assay.
Statistical analysis.
Analysis of variance (ANOVA) was performed to determine differences in mean levels of virus in samples and in virus retention and recovery from different environmental matrices. In all cases, significance was determined at the 95% confidence level. Additional tests, such as Tukey's honestly significant difference test, were performed if a significant difference was obtained by ANOVA. Statistical analysis was performed using SAS software (SAS, Cary, NC).
RESULTS
Wastewater.
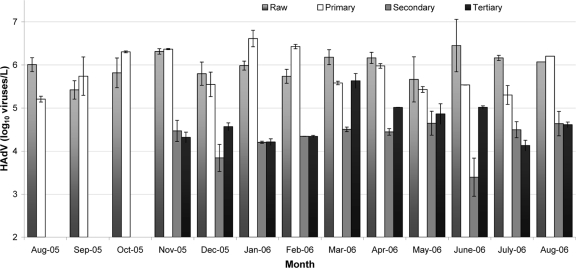
Between August 2005 and August 2006, 46 wastewater samples (13 raw sewage, 13 primary treatment, 10 secondary treatment, and 10 tertiary treatment samples) were collected. Figure 2 and Table 2 show average adenovirus concentrations and log10 removals of human adenovirus at the WWTP. Adenoviruses were consistently isolated in significantly higher concentrations from raw sewage and primary effluent than from secondary and tertiary effluents (P < 0.001). The concentrations of HAdVs ranged between 2.63 × 105 and 2.82 ×106 viruses/liter (mean, 1.15 ×106 viruses/liter) in raw sewage, between 5.37 × 105 and 4.1 × 106 viruses/liter (mean, 1.12 × 106 viruses/liter) in primary effluent, between 1.05 × 103 and 4.42 × 104 viruses/liter (mean, 2.0 × 104 viruses/liter) in secondary effluent, and between 1.35 × 104 and 4.28 × 105 viruses/liter (mean, 8.3 × 104 viruses/liter) in tertiary effluent (Fig. 2). A seasonal trend was not observed for the concentration of adenovirus DNA in wastewater samples. The mean adenovirus removals from raw sewage to secondary and tertiary effluents were 1.77 log10 and 1.14 log10, respectively. Tukey's significant difference test showed that virus concentrations were not significantly different between raw sewage and primary effluent (P = 0.74) and between secondary effluent and tertiary effluent (P = 0.11).
FIG. 2.
Concentrations of HAdV (log10 viruses/liter) in wastewater collected from an advanced WWTP in East Lansing, MI, between August 2005 and August 2006.
TABLE 2.
Number of adenoviruses detected in each treatment step and average log10 removal compared to virus concentration in raw sewage
| Sample type | No. of samples | Mean (range) no. of adenoviruses detected (103 viruses/liter)a | Log removal |
|---|---|---|---|
| Raw | 13 | 1,152* (263-2817) | |
| Primary | 13 | 1,123* (53.7-4094) | 0.01 |
| Secondary | 10 | 20** (1.05-44.2) | 1.77 |
| Tertiary | 10 | 83** (13.5-428) | 1.14 |
*, virus concentrations were not significantly different between raw sewage and primary effluent (P value = 0.74); **, virus concentrations were not significantly different between secondary effluent and tertiary effluent (P value = 0.11).
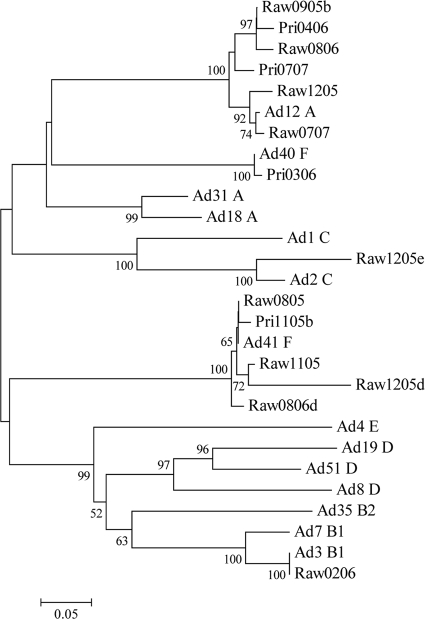
A total of 28 wastewater samples resulted in 35 isolates, and these were amplified by nested PCR for genotyping (Table 3). Samples that produced a high enough concentration of amplified adenoviral DNA were sequenced. Sequences were cropped to approximately 600 bp in length and aligned in web-based ClustalW with reference sequences from GenBank. BLAST analysis of all amplicon sequences identified 92 to 100% sequence identity with the reference prototypes listed below; the number of insertions/deletions between amplicons and the closest prototypes ranged from 0 to 6 (Table 3). Five adenovirus prototypes (adenovirus types 2, 3, 12, 40, and 41) were dominant in raw sewage and primary effluent from wastewater in East Lansing, MI. Adenovirus type 41 (21/35 isolates [60%]) was the most frequently isolated adenovirus, followed by adenovirus type 12 (11/35 isolates [28.5%]), adenovirus type 40 (1/35 isolates [2.9%]), adenovirus type 2 (1/35 isolates [2.9%]), and adenovirus type 3 (1/35 isolates [2.9%]). A neighbor-joining tree was generated from the alignment of the nucleotide sequences of amplicons obtained from this study and hexon gene sequences in GenBank corresponding to adenovirus prototype species A to F (Fig. 3).
TABLE 3.
