Abstract
GCN5 encodes one of the non-essential Histone Acetyl Transferases in Saccharomyces cerevisiae. Extensive evidence has indicated that GCN5 is a key regulator of gene expression and could also be involved in transcriptional elongation, DNA repair and centromere maintenance. Here we show that the deletion of GCN5 decreases the stability of mini-chromosomes; that the tethering of Gcn5p to a crippled origin of replication stimulates its activity; that high dosage of GCN5 suppresses conditional phenotypes caused by mutant alleles of bona fide replication factors, orc2-1, orc5-1 and mcm5-461. Furthermore, Gcn5p physically associates with origins of DNA replication, while its deletion leads to localized condensation of chromatin at origins. Finally, Δgcn5 cells display a deficiency in the assembly of pre-replicative complexes. We propose that GCN5 acts as a positive regulator of DNA replication by counteracting the inhibitory effect of Histone Deacetylases.
Introduction
DNA replication in Saccharomyces cerevisiae initiates at discrete origins referred to as Autonomously Replicating Sequences (ARS). Origins share an essential core element called ACS (ARS Consensus Sequence), which binds the Origin Recognition Complex (ORC). In early G1 phase Cdc6p and six MCM (MiniChromosome Maintenance) proteins (MCM2-7) join ORC to form pre-replicative complexes [1]. In S phase these complexes are activated by protein kinases to initiate DNA replication [1].
It is granted that histone acetylation can play a key role in the regulation of origins of DNA replication [2], [3]. Convincing evidence has shown that in S.cerevisiae two major Histone-DeACetylases (HDAC), RPD3 and SIR2, act as negative regulators of origins [4], [5], [6], [7]. It has been postulated that their inhibitory role is mediated by global deacetylation rather than local recruitment and retention of HDACs to replication foci. However, a recent study suggests that RPD3 may be targeted to multiple non-telomeric origins but the mechanism of recruitment remains elusive [7]. A link between histone deacetylation by Rpd3p and negative impact on origin activity has also been proposed in Drosophila [8]. At the same time, another HDAC, HST1, surprisingly acted as a stimulator of certain origins [9] thus suggesting a more complex cross-talk between histone acetylation and replication. Nevertheless, the prevailing notion is that deacetylation of histones has a negative impact on origin activity [3].
It is conceivable that the opposing activity of Histone Acetyl Transferases (HAT) stimulates origins of DNA replication, but details about the HATs involved are sparse. For example, in S.cerevisiae the tethering of Gcn5p to the late origin ARS1412 causes it to fire earlier [4]. Similarly, the tethering of Chameau (a HAT) to a chromosomal origin in Drosophila stimulates the activity of this origin [8] thus reiterating the general link between histone acetylation and activity of origins. Several recent studies in human cells have demonstrated that the HAT HBO1 is involved in the loading of MCM proteins on chromatin in early G1 phase [10], [11], [12] and in the progression of replication forks in S-phase [13]. Furthermore, HBO1 seems to respond to p53 and FAD24 to halt cell cycle progression [14], [15]. HBO1 has no obvious structural homologue in S.cerevisiae, however the Sas2p-containing HAT complex NuA3 has been proposed as a remote functional homologue [16], [17]. Interestingly, at semi-permissive temperature the deletion of SAS2 exacerbates the low origin activity caused by a mutation in orc2 and delays progression through S-phase in conjunction with orc2 mutations [18]. Another recent study has documented an interaction between Hat1p and ORC and has shown preferential association of Hat1p with origins at the time of their activation [19].
In this study we compared the effects of the deletions or mutations in several HATs including SAS2 and HAT1 on DNA replication in S.cerevisiae. We obtained several lines of evidence indicating that GCN5 is a major positive regulator of origins of DNA replication.
Materials and Methods
The yeast strains used in this study are listed in Table 1. Yeast cells were routinely grown in YPD or SC media at 23°C except in Table 2 as indicated.
Table 1. Yeast strains used in this study.
| Strain | Genotype | Reference |
| W303 | ade2-1 trp1-1 can1-100 leu2-3,112 his3-11,15 ura3-1 MAT a | |
| mcm5-461 | mcm5-461 ade2-1 trp1-1 can1-100 leu2-3,112 his3-11,15 ura3-1 MAT α , isogenic to W303 | [29] |
| orc2-1 | orc2-1 ade2-1 trp1-1 can1-100 leu2-3,112 his3-11,15 ura3-1 MAT a , isogenic to W303 | [31] |
| orc5-1 | orc5-1 ade2-1 trp1-1 can1-100 leu2-3,112 his3-11,15 ura3-1MAT a , isogenic to W303 | [32] |
| esa1-414 | ade2-1 trp1-1 can1-100 leu2-3,112 his3-11,15 ura3-1 esa1-1-414::URA3 MAT a, isogenic to W303 | LPY4679 [27] |
| esa1-L254P | ade2-1 trp1-1 can1-100 leu2-3,112 his3-11,15 ura3-1 esa1-L254P::URA3 MAT a, isogenic to W303 | LPY5001 [27] |
| BY4742 | his3Δ1 leu2Δ0 met 15Δ0 ura3Δ0 MATa | Euroscarf |
| Δgcn5 | gcn5::KanMX his3Δ1 leu2Δ0 met 15Δ0 ura3Δ0 MATa, isogenic to BY4742 | Euroscarf |
| Δsas3 | sas3::KanMX his3Δ1 leu2Δ0 met 15Δ0 ura3Δ0 MATa, isogenic to BY4742 | Euroscarf |
| Δsas2 | sas2::KanMX his3Δ1 leu2Δ0 met 15Δ0 ura3Δ0 MATa, isogenic to BY4742 | Euroscarf |
| Δhat11 | hat1::KanMX his3Δ1 leu2Δ0 met 15Δ0 ura3Δ0 MATa, isogenic to BY4742 | Euroscarf |
| ΔRtt109 | rtt109::KanMX his3Δ1 leu2Δ0 met 15Δ0 ura3Δ0 MATa, isogenic to BY4742 | Euroscarf |
| ADA3-MYC | ADA3-MYC9::HIS3 ura3-1 leu2-3,112 his3-200 lys5 trp1 ada3::TRP1 MATa, isogenic to W303 | [35] |
| GCN5-MYC18 | MATa GCN5-MYC::TRP1 ade2-1 can1-100 his3-11,15 leu2-3,112 his3-11, 15 ura3-1, isogenic to W303 | [36] |
| DF5gal4Δ | gal4Δ gal80Δ ura3-52 trp1-901 leu2-3,112 his3-200 MAT a | [53] |
Table 2. High dosage of GCN5 and SPT8 partially suppress the Ts− phenotypes of replication factor mutants.
| High dosage gene | Growth at 23°C | Growth at 37°C | |
| orc2-1 | No episome | ++++ | − |
| Control | ++++ | − | |
| GCN5 | ++++ | ++ | |
| SPT8 | ++++ | ++ | |
| SPT16 | ++++ | − | |
| POB3 | ++++ | − | |
| orc5-1 | No episome | ++++ | − |
| Control | ++++ | − | |
| GCN5 | ++++ | ++ | |
| SPT8 | ++++ | ++ | |
| SPT16 | ++++ | − | |
| POB3 | ++++ | − | |
| mcm5-461 | No episome | ++++ | +/− |
| Control | ++++ | +/− | |
| GCN5 | ++++ | +++ | |
| SPT8 | ++++ | +++ | |
| SPT16 | ++++ | +/− | |
| POB3 | ++++ | +/− |
orc2-1, orc5-1 and mcm5-461 cell were transformed with high copy episomes (2 µm) carrying the shown genes and then grown at 23°C or 37°C. Growth was assessed by size of colony after 3 days at 23°C or after 5 days at 37°C.
