Summary
The stress response in eukaryotic cells often inhibits translation initiation and leads to the formation of cytoplasmic RNA-protein complexes referred to as stress granules. Stress granules contain non-translating mRNAs, translation initiation components, and many additional proteins affecting mRNA function. Stress granules have been proposed to affect mRNA translation and stability, as well as being linked to apoptosis and nuclear processes. Stress granules also interact with P-bodies, another cytoplasmic RNP granule containing non-translating mRNA, translation repressors and some mRNA degradation machinery. Together, stress granules and P-bodies reveal a dynamic cycle of distinct biochemical and compartmentalized mRNPs in the cytosol, with implications for the control of mRNA function.
Introduction
A key aspect of the control of gene expression is the modulation of cytoplasmic mRNA function. Cytoplasmic mRNAs are controlled by the regulation of mRNA translation, stability, and subcellular location, processes that are often inter-connected. For example, mRNAs are often localized prior to translation (Martin and Ephrussi, 2009), translation initiation and mRNA degradation are often inversely related (Coller and Parker, 2004), and mRNA decay and translation repression mechanisms share similar proteins (Holmes et al, 2004; Coller and Parker, 2005). In eukaryotic cells, non-translating mRNAs can accumulate in two types of cytoplasmic mRNP granules: P-bodies, which generally contain the mRNA decay machinery (reviewed in Anderson and Kedersha, 2006; Parker and Sheth, 2007; Franks and Lykke-Andersen, 2008), and stress granules, which contain many translation initiation components (see below).
The presence of stress granules and P-bodies reveals a dynamic organization of cytoplasmic mRNPs. Moreover, stress granules and P-bodies are related to neuronal RNA granules and germ granules, which play important roles in the localization and control of mRNAs in neurons and embryos, respectively (Figure 1) (Seydoux and Braun, 2006; Kiebler and Bassell, 2006). Herein, we review what is known about the assembly, composition, and possible function of stress granules, and how these roles might fit into the larger picture of cytoplasmic mRNA metabolism.
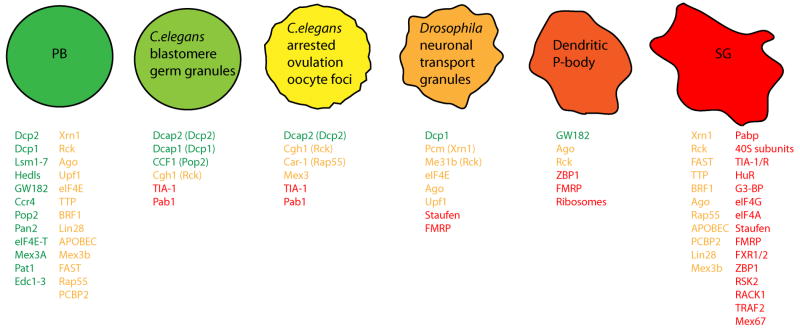
Figure 1. A continuum of mRNP granules.
Select examples of mRNP granules with compositional similarities to both stress granules and P-bodies: C.elegans blastomere germ granules, Gallo et al, 2008; C.elegans arrested ovulation oocyte foci, Jud et al, 2008; Drosophila neuronal transport granules, Barbee et al, 2006; Dendritic P-body, Cougot et al, 2008. Components observed solely in stress granules are highlighted in red, those solely in P-bodies in green, and those seen in both foci are highlighted in yellow. Lists are not necessarily exhaustive, and with specific experimental manipulation, some P-body/stress granule ‘distinct’ components have been observed in both structures.
What are Stress Granules?
Stress granules are cytoplasmic mRNPs that form when translation initiation is impaired, either due to decreased translation initiation rates during a stress response (Kedersha et al, 1999), the addition of drugs blocking translation initiation (Dang et al, 2006; Mazroui et al, 2006; Mokas et al, 2009), knockdown of specific initiation factors (Mokas et al, 2009), or overexpression of RNA binding proteins that repress translation (Mazroui et al, 2002; Gilks et al, 2004; Kedersha et al, 2005; Wilczynska et al, 2005; de Leeuw et al, 2007). Similarly, inducing ribosome-mRNA dissociation with puromycin stimulates stress granule formation, whereas trapping mRNAs in polysomes with drugs that block ribosome elongation inhibits stress granule formation (Kedersha et al, 2000; Buchan et al, 2008). Interestingly, not all initiation blocks induce stress granules. Most notably, knockdown of eukaryotic initiation factor 3 (eIF3) subunits or impairing 60S joining fail to induce stress granules (Ohn et al, 2008; Mokas et al, 2009). This suggests that stress granule formation occurs only when mRNAs are stalled within a defined window of the translation initiation process, and/or that some initiation factors function in stress granule assembly.
The formation of stress granules when translation initiation is inhibited suggests that these granules contain mRNAs stalled in the process of translation initiation, which is consistent with their composition. Stress granules typically contain poly(A)+ mRNA, 40S ribosomal subunits, eIF4E, eIF4G, eIF4A, eIF4B, Poly(A) binding protein (Pabp), eIF3, and eIF2 (Kedersha et al, 1999; 2002; Kimball et al, 2003; Mazroui et al, 2006; Anderson and Kedersha 2006), although the composition can vary. For example, in Saccharomyces cerevisiae, heat shock induced stress granules contain eIF3, whereas glucose deprivation induced stress granules do not (Grousl et al, 2009; Hoyle et al, 2007; Buchan et al, 2008). Depending on experimental conditions, stress granules can also harbor many other protein components including RNA helicases, translation and stability regulators, as well as factors involved in cell signaling (Table S1).
An unresolved issue is the nature of the mRNP complex within stress granules. One possibility is that the mRNAs, translation initiation factors, and 40S ribosomal subunits within stress granules are assembled into a 48S pre-initiation complex. However, many stress responses inhibit translation upstream of 48S complex formation by impairing eIF4E function or via phosphorylation of eIF2, which then limits the formation of a 43S complex containing eIF2, the initiator tRNA, eIF3 and the 40S subunit (Sonenberg and Hinnebusch, 2009). Thus, during most stress responses, mRNAs would stall in translation as mRNPs lacking the 48S complex, and since there is no current evidence for a stress induced block to 60S joining, any mRNAs with a 48S subunit would be expected to join a 60S subunit and complete translation initiation. Moreover, eIF5 is absent from stress granules (Kedersha et al, 2002). Genetic and biochemical studies have identified eIF5 as a component of 48S complexes (Sonenberg and Hinnebusch, 2009), and eIF5 is required for 48S complex assembly, at least in yeast (Asano et al, 2000). Non-phosphorylated eIF2 may also be absent or limiting in stress granules (Kedersha et al, 2002; Kimbal et al, 2003; Kedersha et al, 2005). Taken together, these observations argue that not all mRNAs within stress granules are stalled in translation as a classical 48S complex and suggest three possibilities. First, mRNPs within stress granules may form non-canonical “48S” initiation complexes due to alternative assembly events in initiation being kinetically favored when the “normal” initiation pathway is inhibited. Second, it may be that mRNAs and some translation factors are concentrated in stress granules by distinct mechanisms and not as preassembled “48S” complexes. Finally, stress granules may contain a mix of mRNPs, some forming canonical 48S complexes whilst others lack 40S association.