Comparison of human adenovirus isolates detected in raw sewage and primary effluent of the East Lansing WWTP and their closest prototypes in GenBanka
| Date (mo-yr) | Sample type | Highest scoring prototype |
No. of insertions/ deletions | |
|---|---|---|---|---|
| Serotypeb | % Identity | |||
| Aug-2005 | Raw | 41** | 100 | 0 |
| Aug-2005 | Primary | 41** | 100 | 0 |
| Sep-2005 | Raw | 12* | 93 | 3 |
| 41** | 100 | 0 | ||
| Sep-2005 | Primary | 41** | 100 | 0 |
| Oct-2005 | Raw | 12* | 93 | 3 |
| 41** | 100 | 0 | ||
| Oct-2005 | Primary | 41*** | 99 | 0 |
| Nov-2005 | Raw | 41 | 98 | 0 |
| Nov-2005 | Primary | 12* | 93 | 3 |
| 41 | 99 | 1 | ||
| Dec-2005 | Raw | 12 | 99 | 1 |
| 2 | 93 | 6 | ||
| 41 | 95 | 2 | ||
| Jan-2006 | Raw | 41** | 100 | 0 |
| Jan-2006 | Primary | 41** | 100 | 0 |
| Feb-2006 | Raw | 3 | 100 | 0 |
| Feb-2006 | Primary | 41** | 100 | 0 |
| Mar-2006 | Raw | 41** | 100 | 0 |
| Mar-2006 | Primary | 40 | 99 | 0 |
| Apr-2006 | Raw | 12* | 93 | 3 |
| 41** | 100 | 0 | ||
| Apr-2006 | Primary | 12 | 93 | 3 |
| May-2006 | Raw | 12* | 93 | 3 |
| May-2006 | Primary | 41** | 100 | 0 |
| June-2006 | Raw | 12* | 93 | 3 |
| June-2006 | Primary | 41** | 100 | 0 |
| July-2006 | Raw | 41** | 100 | 0 |
| July-2006 | Primary | 41** | 100 | 0 |
| Aug-2006 | Raw | 12 | 92 | 4 |
| 41*** | 99 | 0 | ||
| Aug-2006 | Primary | 41** | 100 | 0 |
| June-2007 | Raw | 41** | 100 | 0 |
| July-2007 | Raw | 12 | 98 | 0 |
| July-2007 | Primary | 12 | 96 | 0 |
Adenovirus prototypes used in the comparison were as follows: species A, human adenovirus type 12 (AB330093), type 18 (DQ149610), and type 31 (AB330112); species B1, human adenovirus type 3 (EF494650) and type 7 (AC000018); species B2, adenovirus type 35 (AC000019); species C, adenovirus type 1 (AC000017) and type 2 (EU867481); species D, adenovirus type 8 (AB361058), type 19 (AB330133), and type 51 (AB330132); species E, adenovirus type 4 (AB330085); and species F, adenovirus type 40 (AB330121) and type 41 (EF429128).
*, isolates were 100% homologous to each other and were most closely related to prototype HAdV 12; **, isolates were 100% homologous to each other and to prototype HAdV 41; ***, isolates were 100 % homologous to each other and were most closely related to prototype HAdV 41.
FIG. 3.
Neighbor-joining tree of adenovirus hexon amplicons detected in wastewater. The scale indicates the number of nucleotide substitutions per position. Numbers above the branches indicate bootstrap percentages (those above 50%) based on 1,000 replicates. Reference strains of human adenovirus were selected from GenBank, using the accession numbers indicated in the text.
Surface water.
Surface water samples were collected from the lower Grand River during a 1-week intensive study in June 2005 and periodically over a 2-year period between 2005 and 2007. During the intensive study, a total of 16 surface water samples were collected from different sites at the lower Grand River, and human adenovirus was detected in four samples (16%) by conventional PCR (Table 4). Sampling sites that were positive for human adenovirus were Petty's Bayou (i), Deer Creek Park (ii), Kent County (iii), and Sixth Street Park (iv) (as labeled in Fig. 1). Following the intensive study, several sites were chosen for continued monitoring because of high concentrations of bacterial indicators (60) and detection of HAdV DNA at 2 of 16 study sites (Deer Creek Park and Sixth Street Park). Over the 2-year period, 26 samples were collected among the seven sampling sites (North Beach Park, North Shore, Riverside Park, Rosy Mound, Deer Creek Park, Grand River Park, and Sixth Street Park) (Table 4). Adenovirus DNA was detected in four of six samples collected in 2005, through conventional PCR. For the 20 remaining samples, collected in 2006 and 2007, adenovirus concentrations ranged between <10 and 6.6 × 104 viruses/liter (mean, 7.76 × 103 viruses/liter). Six of the 20 samples had an adenovirus DNA concentration above the detection limit.
TABLE 4.
Nested PCR detection of HAdV in surface water samples from the lower Grand River, MI, between 2 November 2005 and 9 August 2007
| Site | Real-time PCR result (no. of viruses/liter) |
|||||
|---|---|---|---|---|---|---|
| 10 January 2006 | 21 March 2006 | 12 June 2006 | 11 July 2006 | 26 July 2007 | 9 August 2007 | |
| Deer Creek Park | <4.0 × 101 | <5.0 × 101 | <1.0 × 101 | <4.0 × 101 | 1.9 × 103 | |
| Grand River Park | 2.7 × 104 | 9.0 × 101 | ||||
| North Beach Park | 8.0 × 101 | <3.0 × 101 | <2.0 × 101 | <2.0 × 101 | ||
| North Shore | 6.0 × 104 | <3.0 × 101 | ||||
| Riverside Park | <3.0 × 101 | <4.0 × 101 | <3.0 × 101 | |||
| Sixth Street Park | <2.0 × 101 | <4.0 × 101 | 6.6 × 104 | <2.0 × 101 | ||
CSO samples.