Plasmids
pGBKT7 (Clontech), pRS305 (SGD), pARS1wt and pARS1/-B23/G24 [20], pARS1wt/linker [21], pGAL4DBD-GCN5, pGAL4DBD-gcn5(KQL), pGAL4DBD-SAS2, pGAL4DBD-SAS3, pGAL4DBD-ESA1 and pGAL4DBD-HAT1 [22] have been described previously. pFW193 (2 µm, URA3, SPT8) is a gift from F. Winston. pTF125 (2 µm, URA3, SPT16) and pTF137 (2 µm, URA3, POB3) are gifts from T. Fromosa. pGBKT7-GCN5 was produced by replacing the large HindIII fragment containing the MCS with a PCR fragment containing GCN5 under its own promoter.
Mini-chromosome stability assays with pARS1wt and pRS305 were performed as in [23], [24]. Suppression of plasmid loss in W303, mcm5-461, orc2-1 and orc5-1 cells was performed by double transformation with pARS1wt and pGBKT7 or pGBKT7-GCN5, respectively. Cells were selected on SC-trp-ura plates. Three individual colonies were inoculated in SC-trp to allow for retention of the pGBKT7 episomes and for loss of pARS1wt and grown for 20 generations. Each culture was serially diluted 1/10, and 5 ul of each dilution were spotted on both SC-trp-ura and on SC-trp plates. Assays with pARS1/-B23/GAL4 were performed by co-transforming in DF5gal4Δ cells with pARS1/-B23/GAL4 together with pGAL4DBD-GCN5, pGAL4DBD-GCN5(KQL), pGAL4DBD-SAS2, pGAL4DBD-SAS3, pGAL4DBD-ESA1 and pGAL4DBD-HAT1, respectively. Cells were selected on SC-his-ura plates, grown for 20 generations in SC-his and then plated on SC-his and SC-his-ura. In all cases plasmid loss was calculated as in [23].
Chromatin Immuno-Precipitation (ChIP)
We used a protocol, which favors weaker protein interactions with chromatin. It includes a second cross-linking agent with a longer arm in the cross-linking step [25]. Briefly, GCN5-MYC or ADA3-MYC cells at OD600 = 1.0 in YPD were cross-linked with 1.6% formaldehyde for 30 minutes at 25°C and then suspended in 10 ml PBS/10 mM DMA(Pierce)/0.2%DMSO for another 30 minutes at 25°C. The cross-linking was quenched with 2.5 ml of 2.5 M glycine, cells were collected and crushed by glass beads in LB/IP buffer (50 mM Tris.HCl, 140 mM NaCl2, 5 mM EDTA, 1x PI cocktail (Sigma)) and sonicated by 12 bursts of 30 seconds at maximum output of Micronix cuphorn to produce fragments of about 500 bp. Chromatin was extracted in LP/IP plus 0.3% TX100 and 0.05% Na deoxycholate and immuno-precipitated overnight with anti-MYC beads (Sigma). The beads were washed three times with 1 ml LB/IP/0.5% TX100, 0.05% Na deoxycholate, one time with 1 ml LB/IP plus 350 mM NaCl, one time with 150 ul TE plus 10 mM NaCl (Final Wash) and eluted/uncrosslinked overnight in 150 ul TE/0.2% SDS at 65°C. After treatment with Proteinase K, DNA was extracted by Phenol/Chloroform and precipitated. The fragments of interest were quantified by PCR (after confirming linear range of amplification with crude genomic DNA) and ImageQuant software. Primer sequences are provided (Suppl. Table S1).
For the analysis of MCM loading on origins, BY4247 and Δgcn5 cells were grown at 23°C in YPD to OD600 = 1.0, arrested in Mitosis with 12 µg/ml Nocodazole for 3.5 hours, then released from mitotic arrest in fresh YPD. Aliquots of 100 ml were cross-linked and processed as above. Immunoprecipitation was conducted with a mixture of 2 ug each of anti-yMCM2 and anti-yMCM5 antibody (Santa Cruz) and Protein G beads (Sigma). Washing, elution, reversal of cross-linking and PCR were as described above.
Restriction Nuclease Accessibility Assay
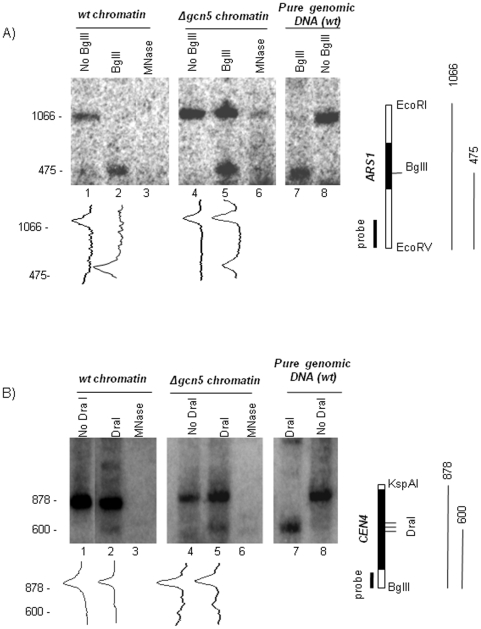
We used the protocol for targeted analysis of loci [26] with slight modifications. BY4247 and Δgcn5 cells (100 ml) were grown in YPD to OD600 = 1.0 and treated with Zymolase and pelleted. Spheroplasts were re-suspended in 5 pellet volumes of nuclease buffer (30 mM Tris.HCl pH 7.5, 10 mM MgCl2, 5 mM CaCl2, 50 mM NaCl, 1/100 volume of PI cocktail (Sigma)), split into 0.7 ml aliquots and incubated for 30 min at 37°C with 50 U of BglII, 50 U of DraI, 200 U of Micrococcal Nuclease (MNase) or no enzyme, respectively. The digestions were stopped with 1/5 volume of 5% SDS/100 mM EDTA. All samples were treated with Proteinase K, extracted with phenol/chloroform and precipitated. Subsequently, samples for the analysis of ARS1 were digested overnight with 0.3 U per µg of DNA of each EcoRI and EcoRV. Samples for the analysis of CEN4 were digested overnight with 0.5 U of KspA1 and 0.2 U of BglII per µg of DNA. After ethanol precipitation, the DNA was resolved on 1.2% agarose gels and analyzed by Southern blot. The probe for ARS1 was a PCR fragment adjacent to the EcoRV site. The probe for CEN4 was adjacent to the BglII site. The actual sequences of the probes are available upon request. Signals were acquired and quantified by Phosphorimager using ImageQuant software.