Interaction of Stress Granules with P-bodies: Evidence for an mRNA Cycle
Several observations demonstrate that stress granules interact with P-bodies and are likely to exchange mRNPs between them. First, mammalian P-bodies and stress granules transiently dock with one another during arsenite treatment and can show prolonged docking when Tristetraprolin (TTP) is overexpressed (Kedersha et al, 2005). Similarly, yeast stress granules often initially form in conjunction with, and partially overlap P-bodies (Brengues and Parker, 2007; Hoyle et al., 2007; Buchan et al., 2008; Grousl et al, 2009). Moreover, due to overexpression or knockdowns of various P-body components, other P-body components re-localize in stress granule-like foci, suggesting these two different mRNPs complexes can now co-associate (Wilczynska et al, 2005; Mollet et al, 2008). Additionally, P-bodies and stress granules share many protein components and the same mRNA species (Kedersha et al., 2005; Hoyle et al., 2007 and Table S1).
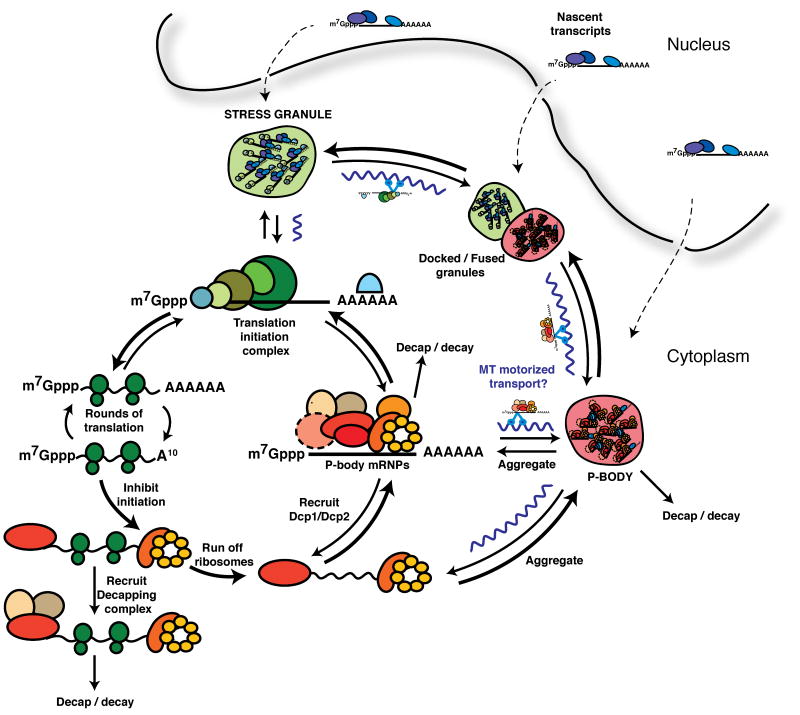
The interaction of P-bodies and stress granules suggests a cytoplasmic mRNP cycle wherein mRNAs exchange between polysomes, P-bodies and stress granules (Figure 2; Parker and Sheth, 2007). Indeed, stress granules are dynamic as FRAP (fluorescent recovery after photobleaching) experiments indicate that the majority of protein and mRNA components examined possess rapid recovery rates (Table S2). Because mRNAs within P-bodies can return to translation (Brengues et al, 2005; Bhattacharyya et al, 2006), one would expect a priori that mRNPs within P-bodies can exchange proteins to form mRNPs competent for translation initiation. These might then accumulate in stress granules, an idea supported by several observations. First, during a glucose deprivation stress response in yeast cells, P-bodies form first, followed by stress granule formation, which initially co-localize with pre-existing P-bodies (Buchan et al., 2008; Hoyle et al., 2007). Similarly, some but not all stress granules in mammalian cells have also been reported to form in association with P-bodies (Mollet et al, 2008) and P-bodies can be induced prior to stress granules (Buchan et al., 2008), although other reports suggest stress granules and P-bodies may form with similar kinetics (Kedersha et al, 2005; Ohn et al, 2008). Second, multiple proteins observed in P-bodies prior to stress, later accumulate in stress granules during stress, with the RNA helicase and translational repressor Rck exhibiting a delayed transition (Mollet et al, 2008; Buchan et al, 2008). No such relocalization of stress granule factors to P-bodies has been observed (Table S1). Finally, mutations decreasing P-body formation in yeast also inhibit stress granule formation during glucose deprivation (but not heat-shock), while mutations increasing P-bodies correspondingly increase stress granules (Buchan et al, 2008; Grousl et al, 2009). Moreover, blocking stress granule assembly in yeast or mammalian cells does not affect P-body assembly (Buchan et al, 2008; Ohn et al, 2008). Taken together, these observations argue that mRNPs within P-bodies can be remodeled and then accumulate within a stress granule, which would imply an important role of mRNP sorting within P-bodies to determine whether an mRNA is stored, degraded, or returns to translation. A precedent for this type of sorting has been described in yeast, as Upf1 targets both normal and nonsense-mediated decay (NMD) target mRNAs to P-bodies, where Upf2 and Upf3 promote decay of NMD targets, whilst Upf1, dependent upon its ATPase activity, aids recycling of normal mRNAs back out of P-bodies (Sheth and Parker, 2006).
Figure 2. Model integrating Stress Granules, and P-bodies, into an mRNP cycle.
A speculative model for mRNP transitions, particularly during stress. Dashed arrows indicate possible destination of exported nascent transcripts. Wavy purple lines represent microtubules, and their possible contribution of dynein/kinesin-mediated motorized transport to granule aggregation, and/or movement of mRNPs between different mRNP states.
mRNA may also be targeted to stress granules directly from polysomes (Figure 2). This is suggested by the observations that stress granules can form spatially independent of P-bodies (Kedersha et al, 2005; Mollet et al, 2008) and that knockdowns of some mammalian proteins leads to decreases in P-bodies without corresponding decreases in stress granules (Ohn et al., 2008). One possibility is that the relative rates of exchange of translation factors for P-body components versus the aggregation of stalled translation initiation complexes determines whether mRNAs accumulate in P-bodies or stress granules when not translating.
Nascent mRNPs exported from the nucleus may also be targeted directly to stress granules or P-bodies. Many nuclear mRNP factors localize in stress granules and P-bodies during stress, including factors involved in transcription, 3′end processing, splicing and export (Table S1), which might affect nuclear events. For example, relocalization of mammalian hnRNP A1 to stress granules during stress (Guil et al, 2006) may explain changes in alternative splicing (van der Houven van Oordt et al, 2000). Stress granules and P-bodies could also modulate, or participate in mRNP export remodeling, given that the RNA helicases Dbp5 and DDX3, which are implicated in this process (Lund and Guthrie, 2005; Yedavalli et al, 2004) are known or likely stress granule and P-body components (Beckham et al, 2008; Scarcelli et al, 2008; Lai et al, 2008). Finally, impairing stress granule or P-body assembly can affect movement of various stress granule or P-body factors to the nucleus (Eisinger-Mathason et al, 2008; Grousl et al, 2009). These observations suggest a possible role for stress granules and P-bodies in remodeling, storage or decay of exported mRNAs, and/or a coupling of cytoplasmic and nuclear gene expression control mechanisms.
The interaction and likely exchange of mRNA between P-bodies and stress granules suggests a continuum of mRNP states between polysomes, P-bodies and stress granules, reflecting different protein compositions in the mRNP due to remodeling events. Thus, the observed composition of each granule, and the location of any given mRNA, will reflect the rate limiting steps in mRNP exchanges; for example, stalls at different initiation steps may lead to mRNAs localizing in different granules. Supporting the idea of an mRNP continuum, the composition and morphology of stress granules and P-bodies, in both yeast and mammals varies under different stresses and experimental conditions, with ‘hybrid’ granules not uncommon (Kedersha et al, 1999; Stoecklin et al, 2004; Serman et al, 2007; Buchan et al 2008; Grousl et al, 2009; Thomas et al, 2009). Granule composition can also vary over time (Mollet et al, 2008, Buchan et al, 2008; Kedersha et al, 2005), and can be heterogeneous i.e. certain proteins only show partial co-localization with other stress granule / P-body marker proteins (Touierre et al, 2003; Tsai et al, 2008). Finally, other related mRNP foci such as germ granules or neuronal transport granules often appear as compositional hybrids of stress granules and P-bodies (Figure 1), possibly reflecting regulation of function at an intermediate stage in the remodeling process.