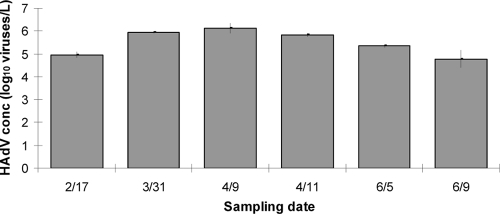
Six CSO samples were collected from a wastewater retention basin in Grand Rapids, MI, between February and June 2008. Adenovirus DNA was detected in 100% of CSO samples, with concentrations ranging from 6.02 ×104 viruses/liter to 1.32 ×106 viruses/liter (average, 5.35 ×105 viruses/liter; standard deviation, 500.81 viruses/liter) (Fig. 4). The concentrations of adenovirus DNA in the CSO discharge were not significantly different from adenovirus DNA concentrations in raw sewage and primary treatment samples (P = 0.39).
FIG. 4.
Human adenovirus concentrations (log10 viruses/liter) in CSO discharge samples collected from Market Avenue retention basin in Grand Rapids, MI.
Virus detection limits and recovery efficiency.
The virus recovery efficiency of the adsorption-elution method of Haramoto et al. (25) was evaluated. Raw sewage was seeded into MilliQ water at ratios of 1:10 to 1:100,000, and conventional nested PCR was able to detect adenovirus DNA at a 1:10,000 dilution for the larger volume tested (3.8 liters) but only up to a 1:1,000 dilution for the smaller volume tested (700 ml) (Table 5). For the 1:10,000 dilution, real-time PCR gave adenovirus concentrations between 4 × 101 and 8 × 101 viruses/liter (mean, 5 × 101 viruses/liter; n = 5), which converted to between 4 × 105 and 8 × 105 viruses/liter in the original sewage samples.
TABLE 5.
Detection of human adenovirus in sewage-inoculated MilliQ water by conventional nested PCR
| Sewage/MilliQ water ratio | Vol of water filtered (ml) | Total no. of samples | No. of positive samples |
|---|---|---|---|
| 1:100,000 | 3,800 | 5 | 0 |
| 1:10,000 | 3,500 | 10 | 4 |
| 1:1,000 | 700 | 5 | 2 |
| 1:1,000 | 3,500 | 6 | 6 |
| 1:100 | 700 | 5 | 4 |
| 1:10 | 700 | 3 | 3 |
In a seeded experiment, known concentrations of adenovirus type 40 were inoculated into MilliQ water and river water and filtered through a cation-coated HA membrane as described previously. Adenovirus was eluted from the membrane in 20 ml of elution buffer (NaOH) as described for the second seeded experiment. Quantitative PCR gave adenovirus recovery rates of 0.32 to 6.98% (Table 6).
TABLE 6.
Recovery of adenovirus DNA from virus-inoculated MilliQ water and river water with a modified cation-coated virus filter methoda
| Sample type | Vol of water filtered (ml) | No. of samples | No. of viruses seeded | No. of viruses recovered | Mean recovery rate (%) | Recovery range (%) |
|---|---|---|---|---|---|---|
| MilliQ water | 1,300 | 8 | 7.63E+04 | 9.97E+02 | 1.34 | 0.17-3.43 |
| 3,900 | 3 | 2.19E+05 | 6.19E+03 | 2.83 | 1.35-4.32 | |
| 1,300 | 3 | 7.29E+05 | 2.41E+04 | 3.31 | 0.19-6.98 | |
| River water | 1,300 | 3 | 7.16E+04 | 7.32E+02 | 1.03 | 0.49-1.38 |
| 1,300 | 3 | 7.24E+05 | 6.65E+03 | 0.92 | 0.32-1.47 |
Method of Haramoto et al. (25).
DISCUSSION
The concentrations of HAdVs in sewage (mean, 1.15 ×106 viruses/liter) and effluent (mean, 8.3 × 104 viruses/liter) samples in this study were similar to adenovirus concentrations found in wastewater in other studies (4, 24, 38). The slightly higher concentration of adenovirus in tertiary effluent than in secondary effluent may be an artifact of real-time PCR caused by the presence of a higher level of inhibitory substances in samples post-secondary treatment. This observation agrees with the findings of He and Jiang (29), in which higher concentrations of adenovirus DNA were detected in secondary chlorinated effluent than in primary effluent. Likewise, using synthetic short RNA fragments as internal controls, da Silva et al. (12) observed that poor real-time reverse transcription (RT-PCR) efficiency occurred more frequently in waste stabilization pond samples than in activated sludge and membrane bioreactor samples.
In addition, the adenovirus concentration in wastewater in Michigan remained high year-round, without a seasonal trend, maybe because the water temperature in wastewater in Michigan was within the range of optimal virus survival throughout the year (<23°C) (43). The mean HAdV removal levels in secondary effluent (1.77 log10) and tertiary effluent (1.14 log10) were lower than the mean HAdV (2.35 log10) and norovirus G1 (2.27 log10) removal levels in WWTPs in Japan (23, 24). In a WWTP in France, an astrovirus removal level of 2.00 log10 was determined by real-time PCR (41). The data suggest that HAdV removal in this WWTP in Michigan is not efficient, and considering that HAdVs comprise one of the most abundant enteric virus groups in raw sewage, the fate of HAdVs in wastewater treatment processes needs further evaluation, with the goals to improve virus removal and to document that the disinfection process can be used to inactivate the viruses.