Results
Deletion of GCN5 Impairs Mini-Chromosome Stability
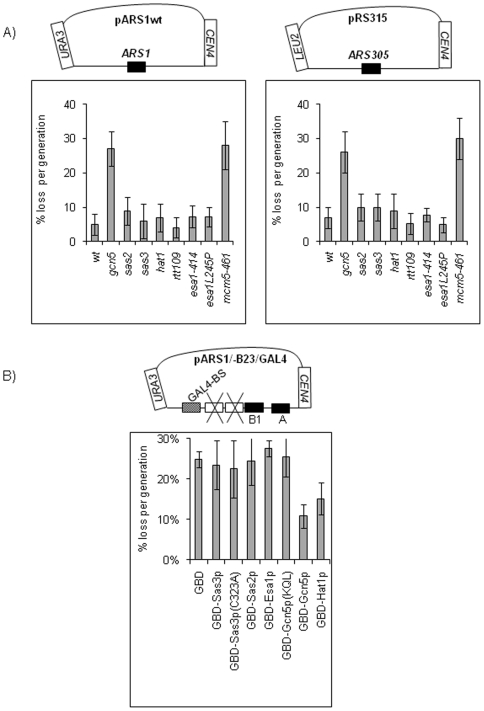
Mini-chromosomes are stably inherited (3–5% loss per generation) plasmids, which contain an ARS, a centromere (CEN) and a selection marker gene. Increased loss rate in non-selective medium is most frequently associated with flawed replication of the mini-chromosome independently of effects on other processes [24]. We used the mini-chromosome stability assay to test the involvement of several HATs in DNA replication. Strains with disrupted non-essential HAT genes (Δgcn5, Δhat1, Δsas2, Δsas3 and Δrtt109) or mutations in the essential ESA1 gene (esa1-414, esa1L245P [27]) were transformed with two different mini-chromosomes carrying ARS1 (pARS1wt) or ARS305 (pRS315). In Fig. 1A we show that the disruption of GCN5 dramatically decreased the stability of both mini-chromosomes, while the deletion of the other HATs or the mutations in esa1 had marginal effects. The level of mini-chromosome instability in Δgcn5 cells was comparable to that of the mcm5-461 strain, which harbors a mutation in one of the essential origin licensing factors, MCM5 [28], [29].
Figure 1. GCN5 regulates mini-chromosome stability.
A. Disruption of GCN5 increases the loss rate of ARS1- or ARS305- mini-chromosomes. Maps of pARS1wt (ARS1, CEN4, URA3) and pRS315 (ARS305, CEN4, LEU2) mini-chromosomes are shown on the top of the graphs. The mini-chromosomes were transformed in the strains shown at the bottom of the graphs. After growth on selective plates (SC-ura or SC-leu, respectively) three colonies were transferred for 20 generations in rich medium (YPD) and plated on YPD and SC-ura or SC-leu, respectively. The proportions of cells with URA+ or LEU+ phenotypes were assessed and the average loss rate per generation was calculated (%) and plotted. Error bars represent standard deviations between the three parallel counts in the experiment. A typical outcome of one of three independent experiments is shown. B. Tethering of Gcn5p increases the stability of pARS1/-B23/GAL4. A diagram of pARS1/-B23/GAL4 (ARS1-B2-B3+GAL4, CEN4, URA3) is shown on the top. The B2 and B3 elements of ARS1 are destroyed and a GAL4 binding site (GAL4-BS) is inserted next to B3. DF5gal4Δ cells were co-transformed with pARS1/-B23/GAL4 and episomes (2 µm/HIS3) expressing the GAL4 DNA binding domain fused to the proteins indicated below the graph. Gcn5p(KQL) lacks HAT activity. Average pARS1/-B23/GAL4 loss per generation and standard deviation were calculated as in Figure 1a. A typical outcome of one of two independent experiments is shown.
Tethering of Gcn5p Stimulates the Activity of an Impaired ARS1
The tethering of Gcn5p to a late origin (ARS1412) causes it to fire earlier [4]. We designed a similar tethering assay to directly compare the overall activity of several HATs on a mini-chromosomal origin. The mini-chromosome pARS1/-B23/G24 (Fig. 1B) carries ARS1, in which two of the auxiliary elements, B2 and B3, are destroyed and a GAL4 binding site is inserted instead of B3 [20]. These ablations render pARS1/-B23/G24 unstable to a point where its maintenance is critically dependent on the tethering of stimulators via the GAL4 binding site [20], [30]. To test the roles of different HATs on this crippled origin, we co-transformed DFΔgal4 cells with pARS1/-B23/G24 plus a set of constructs expressing GAL4-fusions of Gcn5p, gcn5p(KQL) (a GCN5 mutant lacking HAT activity), Sas2p, Sas3p, Esa1p and Hat1p, respectively, and measured the stability of pARS1/-B23/G24. In Fig. 1B we show that Gcn5p and Hat1p reduced the loss rate of pARS1/-B23/G24 from 25% to 10 and 12%, respectively. Since the theoretical maximum loss measurable by this assay is 50% for non-replicating plasmids and is in the range of 30% for very poorly replicating plasmids [24], these effects were considered significant. In contrast, Gcn5p(KQL), Sas2p, Sas3p and Esa1p did not reduce the loss rate of pARS1/-B23/G24 (Fig. 1B). We note that all GAL4-fusion constructs express active HAT proteins in vivo as demonstrated elsewhere [22].
High Dosage of GCN5 Suppresses Mini-Chromosome Instability in Replication Factor Mutants
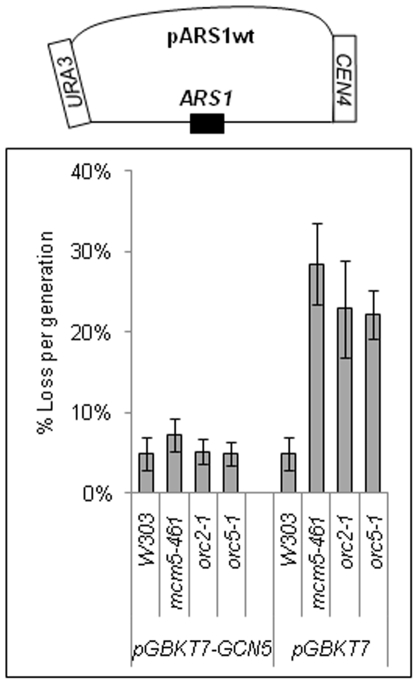
In an earlier study we have shown that high dosage of TRA1 (a component of the HAT complexes SAGA and NuA4) can suppress the Ts− phenotype of a strain with profound replication deficiency, mcm5-461 [29]. We reasoned that if GCN5 works in DNA replication, it may have a similar effect in this and other replication factor mutants. We supplemented the mcm5-461 [29], orc2-1 [31] and orc5-1 [32] strains with stable high copy 2 µm episomes, which carry GCN5 or a non-HAT component of the SAGA/ADA complex, SPT8. As controls, we used two genes, which encode components of the FACT complex, SPT16 and POB3. FACT has been implicated in pol II transcription and in the elongation step of DNA replication, but not in origin activation [33], [34]. In Table 2 we show that high copy of GCN5 and SPT8 suppressed the conditional Ts− phenotypes of mcm5-461, orc2-1 and orc5-1. In contrast, SPT16 and POB3 had no effect. Next, we tested if the high dosage of GCN5 in these mutants also suppressed the severe instability of mini-chromosomes. We co-transformed these strains with a test mini-chromosome (pARS1wt) and the GCN5-carrying episome and assessed the level of loss of pARS1wt. The high dosage of GCN5 significantly improved the stability of pARS1wt relative to the control (Fig. 2). Hence, high dosage of GCN5 specifically suppressed the replication deficiencies of mcm5-461, orc2-1 and orc5-1.
Figure 2. High copy number of GCN5 suppresses mini-chromosome instability in mcm5-461, orc2-1 and orc5-1.