Stress Granule Assembly
In addition to a pool of mRNAs stalled at a step in translation initiation, stress granule assembly is affected by at least three other factors. The first of these is protein modification, which also regulates the interaction and function of many stress granule mRNP components. For example, phosphorylation of eIF2α underlies the decrease in translation initiation required for stress granule assembly in many stress responses (Wek et al, 2006), while phosphorylation of TTP, BRF1, and G3BP reduces their accumulation in stress granules (Stoecklin et al, 2004; Schmidlin et al, 2004; Gallouzi et al. 1998; Tourriere et al. 2001, 2003). Acetylation also affects stress granules. HDAC6 deacetylase mutants are impaired in stress granule formation, and although the target of this activity is unclear, two feasible candidates include Hsp90 and microtubules (Kwon et al, 2007). Stress granules contain ubiquitin-modified proteins (Kwon et al, 2007) and mutations in HDAC6's ubiquitin binding domain and knockdowns of several factors implicated in ubiquitin metabolism affect stress granule formation (Ohn et al, 2008). Modification of proteins with O-Glc-NAc also enhances stress granule formation (Ohn et al, 2008). Finally, methylation, or the ability to bind methyl groups via Tudor domains, is necessary for localization of specific stress granule components (de Leeuw et al, 2007; Goulet et al, 2008), or their ability to drive stress granule formation when overexpressed (Hua and Zhou, 2004). Methylation and Tudor domains have also been implicated in the assembly of other RNA granules (Thomson and Lasko, 2004; Arkov et al, 2006; Chuma et al, 2006).
Post-translational modification of mRNP components is an ideal mechanism to modulate mRNA function during a stress, where rapid and reversible protein modifications allow adaptation to stress without new protein synthesis. Elucidating the key physiological targets of various modifications, and the mechanisms underlying their effects, will therefore be an important future goal.
A second aspect of stress granule assembly are protein-protein interaction domains present on numerous RNA binding proteins. For example, the G3BP protein has a dimerization domain that contributes to stress granule formation during arsenite stress (Tourriere et al, 2003). Moreover, several proteins involved in RNA metabolism contain QN-rich prion-like domains and the ability of those domains to self-aggregate can promote stress granule assembly. For example, the RNA binding proteins TIA-1 and TIA-R, and their orthologs, are found in stress granules and contain a conserved QN-rich domain. Moreover, TIA-1 lacking its QN-rich domain cannot support stress granule formation, though fusion of the yeast SUP35 prion domain in its place recovers TIA-1 stress granule assembly function (Gilks et al, 2004). Conversely, overexpression of TIA-1's QN-rich domain inhibits normal stress granule assembly by generating constitutive micro-aggregates that sequester endogenous TIA proteins (Kedersha et al, 1999; Kedersha et al, 2000; Gilks et al, 2004). The role of QN-rich domains in organizing mRNA metabolism may be quite broad since QN-rich domains also aid P-body assembly and almost half of the 107 QN-rich domain containing proteins in yeast function in RNA-related processes such as transport, translation or degradation (Decker et al, 2007; Reijins et al, 2008). Additionally, stress granule assembly is modulated by heat shock proteins, which disassemble prion aggregates (Rikhvanov et al, 2007) and inhibit stress granule formation when overexpressed (Gilks et al, 2004; Mazroui et al, 2007). Since aggregation of QN-rich prion domains is reversed by specific heat shock protein function (Rikhvanov et al, 2007), stress granule assembly may be promoted during stress due to accumulation of unfolded proteins, which may titrate heat shock proteins, thus driving the equilibrium of QN-rich domains towards an aggregated state.
The microtubule network is the third contributor to stress granule assembly. Microtubule depolymerizing drugs such as nocodazole inhibit stress granule formation, although smaller stress granules generally still form (Ivanov et al, 2003; Kwon et al, 2007; Kolobova et al, 2009; Fujimura et al, 2009; Loschi et al, 2009). Dynein and Kinesin motor proteins are also observed in stress granules, and knockdown experiments have suggested roles in stimulating assembly and disassembly of large stress granules respectively (Loschi et al, 2009). Dynein inhibition or knockdown also increases protease sensitivity of TIA-1 aggregates providing additional evidence for a role in stress granule formation (Kwon et al, 2007; Tsai et al, 2009). Once assembled, stress granules do not require microtubules for their persistence (Fujimura et al, 2009). Interestingly, mobile P-bodies associate with microtubules and their movement is dependent on intact microtubules (Aizer et al., 2008). However, disruption of microtubules increases P-body formation in yeast and mammals (Aizer et al, 2008; Sweet et al., 2007), which argues that microtubules serve different roles in the formation of stress granules and P-bodies. Curiously, P-bodies appear unaffected by dynein knockdown under non-stress conditions, but stress-induced increases are attenuated, suggesting different assembly mechanisms operate under different cellular conditions (Loschi et al, 2009).
The exact role of microtubules in stress granule assembly remains unclear. Since stress granules are relatively non-mobile compared to P-bodies (Kedersha et al, 2005), the assembly defects caused by microtubule disruption may partially reflect impaired mRNP transport in and out of stress granules. One possible model is that when mRNAs exit polysomes, they can assemble directly into P-bodies, which are often associated with microtubules, and then movement of mRNPs from P-bodies to stress granules occurs in conjunction with microtubules. Another possible model is that microtubules provide a surface to concentrate both the translationally inactive mRNPs and translation initiation factors that then facilitate stress granule formation. In this light, it is striking that eIF3 is required for stress granule formation in mammals (Ohn et al, 2008), contains a microtubule binding protein (Hasek et al, 2000) and co-localizes and co-IPs with microtubule proteins (Hasek et al, 2000; Kwon et al, 2007). This suggests a possible model whereby microtubules may play an important role in forming stress granules by independently concentrating untranslating mRNAs (either within or outside of P-bodies), and translation initiation factors. Such a role might then promote translation initiation during stress (see below).
Several observations suggest that stress granule assembly is variable and dynamic and depends on the types of mRNPs present within the granule, whose components likely interact in multiple and stress specific manners. First, stress granule formation can be driven by overexpression of multiple factors, and conversely impaired by depletion of many factors (Ohn et al, 2008; Table S1). Additionally, most factors localize very transiently in stress granules (Table S2), arguing against a rigid structural compartment. Moreover, stress granule shape and size varies significantly over time, and lacks any obvious structural organization as assessed by electron microscopy (Gilks et al, 2004). Finally, assembly factors important under one stress condition are frequently unimportant during other stresses. For example, TIA-1, and its yeast homolog Pub1, facilitate stress granule assembly in response to arsenite and glucose deprivation respectively (Gilks et al, 2004; Buchan et al, 2008), but not in response to other stresses such as heatshock (Lopez de Silanes, 2005; Grousl et al, 2009). Therefore, the nature of the stress, which shapes the non-translating mRNP pool, likely defines the assembly rules for stress granules.