The presence of adenovirus genotypes in wastewater may represent a public health risk. It has been shown that there is a connection between environmental and clinical virus isolates in a given year within specific geographic areas (57). Sedmak et al. (57) compared clinical and sewage isolates of enteroviruses from Milwaukee, WI, collected between 1994 and 2002, and found that the predominant clinical serotype was most often the predominant sewage serotype for that year. Typing of isolates from sewage and environmental water may help to alert community health professionals to predominant and new serotypes that are in circulation in the population, providing new insights into vaccine development and other prevention strategies.
In this study, adenovirus type 41 was identified as the predominant serotype in sewage, followed by adenovirus types 12, 40, 2, and 3. This trend agrees with results from other environmental studies (56, 66). Santos et al. (56) isolated adenoviruses 40 and 41 from 62 of 69 sewage and surface water samples collected in São Paulo, Brazil, over a 3-year period. In South Africa, adenovirus types 2, 40, and 41 and species D HAdVs were isolated from treated drinking water and river water (66). However, even though enteric adenoviruses (serotypes 40 and 41) were most frequently identified in river water, human adenovirus species D isolates were predominant in treated drinking water (66). In addition, Gray et al. (18) reported that the most prevalent serotypes among civilians in the United States (n = 1,608) were types 3 (34.6%), 2 (24.3%), 1 (17.7%), and 5 (5.3%). In the same study, among specimens collected from the gastrointestinal tract and urine, only 26 of 234 (11.1%) and 0 of 44 (0%) isolates were adenovirus species F (i.e., adenovirus type 41). This suggests that adenovirus type 41 may have greater persistence in natural environments than other adenovirus serotypes. Adenovirus species F (types 40 and 41) has been identified as one of the most prevalent viruses globally in the etiology of childhood gastroenteritis (20, 51, 58, 62). Several researchers claimed that adenovirus type 41, the most prevalent serotype isolated in the current study, had gradually replaced serotype 40 as the predominant serotype isolated from gastroenteritis patients globally, starting in the 1980s, and was currently identified as the most prevalent serotype in wastewater (20, 44, 58, 59, 70). These findings were consistent with the fact that in the current study adenovirus type 40 was detected in only 1 of 35 isolates. Adenovirus type 12, the second most prevalent adenovirus isolated in this study (11/35 isolates), has often been associated with meningoencephalitis, and no waterborne outbreak related to it has been identified to date. However, it was recently identified as the etiologic agent of a diarrhea outbreak in a hematology hospital ward in London (32).
Adenovirus types 2 and 3 were both isolated from one wastewater sample. These types are generally associated with pneumonia and childhood respiratory diseases (31). Adenovirus type 3 was also identified as the etiologic agent of pharyngoconjunctival fever and conjunctivitis in several outbreaks associated with contaminated recreational water (16, 47, 48). Gray et al. (18) identified adenovirus type 3 and type 2 as the two most prevalent adenovirus types in civilian specimens (15.4% and 27.8%, respectively). Adenovirus type 3 is also the second most prevalent type (2.6%) in specimens collected from military trainees (18).
Adenovirus levels in CSO samples were not significantly different from those in raw sewage and primary effluents; adenovirus was detected at levels between 6.02 ×104 viruses/liter and 1.32 ×106 viruses/liter (mean, 5.35 ×105 viruses/liter; n = 6) in CSO samples collected from a retention basin in Grand Rapids, MI. This suggests that in cases where the virus concentration in CSO samples is unknown, virus concentrations in raw sewage or primary effluent samples may be used as indicators for estimating total virus discharge during a CSO event. In addition, high adenovirus concentrations in CSOs indicate that recreational waters that receive CSO input are unfit for bodily contact via recreational activities such as swimming.
Virus detection in surface water generally requires filtration and concentration of large volumes of water, which at the same time also concentrates high levels of PCR inhibitors. Katayama et al. (39) and Haramoto et al. (25) developed a virus concentration method that consists of adding salts for adsorption of viruses to a negatively charged filter membrane and an acid rinse step, as well as inorganic eluent, to remove PCR inhibitors. This virus filtration and concentration method was modified and used in several recent studies, with various virus recovery efficiencies, ranging from <1% to >100% (13, 21, 22, 26, 52, 67). In the adenovirus seeding experiment, virus recovery quantified by real-time PCR was low; mean recoveries from the MilliQ water and river water were 2.08% (range, 0.17 to 6.98%) and 0.98% (range, 0.32 and 1.47%), respectively. The low recovery rates in this study may be due to a loss of viruses during the elution step or to incomplete elution of viruses from the membrane. Lukasik et al. (46) suggested that AlCl3 forms flocs that trap viruses and thus inhibit virus elution from the membrane. In addition, Hamza et al. (22) tested the recovery of various viruses (i.e., echovirus 11, norovirus G1, HAdV 5, JC polyomavirus, and bacteriophage ΦX174) from negatively charged HA membrane filters and observed that virus retention and recovery were dependent on virus genetic structure and water quality factors, such as turbidity and dissolved and colloidal organic matter. Hamza et al. (22) also found that viral nucleic acid detection was 2.5- to 6-fold less efficient when beef extract was used as an elution buffer.
