Abstract
NSC 676914 has been identified as a selective NF-κB inhibitor that does not inhibit cell proliferation. This compound was originally identified in a high-throughput cell based assay for AP-1 inhibitors using synthetic compound libraries and the NCI natural product repository. NSC 676914 shows activity against NF-κB (Nuclear factor kappa B) in luciferase reporter assays at concentrations much less than the IC50 for AP-1. An SRE (Serum response element) reporter used as a specificity control and indicator of cell proliferation was relatively insensitive to the compound. Pretreatment with NSC 676914 is here shown to repress TPA-induced IκB-α phosphorylation and translocation of p65/50 to the nucleus, but not the processing of p52 from p100, suggesting inhibition of NF-κB regulator IKKβ rather than IKKα. Inhibition of NF-κB activation occurred as a consequence of blocking phosphorylation of IKK. Induction of IκB-α phosphorylation by TPA was diminished by pretreatment of NSC 676914 even at 1.1 µM. In contrast, kinases JNK and ERK1&2, important for AP-1 activation, showed no significant repression by this compound. Furthermore, a matrigel invasion assay with breast cancer cell lines and a transformation assay in mouse JB6 cells revealed that TPA-induced invasion and transformation responses were completely repressed by this compound. These results suggest that NSC 676914 could be a novel inhibitor having potential therapeutic activity to target NF-κB for cancer treatment or prevention.
Keywords: AP-1, NF-κB, IKK, Inhibitor, Screening
Introduction
AP-1 activation is important in regulating genes involved in normal cell proliferation, differentiation, development and responses to external stimuli. Environmentally induced AP-1 dependent transcription can contribute not only to normal physiological processes but also to tumor promotion and progression. AP-1 activity is often elevated in human and mouse cancers (via elevated Ras-MAPK signaling) and this elevation is required both to induce and to maintain tumor phenotype (1–4). AP-1 activity therefore appears to be a promising target for cancer prevention. Tumor promoter induced AP-1 activation can be repressed by a dominant-negative c-Jun, TAM67 in a transgenic mouse model, inhibiting tumorigenesis and tumor progression without inhibiting cell proliferation or cell survival (4–8). This specificity of TAM67 has enabled the identification of genes the expression of which are critical for tumorigenesis and tumor progression and might be targeted for cancer prevention. Cyclooxygenase-2 (COX-2), Osteopontin, HMGA1, and Vascular endothelial growth factor (VEGF) have been identified as functionally significant targets of TAM67 (9–11). Targeted inhibition of AP-1 activity without cell growth inhibition has identified TAM67 as an efficient molecular tool to prevent cancer development and progression (12). The genes whose expression is inhibited by TAM67 are those associated with inflammation, invasion and metastasis rather than those associated with cell proliferation or cell survival (9). Exploiting TAM67 for therapeutic intervention however calls for using it as a gene therapy reagent, something that awaits further development of delivery technology.
As an alternative approach to targeting AP-1, we sought small molecules that interfere with the activation of AP-1 and that might recapitulate the specificity of TAM67. We established a cell based high throughput screen (HTS) to identify specific inhibitors of AP-1 using HEK293-T cells (Invitrogen) stably expressing a 3× AP-1 promoter driving a beta lactamase reporter (13). Synthetic compound libraries and the NCI natural product repository were screened to identify AP-1 Hits. The optimized HTS was validated and coupled with an XTT assay in the same wells to exclude inhibitors of cell proliferation. Several compounds passed the HTS for AP-1 inhibitors; an inhibitor was defined as a compound inhibiting TPA-induced AP-1 activity by more than 50% and reducing cell viability (XTT assay) by less than 40% (13).
Because TAM67 targets transcription factor Nuclear factor kappa B (NF-κB) in addition to AP-1 (2, 12, 14, 15), and because both AP-1 and NF-κB can be drivers of carcinogenesis (4, 16–18), we measured NF-κB activation as a secondary assay. NF-κB-regulated genes are important for cell differentiation, embryonic development, immune response and inflammation, the latter contributing to carcinogenesis in several cancer sites (19–21). Although NF-κB and AP-1 are regulated by different signaling pathways, cross-talk between these pathways occurs, mediated in part by the ability of certain of the Jun and Fos family proteins to interact with NF-κB p65 (22, 23). Therefore, the HTS for AP-1 inhibitors might also identify NF-κB inhibitors.
NSC 676914 is one of the active compounds from a synthetic library, determined to have an IC50 value of 17 µM for inhibition of TPA induced AP-1. Characterization of NSC 676914 revealed that at lower concentrations it selectively inhibits TPA induced NF-κB activation. The mechanism involves inhibition of protein kinase IKKβ.
Materials and Methods
Compound
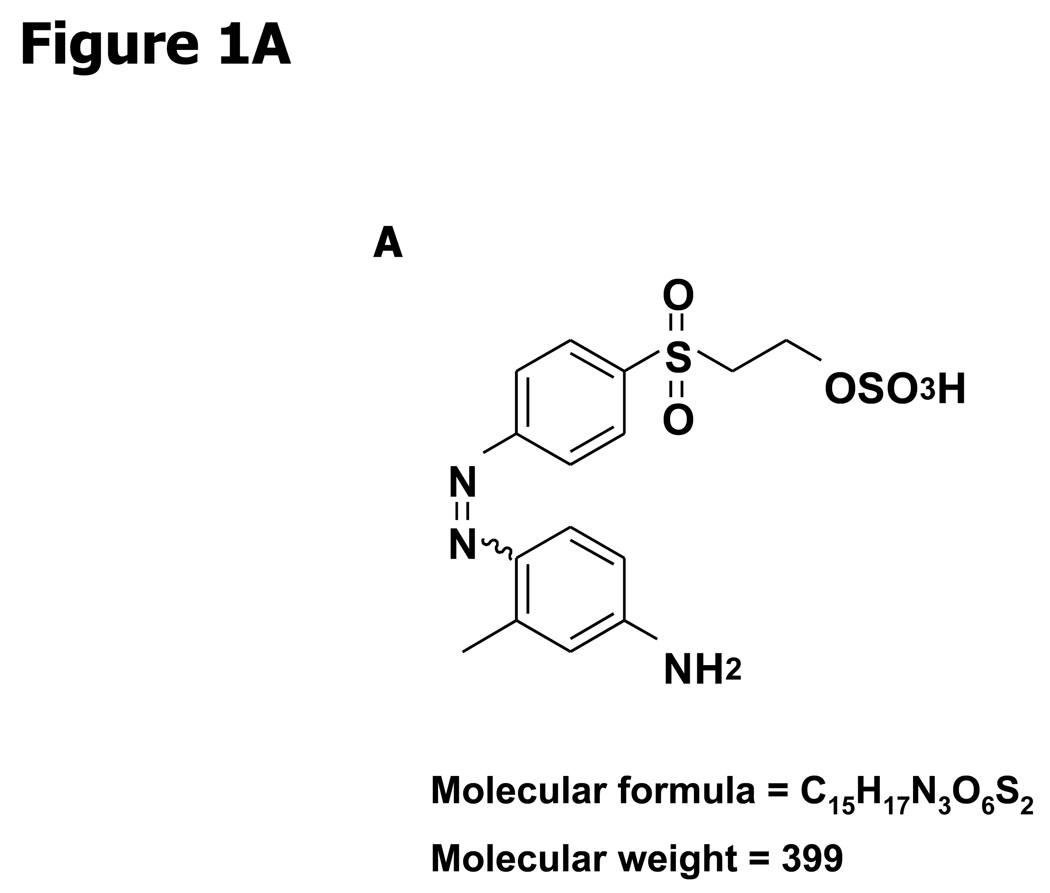
The synthetic compound was provided by the Drug Synthesis and Chemistry Branch, Developmental Therapeutics Program, Division of Cancer Treatment and Diagnosis, National Cancer Institute (Frederick, MD). NSC 676914 was originally identified as 1-[m-methylanilinomalonyl]-3,5-dimethyl-4-[4'.beta.-sulphato ethyl sulphonyl phenyl azo]pyrazole, with a molecular weight of 563.6. Spectroscopic examination of the compound by NMR and LC-MS showed that the sample did not match the published structure. It was subsequently purified by HPLC on Rainin Dynamax C18 (2 × 25 cm) reversed-phase HPLC eluting with a gradient of 20–60% acetonitrile in 0.05% aqueous TFA in 40 min at a flow rate of 10 ml/min to provide (Z)-2-(4-((4-amino-2-methylphenyl)diazenyl)phenylsulfonyl)ethanesulfonoperoxoic acid (NSC 676914). The structure of NSC 676914 (Fig. 1A) was assigned based on spectroscopic characterization and comparison to literature data of structurally related compounds (24). This compound has been previously patented (25) but no NMR data has been published. 1H NMR (DMSO, 500 MHz) δ 7.97* (2H, d, J = 7.5 Hz, H-2′ and H-6′), 7.87* (2H, d, J = 8.0 Hz, H-3′ and H-5′), 7.61 (1H, d, J = 9.0 Hz, H-5), 6.52 (1H, s, H-3), 6.48 (1H, d, J = 7.5 Hz, H-6), 3.96 (2H, d, J = 6.5 Hz, H-8′), 3.65 (2H, d, J = 6.5 Hz, H-7′), 2.55 (3H, s, H-7)). 13C NMR (DMSO, 125 MHz) δ 155.98 (C-1′), 154.21 (C-4), 142.82 (C-1), 141.10 (C-2), 138.32 (C-4′), 129.18 (C-2′ and C-6′), 122.18 (C-3′ and H-5′), 117.49 (C-6), 113.99 (C-3), 112.34 (C-5), 59.25 (C-8′), 54.87 (C-7′), 20.67 (C-7).
Figure 1.