Disassembly of Stress Granules
During recovery from stress, stress granules disassemble in a manner that roughly correlates with the recovery of bulk protein synthesis (Mazroui et al, 2007), as well as translation of individual mRNAs (Lian and Gallouzi, 2009; Tsai et al, 2008), although complete disassembly of stress granules may not be required for translational recovery (Loschi et al, 2009). In principle, stress granules could be disassembled by dissociation of the interactions creating the larger aggregate, by degrading the stress granule mRNA pool in situ or after transfer to a P-body, or by removal of mRNAs from stress granules by entry into polysomes. Interestingly, the rate of stress granule disassembly is increased by treatment with emetine or cycloheximide (Kedersha et al, 2000; Mazroui et al, 2002; Mollet et al, 2008), which suggests that even during recovery from stress some mRNAs are still entering stress granules and there is a dynamic exchange between translating and stress granule pools of mRNAs.
A few RNA binding proteins promote stress granule disassembly. Staufen is a stress granule/neuronal granule/germ granule component (Table S1), whose knockdown facilitates stress granule formation (Thomas et al, 2009), while moderate overexpression of Staufen inhibits stress granule formation. Staufen appears to stabilize mRNA-polysome association (Thomas et al, 2009), though whether it prevents entry into stress granules, or facilitates exit into polysomes is unclear. In another example, phosphorylation of Grb7 by Focal adhesion kinase (FAK) during stress recovery is necessary to weaken interactions with other stress granule components such as HuR and TIA-1, as well as binding to specific mRNAs (Tsai et al, 2008). The inability to phosphorylate Grb7 during stress recovery impairs stress granule disassembly, suggesting maintenance of these interactions may underlie this disassembly defect (Tsai et al, 2008).
Assembly and disassembly of stress granules may also be influenced by complex autoregulatory loops of the key factors. For example, despite TIA-R being a recognized translational repressor (Gueydan et al, 1999; Mazam-Mamczarz et al, 2006), both its overexpression and knockdown increase stress granule formation (Gilks et al, 2004; de Leeuw et al, 2007). Such positive and negative assembly roles for TIA-R could be determined by cellular conditions, by its overall concentration, or be a consequence of altered regulation of other stress granule assembly factors, such as TIA-1, whose translation is repressed by TIA-R (Pullman et al., 2007). In contrast, the RNA binding protein HuR, which also accumulates in stress granules, enhances TIA-1 expression (Pullman et al., 2007). Given these types of regulatory circuits, care must be taken in interpreting mutant phenotypes without a clear understanding of the underlying mechanism.
Stress Granule Function
Stress granules have been hypothesized to function in repression, given that numerous stress granule components are translational repressors, and their formation correlates with decreased global translation (Anderson and Kedersha, 2009). However, formation of stress granules is clearly not required for global translation repression (Kwon et al, 2007; Buchan et al 2008; Ohn et al 2008; Mokas et al, 2009; Fujimura et al, 2009; Loschi et al, 2009). Some specific mRNAs are inefficiently repressed when RNA binding proteins that contribute to stress granule formation are altered (Moeller et al, 2004; Kedersha et al, 2000; Gilks et al 2004; Tsai et al, 2008; Mazroui et al, 2007), but these effects may simply reflect loss of a specific mRNP regulatory component rather than failure to assemble a granule per se. Thus, at the current time, the available evidence suggests that the majority of the translation status of an mRNA is determined by its specific mRNP, and not by its aggregation into stress granules.
Stress granules have also been proposed to function to stabilize mRNAs. During a wide variety of stress responses, mRNA deadenylation, which is a prerequisite for most mRNA degradation, is broadly inhibited (Laroia et al., 1999; Hilgers et al, 2006; Gowrishankar et al, 2006). However, at least in yeast, mutations that prevent stress granule formation do not affect the stabilization of mRNAs during stress (Buchan et al., 2008). Moreover, deadenylation is inhibited during stress even when the mRNA is trapped in polysomes (Hilgers et al., 2006). These results suggest that stress granules are not required for the global stabilization of mRNAs that occur during stress.
Why then do mRNPs aggregate into stress granules? A key point is that the formation of stress granules will lead to a higher local concentration of their components in stress granules and a corresponding lower concentration in the remainder of the cytosol, which has two general affects. First, the concentration of mRNAs and associated proteins into stress granules will reduce the concentration of those molecules in the cytosol thereby altering the interactions and rates of biochemical reactions. For example, as discussed below, the sequestration of the RACK protein into stress granules alters the activation of the MTK1 kinase during stress and thereby affects whether cells enter apoptosis. Similarly, although global control of translational repression or mRNA stability does not depend upon stress granule assembly (see earlier), aggregation of a subset of mRNAs, or mRNP components within stress granules might in principle limit the interaction of some mRNAs with degradation enzymes or polysomes.
A second consequence of stress granules formation is that the higher local concentration of components in a stress granule is likely to increase the rates of mRNP assembly or remodeling driven by these factors. A precedent for this possibility comes from the study of Cajal bodies, which are nuclear structures involved in the assembly and biogenesis of small nuclear ribonucleoproteins (snRNPs), and whose presence is suggested to increase the rate of snRNP assembly by 10 fold (Klingauf et al., 2006). This raises the possibility that stress granules form to promote assembly of translation initiation complexes by increasing the local concentration of mRNAs and translation factors, though translation itself is unlikely to occur in stress granules given the absence of 60S subunits, and mRNA species which are translated during stress (Kedersha and Anderson, 2002). Assembly of initiation complexes may be especially important during stress when certain translational resources are limiting. Additionally, concentration of various mRNP regulators may also be important in promoting the translation of specific mRNAs that are preferentially translated during stress. Consistent with that model, stress granules contain several factors that often promote translation in a stress-specific manner (e.g. HuR, Lin28, DDX3/Ded1, Pbp1 – Table S1). In a larger sense, the dynamic concentration of molecules to enhance reaction rates may be a more general property of cells as many metabolic enzymes form complexes under nutrient starvation that may affect the rates of biochemical reactions within the cell (Narayanaswamy et al., 2009).
mRNAs that are preferentially translated during stress tend to initiate translation by non-canonical mechanisms. For example, mRNAs that contain internal ribosome entry sites (IRES), which recruit translation factors and the ribosome in a cap independent manner, are often preferentially translated during stress (Spriggs et al, 2008). Recent estimates suggest that 10-15% of cellular transcripts in cell lines possess IRES activity, whose translation may vary depending on the stress condition (Spriggs et al, 2008). IRESs rely on various trans-acting factors for ribosomal recruitment, such as stress granule components hnRNP A1 (Guil et al, 2006) and PCBP2 (Fujimura et al, 2008), which bind to IRESs and promote translation (Bonnal et al, 2004; Bedard et al, 2004). Thus, an interesting possibility is that formation of stress granules is required to allow optimal translation of stress responsive mRNAs.
Stress Granules and Apoptosis
Stress granule formation appears to play a role during stress responses in the decision of whether to enter apoptosis, which occurs when a stress is too extreme and the cell is unable to recover. Stress granules, which harbor several apoptosis regulatory factors (Table S1), seem to provide a protective role during stress since impairing stress granule assembly often leads to poorer cell survival rates following stress exposure (Baguet et al, 2007; Kwon et al, 2007; Eisinger-Mathason et al, 2008).