W303, mcm5-461, orc2-1 and orc5-1 cells carrying pARS1wt (ARS1, CEN4, URA3) were transformed with pGBKT7 (2 µm/TRP1) or pGBKT7-GCN5. After selection on SC-trp-ura plates three colonies were transferred for 20 generations in SC-trp and then plated on SC-trp and SC-trp-ura, respectively. The proportion of cells with URA+ phenotype was estimated and the average loss per generation of pARS1wt was calculated (%) as in Fig. 1A and plotted. A typical outcome of one of two independent experiments is shown.
Gcn5p Associates with Origins of DNA Replication
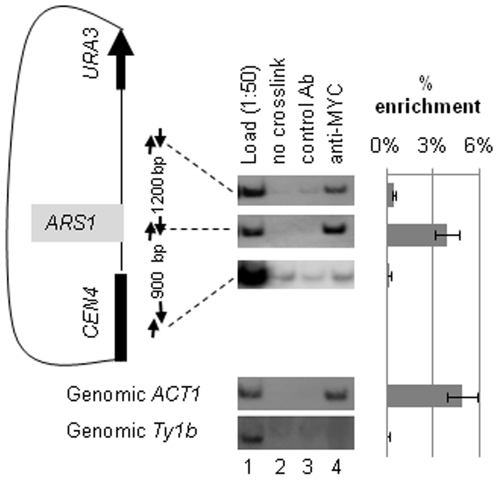
Next, we used ChIP assays to test if Gcn5p directly associates with origins of DNA replication. In the first set of experiments we used a modified pARS1wt/link mini-chromosome, which has been designed to minimize overlap of ChIP signals from URA3 and ARS1 ([21], see Fig. 3). It contains a 2.2 kb fragment of non-yeast DNA between ARS1 and the URA3 reporter. pARS1wt/link was introduced in the ADA3-MYC strain [35] and ChIP was performed with anti-MYC antibodies. Ada3p, Ada2p and Gcn5p build up a stable complex, which forms the catalytic core of all Gcn5p-containing HATs [35]. The amounts of precipitated mini-chromosomal ARS1 only (one of the primers anneals to the plasmid backbone) and two flanking fragments that are positioned about 1 kb away from it were quantified by triplicate PCR. The data showed that about 4% of the mini-chromosomal ARS1, 0.6% of the fragment between ARS1 and URA3 and 0.1% of the fragment next to CEN4 were immunoprecipitated by anti-MYC antibodies (Fig. 3, lane 4). In comparison, the anti-MYC antibodies precipitated none of the genomic Ty1b transposon (negative control) (Fig. 3, lower panel) and about 5% of the genomic ACT1 (positive control). The non-crosslinked samples (Fig. 3, lane 2) and the samples without antibody (Fig. 3, lane 3) uniformly produced negligible signals with all loci tested. We concluded that Ada2p/Ada3p/Gcn5p specifically associates with the mini-chromosomal ARS1.
Figure 3. Gcn5p associates with mini-chromosomal ARS1.
A map of the test mini-chromosome (pARS1wt/linker) is shown on the left. The distance between ARS1 and URA3 is 2.2 kb. The distance between the PCR fragments is shown between the pairs of arrows. MYC-ADA3 cells harboring this mini-chromosome were cross-linked with formaldehyde and immunoprecipitated with anti-MYC antibodies. The precipitated DNA was quantified by PCR in triplicate. The per cent of signal from each fragment in the anti-MYC immuneprecipitate versus Load (diluted 1∶50) was calculated and plotted in the graph on the right together with the standard deviation in the three PCR reactions. Controls with genomic ACT1 and Ty1b DNAs are shown below the mini-chromosome diagram. A typical outcome of one of two independent experiments is shown.
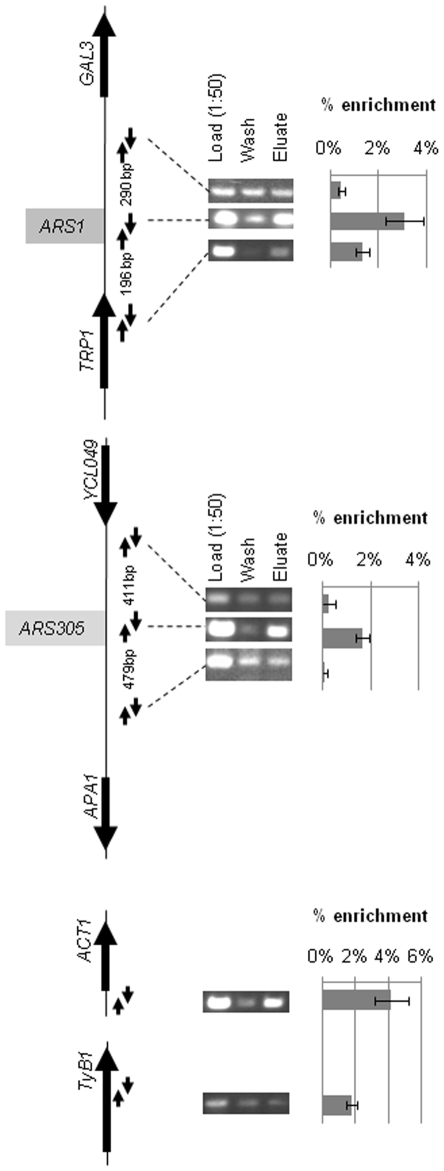
In another set of experiments we analyzed the association of Gcn5p with chromosomal origins. We must note that the high gene density in the yeast genome does not provide the comfortable distances between genes and origins that were available on pARSwt/link. Because of this concern we focused on two extensively characterized origins with finely mapped ACS and auxiliary elements, ARS1 and ARS305, in a GCN5-MYC18 strain [36]. The ACS elements of these origins are located more than 500 base pairs (the average resolution of ChIP) away from the nearest promoter. In these assays we compared the signal from the core ACS to the signals from two flanking elements positioned between 200 and 500 bases away from the ACSs (see maps in Fig. 4). The relative amounts of each of the analyzed fragments in the Load, Wash and anti-MYC-Eluate fractions were quantified by PCR (triplicate). The per cent of signal in Eluate versus Load was calculated and plotted next the maps of ARS1 and ARS305 (Fig. 4). In addition, we tested association of Gcn5-MYCp to two control loci: the ACT1 gene and the repressed Tyb1 transposon (Fig. 4).
Figure 4. Gcn5p associates with chromosomal origins of DNA replication.
Maps of the four genomic loci analyzed are shown on the left. Exponentially growing MYC18-GCN5 cells were cross-linked with formaldehyde and immunoprecipitated with anti-MYC antibodies and the precipitated DNA was quantified by PCR in triplicate. The per cent of signal from each fragment in the anti-MYC immunoprecipitate versus Load (diluted 1∶50) was calculated and plotted in the graphs on the right together with the standard deviation in the three PCR reactions. The arrows in the diagrams represent the pairs of PCR primers used. The distance between the PCR fragments is shown between the pairs of arrows. A typical outcome of one of two independent experiments is shown.