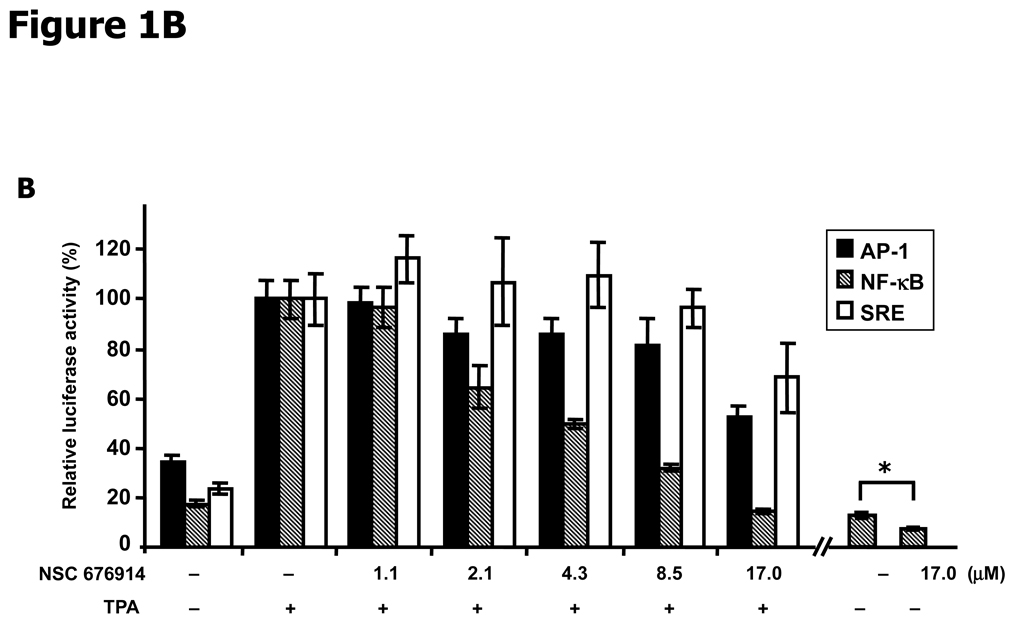
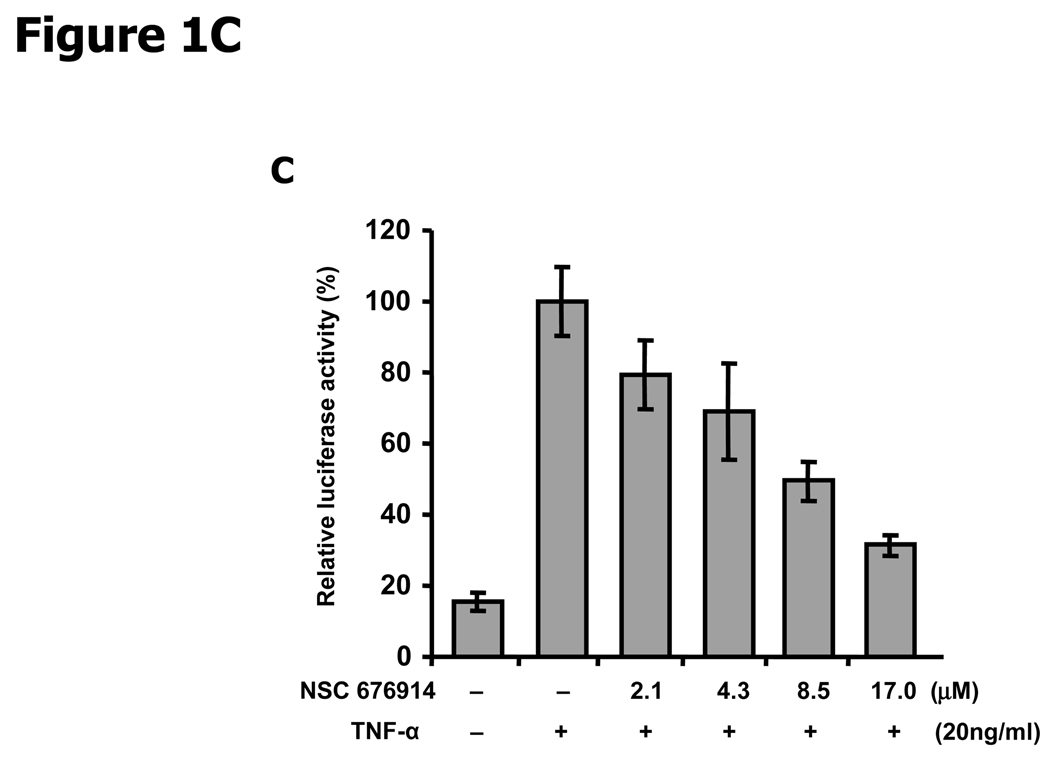
NF-κB is more sensitive to NSC 676914 inhibition than AP-1 or SRE-dependent transcription. A, Molecular structure of NSC 676914. B, TPA-induced and basal Luciferase reporter activity. HEK293 cells were transfected with AP-1, NF-κB, or SRE promoted luciferase reporter. The transfected cells were pre-treated with varied concentrations of NSC 676914 or DMSO for 1 hour before 18 hours stimulation by TPA (10 ng/ml) or DMSO as vehicle. * A separate experiment with NF-κB dependent luciferase only. C, NSC 676914 inhibits TNFα-induced NF-κB-luciferase activity. Luciferase activities with TPA or TNFα induction alone were set at 100% and the relative activities with TPA or TNFα in the presence of NSC 676914 are shown. Assays were performed with two independent experiments each in triplicate (C) or quadruplicate (B).
*Assignments may be interchanged.
Transfection and Luciferase Reporter Assay
All expression plasmids were transiently transfected into HEK293 cells in a 96-well plate (0.1 µg/well) by using Effectene transfection reagent (Qiagen) according to the manufacturer's manual. HEK293 cells were maintained in Dulbecco's modified Eagle medium (Sigma) supplemented with 10% fetal bovine serum (Atlanta biologicals) and incubated at 37 °C in a 5% CO2 incubator. After 24 hrs incubation, the medium was changed to DMEM with 0.2% FBS, and the transfectants were exposed to various concentrations of NSC 676914 and TPA (10 ng/ml). Cells were lysed with passive lysis buffer and the luciferase assay was performed using the Firefly-Luciferase reporter assay system (Promega) and MLX Luminometer (Dynex technology).
Plasmids
Three different luciferase reporter genes, AP-1 reporter containing four times TRE consensus originated from GCN4 and SRE reporter purchased from Stratagene respectively are as described previously (26). NF-κB reporter is driven by five times NF-κB responsive element inserted into Cis-reporter backbone (Stratagene). Others include mouse COX-2 promoter region from −203 to +70 relative to the transcription site inserted into pTIS10L vector (27), human uPAR promoter region from −141 to +47 relative to transcription start site in pGL3 vector (28), and human VEGF promoter sequence from −2274 to +379 inserted into the pGL2 vector (11).
Immunoblot Assay and Cytoplasmic and Nuclear preparation
To measure endogenous level of several proteins in response to TPA induction and NSC 676914, HEK293 cells were lysed with RIPA buffer (10 mM Tris-HCl pH 7.5, 1 mM EDTA, 0.1% sodium dodecyl sulfate, 0.1% deoxycholate, and protease inhibitor). Whole cell extracts were subjected to immunoblot analysis with the following antibodies; p65 (F-6), p50 (E-10), p52 (C-5), RelB (C-19), IκB-α (C-21), PCNA (F-2) were obtained from Santa Cruz. IKKα, IKKβ, IKKγ, phospho-IκB-α, phospho-IKKα/β, phospho-IKKγ, pohospho-Stat3 (Tyr 705 and Ser 727), phospho-p65 (Ser536 and Ser276), phospho-p44/42, phospho-JNKs were purchased from Cell Signaling, and α-tubulin from Sigma. Nuclear and cytoplasmic extracts were prepared with HEK293 cells by NE-PER Nuclear and Cytoplasmic Extraction Reagents (Pierce Biotechnology) according to the manufacturer’s protocol.
Quantitative real-time PCR
Total RNA was isolated from HEK293 cells after pretreatment with varying NSC 676914 and/or TPA stimulation by ToTALLY RNA kit (Ambion), and first-strand cDNA was synthesized from 1µg of the isolated total RNA using the SuperScript III First-Strand Synthesis System (Invitrogen) with random hexamers, oligonucleotide primers. cDNA was purified by QIAquick PCR purification kit (Qiagen) according to the manufacturer's manual. Real-time PCR was performed with iQ SYBR Green Supermix (Bio-Rad) by iQ5 Multicolor RT-PCR detection system (Bio-Rad). Primers for endogenous human IL-6 were designed by Primer 3 program (http://frodo.wi.mit.edu/). Human glyceraldehyde-3-phosphate dehydrogenase (hGAPDH) was used as a control. The sequences for RT-PCR are as follows: IL-6 F: 5’-TACCCCCAGGAGAAGATTCC-3’, R: 5’-TTTTCTGCCAGTGCCTCTTT-3’ and hGAPDH F: 5’-TGCACCACCACCTGCTTAGC-3’, R: 5’-GGCATGGACTGTGGTCATGAG-3’.
Electrophoretic mobility shift assay (EMSA)
EMSA was performed with the nuclear fraction of HEK293 cell using LightShift Chemiluminescent EMSA Kit (Pierce Biotechnology) according to the manufacture’s instruction. Oligonucleotides for NF-κB, NF-1, and AP-1 were labeled with biotin by Biotin 3’ End DNA labeling Kit (Pierce Biotechnology). The quantified 15 µg of nuclear extracts were incubated for 20 mins with 1µg/ul Poly (dI·dC), biotin end-label target nucleotides in 20 µl binding buffer DNA-protein complexes were fractionated on 6% a polyacrylamide gel and transferred to a nylon membrane. The membranes were processed to detect biotin-label DNA by cross-linking, blocking, and detection incubations. Results were obtained by X-ray film exposure for 10 sec and development. The target sequences of oligonucleotides for NF-κB, NF-1, and AP-1 are as follows; NF-κB F: 5’-GGTTACAAGGGACTTTCCGCTG-3’, R: 5’-CAGCGGAAAGTCCCTTGTAACC-3’, NF-1 F: 5’-TTTTGGATTGAAGCCAATATGATA-3’, R: 5’-TATCATATTGGCTTCAATCCAAAA-3’, AP-1 F: 5’-CTAGAGGTGTCTGACTCATGCTTTA-3’, R: 5’-AGCTTAAAGCATGAGTCAGACACCT-3’.
Invasion assay
Breast cancer cell lines, MCF-7, T47D, and MDA-MB-231 were used to measure the effect of NSC 676914 on TPA-induced invasion activity using Matrigel Invasion Chamber (BD Bioscience) according to the manufacturer’s protocol. Matrigel-coated chambers containing a 8 µm pore size filter were fitted into a 24-well tissue culture plate. Briefly, breast cancer cells (1 × 105 cells/ml) of each cell line were seeded into 6-well Matrigel-coated chambers with DMSO, TPA, or TPA plus NSC 676914 conditions and incubated for 24 hrs. The invaded cells on the bottom side were stained using Diff-Quik staining (Dade Behring) and photographed. Three independent results were very similar.
Transformation assay
JB6 cells (clone 41) were used to assess the inhibitory activity of NSC 676914 using CytoSelect 96-well Cell Transformation Assay (Cell Biolabs) as Soft Agar Assay according to manufacturer’s instructions. Cells were incubated for 10 days in 0.4% agar medium over 0.6% agar containing TPA or TPA with NSC 676914 or DMSO as a control. To measure anchorage-independent growth, agar-layers were dissolved and lysed. 10 µl of the lysed solutions of each well were mixed with CyQuant and the fluorescence read by iQ5 with a FAM filter of 485/520 nm. As an alternative analysis, cell dose curves of JB6 cells were quantified by the End point using CyQuant to indicate relative anchorage-independent colony number.
Results
NSC 676914 prevents activation of NF-κB
To evaluate whether the NSC 676914 biological activity can imitate TAM67 specificity we used three different transcriptional promoter reporters, AP-1, NF-κB, and SRE (Serum response element) in luciferase reporter assays (Fig. 1B). All three activities were induced by TPA but only induction of AP-1 and NF-κB were inhibited by TAM67. The SRE reporter was used as a specificity control and indicator of cell proliferation. TPA-induced SRE activity was not repressed by TAM67 when coexpressed with the SRE-Luc reporter in HEK293 cells (Data not shown). In this study, we used the same concentration (10 ng/ml, 16.2 nM) and incubation time (18 hrs) for TPA induction as previously used for the HTS (13) along with the presence of NSC 676914 for 18 hrs incubation in addition to 1 h pretreatment. The screening results had tested concentrations as high as 40 µM NSC 676914 with cell numbers showing 95.5 +/− 6.2% of DMSO control (ave +/− sd, n = 3) by the XTT assay in HEK293-T cells.