Sequestration of apoptotic regulatory proteins in stress granules can prevent interactions with other factors that would otherwise promote apoptosis in response to a given stress. For example, severe apoptosis-inducing stress strongly activates MTK1 kinase. This activation process is facilitated by interaction with RACK1. However, during modest stress, from which cells can recover, RACK1 is sequestered in stress granules, dependent on its ability to bind 40S subunits. This limits MTK1 activation and apoptosis is avoided (Arimoto et al, 2008). Driving stress granule assembly by G3BP overexpression also inhibits MTK1 activation and increases apoptotic resistance (Arimoto et al, 2008). Similarly, sequestration of TNF-α receptor associated factor 2 (TRAF2) in heatshock-induced stress granules, via eIF4GI interaction, impairs TNF-α-mediated activation of NF-κB, a key transcriptional regulator of inflammatory responses and apoptosis (Kim et al, 2005).
Apoptosis regulation involving stress granule assembly may also link and directly impact upon mRNP regulation. For example, Ribosomal S6 kinase 2 (RSK2) and FAST kinase are both anti-apoptotic factors that localize in stress granules, and that directly bind the QN-rich domain of TIA-1. RSK2 additionally regulated the localization and concentration of both TIA-1 and Pabp in stress granules (Eisinger-Mathason et al, 2008). Interestingly, a domain present in FAST that inhibits caspase-3 activation is nullified upon TIA-1 binding, which may partly underlie the pro-apoptotic nature of TIA-1 (Li et al, 2004). Conversely, overexpression or increased release of FAST from the mitochondrial membrane promotes expression of TIA-1-repressed mRNA reporters and stimulates expression of anti-apoptotic factors. This too depends on the ability to bind TIA-1, suggesting these two proteins are apoptosis antagonists (Li et al, 2004). Binding by FAST could antagonize TIA-1 promoted apoptosis via phosphorylation (Tian et al, 1995), or it could impair mRNA binding (Yu et al, 2007), alter mRNP composition, or negate TIA-1 QN-rich domain activity, thus affecting stress granule localization or assembly. Interestingly, RSK2 and FAST are two rare examples of near-static stress granule components (Table S2), which combined with interactions with other stress granule factors, suggests a possible scaffolding role in stress granule assembly (Eisinger-Mathason et al, 2008; Kedersha et al, 2005).
In summary, the role of stress granules in controlling apoptosis could be to sequester and nullify apoptosis-promoting factors, and simultaneously link appropriate mRNP regulation to this decision process.
Concluding Remarks: A Working Model for Stress Granule Function
Taken together, the above observations suggest a working model for stress granule formation, function, and disassembly with the following key points. First, when steps in translation initiation are compromised, the resulting mRNPs that form can aggregate into stress granules. The mRNAs that contribute to stress granule formation can be mRNAs re-entering translation from P-bodies, and may also be nascent transcripts or those directly exiting polysomes. Second, the assembly and disassembly of stress granules is partially reliant on microtubules and is mediated by protein-protein interactions on RNA binding proteins, many of which are modulated by stress induced modifications. Third, the formation of stress granules both creates a high local concentration of factors and depletes them from the cytosol, which may preferentially increase, or decrease the rates of specific reactions, mRNP associated or otherwise. A key implication of this type of model is that changes in an mRNP that affects its accumulation within stress granules, or even P-bodies, have the potential to alter the translation and/or degradation of the mRNA. However, the ability to form aggregated stress granules per se, despite harboring a diverse array of mRNP regulators, does not play a global role in causing translational repression or mRNA stabilization. Possible roles in controlling the translation and degradation of specific mRNAs, as well as possibly enhancing the assembly of translation initiation complexes remain possible functions for stress granule formation, which are yet to be carefully addressed.
There are many other unresolved and interesting issues with regards to stress granules. For example, stress granules, and their components, have been implicated in viral infection (Beckham and Parker, 2008), inflammatory disease (Anderson, 2008), cancer (Arimoto et al, 2008; Moeller et al, 2004) and multiple neurological diseases including Fragile X syndrome (Vanderklish and Edlemann, 2005), Spinal Muscular Atrophy (McWhorter et al, 2003), Spinocerebellar Ataxia 2 (Nonhoff et al, 2007), and Myotonic Dystrophy (Ranum and Cooper 2006. Determining any role of stress granules in these pathologies will be important. It will also be important, perhaps by following single mRNA molecules, to determine the pathways that mRNAs follow between these different subcellular compartments and how those transitions are modulated in an mRNA specific manner to affect either translation or degradation of the mRNA. Finally, understanding the nature of the mRNP complexes that form within stress granules is likely to provide insight into their function.
Supplementary Material
Acknowledgments
The authors thank members of the Parker lab, particularly Angie Hilliker and Carolyn Decker, for critical review of the manuscript, as well as Mark Ashe, and the reviewers for helpful comments. We apologize for not being able to cite all relevant works due to space constraints.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference list
- Aizer A, Brody Y, Ler LW, Sonenberg N, Singer RH, Shav-Tal Y. Mol Biol Cell. 2008;19:4154–4166. doi: 10.1091/mbc.E08-05-0513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P. Post-transcriptional control of cytokine production. Nat Immunol. 2008;9:353–359. doi: 10.1038/ni1584. [DOI] [PubMed] [Google Scholar]
- Anderson P, Kedersha N. RNA granules. J Cell Biol. 2006;172:803–808. doi: 10.1083/jcb.200512082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P, Kedersha N. RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat Rev Mol Cell Biol. 2009;10:430–436. doi: 10.1038/nrm2694. [DOI] [PubMed] [Google Scholar]
- Arimoto K, Fukuda H, Imajoh-Ohmi S, Saito H, Takekawa M. Formation of stress granules inhibits apoptosis by suppressing stress-responsive MAPK pathways. Nat Cell Biol. 2008;10:1324–1332. doi: 10.1038/ncb1791. [DOI] [PubMed] [Google Scholar]
- Arkov AL, Wang JY, Ramos A, Lehmann R. The role of Tudor domains in germline development and polar granule architecture. Development. 2006;133:4053–4062. doi: 10.1242/dev.02572. [DOI] [PubMed] [Google Scholar]
- Asano K, Clayton J, Shalev A, Hinnebusch AG. A multifactor complex of eukaryotic initiation factors, eIF1, eIF2, eIF3, eIF5, and initiator tRNA(Met) is an important translation initiation intermediate in vivo. Genes Dev. 2000;14:2534–2546. doi: 10.1101/gad.831800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baguet A, Degot S, Cougot N, Bertrand E, Chenard MP, Wendling C, Kessler P, Le Hir H, Rio MC, Tomasetto C. The exon-junction-complex-component metastatic lymph node 51 functions in stress-granule assembly. J Cell Sci. 2007;120:2774–2784. doi: 10.1242/jcs.009225. [DOI] [PubMed] [Google Scholar]