In conclusion, this research demonstrates that human adenoviruses are consistently present in sewage and were occasionally detected in rivers receiving sewage effluents in Michigan. The real-time PCR assay used in the current study was not able to evaluate the viability of viruses detected after disinfection; however, the virus concentration in tertiary effluent showed little physical removal. In general, there is not a consistent seasonal trend for the presence of adenoviruses in wastewater and surface water in Michigan, but their isolation from surface water can be used to indicate fecal contamination and a lack of efficient treatment. Overall, wastewater treatment in East Lansing reduces the level of adenoviruses by 95% from untreated to treated sewage. The low adenovirus recovery rate with the cation-coated negatively charged membrane method used in this study warrants future study for improved recovery of viruses. Despite this low recovery rate, the presence of adenoviruses in surface waters may represent a public health risk if disinfection is not shown to be adequate. Quantitative detection of adenoviruses as indicators in surface water may provide a means of better understanding the risk associated with recreation in contaminated water.
Acknowledgments
This study was partly funded by a grant from the National Oceanic and Atmospheric Administration (NA04OAR4600199).
We give special thanks to Rebecca Ives, Marc Verhougstraete, Arun Nayak, Sangeetha Srinivasan, and Shikha Singh for help with sample collection.
Footnotes
Published ahead of print on 30 November 2009.
REFERENCES
- 1.Akihara, S., T. G. Phan, T. A. Nguyen, G. Hansman, S. Okitsu, and H. Ushijima. 2005. Existence of multiple outbreaks of viral gastroenteritis among infants in a day care center in Japan. Arch. Virol. 150:2061-2075. [DOI] [PubMed] [Google Scholar]
- 2.Allard, A., B. Albinsson, and G. Wadell. 1992. Detection of adenoviruses in stools from healthy persons and patients with diarrhea by two-step polymerase chain reaction. J. Med. Virol. 37:149-157. [DOI] [PubMed] [Google Scholar]
- 3.Basu, G., J. Rossouw, T. K. Sebunya, B. A. Gashe, M. De Beer, J. B. Dewar, and A. D. Steele. 2003. Prevalence of rotavirus, adenovirus and astrovirus infection in young children with gastroenteritis in Gaborone, Botswana. East Afr. Med. J. 80:652-655. [DOI] [PubMed] [Google Scholar]
- 4.Bofill-Mas, S., N. Albinana-Gimenez, P. Clemente-Casares, A. Hundesa, J. Rodriguez-Manzano, A. Allard, M. Calvo, and R. Girones. 2006. Quantification and stability of human adenoviruses and polyomavirus JCPyV in wastewater matrices. Appl. Environ. Microbiol. 72:7894-7896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brandt, C., H. Kim, B. J. A. Vargosko, J. Arrobio, B. Rindge, R. Parrott, and R. Chanock. 1969. Infections in 18000 infants and children in a controlled study of respiratory tract disease. I. Adenovirus pathogenicity in relation to serologic type and illness syndrome. Am. J. Epidemiol. 90:484-500. [DOI] [PubMed] [Google Scholar]
- 6.Chaberny, I. F., P. Schnitzler, H. K. Geiss, and C. Wendt. 2003. An outbreak of epidemic keratoconjunctivitis in a pediatric unit due to adenovirus type 8. Infect. Control Hosp. Epidemiol. 24:514-519. [DOI] [PubMed] [Google Scholar]
- 7.Chmielewicz, B., J. Benzler, G. Pauli, G. Krause, F. Bergmann, and B. Schweiger. 2005. Respiratory disease caused by a species B2 adenovirus in a military camp in Turkey. J. Med. Virol. 77:232-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crabtree, K. D., C. P. Gerba, J. B. Rose, and C. N. Haas. 1997. Waterborne adenovirus: a risk assessment. Water Sci. Technol. 35:1-6. [Google Scholar]
- 9.Craun, G. F. 1991. Causes of waterborne outbreaks in the United States. Water Sci. Technol. 24:17-20. [Google Scholar]
- 10.Cruz, J. R., P. Cáceres, F. Cano, J. Flores, A. Bartlett, and B. Torún. 1990. Adenovirus types 40 and 41 and rotaviruses associated with diarrhea in children from Guatemala. J. Clin. Microbiol. 28:1780-1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.D'Ambrosio, E., N. Del Grosso, A. Chicca, and M. Midulla. 1982. Neutralizing antibodies against 33 human adenoviruses in normal children in Rome. J. Hyg. (London) 89:155-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.da Silva, A. K., J.-C. Le Saux, S. Parnaudeau, M. Pommepuy, M. Elimelech, and F. S. Le Guyader. 2007. Evaluation of removal of noroviruses during wastewater treatment, using real-time reverse transcription-PCR: different behaviors of genogroups I and II. Appl. Environ. Microbiol. 73:7891-7897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Paula, V. S., L. Diniz-Mendes, L. M. Villar, S. L. B. Luz, L. A. Silva, M. S. Jesus, N. M. V. S. da Silva, and A. M. C. Gaspar. 2007. Hepatitis A virus in environmental water samples from the Amazon Basin. Water Res. 41:1169-1176. [DOI] [PubMed] [Google Scholar]