AP-1 luciferase activity was repressed to 50% of the activity with TPA alone after pretreatment by NSC 676914 at 17.0 µM (Fig. 1B). SRE activity was also repressed but to a lesser degree than AP-1. Surprisingly, TPA-induced NF-κB luciferase activity was completely blocked to the non-induced level at 17.0 µM NSC 676914. NF-κB luciferase activity was dose-dependently inhibited by NSC 676914 at all concentrations tested. The IC50 of NSC 676914 against TPA-induced NF-κB was around 4 µM or about 1/4 of the 17.0 µM IC50 for AP-1 inhibition. The basal activity for NF-κB was also inhibited, but to a lesser extent, by NSC 676914 without TPA induction (Fig. 1B). In addition, NSC 676914 shows dose-dependent inhibition of NF-κB induced by TNFα (Fig 1C). These results suggest that NSC 676914 may preferentially target events needed for NF-κB activation rather than other transcription factor activations. The magnitude of NF-κB activity remaining in the presence of NSC 676914 apparently suffices to support normal cell survival.
NSC 676914 inhibits phosphorylation of endogenous IκB-α and IKKα/β
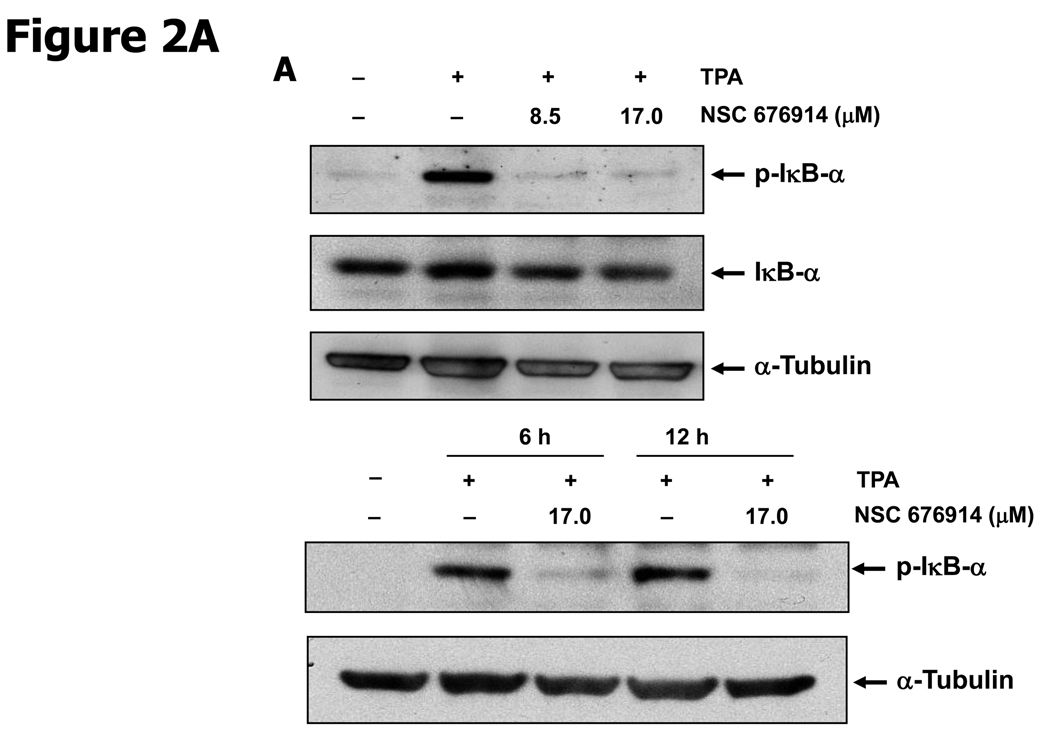
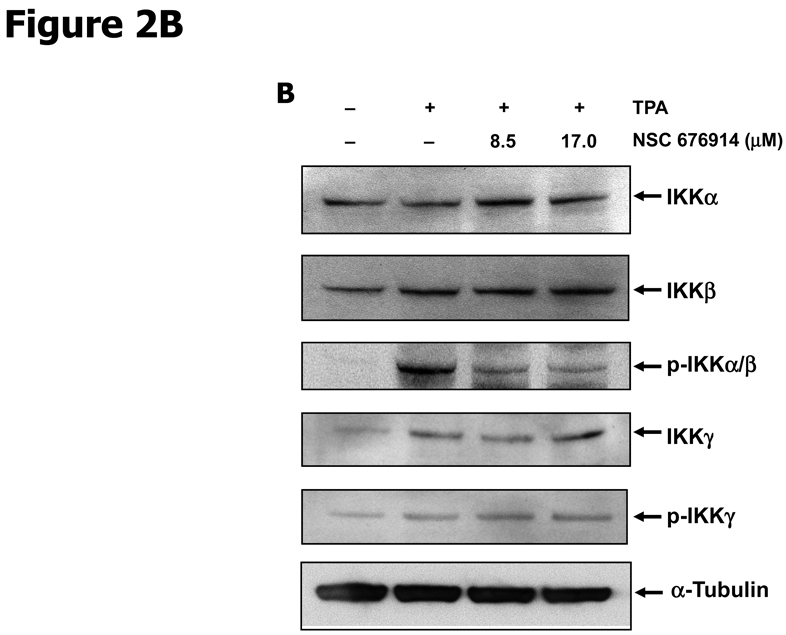
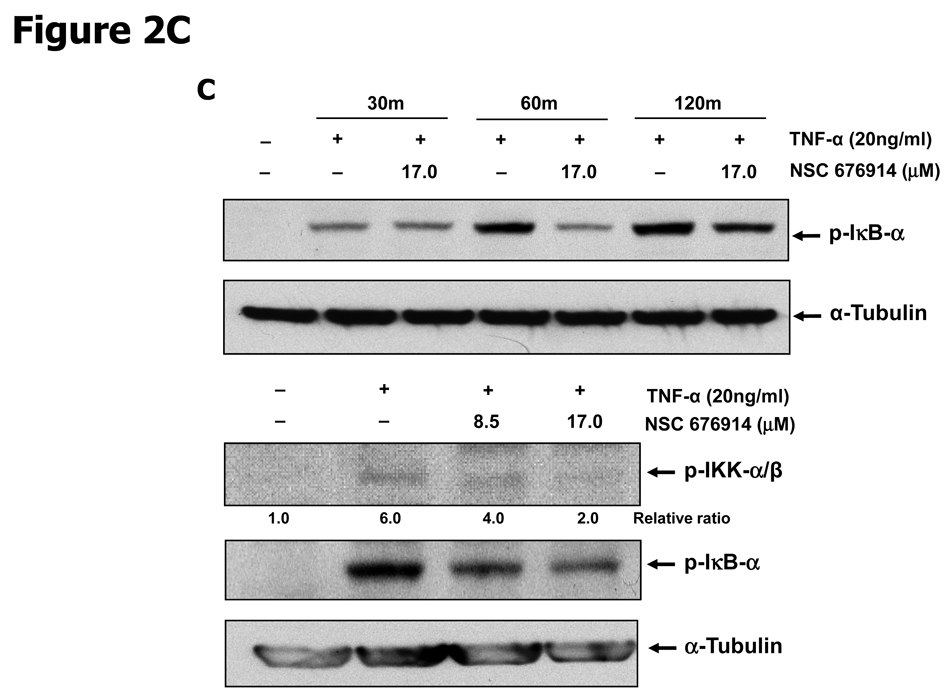
To characterize the activity of NSC 676914 in targeting events upstream of NF-κB, endogenous levels of kinases involved in NF-κB activation were examined by immunoblot analysis (Fig. 2). The well-established critical upstream regulators of NF-κB activation are IKKα, IKKβ, and IκB-α. Stimulation of HEK293 cells with TPA resulted in induced phosphorylation of IκB-α as well as of protein kinase(s) I Kappa B Kinase α and/or β. The induced phosphorylation of IκB-α following TPA stimulation was almost completely blocked after pretreatment with NSC 676914 at 17.0 and 8.5 µM (Fig. 2A upper). A similar potency for inhibition of IKKα/β phosphorylation was seen (Fig. 2B). Moreover, the phosphorylation of IκB-α was inhibited even at 1.1 µM showing dose-dependent decrease by NSC 676914 pretreatment (Supplementary Fig. S1). TPA-induced phosphorylation of IκB-α was also inhibited by the compound at 6 and 12 hours (Fig. 2A bottom), but not at 3 hrs (Data not shown). In addition NSC 676914 inhibits TNFα-induced phosphorylation of IKKα/β (Fig. 2C bottom) and IκB-α at 1 hour (Fig. 2C upper). We were not able to distinguish IKKβ from IKKα in this analysis with antibodies which detect phosphorylated IKKα at Ser176/180 and IKKβ at Ser177/181. TPA-induced phosphorylation of IκB-α is mediated primarily through IKKβ catalytic activity (29, 30); thus it appears that NSC 676914 represses phosphorylation of IKKβ only or both IKKβ and IKKα. Phosphorylation of IKKγ did not change in response to NSC 676914 pretreatment. The total protein levels of IKKα/β, IKKγ, and IκB-α were not affected (Fig. 2A & B). Thus, upstream activation events rather than transcription factor binding to DNA appear to be targeted by NSC 676914 when it inhibits NF-κB induction.
Figure 2.
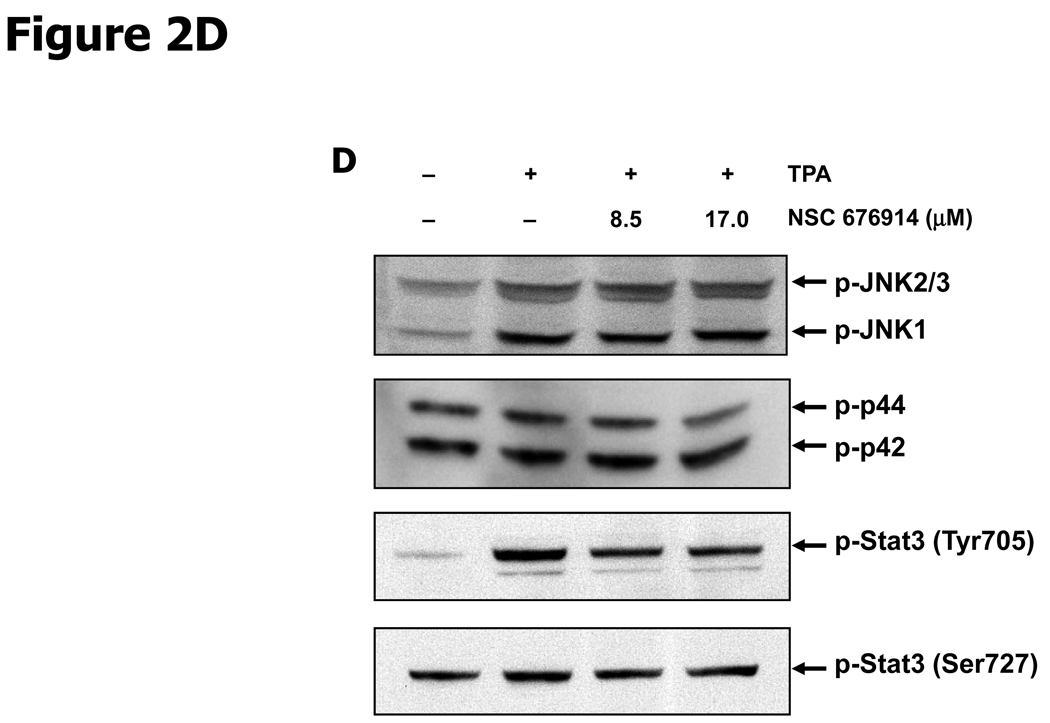
NSC 676914 inhibits phosphorylation of IKKs and IκB-α. HEK293 cells were treated with various concentrations of NSC 676914 or DMSO for 1 hour followed by induction with TPA (10 ng/ml) for 18 hours (A upper, B, D) or 6 and 12 hours (A bottom) with TNFα (20ng/ml) for 30, 60, and 120 mins (C bottom) or 60 mins only (C upper). Endogenous protein expression was measured with whole cell extract by immunoblot analysis. Immunoblots were probed with antibodies for (A, B) IκB-α, phospho-IκB-α, α-tubulin, (C) IKK using antibodies for IKKα, IKKβ, IKKγ, phospho-IKKα/β, phospho-IKKγ, (D) phospho-JNKs, phospho-ERK 1/2 (p42/44), and phospho-STAT3.