The ChIP experiments consistently produced higher signals from the ACS elements of ARS1 and ARS305 as compared to the flanking fragments. The moderately high signal of 1.3% at the ARS1 flanking element towards TRP1 probably reflects its proximity to the origin (196 bp) or strong association of Gcn5p to the upstream TRP1 gene. The recovery of DNAs in the immunoprecipitates represented 2.9% for ARS1 and 1.8% for ARS305 of the total DNA analyzed, while the recovery of ACT1 (positive control) DNA was 4.3% (Fig. 4). As expected, no association of Gcn5p to the repressed Ty1b element was observed (Fig. 4). These weaker but still noticeable chromatin immunioprecipitation signals from origins relative to promoters are in tune with the observations in [36]. In this comprehensive genome-wide mapping of Gcn5p association (data available on http://frodo.wi.mit.edu/cgi-bin/yeast_histones/gbrowse.cgi/yeast_histones) moderately higher signals of Gcn5p binding at ARS1 and ARS305 (and also at ARS501, ARS606, ARS1021, ARS1414, ARS1618) relative to the immediate neighboring regions can be observed. However, these peaks were below the preset threshold of the computational analysis and were not discussed in [36].We should also mention that many ARS reside within short distance from gene promoters thus adding to the problematic interpretation of such weak signals. We completely agree that the genome-wide approach in [36] has not identified association of Gcn5p to ARSs. Nevertheless, their data are in tune with our focused approach and support the notion of weak association of Gcn5p to origins. We also need to note that we used a protocol with two cross-linkers (formaldehyde and DMA, [25]) that enhances weak ChIP signals as compared to a single cross-linker (formaldehyde) in [36]. In summary, the results in Figure 4 showed specific, but weaker association of Gcn5p to chromosomal origins relative to the better isolated mini-chromosomal ARS1 origin (Fig. 3). This outcome could be a consequence of the expected higher background signal on chromosomal origins. The alternatively explanation is that for some unknown reason the association of Gcn5p to the mini-chromosomal ARS1 origin is intrinsically stronger.
Deletion of GCN5 Perturbs Chromatin Structure of a Chromosomal Origin
We reasoned that if the weak association of Gcn5p to chromosomal origins is functionally relevant, the deletion of GCN5 would affect their chromatin structure. Initial analyses by limited MNase digestion showed the existence of hypersensitivity sites in the vicinity of ARS1, ARS305 and ARS501 (not shown). However, the differences between Δgcn5 and wt cells were hard to assess because this enzyme also cleaves in the broader area of the analyzed loci. It is noteworthy that a similar DNAase accessibility assay has failed to detect variations in the structure of ARS1 between wt and orc2-1 cells [18], yet it is known that ARS1 activity is severely impaired in orc2-1 cells [31]. We concluded that the MNase or DNAase sensitivity assays are inadequate for analysis of ARS. Therefore, we tailored a focused restriction enzyme (BglII) accessibility assay [26] for the analysis of ARS1 on chromosome IV. BglII cleaves ARS1 between the B2 and B3 elements [20]. Briefly, equal amount of spheroplasts from exponentially growing wt or Δgcn5 cells, respectively, were lysed and exposed to excess of BglII. Under these conditions the cleavage of sites depends on site accessibility thus representing the level of chromatin compactness of the tested locus. Control samples were incubated without BglII to monitor for endogenous nuclease activity. In addition, to distinguish specific signals during the indirect end-labeling, excess of MNase was added in parallel samples of lysed spheroplasts. Subsequently, total DNA from all samples was purified and a second complete digestion was performed with EcoRI and EcoRV, which cut on either side of ARS1. Finally, the produced DNA fragments were analyzed by indirect end-labeling with a probe, which anneals 475 bp away from the BglII site. The ratio of un-cleaved EcoRI-EcoRV versus BglII-cleaved fragment was used as a measure for the accessibility to ARS1 and therefore for chromatin compactness of the analyzed locus.
Using this assay we reproducibly observed high accessibility to the BglII site in ARS1 in wt cells (Fig. 5A, lane 2), while the parallel samples from Δgcn5 cells (Fig. 5A, lane 5) consistently showed moderate accessibility. The bands corresponding the BglII-cleaved (475 bp) and un-cleaved (1066 bp) fragment were abolished by treatment with MNase. These results clearly show that in Δgcn5 cells ARS1 acquires a compact structure resistant to nuclease digestion.
Figure 5. Chromatin structure of ARS1 and CEN4 in Δgcn5 cells.
Spheroplasts from Δgcn5 and the isogenic wild type cells (BY4741) were lysed and exposed to excess of BglII, DraI or MNase as indicated on top of each lane. After purification and complete digestion with restriction enzymes that cut at each side of the analyzed loci (shown on the right), DNA fragments were analyzed by Southern blot with the probes shown on the right. A typical outcome of one of three independent experiments is shown. A. Analysis of ARS1 . Diagram of the ARS1 locus on chromosome IV with the positions of ARS1, of the probe for Southern blot and of the BglII, EcoRV and EcoRI sites is shown on the right. The BglII-cleaved (475 bp) and uncleaved (1066 bp) fragments are also shown. Density graphs of lanes 1, 2, 4 and 5 are shown underneath. B. Analysis of CEN4 . Diagram of the CEN4 locus on chromosome IV with the positions of CEN4, of the probe for Southern blot, of the BglII and KspAI sites and the three juxtaposed DraI sites is shown on the right. The DraI-cleaved (600 bp) and uncleaved (878 bp) fragments are also shown. Density graphs of lanes 1, 2, 4 and 5 are shown underneath.
Deletion of GCN5 Has a Relatively Minor Effect on the Structure of CEN4
An earlier study has implicated GCN5 in the regulation of centromeres [37]. More specifically, the study has shown increased accessibility of DraI sites in centromeres in Δgcn5 cells. We attempted to evaluate the extent of chromatin modifications at origins and centromeres in Δgcn5 cells using a similar DraI-accessibility assay for the analysis of CEN4. Samples from wt and Δgcn5 cells prepared in parallel with the samples for Fig. 5A were digested with DraI and processed accordingly with KspI and BglII. Next, indirect end-labeling with a probe annealing 600 bp from the three juxtaposed DraI sites in CEN4 was performed (Fig. 5B). In these experiments we consistently observed fairly similar levels of DraI cleavage in both wt and Δgcn5 cells (Fig. 5B, lanes 2 and 5). Calculation of the intensity of the cleaved (600 bp) and uncleaved (878 bp) CEN4 fragments in Fig. 5B showed that 14% and 18% of the DraI sites were digested in wt cells or Δgcn5 cells, respectively. In agreement, the accessibility to DraI in [37] was similarly estimated at 9% in wt cells and 13% in Δgcn5 cells.
In summary, our assays have demonstrated that the deletion of GCN5 produces minor alterations in the chromatin structure of CEN4 and major alterations of ARS1.
Deletion of GCN5 Impairs the Assembly of Pre-Replicative Complexes on Origins in G1
Earlier studies have shown that the lack of GCN5 delays progression through G2 and Mitosis after synchronization with α-factor [37], [38]. When cells are synchronized in Mitosis, they also progress slower through G1/early S phase and display later appearance of buds as compared to wild type cells (Figure S1). We considered that an event in Mitosis or G1 that is important for regulation of origins could be impaired in Δgcn5 cells.