- 14.Fong, T.-T., D. W. Griffin, and E. K. Lipp. 2005. Molecular assays for targeting human and bovine enteric viruses in coastal waters and their application for library-independent source tracking. Appl. Environ. Microbiol. 71:2070-2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fong, T., L. S. Mansfield, D. L. Wilson, D. J. Schwab, S. L. Molloy, and J. B. Rose. 2007. Massive microbiological groundwater contamination associated with a waterborne outbreak in Lake Erie, South Bass Island, OH. Environ. Health Perspect. 115:856-864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Foy, H. M., M. K. Cooney, and J. B. Hatlen. 1968. Adenovirus type 3 epidemic associated with intermittent chlorination of a swimming pool. Arch. Environ. Health 17:795-802. [DOI] [PubMed] [Google Scholar]
- 17.Foy, H. M., M. K. Cooney, R. McMahan, and J. T. Grayston. 1973. Viral and mycoplasmal pneumonia in a prepaid medical care group during an eight-year period. Am. J. Epidemiol. 97:93-102. [DOI] [PubMed] [Google Scholar]
- 18.Gray, G. C., T. McCarthy, M. G. Lebeck, D. P. Schnurr, K. L. Russell, A. E. Kajon, M. L. Landry, D. S. Leland, G. A. Storch, C. C. Ginocchio, C. C. Robinson, G. J. Demmler, M. A. Saubolle, S. C. Kehl, R. Selvarangan, M. B. Miller, J. D. Chappell, D. M. Zerr, D. L. Kiska, D. C. Halstead, A. W. Capuano, S. F. Setterquist, M. L. Chorazy, J. D. Dawson, and D. D. Erdman. 2007. Genotype prevalence and risk factors for severe clinical adenovirus infection, United States 2004-2006. Clin. Infect. Dis. 45:1120-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Griffin, D. W., E. K. Lipp, M. R. McLaughlin, and J. B. Rose. 2001. Marine recreation and public health microbiology: quest for the ideal indicator. Bioscience 51:817-825. [Google Scholar]
- 20.Grimwood, K., R. Carzino, G. L. Narnes, and R. F. Bishop. 1995. Patients with enteric adenovirus gastroenteritis admitted to an Australian pediatric teaching hospital from 1981 to 1992. J. Clin. Microbiol. 33:131-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guimarães, F. R., F. F. M. Ferreira, C. B. Vieira, T. M. Fumian, T. Shubo, J. P. G. Leite, and M. P. Miagostovich. 2008. Molecular detection of human astrovirus in an urban sewage treatment plant in Rio de Janeiro, Brazil. Mem. Inst. Oswaldo Cruz 103:819-823. [DOI] [PubMed] [Google Scholar]
- 22.Hamza, I. A., L. Jurzik, A. Stang, K. Sure, K. Überla, and M. Wilhelm. 2009. Detection of human viruses in rivers of a densely-populated area in Germany using a virus adsorption elution method optimized for PCR analyses. Water Res. 43:2657-2668. [DOI] [PubMed] [Google Scholar]
- 23.Haramoto, E., E. Katayama, K. Oguma, H. Yamashita, A. Tajima, H. Nakajima, and S. Ohgaki. 2006. Seasonal profiles of human noroviruses and indicator bacteria in wastewater treatment plant in Tokyo, Japan. Water Sci. Technol. 54:301-308. [DOI] [PubMed] [Google Scholar]
- 24.Haramoto, E., H. Katayama, K. Oguma, and S. Ohgaki. 2007. Quantitative analysis of human enteric adenoviruses in aquatic environments. J. Appl. Microbiol. 103:2153-2159. [DOI] [PubMed] [Google Scholar]
- 25.Haramoto, E., H. Katayama, K. Oguma, and S. Ohgaki. 2005. Application of cation-coated filter method to detection of noroviruses, enteroviruses, adenoviruses, and torque teno viruses in the Tamagawa River in Japan. Appl. Environ. Microbiol. 71:2403-2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haramoto, E., M. Kitajima, H. Katayama, T. Ito, and S. Ohgaki. 2009. Development of virus concentration methods for detection of koi herpesvirus in water. J. Fish Dis. 32:297-300. [DOI] [PubMed] [Google Scholar]
- 27.Harley, D., B. Harrower, M. Lyon, and A. Dick. 2001. A primary school outbreak of pharyngoconjunctival fever caused by adenovirus type 3. Commun. Dis. Intell. 25:9-12. [PubMed] [Google Scholar]
- 28.Hatherill, M., M. Levin, J. Lawrenson, N. Y. Hsiao, L. Reynolds, and A. Argent. 2004. Evolution of an adenovirus outbreak in a multidisciplinary children's hospital. J. Paediatr. Child. Health 40:449-454. [DOI] [PubMed] [Google Scholar]
- 29.He, J.-W., and S. Jiang. 2005. Quantification of enterococci and human adenoviruses in environmental samples by real-time PCR. Appl. Environ. Microbiol. 71:2250-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hilleman, M. R., and J. H. Werner. 1954. Recovery of new agents from patients with acute respiratory illness. Proc. Soc. Exp. Biol. Med. 85:183-188. [DOI] [PubMed] [Google Scholar]