We next evaluated the selectivity of NSC 676914 by measuring phosphorylation of proteins involved in the AP-1 induction pathway (Fig. 2D). AP-1 dependent transcription can be activated by MAP Kinases JNKs, ERK 1/2 (p44/42) (22, 31). Immunoblot assay revealed no repression of TPA-induced phosphorylation of endogenous JNKs, ERK 1/2 (p42/44), or of TPA inducible phosphorylation of an unrelated transcription factor STAT3 by pretreatment with N SC 676914 at 17.0 µM and 8.5 µM. At these concentrations NSC 676914 in contrast, completely repressed major regulators in the NF-κB activation pathway as shown in Figure 2A & B. JNKs are activated by a variety of environmental stresses responsible for AP-1 activation through c-Jun phosphorylation. TPA-induced phosphorylation of JNKs showed no repression by NSC 676914 pretreatment. These results indicate that NSC 676914 does not negatively regulate MAP Kinases that activate AP-1 or kinases that activate STAT3. Instead it suppresses the phosphorylation of IκB-α needed for release from p65/p50 and of IKKs that activate NF-κB. The mechanism by which NSC 676914 at higher concentrations inhibits AP-1- luciferase as shown in Figure 1B is not known, but it can be said that this mechanism is independent of JNK or ERK. The inhibition of AP-1 could be secondary to inhibition of NF-κB, as p65 is known to positively transactivate c-Jun through binding to its b-Zip domain (22).
NSC 676914 did not inhibit either IKKα or β in vitro
In an attempt to determine whether NSC 676914 interferes with ATP binding to the catalytic site, we measured the activity of NSC 676914 with fully activated, phosphorylated IKKα or IKKβ. NSC 676914 showed no activity in an in vitro assay, the off-chip incubation mobility-shift kinase assay measuring the ability to block phosphorylation of a peptide substrate of IKKα or β (Supplementary Fig. S2). This result indicates that NSC 676914 does not inhibit the activity of either IKKα or β by targeting its conserved ATP binding pocket. Whether this compound interacts outside of the ATP binding pocket of IKK as an ATP non-competitive inhibitor similar to BMS-345541 (32) is unknown. However, both NSC 676914 and BMS-345541 at concentrations from 0.1 to 100 µM showed no inhibition of phosphorylation of IκB-α peptide substrate catalyzed by IKKβ incubated with ATP in a vitro assay (Calbiochem; K-LISA™ IKKβ Inhibitor Screening Kit). BMS-345541, showed dose-dependent inhibition of NF-κB-Luciferase at 1 to 8 µM without inhibition of AP-1, similar to NSC 676914 (Fig. 1B and Supplementary Fig. S3A). In contrast, other NF-κB inhibitors ATP competitive inhibitor SC-514 and sulfasalazine showed similar or greater potency for inhibiting AP-1 and no sparing of SRE in the luciferase assay (Supplementary Fig. S3B & C).
NSC 676914 inhibits TPA-induced nuclear accumulation of NF-κB p65/p50 by a mechanism involving inhibition of IKKβ
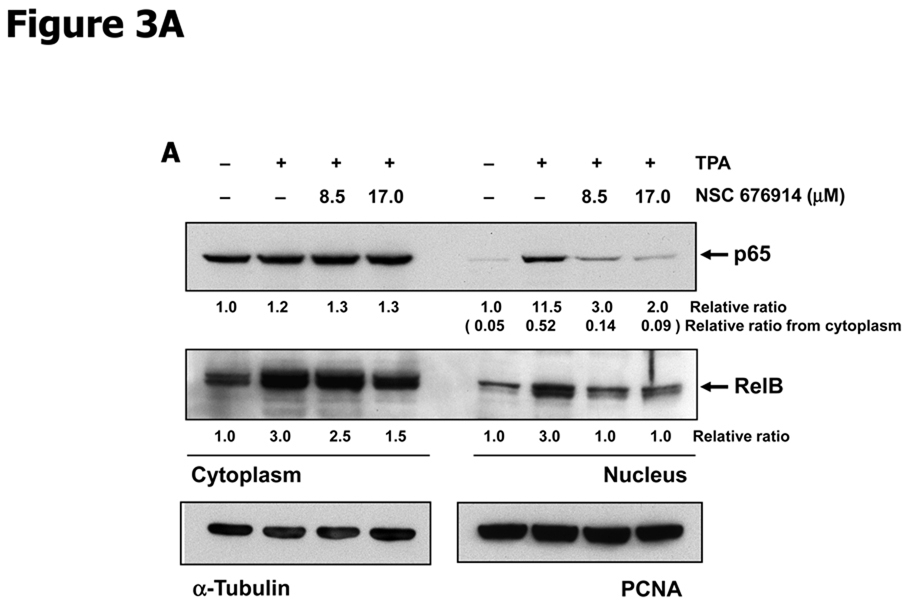
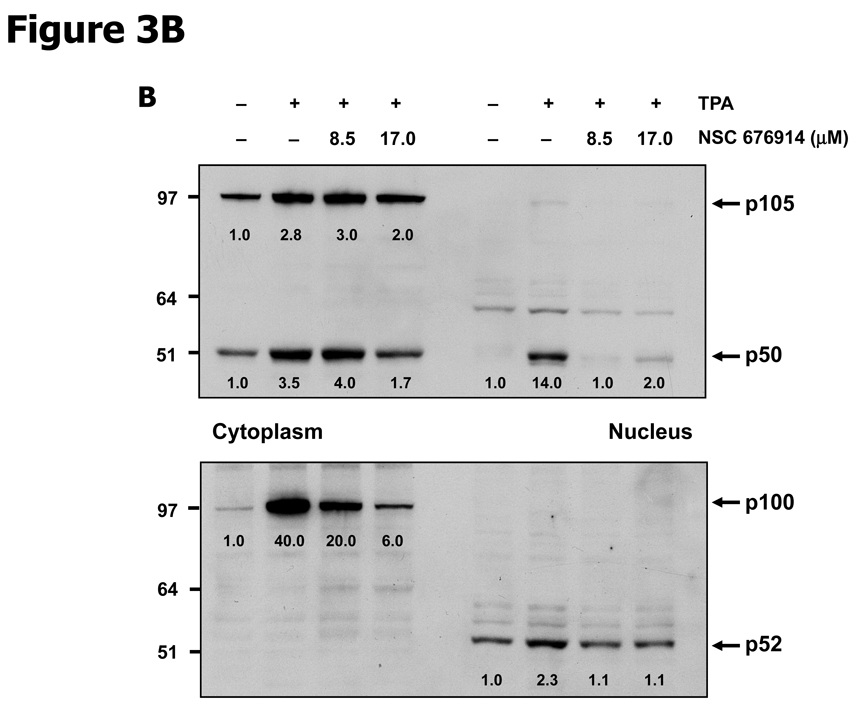
In order to further characterize the mechanism by which NSC 676914 acts as an inhibitor of NF-κB activity, we monitored nuclear translocation of endogenous p65/p50 and RelB/p52 heterodimers. The activation of heterodimers p65/p50 and RelB/p52 occurs by distinct pathways dependent on IKKβ or IKKα catalytic activity. Repression of IKKβ would be expected to influence nuclear translocation of p65/p50 while repression of IKKα would influence processing of p100 to p52 (30). As shown in Figure 3A, nuclear translocation of p65 (also known as RelA) and RelB were induced by TPA and translocation in both cases were remarkably repressed by pretreatment with NSC 676914. Translocation of p65 on the canonical pathway depends on phosphorylation of IκB-α by IKKβ. However translocation of RelB is only dependent on IKKα for processing of its dimer partner p52 from p100. This processing depends on phosphorylation of p100 by IKKα (30, 33). As seen in figure 3B, the processing of p100 to nuclear p52 was not inhibited by NSC 676914 but was proportional to expression levels of p100. NSC 676914 inhibited expression of p100 by an unknown mechanism. Thus because NSC 676914 does not inhibit the processing of p100 to p52, it appears not to inhibit IKKα. Instead NSC 676914 inhibits IKKβ.
Figure 3.
NSC 676914 inhibits translocation o f NF-κB (p65, RelB, p50) from cytoplasm to nucleus. HEK293 cells were stimulated with TPA (10 ng/ml) for 18 hours after pretreatment either with various concentrations of NSC 676914 or DMSO for 1h. Nuclear and cytoplasmic fractions were separated for immunoblot. A, p65 (RelA) and RelB. PCNA was used as loading control for nucleus and α-tubulin for cytoplasm. B, p50 and p52 (antibodies also detect p105 and p100, respectively).
NSC 676914 does not appear to inhibit proteasome-dependent degradation. This is supported by the observation of no interference with processing of p105 to p50 (Fig. 3B). As a control, the processing of p105 to p50 was completely blocked by pretreatment with proteasome inhibitor MG-132 (Supplementary Fig. S4). We also measured the level of p65 phosphorylation at Ser536 known to be required for p65 transactivational activity. Exposure to NSC 676914 attenuated TPA induced phosphorylation at Ser536 and to a lesser extent at Ser276 (Supplementary Fig. S5). The decline in phosphorylation of p65 at Ser536 was expected as p65-S536 is phosphorylated at least in part by IKKβ (34). The decline in Ser276 phosphorylation may be attributable to inhibition by NSC 676914 of another kinase such as MSK1 or PKA (35, 36). The possibility that NSC 676914 targets the upstream MAP kinase kinase kinase TAK1 was not readily testable due to lack of a suitable antibody that can detect endogenous expression. The fact that TAK1 is important for activating JNKs (37) and the observation that activation of JNKs were unaffected suggest that NSC 676914 acts downstream of TAK1. TAK1 cannot however be excluded as a target of NSC 676914 because JNKs might be phosphorylated by a kinase other than TAK1. If NIK (NF-κB inducing kinase) regulates only the non-canonical pathway upstream of IKKα, then NIK targeting by NSC 676914 is unlikely. However if NIK also can regulate the canonical pathway upstream of IKKβ, one cannot exclude the possibility that NIK could be targeted.