- Beckham CJ, Hilliker A, Cziko AM, Noueiry A, Ramaswami M, Parker R. The DEAD-box RNA helicase Ded1p affects and accumulates in Saccharomyces cerevisiae P-bodies. Mol Biol Cell. 2008;19:984–993. doi: 10.1091/mbc.E07-09-0954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckham CJ, Parker R. P-bodies, stress granules, and viral life cycles. Cell Host Microbe. 2008;17:206–212. doi: 10.1016/j.chom.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedard KM, Walter BL, Semler BL. Multimerization of poly(rC) binding protein 2 is required for translation initiation mediated by a viral IRES. RNA. 2004;12:1266–1276. doi: 10.1261/rna.7070304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 125:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- Bonnal S, Pileur F, Orsini C, Parker F, Pujol F, Prats AC, Vagner S. Heterogenous nuclear ribonucleoprotein A1 is a novel internal ribosome entry site trans-acting factor that modulates alternative initiation of translation of the fibroblast growth factor 2 mRNA. J Biol Chem. 2005;280:4144–4153. doi: 10.1074/jbc.M411492200. [DOI] [PubMed] [Google Scholar]
- Brengues M, Sheth U, Parker R. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science. 2005;310:486–489. doi: 10.1126/science.1115791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brengues M, Parker R. Accumulation of polyadenylated mRNA, Pab1p, eIF4E, and eIF4G with P-bodies in Saccharomyces cerevisiae. Mol Biol Cell. 2007;18:2592–2602. doi: 10.1091/mbc.E06-12-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR, Muhlrad D, Parker R. P bodies promote stress granule assembly in Saccharomyces cerevisiae. J Cell Biol. 2008;183:441–455. doi: 10.1083/jcb.200807043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuma S, Hosokawa M, Kitamura K, Kasai S, Fujioka M, Hiyoshi M, Takamune K, Noce T, Nakatsuji N. Tdrd1/Mtr-1, a tudor-related gene, is essential for male germ-cell differentiation and nuage/germinal granule formation in mice. PNAS. 2006;103:15894–15899. doi: 10.1073/pnas.0601878103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller J, Parker R. Eukaryotic mRNA decapping. Annu Rev Biochem. 2004;73:861–890. doi: 10.1146/annurev.biochem.73.011303.074032. [DOI] [PubMed] [Google Scholar]
- Coller J, Parker R. General translational repression by activators of mRNA decapping. Cell. 2005;136:719–730. doi: 10.1016/j.cell.2005.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang Y, Kedersha N, Low WK, Romo D, Gorsope M, Kaufman R, Anderson P, Liu JO. Eukaryotic initiation factor 2alpha-independent pathway of stress granule induction by the natural product pateamine A. J Biol Chem. 2006;281:32870–32878. doi: 10.1074/jbc.M606149200. [DOI] [PubMed] [Google Scholar]
- Decker CJ, Teixeira D, Parker R. Edc3p and a glutamine/asparagines-rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae. J Cell Biol. 2007;179:437–449. doi: 10.1083/jcb.200704147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Leeuw F, Zhang T, Wauquier C, Huez G, Kruys V, Gueydan C. The cold-inducible RNA-binding protein migrates from the nucleus to cytoplasmic stress granules by a methylation-dependent mechanism and acts as a translational repressor. Exp Cell Res. 2007;313:4130–4144. doi: 10.1016/j.yexcr.2007.09.017. [DOI] [PubMed] [Google Scholar]
- Eisinger-Mathason TS, Andrade J, Groehler AL, Clark DE, Muratore-Schroeder TL, Pasic L, Smith JA, Shabanowitz J, Hunt DF, Macara IG, et al. Codependent functions of RSK2 and the apoptosis-promoting factor TIA-1 in stress granule assembly and cell survival. Mol Cell. 2008;31:722–736. doi: 10.1016/j.molcel.2008.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks TM, Lykke-Andersen J. The control of mRNA decapping and P-body formation. Mol Cell. 2008;32:605–615. doi: 10.1016/j.molcel.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimura K, Kano F, Murata M. Identification of PCBP2, a facilitator of IRES-mediated translation, as a novel constituent of stress granules and processing bodies. RNA. 2008;14:425–431. doi: 10.1261/rna.780708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimura K, Katahira J, Kano F, Yoneda Y, Murata M. Microscopic dissection of the process of stress granule assembly. Biochim Biophys Acta. 2009;1793:1728–1737. doi: 10.1016/j.bbamcr.2009.08.010. [DOI] [PubMed] [Google Scholar]
- Gallouzi IE, Parker F, Chebli K, Maurier F, Labourier E, Barlat I, Capony JP, Tocque B, Tazi J. A novel phosphorylation-dependent RNase activity of GAP-SH3 binding protein: a potential link between signal transduction and RNA stability. Mol Cell Biol. 1998;18:3956–3965. doi: 10.1128/mcb.18.7.3956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilks N, Kedersha N, Ayodele M, Shen L, Stoecklin G, Dember LM, Anderson P. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol Biol Cell. 2004;15:5383–5398. doi: 10.1091/mbc.E04-08-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulet I, Boisvenue S, Mokas S, Mazroui R, Côté J. TDRD3, a novel Tudor domain-containing protein, localizes to cytoplasmic stress granules. Hum Mol Genet. 2008;17:3055–3074. doi: 10.1093/hmg/ddn203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowrishankar G, Winzen R, Dittrich-Breiholz O, Redich N, Kracht M, Holtmann H. Inhibition of mRNA deadenylation and degradation by different types of cell stress. Biol Chem. 2006;387:323–327. doi: 10.1515/BC.2006.043. [DOI] [PubMed] [Google Scholar]
- Grousl T, Ivanov P, Frydlova I, Vasicova P, Janda F, Vojtova J, Malinska K, Malcova I, Novakova L, Janoskova D, et al. Robust heat shock induces eIF2alpha-phosphorylation-independent assembly of stress granules containing eIF3 and 40S ribosomal subunits in budding yeast, Saccharomyces cerevisiae. J Cell Sci. 2009;122:2078–2088. doi: 10.1242/jcs.045104. [DOI] [PubMed] [Google Scholar]
- Gueydan C, Droogmans L, Chalon P, Huez G, Caput D, Kruys V. Identification of TIAR as a protein binding to the translational regulatory AU-rich element of tumor necrosis factor alpha mRNA. J Biol Chem. 1999;274:2322–2326. doi: 10.1074/jbc.274.4.2322. [DOI] [PubMed] [Google Scholar]
- Guil S, Long JC, Cáceres JF. hnRNP A1 relocalization to the stress granules reflects a role in the stress response. Mol Cell Biol. 2006;26:5744–5758. doi: 10.1128/MCB.00224-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasek J, Kovarik P, Valásek L, Malínská K, Schneider J, Kohlwein SD, Ruis H. Rpg1, the subunit of the Saccharomyces cerevisiae eIF3 core complex, is a microtubule-interactig protein. Cell Motil Cytoskeleton. 2000;45:235–246. doi: 10.1002/(SICI)1097-0169(200003)45:3<235::AID-CM6>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Hilgers V, Teixeira D, Parker R. Translation-independent inhibition of mRNA deadenylation during stress in Saccharomyces cerevisiae. RNA. 2006;12:1835–1845. doi: 10.1261/rna.241006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes LE, Campbell SG, De Long SK, Sachs AB, Ashe MP. Loss of translational control in yeast compromised for the major mRNA decay pathway. Mol Cell Biol. 2004;24:2998–3010. doi: 10.1128/MCB.24.7.2998-3010.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyle NP, Castelli LM, Campbell SG, Holmes LE, Ashe MP. Stress-dependent relocalization of translationally primed mRNPs to cytoplasmic granules that are kinetically and spatially distinct from P-bodies. J Cell Biol. 2007;179:65–74. doi: 10.1083/jcb.200707010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Y, Zhou J. Survival motor neuron protein facilitates assembly of stress granules. FEBS Lett. 2004;572:69–74. doi: 10.1016/j.febslet.2004.07.010. [DOI] [PubMed] [Google Scholar]