- 31.Horwitz, M. S. 2001. Adenoviruses, p. 2301-2326. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 32.Jalal, H., D. F. Bibby, J. W. Tang, J. Bennett, C. Kyriakou, K. Peggs, D. Cubitt, N. S. Brink, K. N. Ward, and R. S. Tedder. 2005. First reported outbreak of diarrhea due to adenovirus infection in a hematology unit for adults. J. Clin. Microbiol. 43:2575-2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiang, S., H. Dezfulian, and W. Chu. 2005. Real-time quantitative PCR for enteric adenovirus serotype 40 in environmental waters. Can. J. Microbiol. 51:393-398. [DOI] [PubMed] [Google Scholar]
- 34.Jiang, S., R. Noble, and W. P. Chu. 2001. Human adenoviruses and coliphages in urban runoff-impacted coastal waters of Southern California. Appl. Environ. Microbiol. 67:179-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jothikumar, N., T. L. Cromeans, V. R. Hill, X. Lu, M. D. Sobsey, and D. D. Erdman. 2005. Quantitative real-time PCR assays for detection of human adenoviruses and identification of serotypes 40 and 41. Appl. Environ. Microbiol. 71:3131-3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kajon, A., J. Moseley, D. Metzgar, H. S. Huong, A. Wadleigh, M. K. Ryan, and K. Russell. 2007. Molecular epidemiology of adenovirus type 4 infections in US military recruits in the postvaccination era (1997-2003). J. Infect. Dis. 196:67-75. [DOI] [PubMed] [Google Scholar]
- 37.Kapikian, A. Z., and R. G. Wyatt. 1992. Viral gastrointestinal infections, p. 667-676. In Textbook of pediatric infectious diseases, 3rd ed., vol. 1. W. B. Saunders Co., Philadelphia, PA. [Google Scholar]
- 38.Katayama, H., E. Haramoto, K. Oguma, H. Yamashita, A. Tajima, H. Nakajima, and S. Ohgaki. 2008. One-year monthly quantitative survey of noroviruses, enteroviruses, and adenoviruses in wastewater collected from six plants in Japan. Water Res. 42:1441-1448. [DOI] [PubMed] [Google Scholar]
- 39.Katayama, H., A. Shimasaki, and S. Ohgaki. 2002. Development of a virus concentration method and its application to detection of enterovirus and Norwalk virus from coastal seawater. Appl. Environ. Microbiol. 68:1033-1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kolavic-Gray, S., L. Binn, J. Sanchez, S. Cersovsky, C. Polyak, F. Mitchell-Raymundo, L. Asher, D. Vaughn, B. Feighner, and B. Innis. 2002. Large epidemic of adenovirus type 4 infection among military trainees: epidemiological, clinical, and laboratory studies. Clin. Infect. Dis. 35:808-818. [DOI] [PubMed] [Google Scholar]
- 41.Le Cann, P., S. Ranarijaona, S. Monpoeho, F. Le Guyader, and V. Ferre. 2004. Quantification of human astroviruses in sewage using real-time RT-PCR. Res. Microbiol. 155:11-15. [DOI] [PubMed] [Google Scholar]
- 42.Lee, S. H., and S. J. Kim. 2002. Detection of infectious enteroviruses and adenoviruses in tap water in urban areas in Korea. Water Res. 36:248-256. [DOI] [PubMed] [Google Scholar]
- 43.Lipp, E. K., R. Kurz, R. Vincent, C. Rodriguez-Palacios, S. R. Farrah, and J. B. Rose. 2001. The effects of seasonal variability and weather on microbial fecal pollution and enteric pathogens in a subtropical estuary. Estuaries 24:266-276. [Google Scholar]
- 44.Logan, C., J. J. O'Leary, and N. O'Sullivan. 2006. Real-time reverse transcription-PCR for detection of rotavirus and adenovirus as causative agents of acute viral gastroenteritis in children. J. Clin. Microbiol. 44:3189-3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu, X., and D. D. Erdman. 2006. Molecular typing of human adenoviruses by PCR and sequencing of a partial region of the hexon gene. Arch. Virol. 151:1587-1602. [DOI] [PubMed] [Google Scholar]
- 46.Lukasik, J., T. M. Scott, D. Andryshak, and S. R. Farrah. 2000. Influence of salts on virus adsorption to microporous filters. Appl. Environ. Microbiol. 66:2914-2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martone, W. J., J. C. Hierholzer, R. A. Keenlyside, D. W. Fraser, L. J. D' Angelo, and W. G. Winkler. 1980. An outbreak of adenovirus type 3 disease at a private recreation center swimming pool. Am. J. Epidemiol. 111:229-237. [DOI] [PubMed] [Google Scholar]
- 48.McMillan, N. S., S. A. Martin, M. D. Sobsey, D. A. Wait, R. A. Meriwether, and J. N. MacCormack. 1992. Outbreak of pharyngoconjunctival fever at a summer camp: North Carolina. MMWR Morb. Mortal. Wkly. Rep. 41:342-347. [PubMed] [Google Scholar]
- 49.McNeil, K. M., R. M. Hendrix, J. L. Lindner, R. R. Benton, S. C. Monteith, M. A. Tuchscherer, G. C. Gray, and J. C. Gaydos. 1999. Large, persistent epidemic of adenovirus type 4-associated acute respiratory disease in U.S. army trainees. Emerg. Infect. Dis. 5:798-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.MDEQ. 2008. Combined sewer overflow (CSO) & sanitary sewer overflow (SSO) 2007 annual report (January 1, 2007-December 31, 2007). Michigan Department of Environmental Quality, Lansing, MI. http://www.michigan.gov/documents/deq/deq-wb-csossoreport07_243725_7.pdf.