NSC 676914 inhibits DNA binding of NF-κB
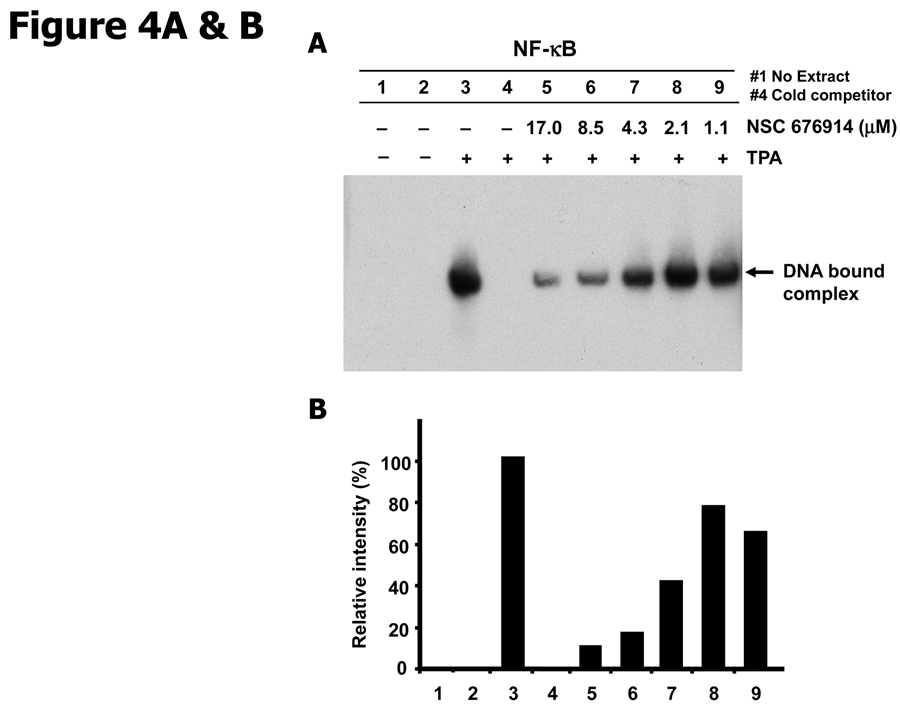
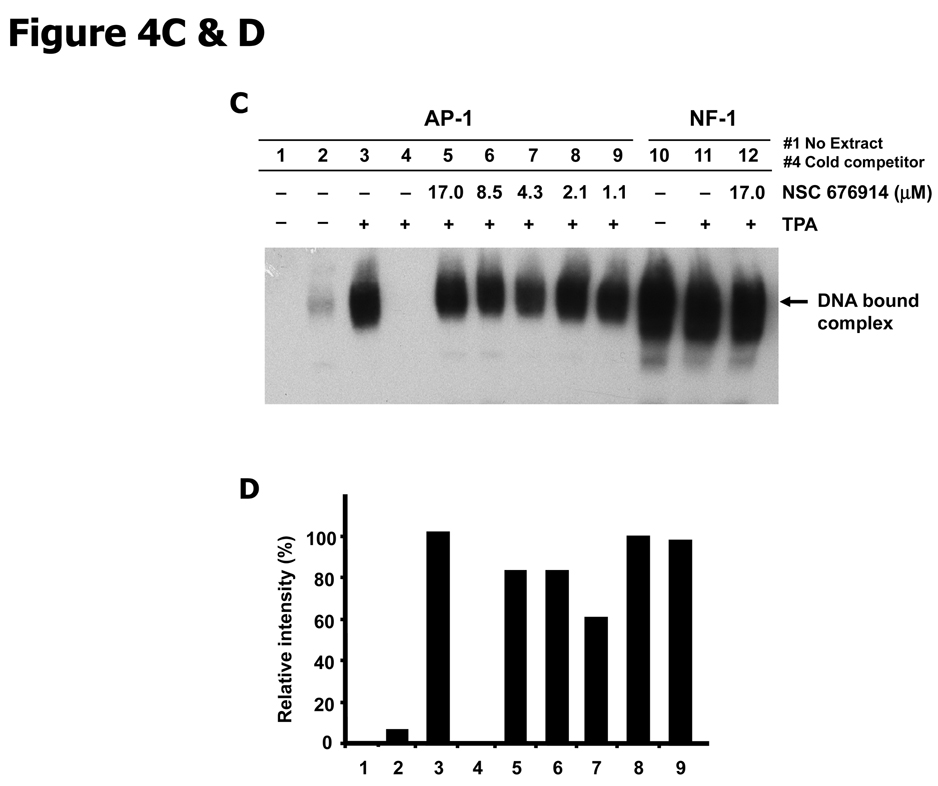
To determine whether the effect of NSC 676914 on nuclear translocation of NF-κB leads to inhibited binding of NF-κB proteins to DNA, we analyzed endogenous NF-κB DNA binding activity by EMSA with a probe representing an NF-κB-dependent promoter (Fig. 4A & B). The intensity of NF-κB probe and protein complex was increased by TPA stimulation (lane 3) and pretreatment of NSC 676914 at 17 µM resulted in marked loss of binding complex. TPA-induced binding complex was attenuated by pretreatment with NSC 676914 in a dose-dependent manner (lanes 5–9). This binding inhibition parallels the inhibition of luciferase reporter transactivation and inhibition of nuclear translocation of NF-κB proteins (Fig. 3A). The TPA-induced binding complex disappeared when the labeled NF-κB oligonucleotide was competed with an excess of unlabelled probe (90× fold). Specificity controls showed that NSC 676914 at 17 µM had little or no effect on NF-1 and AP-1 binding activity (Fig. 4C & D). Taken together, these results suggest that NSC 676914 inhibits DNA binding as a consequence of interference with nuclear localization of NF-κB proteins, a process that is regulated by IKK activity.
Figure 4.
NSC 676914 inhibits DNA binding of NF-κB. A, Inhibition of NF-κB binding. EMSA analysis of basal and TPA-induced NF-κB DNA-binding was performed with NF-κB oligonucleotide probe on the nuclear fractions of HEK293 cells. HEK293 cells were treated with TPA (10 ng/ml) for 18 hours following pretreatment with NSC 676914 or DMSO for 1 hour. The lane 1 was mixed with no extract of nuclear fractions of HEK293 cells. B, Semiquantitiative analysis of EMSA using densitometry of the band in each lane from A. NF-κB DNA binding activities were converted to the value of the maximal activity with TPA induction alone without NSC 676914. C, Lack of NSC 676914 effect on AP-1 and NF-1 binding. EMSA analysis of basal and TPA-induced AP-1 (lane 1–9) and NF-1 (lane 10–12) DNA-binding was performed with AP-1 and NF-1 consensus probes. D, Semiquantitiative analysis of EMSA was shown using densitometry of the band in each lane from C.
TPA-induced transcriptional targets of NF-κB were repressed by NSC 676914 pretreatment
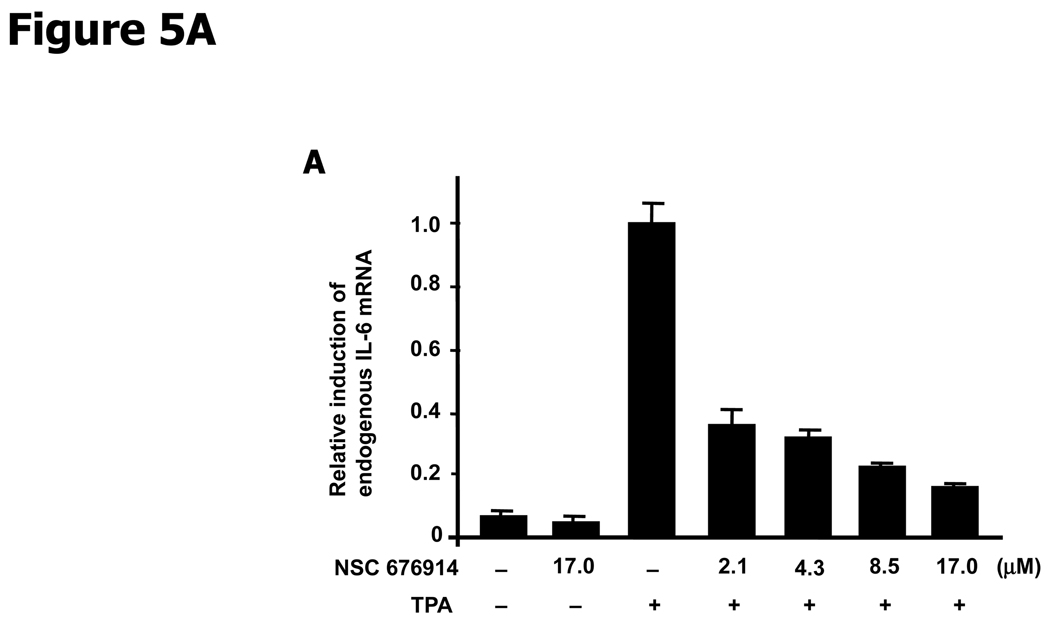
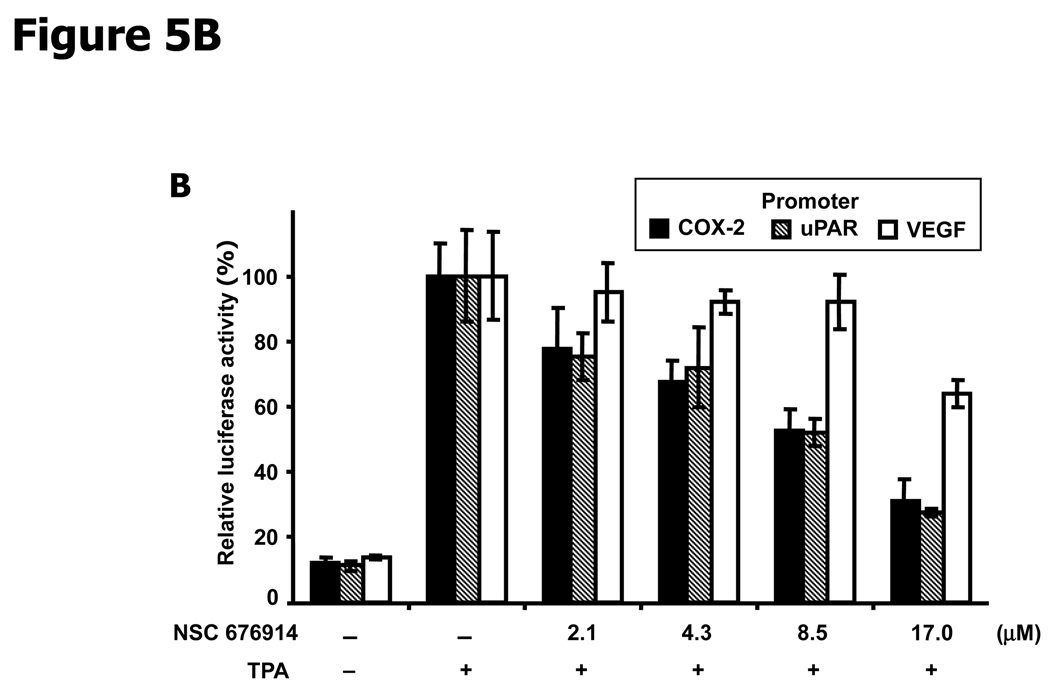
Inhibition of NF-κB transactivation would be expected to result in suppressed expression of genes whose transcription depends on NF-κB. Quantitative RT-PCR was used to measure endogenous levels of IL-6 mRNA, transcribed from a gene whose transcription depends on NF-κB but not AP-1. While TPA induction increased the IL-6 mRNA by more than 10 fold, pretreatment with NSC 676914 prevented more than half of the induction at concentrations as low as 2.1 µM (Fig. 5A). We also examined the effect of NSC 676914 on several well-known transcriptional targets of NF-κB, also regulatable by AP-1, using luciferase reporter assays in HEK293 cells. Plasmids expressing luciferase reporters were driven by promoters for COX-2, uPAR (urokinase-type plasminogen activator receptor), or VEGF. COX-2 and uPAR luciferase activities after treatments with NSC 676914 showed similar dose-dependent inhibition (Fig. 5B). In contrast, NSC 676914 did not significantly affect the luciferase activity when driven by VEGF promoter. One NF-IL6 and two AP-l-like sequences are located in the promoter region of COX-2. uPAR promoter region contains one NF-κB-like sequence and two AP-1 binding sites, and VEGF promoter region has multiple binding sites for AP-1, HIF-1α, and NF-κB-like. These mixed promoter regions of AP-1 and NF-κB in the COX-2 and uPAR are known to be important sequences for their expression. However, VEGF has not shown direct dependency on an NF-κB consensus sequence in the promoter region of mouse macrophages, whereas several AP-1 and HIF-1α sites are required. Deletion of putative NF-κB binding sites from the VEGF promoter reporter showed no effect, suggesting that NF-κB is not directly involved in VEGF transcription (38). The significant differential in sensitivity to NSC 676914 between VEGF versus COX-2 and uPAR as shown in Figure 5B, might be explained by the relative importance of NF-κB binding sites compared to AP-1 sites in the promoter region. These results establish that genes known to be dependent on NF-κB are transcriptionally inhibited by NSC 676914.