- Ivanov PA, Chudinova EM, Nadezhdina ES. Disruption of microtubules inhibits cytoplasmic ribonucleoprotein stress granule formation. Exp Cell Res. 2003;290:227–233. doi: 10.1016/s0014-4827(03)00290-8. [DOI] [PubMed] [Google Scholar]
- Kedersha NL, Gupta M, Li W, Miller I, Anderson P. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J Cell Biol. 1999;147:1431–1342. doi: 10.1083/jcb.147.7.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Cho MR, Li W, Yacono PW, Chen S, Gilks N, Golan DE, Anderson P. Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. J Cell Biol. 2000;151:1257–1268. doi: 10.1083/jcb.151.6.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Anderson P. Stress granules: Sites of mRNA triage that regulate mRNA stability and translatability. Biochem Soc Trans. 2002;30:963–969. doi: 10.1042/bst0300963. [DOI] [PubMed] [Google Scholar]
- Kedersha N, Chen S, Gilks N, Li W, Miller IJ, Stahl J, Anderson P. Evidence that ternary complex (eIF2-GTP-tRNA(i)(Met))-deficient preinitiation complexes are core constituents of mammalian stress granules. Mol Biol Cell. 2002;13:195–210. doi: 10.1091/mbc.01-05-0221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Stoecklin G, Ayodele M, Yacono P, Lykke-Andersen J, Fritzler MJ, Scheuner D, Kaufman RJ, Golan DE, Anderson P. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J Cell Biol. 2005;169:871–884. doi: 10.1083/jcb.200502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiebler MA, Bassel GJ. Neuronal granules: Movers and makers. Neuron. 2006;51:685–690. doi: 10.1016/j.neuron.2006.08.021. [DOI] [PubMed] [Google Scholar]
- Kim WJ, Back SH, Kim V, Ryu I, Jang SK. Sequestration of TRAF2 into stress granules interrupts tumor necrosis factor signaling under stress condition. Mol Cell Biol. 2005;25:2450–2462. doi: 10.1128/MCB.25.6.2450-2462.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimball SR, Horetsky RL, Ron D, Jefferson LS, Harding HP. Mammalian stress granules represent sites of accumulation of stalled translation initiation complexes. Am J Physiol Cell Physiol. 2003;284:273–284. doi: 10.1152/ajpcell.00314.2002. [DOI] [PubMed] [Google Scholar]
- Klingauf M, Stanek D, Neugebauer KM. Enhancement of U4/U6 small nuclear ribonucleoprotein particle association in Cajal bodies predicted by mathematical modeling. Mol Biol Cell. 2006;17:4972–4981. doi: 10.1091/mbc.E06-06-0513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolobova E, Efimov A, Kaverina I, Rishi AK, Schrader JW, Ham AJ, Larocca MC, Goldenring JR. Microtubule-dependent association of AKAP350A and CCAR1 with RNA stress granules. Exp Cell Res. 2009;315:542–555. doi: 10.1016/j.yexcr.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon S, Zhang Y, Matthias P. The deacetylase HDAC6 is a novel critical component of stress granules involved in the stress response. Genes Dev. 2007;15:3381–3394. doi: 10.1101/gad.461107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai MC, Lee YH, Tarn WY. The DEAD-box RNA helicase DDX3 associates with export messenger ribonucleoproteins as well as tip-associated protein and participates in translational control. Mol Biol Cell. 2008;19:3847–3858. doi: 10.1091/mbc.E07-12-1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laroia G, Cuesta R, Brewer G, Schneider RJ. Control of mRNA decay by heat shock-ubiquitin-proteasome pathway. Science. 1999;284:499–502. doi: 10.1126/science.284.5413.499. [DOI] [PubMed] [Google Scholar]
- Li W, Simarro M, Kedersha N, Anderson P. FAST is a survival protein that senses mitochondrial stress and modulates TIA-1-regulated changes in protein expression. Mol Cell Biol. 2004;24:10178–10732. doi: 10.1128/MCB.24.24.10718-10732.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian XJ, Gallouzi IE. Oxidative stress increases the number of stress granules in senescent cells and triggers a rapid decrease in p21waf1/cip1 translation. J Biol Chem. 2009;284:8877–8887. doi: 10.1074/jbc.M806372200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López de Silanes I, Galbán S, Martindale JL, Yang X, Mazan-Mamczarz K, Indig FE, Falco G, Zhan M, Gorospe M. Identification and functional outcome of mRNAs associated with RNA-binding protein TIA-1. Mol Cell Biol. 2005;25:9520–9531. doi: 10.1128/MCB.25.21.9520-9531.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loschi M, Leishman CC, Beradone N, Boccaccio GL. Dynein and kinesin regulate stress-granule and P-body dynamics. J Cell Sci. 2009;122:3973–3982. doi: 10.1242/jcs.051383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund MK, Guthrie C. The DEAD-box protein Dbp5 is required to dissociate Mex67p from exported mRNPs at the nuclear rim. Mol Cell. 2005;23:645–651. doi: 10.1016/j.molcel.2005.10.005. [DOI] [PubMed] [Google Scholar]
- Martin KC, Ephrussi A. mRNA localization: gene expression in the spatial dimension. Cell. 2009;136:719–730. doi: 10.1016/j.cell.2009.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazan-Mamczarz K, Lal A, Martindale JL, Kawai T, Gorospe M. Translational repression by RNA-binding protein TIAR. Mol Cell Biol. 2006;26:2716–2727. doi: 10.1128/MCB.26.7.2716-2727.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazroui R, Huot ME, Tremblay S, Filion C, Labelle Y, Khandjian EW. Trapping of messenger RNA by Fragile X Mental Retardation protein into cytoplasmic granules induces translation repression. Hum Mol Gen. 2002;15:3007–3017. doi: 10.1093/hmg/11.24.3007. [DOI] [PubMed] [Google Scholar]
- Mazroui R, Sukarieh R, Bordeleau ME, Kaufamn RJ, Northcote P, Tanaka J, Gallouzi I, Pelletier J. Inhibition of ribosome recruitment induces stress granule formation independently of eukaryotic initiation factor 2alpha phosphorylation. Mol Biol Cell. 2006;17:4212–4219. doi: 10.1091/mbc.E06-04-0318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazroui R, Di Marco S, Kaufman RJ, Gallouzi IE. Inhibition of the ubiquitin-proteasome system induces stress granule formation. Mol Biol Cell. 2007;18:2603–2618. doi: 10.1091/mbc.E06-12-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWhorter ML, Monani UR, Burghes AH, Beattie CE. Knockdown of the survival motor neuron (Smn) protein in zebrafish causes defects in motor axon outgrowth and pathfinding. J Cell Biol. 2003;162:919–931. doi: 10.1083/jcb.200303168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moeller BJ, Cao Y, Li CY, Dewhirst MW. Radiation activates HIF-1 to regulate vascular radiosensitivity in tumors: role of reoxygenation, free radicals, and stress granules. Cancer Cell. 2004;5:429–441. doi: 10.1016/s1535-6108(04)00115-1. [DOI] [PubMed] [Google Scholar]