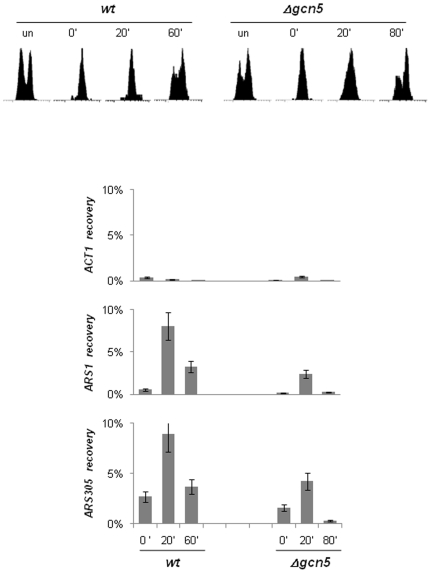
Shortly after exit from Mitosis the origins assemble and “license” pre-replicative complexes. This “licensing” is highlighted by the loading of the MCM proteins [1]. Consequently, the binding of MCM proteins to origins is used as an assessment for the efficiency of assembly of pre-replicative complexes [39]. We tested the hypothesis that the negative effect on DNA replication in Δgcn5 cells is linked to impaired loading of MCM proteins. We synchronized wt and Δgcn5 cells in Mitosis by Nocodazole and then released them in YPD. Cells were cross-linked at 20 min past release when both cell strains are in G1 and show no buds (Fig. 6 and Suppl. Fig. S1). Another sample was collected and cross-linked 60 or 80 min past release for wild type or Δgcn5 cells, respectively. Cross-linked chromatin was sheared, extracted and immunoprecipitated with a mixture of anti-MCM2 and anti-MCM5 antibodies (Santa Cruz). We have found that this mixture of anti-peptide antibodies is more efficient in recovery of origin DNA as compared to the individual antibodies used alone [40], [41]. Finally, the immunoprecipitated DNA was analyzed by quantitative PCR with primers for ARS1 and ARS305. Consistent with many previous reports (reviewed in [1]), in wt cells we saw significant enrichment of anti-MCM immunoprecipitated ARS1 and ARS305 DNA at the 20th min past release (G1) with significant decrease of the signals at the 60th min (S) (Fig. 6). In comparison, ACT1 showed very little association with MCM proteins at the time points of our experiment. A similar pattern of MCM binding to origins was observed in Δgcn5 cells except that the S phase sample was collected at the 80th minute to compensate for the slower progression through the cell cycle. Importantly, in Δgcn5 cells the recovery of ARS1 and ARS305 DNA in G1 was two-three fold lower as compared to wt cells (Fig. 6). These results demonstrate significant deficiency in the assembly of pre-replicative complexes in Δgcn5 cells.
Figure 6. Reduced association of MCM proteins to ARS1 and ARS305.
Wild type (BY4742) and Δgcn5 cells were arrested in Mitosis by12 µg/ml Nocodazole for 3.5 hours and released in YPD. Wild type cells were harvested at 0, 20 and 60 min past Nocodazole release. Δgcn5 cells were harvested at 0, 20 and 60 min past Nocodazole release. Progression through the cell cycle as monitored by Propidium Iodide staining/FACS is shown on the top. Crosslinked chromatin was isolated and immunoprecipitated with anti-MCM2 and anti-MCM5 antibodies. The ACS fragments of ARS1 and ARS305 and a fragment from ACT1 were amplified by PCR (triplicate) and quantified. The per cent of signal from each fragment in the anti-MYC immunoprecipitate versus Load was calculated and plotted. A typical outcome of one of two independent experiments is shown.
Discussion
In S.cerevisiae, ten HATs have been described to date [42], [43], [44]. Several genome-wide studies on gene expression [45], [46], [47], [48] and chromatin association [36], [49] have provided significant details on the engagements of the HAT complexes SAGA, TFIID, Mediator and NuA4 at gene promoters. However, these papers have not suggested the participation of any of these HATs in DNA replication. On the other hand, focused studies of origins or studies on protein interactions have tentatively linked HAT1 [19] and SAS2 [18] to DNA replication. In the present study we have compared the effects on DNA replication of deletions of HAT1, SAS2, SAS3, GCN5 and RTT109 and mutations in the essential ESA1 gene. These assays clearly outlined a major replication deficiency in the Δgcn5 strain with minor effects in the others (Figures 1 and 2). All subsequent analyses supported the notion that GCN5 is a positive regulator of origins. We postulate that GCN5 counteracts the negative effects of HDACs such as SIR2 and RPD3 [4], [5], [6]. Our observations expand the functional repertoire of GCN5, which has already been implicated in gene expression [43] and also in centromere function [37].
GCN5 has been extensively characterized as a global regulator of gene expression [36], [47], [50], [51]. Is it then possible that all effects we have observed are indirect via deregulated gene expression? Several arguments reduce the likelihood of this possibility. Firstly, we have detected credible signals of direct association of Gcn5p with a mini-chromosomal ARS1 (Fig. 3). Whereas the chromosomal ARS1 and ARS305 do not provide the luxury arrangement and distances between the genes and origins on the mini-chromosome, signals from direct Gcn5p association at these loci have been detected by us (Fig. 4) and others [36]. Finally, transcriptome analysis in Δgcn5 cells has revealed a bias towards the regulation of stress induced genes, but not towards genes responsible for DNA replication factors [47]. We therefore favor the idea that GCN5 directly regulates DNA replication.
The fact that Gcn5p weakly and possibly only transiently associates with origins (Fig. 4) suggests that its effects on origins are produced by global activity rather than targeted recruitment as is the case on promoters [36]. This idea is in tune with the notion that the negative origin regulators SIR2 and RPD3 are likely to act by global deacetylation [4], [5], [6]. A recent study suggests that at least in the case of RPD3 some targeted recruitment to origins is possible, but the mechanism remains unknown [7]. Regardless of whether the mode of action of Gcn5p is targeted or global, the effect of GCN5 on the chromatin structure of origins is substantial as shown in Fig. 5A. Another point of consideration regarding the mechanism of action of GCN5 is the observed deficiency of assembly of pre-replicative complexes in Δgcn5 cells (Fig. 6). This result suggests that GCN5 could have a major role in chromatin de-condensation, which in turn affects the licensing of origins. Studies in human cells have allocated a similar role in G1 for HBO1 [10], [11], [12]. At this point it is not clear if in human cells HBO1 is the only HAT engaged in G1 or if GCN5 also participates. As for S.cerevisiae, the remote functional homologue of HBO1, SAS2, shows little activity consistent with a major role in replication (Fig. 1). On the other hand, the deletion of HAT1 seems to have only minor effect on mini-chromosome stability (Fig. 1A), but strongly stimulates an origin when tethered to it (Fig. 1B). This observation and the data in [19] indicate that HAT1 could be involved in replication but the assays employed by us did not properly reveal such function. Finally, the proposed role of GCN5 in chromatin de-condensation and facilitation of pre-replicative complexes in G1 does not at all exclude additional roles in S phase as activator of origins or during elongation.
A very recent study has shown that in human cells GCN5 acetylates the key component of pre-replicative complexes CDC6 thus modulating its subsequent phosphorylation by Cyclin A-CDKs [52]. It is not clear if similar acetylation of Cdc6p by Gcn5p takes place in S. cerevisiae. It is tempting to speculate that at least some of the effects we observe here are consequence of not only histone acetylation, but of acetylation of Cdc6p as well. This question remains open until clear link between GCN5 and CDC6 is established.
Our data suggests that GCN5 has a major role in DNA replication. Nevertheless, GCN5 is not an essential gene so it is obviously not the only HAT involved. In support, the deletion of sas2, sas3 and hat1 and the esa1 mutants displayed slightly higher mini-chromosome instability as compared to wild type cells, but significantly lower as compared to gcn5 (Fig. 1A). It is conceivable that these HATs also play roles in the same steps, which are regulated by GCN5. In particular, it is not clear to what extent the mutations in ESA1 impair its function thus rendering its role in DNA replication far for from being excluded. It is noteworthy that ESA1 and GCN5 show a significant level of functional overlap on gene promoters [36].