Figure 5.
NSC 676914 inhibits TPA-induction of NF-κB dependent genes. A, IL-6 mRNA expression. Detection by quantitative RT-PCR showed TPA induced IL-6 mRNA was repressed by NSC 676914 pretreatment. Total RNA was isolated and analyzed from 18 hours TPA-treated HEK293 cells pretreated with NSC 676914 or DMSO for 1 hour. Human IL-6 mRNA was normalized by GAPDH. B, Luciferase reporter assays of NF-κB targets. Promoter directed luciferase reporters for COX-2, uPAR, and VEGF were transiently transfected into HEK293 cells separately. The cells were pretreated with NSC 676914 followed by exposure to TPA (10 ng/ml) or DMSO for 18 hours. The luciferase activity attained by TPA induction only was set at 100% and used to compare the activity in the presence of NSC 676914. Assays were performed in triplicate.
NSC 676914 suppresses TPA-induced transformation and invasion
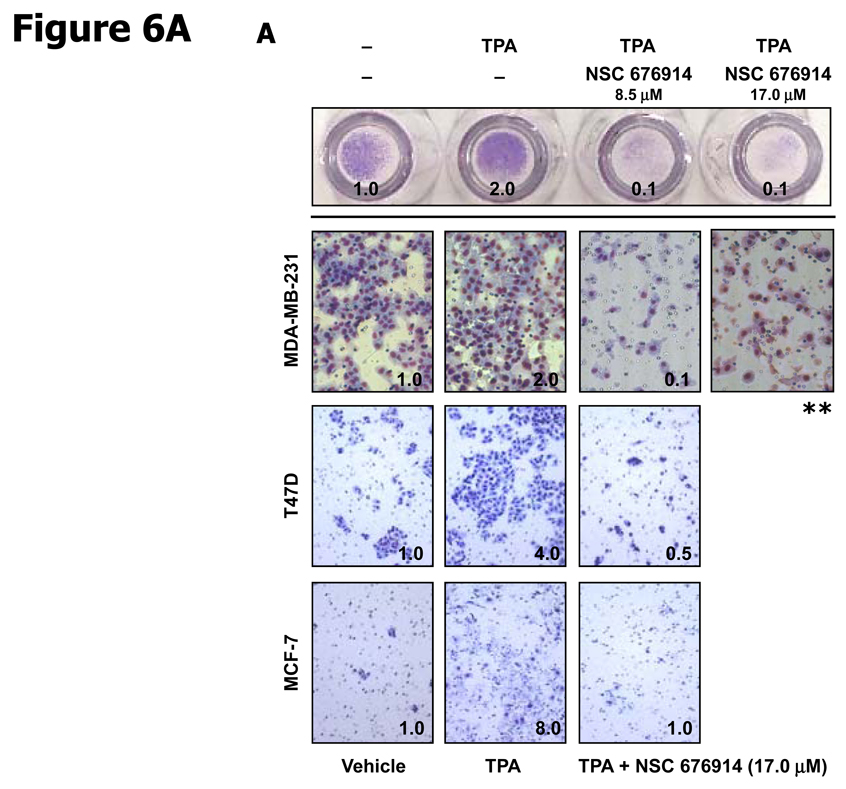
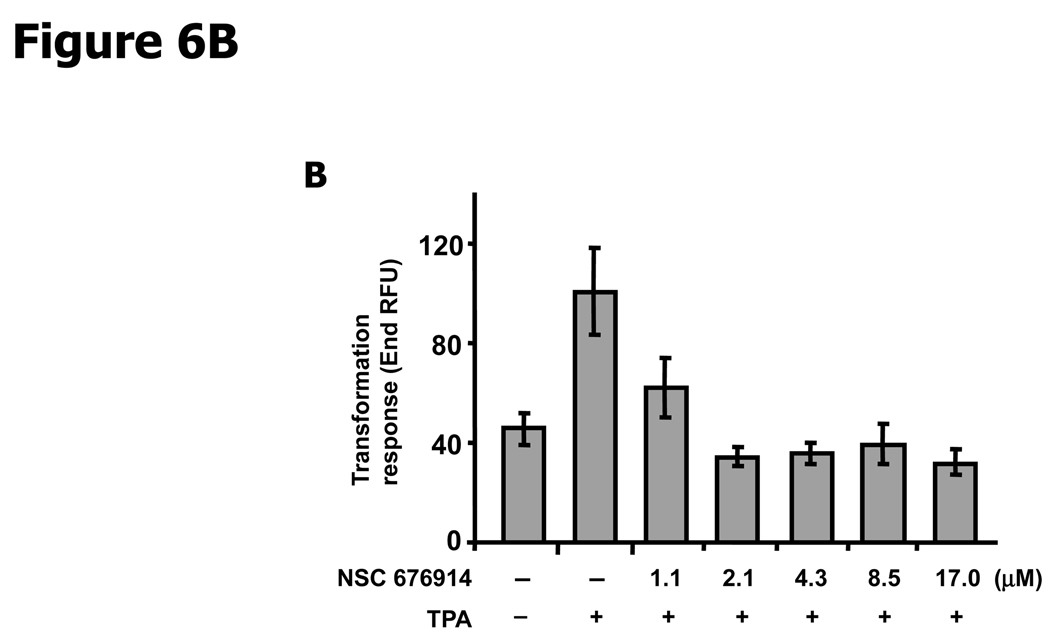
In order to evaluate the potential therapeutic activity of the compound, we considered tests for the biological activity of NSC 676914. The compound is non-toxic to MCF-7, T47D, and MDA-MB-231 breast cancer cells as estimated by XTT assay at 10 µM (Data not shown). To determine the effects of NSC 676914 on invasion by breast cancer cell lines, a matrigel invasion assay was used (Fig. 6A). We first confirmed the levels of phosphorylated IκB-α in TPA treated ER-positive (MCF-7 and T47D) and ER-negative (MDA-MB-231) breast cancer cell lines and compared the levels to those after pretreatment with NSC 676914. Although induced levels of phosphorylated IκB-α varied with the cell line, IκB-α phosphorylation was eliminated by NSC 676914 treatment in all three breast cancer cell lines (Data not shown). NSC 676914 also produced no effect on the estrogen receptor activity in the ER-positive breast cancer cells using ERE promoter luciferase reporter assay (Data not shown). The upper panel in Figure 6A shows the overall picture of MBA-MB-231 cells with the chamber turned upside down and the lower panel shows MBA-MB-231, T47D, and MCF-7 cells invaded through the Matrigel coated filter located on the underside of chambers. Invasion capacity of breast cancer cells was increased by TPA stimulation compared to DMSO treated vehicle control. However, NSC 676914 completely inhibited the TPA-increased invasion in the three breast cancer cell lines as well as basal invasive activity in MBA-MB-231 cells. In another assay, to assess the inhibitory effect of NSC 676914 on TPA-induced transformation, soft agar assays were performed in mouse JB6 cells using the Cytoselect 96-well cell transformation assay. End RFU measuring DNA concentration is proportional to the number and size of anchorage independent JB6 colonies (Fig. 6B). Transformation response to TPA was repressed to below vehicle control level by NSC 676914 treatment at 17.0 µM to 2.1 µM. NSC 676914 even decreased transformation response at 1.1 µM. This result is consistent with the immunoblot analysis showing expression levels of phosphorylated IκB-α inhibited by NSC 676914 treatment at 1.1 µM (Supplementary Fig. S1). Thus, these results demonstrate that NSC 676914, when it blocks phosphorylation of IκB-α, completely blocks the TPA-induced invasion in breast cancer cells and the transformation response in JB6 cells.
Figure 6.
NSC 676914 inhibits TPA induced and basal invasion of breast cancer cells and transformation of JB6 cells. A, A matrigel invasion assay in breast cancer cells. Three human breast cancer cell lines, MDA-MD-231, T47D, and MCF-7 were distributed in BD Biocoat matrigel invasion chamber and invasion activity was stimulated by TPA (10 ng/ml) for 48 hours after pretreatment with NSC 676914 or DMSO for 1h. Only invasive cells can digest the matrix and move through the chambers. (Values, relative ratio compared to vehicle.) ** The MDA-MB-231 cell line was tested with NSC 676914 (17 µM) alone without TPA induction. B, Anchorage-independent transformation response. JB6 cells were suspended in soft agar and incubated with medium containing TPA (10 ng/ml) with NSC 676914 or DMSO for 10 days.
Discussion
NSC 676914, initially identified in a high throughput screen for AP-1 inhibitors, shows greater potency for inhibiting NF-κB activation. This compound was identified from synthetic compound libraries, and was previously not known to function as a selective NF-κB inhibitor. With an IC50 of about 4 µM for inhibition of NF-κB reporter, NSC 676914 reduced phosphorylation of IKKα/β significantly. The repressed phosphorylation of IKK produced complete blocking of phosphorylation on IκB-α at concentrations as low as 8.5 µM. The phosphorylation of IκB-α was reduced even at 1.1 µM NSC 676914 compared to its level with TPA induction alone. Concentrations of NSC 676914 as low as 1.1 µM repressed NF-κB DNA binding and transcriptional activation of NF-κB dependent genes. Moreover NSC 676914 inhibits tumor promoter induced transformation of JB6 cells and invasion of breast cancer cells. These results suggest that single digit micromolar-range treatment is sufficient to inhibit NF-κB transactivation required for tumor promotion and progression.