- Mokas S, Mills JR, Garreau C, Fournier MJ, Robert F, Arya P, Kaufman RJ, Pelletier J, Mazroui R. Uncoupling stress granule assembly and translation initiation inhibition. Mol Biol Cell. 2009;20:2673–2683. doi: 10.1091/mbc.E08-10-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollet S, Cougot N, Wilczynska A, Dautry F, Kress M, Bertrand E, Weil D. Translationally repressed mRNA transiently cycles through stress granules during stress. Mol Biol Cell. 2008;19:4469–4479. doi: 10.1091/mbc.E08-05-0499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanaswamy R, Levy M, Tsechansky M, Stovall GM, O'Connell JD, Mirrielees J, Ellington AD, Marcotte EM. PNAS. 2009;106:10147–10152. doi: 10.1073/pnas.0812771106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonhoff U, Ralser M, Welzel F, Piccini I, Balzereit D, Yaspo ML, Lehrach H, Krobitsch S. Ataxin-2 interacts with the DEAD/H-box RNA helciase DDX6 and interferes with P-bodies and stress granules. Mol Biol Cell. 2007;18:1385–1396. doi: 10.1091/mbc.E06-12-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohn T, Kedersha N, Hickman T, Tisdale S, Anderson P. A functional RNAi screen links O-GlcNAc modification of ribosomal proteins to stress granule and processing body assembly. Nat Cell Biol. 2008;10:1224–1231. doi: 10.1038/ncb1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R, Sheth U. P bodies and the control of mRNA translation and degradation. Mol Cell. 2007;25:635–646. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Pullmann R, Jr, Kim HH, Abdelmohsen K, Lal A, Martindale JL, Yang X, Gorospe M. Analysis of turnover and translation regulatory RNA-binding protein expression through binding to cognate mRNAs. Mol Cell Biol. 2007;27:6265–6278. doi: 10.1128/MCB.00500-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranum LP, Cooper TA. RNA-mediated neuromusucular disorders. Annu Rev Neurosci. 2006;29:259–277. doi: 10.1146/annurev.neuro.29.051605.113014. [DOI] [PubMed] [Google Scholar]
- Reijns MA, Alexander RD, Spiller MP, Beggs JD. A role for Q/N-rich aggregation-prone domains in P-body localization. J Cell Sci. 2008;121:2463–2472. doi: 10.1242/jcs.024976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rikhvanov EG, Romanova NV, Chernoff YO. Chaperone effects on prion and nonprion aggregates. Prion. 2007;1:217–222. doi: 10.4161/pri.1.4.5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarcelli JJ, Viggiano S, Hodge CA, Heath CV, Amberg DC, Cole CN. Synthetic genetic array analysis in Saccharomyces cerevisiae provides evidence for an interaction between RAT8/DBP5 and genes encoding P-body components. Genetics. 2008;179:1945–1955. doi: 10.1534/genetics.108.091256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidlin M, Lu M, Leuenberger SA, Stoecklin G, Mallaun M, Gross B, Gherzi R, Hess D, Hemmings BA, Moroni C. The ARE-dependent mRNA-destabilizing activity of BRF1 is regulated by protein kinase B. EMBO J. 2004;23:4760–4769. doi: 10.1038/sj.emboj.7600477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serman A, Le Roy F, Aigueperse C, Kress M, Dautry F, Weil D. GW body disassembly triggered by siRNAs independently of their silencing activity. Nucleic Acids Res. 2007;35:4715–4727. doi: 10.1093/nar/gkm491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seydoux G, Braun RE. Pathway to totipotency: lessons from germ cells. Cell. 2006;127:891–904. doi: 10.1016/j.cell.2006.11.016. [DOI] [PubMed] [Google Scholar]
- Sheth U, Parker R. Targeting of aberrant mRNA to cytoplasmic processing bodies. Cell. 2006;125:1095–1109. doi: 10.1016/j.cell.2006.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spriggs KA, Stoneley M, Bushell M, Willis AE. Re-programming of translation following cell stress allows IRES-mediated translation to predominate. Biol Cell. 2008;100:27–38. doi: 10.1042/BC20070098. [DOI] [PubMed] [Google Scholar]
- Stoecklin G, Stubbs T, Kedersha N, Wax S, Rigby WF, Blackwell TK, Anderson P. MK2-induced tristetraprolin:14-3-3 complexes prevent stress granule association and ARE-mRNA decay. EMBO J. 2004;23:1313–1324. doi: 10.1038/sj.emboj.7600163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweet TJ, Boyer B, Hu W, Baker KE, Coller J. Microtubule disruption stimulates P-body formation. RNA. 2007;13:493–502. doi: 10.1261/rna.355807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadesse H, Deschênes-Furry J, Boisvenue S, Côté J. KH-type splicing regulatory protein interacts with survival motor neuron protein and is misregulated in spinal muscular atrophy. Hum Mol Genet. 2008;17:506–524. doi: 10.1093/hmg/ddm327. [DOI] [PubMed] [Google Scholar]
- Thomson T, Lasko P. Drosophila tudor is essential for polar granule assembly and pole cell specification, but not for posterior patterning. Genesis. 2004;40:164–170. doi: 10.1002/gene.20079. [DOI] [PubMed] [Google Scholar]
- Tian Q, Taupin J, Elledge S, Robertson M, Anderson P. Fas-activated serine/threonine kinase (FAST) phosphorylates TIA-1 during Fas-mediated apoptosis. J Exp Med. 1995;182:865–874. doi: 10.1084/jem.182.3.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourrière H, Gallouzi IE, Chebli K, Capony JP, Mouaikel J, van der Geer P, Tazi J. RasGAP-associated endoribonuclease G3BP: selective RNA degradation and phosphorylation-dependent localization. Mol Cell Biol. 2001;21:7747–7760. doi: 10.1128/MCB.21.22.7747-7760.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourrière H, Chebli K, Zekri L, Courselaud B, Blanchard JM, Bertrand E, Tazi J. The RasGAP-associated endoribonucleae G3BP assembles stress granules. J Cell Biol. 2003;160:823–831. doi: 10.1083/jcb.200212128. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Thomas MG, Martinez Tosar LJ, Desbats MA, Leishman CC, Boccaccio GL. Mammalian Staufen 1 is recruited to stress granules and impairs their assembly. J Cell Sci. 2009;155:563–573. doi: 10.1242/jcs.038208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai NP, Ho PC, Wei LN. Regulation of stress granule dynamics by Grb7 and FAK signaling pathway. EMBO J. 2008;27:715–726. doi: 10.1038/emboj.2008.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai NP, Tsui YC, Wei LN. Dynein motor contributes to stress granule dynamics in primary neurons. Neuron. 2009;159:647–656. doi: 10.1016/j.neuroscience.2008.12.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Houven van Oordt W, Diaz-Meco MT, Lozano J, Krainer AR, Moscat J, Cáceres JF. The MKK(3/6)-p38-signaling cascade alters the subcellular distribution of hnRNP A1 and modulates alternative splicing regulation. J Cell Biol. 2000;149:307–316. doi: 10.1083/jcb.149.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderklish PW, Edelman GM. Differential translation and fragile X syndrome. Genes Brain Behav. 2005;4:360–384. doi: 10.1111/j.1601-183X.2005.00134.x. [DOI] [PubMed] [Google Scholar]
- Wek RC, Jiang HY, Anthony TG. Coping with stress: eIF2 kinases and translational control. Biochem Soc Trans. 2006;34:7–11. doi: 10.1042/BST20060007. [DOI] [PubMed] [Google Scholar]
- Wilczynska A, Aigueperse C, Kress M, Dautry F, Weil D. The translational regulator CPEB1 provides a link between dcp1 bodies and stress granules. J Cell Sci. 2005;118:981–992. doi: 10.1242/jcs.01692. [DOI] [PubMed] [Google Scholar]
- Yedavalli VS, Neuveut C, Chi YH, Kleiman L, Jeang KT. Requirement of DDX3 DEAD box RNA helicase for HIV-1 Rev-RRE export function. Cell. 2004;119:381–392. doi: 10.1016/j.cell.2004.09.029. [DOI] [PubMed] [Google Scholar]
- Yu C, York B, Wang S, Feng Q, Xu J, O'Malley BW. An essential function of the SRC-3 coactivator in suppression of cytokine mRNA translation and inflammatory response. Mol Cell. 2007;25:765–778. doi: 10.1016/j.molcel.2007.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.