Another issue stemming from our study is the possible role of GCN5 on centromeres [37]. As mentioned earlier, GCN5 has been implicated as a regulator of chromatin structure and in the function of centromeres [37]. Indeed, double deficiency of the ARS and CEN elements can explain the magnitude of Δgcn5 effect in Fig. 1. Whereas such scenario is not unlikely, our data on two chromosomal origins and a centromere shows that the alteration in chromatin structure is significant at origins and moderate to low on centromeres (Fig. 5). Even more, GCN5 strongly suppresses the replication deficiency phenotype of the mcm5-461, orc2-1 and orc5-1strains (Fig. 2). Hence, we suspect that GCN5 is a major positive replication factor independently of its activity on centromeres.
In summary, we have assigned a new role for GCN5. We have also introduced a new HAT in the regulation of DNA replication. Future studies should focus on the interplay between GCN5 and other HATs in modulating the activity of origins and the subsequent steps in DNA replication.
Supporting Information
(0.06 MB TIF)
(0.03 MB DOC)
Acknowledgments
We thank Dr. F. Robert for GCN5-MYC strain; Dr. C. Brandl for the ADA3-MYC strain; Dr. F. Winston for the SPT8 episome; Dr. T. Formosa for the SPT16 and the POB3 episomes; Dr. L. Pillus for the GAL4-HAT constructs and for the esa1 mutant strains; Dr. R. Li for the pARS1wt and pARS1/-B23/GAL4 mini-chromosomes.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This study was supported by a grant from Natural Sciences and Engineering Research Council (NSERC) (043230) to KY. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Blow JJ, Dutta A. Preventing re-replication of chromosomal DNA. Nat Rev Mol Cell Biol. 2005;6:476–486. doi: 10.1038/nrm1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Groth A, Rocha W, Verreault A, Almouzni G. Chromatin challenges during DNA replication and repair. Cell. 2007;128:721–733. doi: 10.1016/j.cell.2007.01.030. [DOI] [PubMed] [Google Scholar]
- 3.Knott SR, Viggiani CJ, Aparicio OM. To promote and protect: coordinating DNA replication and transcription for genome stability. Epigenetics. 2009;4:362–365. doi: 10.4161/epi.4.6.9712. [DOI] [PubMed] [Google Scholar]
- 4.Vogelauer M, Rubbi L, Lucas I, Brewer BJ, Grunstein M. Histone acetylation regulates the time of replication origin firing. Mol Cell. 2002;10:1223–1233. doi: 10.1016/s1097-2765(02)00702-5. [DOI] [PubMed] [Google Scholar]
- 5.Aparicio JG, Viggiani CJ, Gibson DG, Aparicio OM. The Rpd3-Sin3 histone deacetylase regulates replication timing and enables intra-S origin control in Saccharomyces cerevisiae. Mol Cell Biol. 2004;24:4769–4780. doi: 10.1128/MCB.24.11.4769-4780.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pappas DL, Jr, Frisch R, Weinreich M. The NAD(+)-dependent Sir2p histone deacetylase is a negative regulator of chromosomal DNA replication. Genes Dev. 2004;18:769–781. doi: 10.1101/gad.1173204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Knott SR, Viggiani CJ, Tavare S, Aparicio OM. Genome-wide replication profiles indicate an expansive role for Rpd3L in regulating replication initiation timing or efficiency, and reveal genomic loci of Rpd3 function in Saccharomyces cerevisiae. Genes Dev. 2009;23:1077–1090. doi: 10.1101/gad.1784309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aggarwal BD, Calvi BR. Chromatin regulates origin activity in Drosophila follicle cells. Nature. 2004;430:372–376. doi: 10.1038/nature02694. [DOI] [PubMed] [Google Scholar]
- 9.Irlbacher H, Franke J, Manke T, Vingron M, Ehrenhofer-Murray AE. Control of replication initiation and heterochromatin formation in Saccharomyces cerevisiae by a regulator of meiotic gene expression. Genes Dev. 2005;19:1811–1822. doi: 10.1101/gad.334805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iizuka M, Matsui T, Takisawa H, Smith MM. Regulation of replication licensing by acetyltransferase Hbo1. Mol Cell Biol. 2006;26:1098–1108. doi: 10.1128/MCB.26.3.1098-1108.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miotto B, Struhl K. HBO1 histone acetylase is a coactivator of the replication licensing factor Cdt1. Genes Dev. 2008;22:2633–2638. doi: 10.1101/gad.1674108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu ZQ, Liu X. Role for Plk1 phosphorylation of Hbo1 in regulation of replication licensing. Proc Natl Acad Sci U S A. 2008;105:1919–1924. doi: 10.1073/pnas.0712063105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doyon Y, Cayrou C, Ullah M, Landry AJ, Cote V, et al. ING tumor suppressor proteins are critical regulators of chromatin acetylation required for genome expression and perpetuation. Mol Cell. 2006;21:51–64. doi: 10.1016/j.molcel.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 14.Iizuka M, Sarmento OF, Sekiya T, Scrable H, Allis CD, et al. Hbo1 Links p53-dependent stress signaling to DNA replication licensing. Mol Cell Biol. 2008;28:140–153. doi: 10.1128/MCB.00662-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johmura Y, Osada S, Nishizuka M, Imagawa M. FAD24 acts in concert with histone acetyltransferase HBO1 to promote adipogenesis by controlling DNA replication. J Biol Chem. 2008;283:2265–2274. doi: 10.1074/jbc.M707880200. [DOI] [PubMed] [Google Scholar]
- 16.Avvakumov N, Cote J. The MYST family of histone acetyltransferases and their intimate links to cancer. Oncogene. 2007;26:5395–5407. doi: 10.1038/sj.onc.1210608. [DOI] [PubMed] [Google Scholar]
- 17.Lafon A, Chang CS, Scott EM, Jacobson SJ, Pillus L. MYST opportunities for growth control: yeast genes illuminate human cancer gene functions. Oncogene. 2007;26:5373–5384. doi: 10.1038/sj.onc.1210606. [DOI] [PubMed] [Google Scholar]
- 18.Zou Y, Bi X. Positive roles of SAS2 in DNA replication and transcriptional silencing in yeast. Nucleic Acids Res. 2008;36:5189–5200. doi: 10.1093/nar/gkn465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suter B, Pogoutse O, Guo X, Krogan N, Lewis P, et al. Association with the origin recognition complex suggests a novel role for histone acetyltransferase Hat1p/Hat2p. BMC Biol. 2007;5:38. doi: 10.1186/1741-7007-5-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marahrens Y, Stillman B. A yeast chromosomal origin of DNA replication defined by multiple functional elements. Science. 1992;255:817–823. doi: 10.1126/science.1536007. [DOI] [PubMed] [Google Scholar]
- 21.Gauthier L, Dziak R, Kramer DJ, Leishman D, Song X, et al. The role of the carboxyterminal domain of RNA polymerase II in regulating origins of DNA replication in Saccharomyces cerevisiae. Genetics. 2002;162:1117–1129. doi: 10.1093/genetics/162.3.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jacobson S, Pillus L. Molecular requirements for gene expression mediated by targeted histone acetyltransferases. Mol Cell Biol. 2004;24:6029–6039. doi: 10.1128/MCB.24.13.6029-6039.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kramer DJ, Gauthier L, Yankulov K. Higher-accuracy method for measuring minichromosome stability in Saccharomyces cerevisiae. Biotechniques. 2002;32:1036, 1038, 1040 passim. [PubMed] [Google Scholar]