High selectivity and low toxicity can be valuable characteristics of a potential chemopreventive agent against its target. These are characteristics that were sought in the original screen for a TAM67 (dominant negative c-Jun) mimetic. Although inhibition of cell proliferation or cell survival can be useful features for cancer treatment and can be tolerated for prevention in high risk groups, prevention generally calls for selectivity and minimal toxicity (18). For prevention utility, more than one screen for toxicity is needed. In addition to the previously reported XTT assays, we used a Serum Response Element (SRE)-driven luciferase reporter to establish lack of inhibition of cell proliferation by NSC 676914.
NSC 676914 inhibits IKKβ without targeting MAP kinases or JAKs that activate STAT3. As a consequence of targeting IKKβ, NSC 676914 regulates the canonical pathway of NF-κB activation. This results in interception of nuclear translocation of p65/50. Although the expression of cytoplasmic p100 and of nuclear p52 was reduced, NSC 676914 did not inhibit the processing of p100 to p52 that is known to depend on IKKα. The decrease in nuclear p52 can be attributed to inhibited expression of the p100 precursor, a process not known to involve IKKα. The inhibitory activity and pharmacokinetic properties of the compound need further testing in vitro and in vivo to determine IC50 against IKKβ and to identify any direct binding targets of NSC676914 that may act upstream of IKKβ activation.
The mouse epidermal JB6 model of transformation response variants has proven to be a unique system for identifying molecular events that drive or inhibit tumor promotion (12). Transformation sensitive (P+) JB6 cells, used in this study, respond to tumor promoter by irreversibly acquiring a tumor phenotype as measured by anchorage-independence and tumorigenicity. NSC 676914 inhibited TPA-induced transformation. NSC 676914 substantially inhibits the TPA-induced invasion of all breast cancer cell lines tested. The inhibition of invasion by NSC 676914 is correlated with the repression of uPAR expression required for invasion (39). These results suggest that NSC 676914 could be a novel inhibitor having potential therapeutic activity to target NF-κB for cancer treatment and prevention. Assays for in vivo activity of NSC 676914 against tumorigenesis and tumor progression will of course be important.
The NCI cancer screen data for NSC 676914 in the NCI-60 panel of human cancer cell lines showed that leukemia and breast cancer cell lines are more sensitive to NSC 676914 for growth inhibition than epithelial lines from other cancer sites. NSC 676914 at 1 µM inhibits the growth of leukemia lines more than 50% but does not kill these leukemia lines at concentrations up to 100 µM. Most of the NCI-60 human cancer cell lines showed growth inhibition at greater than 10 µM. Thus in addition to inhibiting tumorigenesis (transformation) and tumor progression (invasion), NSC 676914 may have potential for tumor growth inhibition in some cancer sites.
NF-κB selective inhibitors are important for cancer prevention and treatment. Obligatory relationships between NF-κB activation and inflammation-associated cancer have been demonstrated using several mouse models. NF-κB activation has been implicated in inflammation associated liver, prostate and colon cancer induction in humans and mouse models (20–22). Several antioxidants having electrophilic capacity such as cyclopentenone prostaglandins, dimethoxylsulfoxide, glutathione and non-steroidal anti-inflammatory drugs (NSAIDs) Ibuprofen, sulindac, as well as curcumin inhibit NF-κB activity but do not show high selectivity. Aspirin, sulfasalazine, SC-514, and PS-1145 also inhibit NF-κB by interrupting phosphorylation of IKK. Some of these agents are competitive inhibitors of the ATP-binding site of IKKβ (30, 40, 41). However, others such as BMS-345541 do not interfere with the ATP-binding site but target amino acids on the kinase activation loop of IKK. These non-ATP competitive inhibitors are interesting because they may act with greater specificity than can be achieved when competing with ATP for binding to highly conserved sites (42). A new IKKβ inhibitor MLN120B acts as an ATP competitive inhibitor showing an IC50 of 0.06 µM for IKKβ (43, 44). NSC 676914 shows a similar specificity as an inhibitor of NF-κB and IKKβ. NSC 676914 shows no activity to bind to the ATP binding pocket of IKKβ. Unlike MLN120B, NSC 676914 does not suppress cell proliferation. NSC 676914 appears to be novel in completely inhibiting transformation of epithelial cells and invasion of breast cancer cells at low concentrations. In conclusion, our findings suggest that NSC 676914 selectively inhibits NF-κB activation by blocking IKKβ phosphorylation in extracellular stimulus-induced environments. NSC 676914 may offer promise as a nontoxic inhibitor for cancer prevention.
Supplementary Material
Supplementary Figure S1. NSC 676914 blocks phosphorylation of IκB-α at 1.1 µM. HEK293 cells were treated with various concentrations of NSC 676914 or DMSO for 1 hour followed by induction with TPA for 18 hours. Endogenous phospho-IκB-α expression was measured by immunoblot analysis.
Supplementary Figure S2. In vitro kinase inhibition assay by the off-chip incubation mobility shift. NSC 676914 was tested against 3 kinase targets, JNK2a2, IKKβ, and IKKα. Assay consisted of testing the compound in duplicate at concentrations of 1.25, 2.50, 5.00, and 10.00 µM. Less than 20% inhibition of binding or enzyme activity is considered inactive.
Supplementary Figure S3. Comparison of NSC 676914 with other IKK inhibitors for kinase and transcription factor inhibition. AP-1, NF-κB, and SRE promoted luciferase inhibition by (A) BMS-345541, (B) SC-514 and (C) sulfasalazine. HEK293 cells were transfected with AP-1, NF-κB, or SRE promoted luciferase reporter. The transfected cells were pre-treated with varied concentrations of BMS-345541, SC-514, sulfasalazine or DMSO for 1 hour before 18 hours stimulation by TPA (10 ng/ml) or DMSO as vehicle. Luciferase activities with TPA induction alone were set at 100% and the relative activities with TPA in the presence of compounds are shown. Reporter assays were performed with two independent experiments each in quadruplicate.
Supplementary Figure S4. Control for NSC 676914 shows little anti-proteolysis activity (Fig 3B). HEK293 cells were treated with Mg-132 (10 µM) for 1 hour before treatment with various concentrations of NSC 676914 or DMSO for 1 hour followed by induction with TPA (10 ng/ml) for 18 hours. Endogenous p105, p50 and phospho-IκB-α expression were measured by immunoblot analysis.
Supplementary Figure S5. NSC 676914 decreased the phosphorylation of p65 at Ser536 and Ser276. HEK293 cells were treated with NSC 676914 or DMSO for 1 hour followed by induction with TPA (10 ng/ml) for 18 hours. Endogenous protein expression was measured with whole cell extract by immunoblot analysis. Immunoblots were probed with antibodies for (A) p65, phospho-p65 at Ser536, phospho-p65 at Ser276. B, Semiquantitiative analysis of the phosphorylated p65 using densitometry of the band in each lane from A.
Acknowledgments
We thank members of the Gene Regulation Section, LCP and the MTDP for helpful discussions.
Grant supports: This research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract N01-CO-12400.
Abbreviations
- NSC
National Cancer Institute screen compound
- AP-1
activator protein 1
- JNK
c-Jun N-terminal kinase
- MAPK
mitogen-activated protein kinase
- TPA
12-O-tetradecanoylphorbol-13-acetate
- XTT
2,3-bis (2-methoxy-4-nitro-5-sulfophenyl)-5-[(phenylamino) carbonyl]-2H-tetrazolium hydroxide
Footnotes
Note: The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Silberman S, Janulis M, Schultz RM. Characterization of downstream Ras signals that induce alternative protease-dependent invasive phenotypes. J Biol Chem. 1997;272:5927–5935. doi: 10.1074/jbc.272.9.5927. [DOI] [PubMed] [Google Scholar]
- 2.Dong Z, Crawford HC, Lavrovsky V, et al. A dominant negative mutant of jun blocking 12-O-tetradecanoylphorbol-13-acetate-induced invasion in mouse keratinocytes. Mol Carcinog. 1997;19:204–212. doi: 10.1002/(sici)1098-2744(199707)19:3<204::aid-mc8>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 3.Dong Z, Birrer MJ, Watts RG, Matrisian LM, Colburn NH. Blocking of tumor promoter-induced AP-1 activity inhibits induced transformation in JB6 mouse epidermal cells. Proc Natl Acad Sci U S A. 1994;91:609–613. doi: 10.1073/pnas.91.2.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Young MR, Li JJ, Rincón M, et al. Transgenic mice demonstrate AP-1 (activator protein-1) transactivation is required for tumor promotion. Proc Natl Acad Sci U S A. 1999;96:9827–9832. doi: 10.1073/pnas.96.17.9827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thompson EJ, MacGowan J, Young MR, Colburn N, Bowden GT. A dominant negative c-jun specifically blocks okadaic acid-induced skin tumor promotion. Cancer Res. 2002;62:3044–3047. [PubMed] [Google Scholar]
- 6.Young MR, Farrell L, Lambert P, Awasthi P, Colburn NH. Protection against human papillomavirus type 16-E7 oncogene-induced tumorigenesis by in vivo expression of dominant-negative c-jun. Mol Carcinog. 2002;34:72–77. doi: 10.1002/mc.10050. [DOI] [PubMed] [Google Scholar]
- 7.Cooper SJ, MacGowan J, Ranger-Moore J, Young MR, Colburn NH, Bowden GT. Expression of dominant negative c-jun inhibits ultraviolet B-induced squamous cell carcinoma number and size in an SKH-1 hairless mouse model. Mol Cancer Res. 2003;1:848–854. [PubMed] [Google Scholar]
- 8.Shen Qiang, Uray Ivan P., Li Yuxin, et al. Targeting the Activator Protein 1 Transcription Factor for the Prevention of Estrogen Receptor-Negative Mammary Tumors. Cancer Pre Res. 2008;1:45–55. doi: 10.1158/1940-6207.CAPR-08-0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matthews CP, Birkholz AM, Baker AR, et al. Dominant-negative activator protein 1 (TAM67) targets cyclooxygenase-2 and osteopontin under conditions in which it specifically inhibits tumorigenesis. Cancer Res. 2007;67:2430–2438. doi: 10.1158/0008-5472.CAN-06-0522. [DOI] [PubMed] [Google Scholar]
- 10.Dhar A, Hu J, Reeves R, Resar LM, Colburn NH. Dominant-negative c-Jun (TAM67) target genes: HMGA1 is required for tumor promoter-induced transformation. Oncogene. 2004;23:4466–4476. doi: 10.1038/sj.onc.1207581. [DOI] [PubMed] [Google Scholar]