- 51.Meqdam, M. M., and I. R. Thwiny. 2007. Prevalence of group A rotavirus, enteric adenovirus, norovirus and astrovirus infections among children with acute gastroenteritis in Al-Qassim, Saudi Arabia. Pak. J. Med. Sci. 23:551-555. [Google Scholar]
- 52.Miagostovich, M. P., F. F. M. Ferreira, F. R. Guimaraes, T. M. Fumian, L. Diniz-Mendes, S. L. B. Luz, L. A. Silva, and J. P. G. Leite. 2008. Molecular detection and characterization of gastroenteritis viruses occurring naturally in the stream waters of Manaus, Central Amazonia, Brazil. Appl. Environ. Microbiol. 74:375-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Papapetropoulou, M., and A. C. Vantarakis. 1998. Detection of adenovirus outbreak at a municipal swimming pool by nested PCR amplification. J. Infect. 36:101-103. [DOI] [PubMed] [Google Scholar]
- 54.Pina, S., M. Puig, F. Lucena, J. Jofre, and R. Girones. 1998. Viral pollution in the environment and in shellfish: human adenovirus detection by PCR as an index of human viruses. Appl. Environ. Microbiol. 64:3376-3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rowe, W. P., R. J. Huebner, L. K. Gilmore, R. H. Parrot, and T. G. Ward. 1953. Isolation of a cytopathic agent from human adenoids undergoing spontaneous degradation in tissue culture. Proc. Soc. Exp. Biol. Med. 84:570-573. [DOI] [PubMed] [Google Scholar]
- 56.Santos, F. M., M. J. Vieira, P. Garrafa, T. A. Monezi, V. H. Pellizari, C. M. Harsi, and D. U. Mehnert. 2004. Discrimination of adenovirus types circulating in urban sewage and surface polluted waters in Sao Paulo city, Brazil. Water Sci. Technol. 4:79-85. [Google Scholar]
- 57.Sedmak, G., D. Bina, and J. MacDonald. 2003. Assessment of an enterovirus sewage surveillance system by comparison of clinical isolates with sewage isolates from Milwaukee, Wisconsin, collected August 1994 to December 2002. Appl. Environ. Microbiol. 69:7181-7187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shimizua, H., T. G. Phana, S. Nishimuraa, S. Okitsua, N. Maneekarnb, and H. Ushijima. 2007. An outbreak of adenovirus serotype 41 infection in infants and children with acute gastroenteritis in Maizuru City, Japan. Infect. Genet. Evol. 7:279-284. [DOI] [PubMed] [Google Scholar]
- 59.Shinozaki, T., K. Araki, Y. Fujita, M. Kobayashi, T. Tajima, and T. Abe. 1991. Epidemiology of enteric adenoviruses 40 and 41 in acute gastroenteritis in infants and young children in the Tokyo area. Scand. J. Infect. Dis. 23:543-547. [DOI] [PubMed] [Google Scholar]
- 60.Singh, S. 2007. Investigation of bacterial fecal indicators and coliphage virus in sediment and surface water of parks and beaches along the Grand River (MI) and Lake Michigan (MI). Master's thesis. Michigan State University, East Lansing, MI.
- 61.Tani, N., Y. Dohi, N. Jurumatani, and K. Yonemasu. 1995. Seasonal distribution of adenoviruses, enteroviruses and reoviruses in urban river water. Microbiol. Immunol. 39:577-580. [DOI] [PubMed] [Google Scholar]
- 62.Topkaya, A. E., B. Aksungar, F. Özakkafl, and N. Çapan. 2006. Examination of rotavirus and enteric adenovirus in children with acute gastroenteritis. Türk Mikrobiyol. Cem. Derg. 36:210-213. [Google Scholar]
- 63.Reference deleted.
- 64.Turner, M., G. R. Istre, H. Beauchamp, M. Baum, and S. Arnold. 1987. Community outbreak of adenovirus type 7a infections associated with a swimming pool. South. Med. J. 80:712-715. [DOI] [PubMed] [Google Scholar]
- 65.Uhnoo, I., G. Wadell, L. Svensson, and M. E. Johansson. 1984. Importance of enteric adenoviruses 40 and 41 in acute gastroenteritis in infants and young children. J. Clin. Microbiol. 20:365-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.van Heerden, J., M. M. Ehlers, A. Heim, and W. O. K. Grabow. 2005. Prevalence, quantification and typing of adenoviruses detected in river and treated drinking water in South Africa. J. Appl. Microbiol. 99:234-242. [DOI] [PubMed] [Google Scholar]
- 67.Victoria, M., F. Guimarães, T. Fumian, F. Ferreira, C. Vieira, J. P. Leite, and M. Miagostovich. 2009. Evaluation of an adsorption-elution method for detection of astrovirus and norovirus in environmental waters. J. Virol. Methods 156:73-76. [DOI] [PubMed] [Google Scholar]
- 68.Wadell, G. A., A. Allard, M. Johansson, L. Svensson, and I. Uhnoo. 1987. Enteric adenoviruses, p. 73-91. In Enteric adenoviruses. John Wiley & Son, Inc., Chichester, United Kingdom.
- 69.Xagoraraki, I., D. H. W. Kuo, K. Wong, M. Wong, and J. B. Rose. 2007. Occurrence of human adenoviruses in two Great Lakes recreational beaches. Appl. Environ. Microbiol. 73:7874-7881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yamashita, Y., M. Hattori, M. Oseto, M. Mori, H. Inouye, K. Takagi, Y. Ishimaru, and S. Nakano. 1995. Epidemiological studies on enteric adenovirus gastroenteritis in children. Kansenshogaku Zasshi 69:377-382. [DOI] [PubMed] [Google Scholar]