- 11.Ding J, Li J, Chen J, et al. Effects of polycyclic aromatic hydrocarbons (PAHs) on vascular endothelial growth factor induction through phosphatidylinositol 3-kinase/AP-1-dependent, HIF-1α-independent pathway. J Biol Chem. 2006;281:9093–9100. doi: 10.1074/jbc.M510537200. [DOI] [PubMed] [Google Scholar]
- 12.Young MR, Yang HS, Colburn NH. Promising molecular targets for cancer prevention: AP-1, NF-kappa B and Pdcd4. Trends Mol Med. 2003;9:36–41. doi: 10.1016/s1471-4914(02)00009-6. [DOI] [PubMed] [Google Scholar]
- 13.Ruocco KM, Goncharova EI, Young MR, Colburn NH, McMahon JB, Henrich CJ. A high-throughput cell-based assay to identify specific inhibitors of transcription factor AP-1. J Biomol Screen. 2007;12:133–139. doi: 10.1177/1087057106296686. [DOI] [PubMed] [Google Scholar]
- 14.Li JJ, Rhim JS, Schlegel R, Vousden KH, Colburn NH. Expression of dominant negative Jun inhibits elevated AP-1 and NF-kappaB transactivation and suppresses anchorage independent growth of HPV immortalized human keratinocytes. Oncogene. 1998;16:2711–2721. doi: 10.1038/sj.onc.1201798. [DOI] [PubMed] [Google Scholar]
- 15.Li JJ, Cao Y, Young MR, Colburn NH. Induced expression of dominant-negative c-jun downregulates NFkappaB and AP-1 target genes and suppresses tumor phenotype in human keratinocytes. Mol Carcinog. 2000;29:159–169. doi: 10.1002/1098-2744(200011)29:3<159::aid-mc5>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 16.Karin M, Cao Y, Greten FR, Li ZW. NF-kappaB in cancer: from innocent bystander to major culprit. Nat Rev Cancer. 2002;2:301–310. doi: 10.1038/nrc780. [DOI] [PubMed] [Google Scholar]
- 17.Karin M, Ben-Neriah Y. Phosphorylation meets ubiquitination: the control of NF-kappaB activity. Annu Rev Immunol. 2000;18:621–663. doi: 10.1146/annurev.immunol.18.1.621. [DOI] [PubMed] [Google Scholar]
- 18.Shaulian E, Karin M. AP-1 as a regulator of cell life and death. Nat Cell Biol. 2002;4:E131–E136. doi: 10.1038/ncb0502-e131. [DOI] [PubMed] [Google Scholar]
- 19.Karin M. Nuclear factor-kappaB in cancer development and progression. Nature. 2006;441:431–436. doi: 10.1038/nature04870. [DOI] [PubMed] [Google Scholar]
- 20.Luo JL, Tan W, Ricono JM, et al. Nuclear cytokine-activated IKKalpha controls prostate cancer metastasis by repressing Maspin. Nature. 2007;446:690–694. doi: 10.1038/nature05656. [DOI] [PubMed] [Google Scholar]
- 21.Luo JL, Maeda S, Hsu LC, Yagita H, Karin M. Inhibition of NF-kappaB in cancer cells converts inflammation- induced tumor growth mediated by TNFalpha to TRAIL-mediated tumor regression. Cancer Cell. 2004;6:297–305. doi: 10.1016/j.ccr.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 22.Stein B, Baldwin AS, Jr, Ballard DW, Greene WC, Angel P, Herrlich P. Cross-coupling of the NF-kappa B p65 and Fos/Jun transcription factors produces potentiated biological function. EMBO J. 1993;12:3879–3891. doi: 10.1002/j.1460-2075.1993.tb06066.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fujioka S, Niu J, Schmidt C, et al. NF-kappaB and AP-1 connection: mechanism of NF-kappaB-dependent regulation of AP-1 activity. Mol Cell Biol. 2004;24:7806–7819. doi: 10.1128/MCB.24.17.7806-7819.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lungu ME, Rădiţoiu V, Wagner L, et al. Structural characterization of some dyes derived from 2-(4-aminobenzenesulfonyl)ethanol monosulfate. Revista de Chimie (Bucharest, Romanina) 2004;55:7782–7787. [Google Scholar]
- 25.Eichhorn J, Meier S, Werner R. PCT Int Appl WO2007039573. 2007 [Google Scholar]
- 26.Suzukawa K, Weber TJ, Colburn NH. AP-1, NF-kappa-B, and ERK activation thresholds for promotion of neoplastic transformation in the mouse epidermal JB6 model. Environ Health Perspect. 2002;110:865–870. doi: 10.1289/ehp.02110865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim Y, Fischer SM. Transcriptional regulation of cyclooxygenase-2 in mouse skin carcinoma cells. Regulatory role of CCAAT/enhancer-binding proteins in the differential expression of cyclooxygenase-2 in normal and neoplastic tissues. J Biol Chem. 1998;273:27686–27694. doi: 10.1074/jbc.273.42.27686. [DOI] [PubMed] [Google Scholar]
- 28.Wang Y, Dang J, Wang H, Allgayer H, Murrell GA, Boyd D. Identification of a novel nuclear factor-kappaB sequence involved in expression of urokinase-type plasminogen activator receptor. Eur J Biochem. 2000;267:3248–3254. doi: 10.1046/j.1432-1327.2000.01350.x. [DOI] [PubMed] [Google Scholar]
- 29.Hu Y, Baud V, Delhase M, et al. Abnormal morphogenesis but intact IKK activation in mice lacking the IKKalpha subunit of IkappaB kinase. Science. 1999;284:316–320. doi: 10.1126/science.284.5412.316. [DOI] [PubMed] [Google Scholar]
- 30.Karin M, Yamamoto Y, Wang QM. The IKK NF-kappa B system: a treasure trove for drug development. Nat Rev Drug Discov. 2004;3:17–26. doi: 10.1038/nrd1279. [DOI] [PubMed] [Google Scholar]
- 31.Su B, Karin M. Mitogen-activated protein kinase cascades and regulation of gene expression. Curr Opin Immunol. 1996;8:402–411. doi: 10.1016/s0952-7915(96)80131-2. [DOI] [PubMed] [Google Scholar]
- 32.Burke JR, Pattoli MA, Gregor KR, et al. BMS-345541 is a highly selective inhibitor of I kappa B kinase that binds at an allosteric site of the enzyme and blocks NF-kappa B-dependent transcription in mice. J Biol Chem. 2003;278:1450–1456. doi: 10.1074/jbc.M209677200. [DOI] [PubMed] [Google Scholar]
- 33.Xiao G, Fong A, Sun SC. Induction of p100 processing by NF-kappaB-inducing kinase involves docking IkappaB kinase alpha (IKKalpha) to p100 and IKKalpha-mediated phosphorylation. J Biol Chem. 2004;279:30099–30105. doi: 10.1074/jbc.M401428200. [DOI] [PubMed] [Google Scholar]
- 34.Sakurai H, Chiba H, Miyoshi H, Sugita T, Toriumi W. IkappaB kinases phosphorylate NF-kappaB p65 subunit on serine 536 in the transactivation domain. J Biol Chem. 1999;274:30353–30356. doi: 10.1074/jbc.274.43.30353. [DOI] [PubMed] [Google Scholar]
- 35.Zhong H, SuYang H, Erdjument-Bromage H, Tempst P, Ghosh S. The transcriptional activity of NF-kappaB is regulated by the IkappaB-associated PKAc subunit through a cyclic AMP-independent mechanism. Cell. 1997;89:413–424. doi: 10.1016/s0092-8674(00)80222-6. [DOI] [PubMed] [Google Scholar]
- 36.Vermeulen L, De Wilde G, Van Damme P, Vanden Berghe W, Haegeman G. Transcriptional activation of the NF-kappaB p65 subunit by mitogen- and stress-activated protein kinase-1 (MSK1) EMBO J. 2003;22:1313–1324. doi: 10.1093/emboj/cdg139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang C, Deng L, Hong M, Akkaraju GR, Inoue J, Chen ZJ. TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature. 2001;412:346–351. doi: 10.1038/35085597. [DOI] [PubMed] [Google Scholar]
- 38.Ramanathan M, Pinhal-Enfield G, Hao I, Leibovich SJ. Synergistic up-regulation of vascular endothelial growth factor (VEGF) expression in macrophages by adenosine A2A receptor agonists and endotoxin involves transcriptional regulation via the hypoxia response element in the VEGF promoter. Mol Biol Cell. 2007;18:14–23. doi: 10.1091/mbc.E06-07-0596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Blasi F, Carmeliet P. uPAR: a versatile signalling orchestrator. Nat Rev Mol Cell Biol. 2002;3:932–943. doi: 10.1038/nrm977. [DOI] [PubMed] [Google Scholar]
- 40.Pande V, Ramos MJ. NF-kappaB in human disease: current inhibitors and prospects for de novo structure based design of inhibitors. Curr Med Chem. 2005;12:357–374. doi: 10.2174/0929867053363180. [DOI] [PubMed] [Google Scholar]
- 41.Hideshima T, Chauhan D, Richardson P, et al. NF-kappa B as a therapeutic target in multiple myeloma. J Biol Chem. 2002;277:16639–16647. doi: 10.1074/jbc.M200360200. [DOI] [PubMed] [Google Scholar]
- 42.Bogoyevitch MA, Fairlie DP. A new paradigm for protein kinase inhibition: blocking phosphorylation without directly targeting ATP binding. Drug Discov Today. 2007;12:622–633. doi: 10.1016/j.drudis.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 43.Nagashima K, Sasseville VG, Wen D, et al. Rapid TNFR1-dependent lymphocyte depletion in vivo with a selective chemical inhibitor of IKKbeta. Blood. 2006;107:4266–4273. doi: 10.1182/blood-2005-09-3852. [DOI] [PubMed] [Google Scholar]
- 44.Newton R, Holden NS, Catley MC, et al. Repression of inflammatory gene expression in human pulmonary epithelial cells by small-molecule IkappaB kinase inhibitors. J Pharmacol Exp Ther. 2007;321:734–742. doi: 10.1124/jpet.106.118125. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1. NSC 676914 blocks phosphorylation of IκB-α at 1.1 µM. HEK293 cells were treated with various concentrations of NSC 676914 or DMSO for 1 hour followed by induction with TPA for 18 hours. Endogenous phospho-IκB-α expression was measured by immunoblot analysis.
Supplementary Figure S2. In vitro kinase inhibition assay by the off-chip incubation mobility shift. NSC 676914 was tested against 3 kinase targets, JNK2a2, IKKβ, and IKKα. Assay consisted of testing the compound in duplicate at concentrations of 1.25, 2.50, 5.00, and 10.00 µM. Less than 20% inhibition of binding or enzyme activity is considered inactive.
Supplementary Figure S3. Comparison of NSC 676914 with other IKK inhibitors for kinase and transcription factor inhibition. AP-1, NF-κB, and SRE promoted luciferase inhibition by (A) BMS-345541, (B) SC-514 and (C) sulfasalazine. HEK293 cells were transfected with AP-1, NF-κB, or SRE promoted luciferase reporter. The transfected cells were pre-treated with varied concentrations of BMS-345541, SC-514, sulfasalazine or DMSO for 1 hour before 18 hours stimulation by TPA (10 ng/ml) or DMSO as vehicle. Luciferase activities with TPA induction alone were set at 100% and the relative activities with TPA in the presence of compounds are shown. Reporter assays were performed with two independent experiments each in quadruplicate.
Supplementary Figure S4. Control for NSC 676914 shows little anti-proteolysis activity (Fig 3B). HEK293 cells were treated with Mg-132 (10 µM) for 1 hour before treatment with various concentrations of NSC 676914 or DMSO for 1 hour followed by induction with TPA (10 ng/ml) for 18 hours. Endogenous p105, p50 and phospho-IκB-α expression were measured by immunoblot analysis.
Supplementary Figure S5. NSC 676914 decreased the phosphorylation of p65 at Ser536 and Ser276. HEK293 cells were treated with NSC 676914 or DMSO for 1 hour followed by induction with TPA (10 ng/ml) for 18 hours. Endogenous protein expression was measured with whole cell extract by immunoblot analysis. Immunoblots were probed with antibodies for (A) p65, phospho-p65 at Ser536, phospho-p65 at Ser276. B, Semiquantitiative analysis of the phosphorylated p65 using densitometry of the band in each lane from A.