- 24.Tye BK. Minichromosome maintenance as a genetic assay for defects in DNA replication. Methods. 1999;18:329–334. doi: 10.1006/meth.1999.0793. [DOI] [PubMed] [Google Scholar]
- 25.Kurdistani SK, Grunstein M. In vivo protein-protein and protein-DNA crosslinking for genomewide binding microarray. Methods. 2003;31:90–95. doi: 10.1016/s1046-2023(03)00092-6. [DOI] [PubMed] [Google Scholar]
- 26.Gregory PD, Barbaric S, Horz W. Restriction nucleases as probes for chromatin structure. Methods Mol Biol. 1999;119:417–425. doi: 10.1385/1-59259-681-9:417. [DOI] [PubMed] [Google Scholar]
- 27.Clarke AS, Samal E, Pillus L. Distinct roles for the essential MYST family HAT Esa1p in transcriptional silencing. Mol Biol Cell. 2006;17:1744–1757. doi: 10.1091/mbc.E05-07-0613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maine GT, Sinha P, Tye BK. Mutants of S. cerevisiae defective in the maintenance of minichromosomes. Genetics. 1984;106:365–385. doi: 10.1093/genetics/106.3.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dziak R, Leishman D, Radovic M, Tye BK, Yankulov K. Evidence for a role of MCM (mini-chromosome maintenance)5 in transcriptional repression of sub-telomeric and Ty-proximal genes in Saccharomyces cerevisiae. J Biol Chem. 2003;278:27372–27381. doi: 10.1074/jbc.M301110200. [DOI] [PubMed] [Google Scholar]
- 30.Li R, Yu DS, Tanaka M, Zheng L, Berger SL, et al. Activation of chromosomal DNA replication in Saccharomyces cerevisiae by acidic transcriptional activation domains. Mol Cell Biol. 1998;18:1296–1302. doi: 10.1128/mcb.18.3.1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ehrenhofer-Murray AE, Gossen M, Pak DT, Botchan MR, Rine J. Separation of origin recognition complex functions by cross-species complementation. Science. 1995;270:1671–1674. doi: 10.1126/science.270.5242.1671. [DOI] [PubMed] [Google Scholar]
- 32.Dillin A, Rine J. Separable functions of ORC5 in replication initiation and silencing in Saccharomyces cerevisiae. Genetics. 1997;147:1053–1062. doi: 10.1093/genetics/147.3.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wittmeyer J, Joss L, Formosa T. Spt16 and Pob3 of Saccharomyces cerevisiae form an essential, abundant heterodimer that is nuclear, chromatin-associated, and copurifies with DNA polymerase alpha. Biochemistry. 1999;38:8961–8971. doi: 10.1021/bi982851d. [DOI] [PubMed] [Google Scholar]
- 34.VanDemark AP, Blanksma M, Ferris E, Heroux A, Hill CP, et al. The structure of the yFACT Pob3-M domain, its interaction with the DNA replication factor RPA, and a potential role in nucleosome deposition. Mol Cell. 2006;22:363–374. doi: 10.1016/j.molcel.2006.03.025. [DOI] [PubMed] [Google Scholar]
- 35.Saleh A, Lang V, Cook R, Brandl CJ. Identification of native complexes containing the yeast coactivator/repressor proteins NGG1/ADA3 and ADA2. J Biol Chem. 1997;272:5571–5578. doi: 10.1074/jbc.272.9.5571. [DOI] [PubMed] [Google Scholar]
- 36.Robert F, Pokholok DK, Hannett NM, Rinaldi NJ, Chandy M, et al. Global position and recruitment of HATs and HDACs in the yeast genome. Mol Cell. 2004;16:199–209. doi: 10.1016/j.molcel.2004.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vernarecci S, Ornaghi P, Bagu A, Cundari E, Ballario P, et al. Gcn5p plays an important role in centromere kinetochore function in budding yeast. Mol Cell Biol. 2008;28:988–996. doi: 10.1128/MCB.01366-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adkins MW, Carson JJ, English CM, Ramey CJ, Tyler JK. The histone chaperone anti-silencing function 1 stimulates the acetylation of newly synthesized histone H3 in S-phase. J Biol Chem. 2007;282:1334–1340. doi: 10.1074/jbc.M608025200. [DOI] [PubMed] [Google Scholar]
- 39.Crampton A, Chang F, Pappas DL, Jr, Frisch RL, Weinreich M. An ARS element inhibits DNA replication through a SIR2-dependent mechanism. Mol Cell. 2008;30:156–166. doi: 10.1016/j.molcel.2008.02.019. [DOI] [PubMed] [Google Scholar]
- 40.Rehman MA, Fourel G, Mathews A, Ramdin D, Espinosa M, et al. Differential requirement of DNA replication factors for subtelomeric ARS consensus sequence protosilencers in Saccharomyces cerevisiae. Genetics. 2006;174:1801–1810. doi: 10.1534/genetics.106.063446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rehman MA, Wang D, Fourel G, Gilson E, Yankulov K. Subtelomeric ACS-containing proto-silencers act as antisilencers in replication factors mutants in Saccharomyces cerevisiae. Mol Biol Cell. 2009;20:631–641. doi: 10.1091/mbc.E08-01-0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sterner DE, Berger SL. Acetylation of histones and transcription-related factors. Microbiol Mol Biol Rev. 2000;64:435–459. doi: 10.1128/mmbr.64.2.435-459.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carrozza MJ, Utley RT, Workman JL, Cote J. The diverse functions of histone acetyltransferase complexes. Trends Genet. 2003;19:321–329. doi: 10.1016/S0168-9525(03)00115-X. [DOI] [PubMed] [Google Scholar]
- 44.Yang XJ. The diverse superfamily of lysine acetyltransferases and their roles in leukemia and other diseases. Nucleic Acids Res. 2004;32:959–976. doi: 10.1093/nar/gkh252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee TI, Causton HC, Holstege FC, Shen WC, Hannett N, et al. Redundant roles for the TFIID and SAGA complexes in global transcription. Nature. 2000;405:701–704. doi: 10.1038/35015104. [DOI] [PubMed] [Google Scholar]
- 46.Holstege FC, Jennings EG, Wyrick JJ, Lee TI, Hengartner CJ, et al. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 47.Huisinga KL, Pugh BF. A genome-wide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Mol Cell. 2004;13:573–585. doi: 10.1016/s1097-2765(04)00087-5. [DOI] [PubMed] [Google Scholar]
- 48.Reid JL, Moqtaderi Z, Struhl K. Eaf3 regulates the global pattern of histone acetylation in Saccharomyces cerevisiae. Mol Cell Biol. 2004;24:757–764. doi: 10.1128/MCB.24.2.757-764.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 50.Huisinga KL, Pugh BF. A TATA binding protein regulatory network that governs transcription complex assembly. Genome Biol. 2007;8:R46. doi: 10.1186/gb-2007-8-4-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baker SP, Grant PA. The SAGA continues: expanding the cellular role of a transcriptional co-activator complex. Oncogene. 2007;26:5329–5340. doi: 10.1038/sj.onc.1210603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Paolinelli R, Mendoza-Maldonado R, Cereseto A, Giacca M. Acetylation by GCN5 regulates CDC6 phosphorylation in the S phase of the cell cycle. Nat Struct Mol Biol. 2009;16:412–420. doi: 10.1038/nsmb.1583. [DOI] [PubMed] [Google Scholar]
- 53.Harper JW, Adami GR, Wei N, Keyomarsi K, Elledge SJ. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(0.06 MB TIF)
(0.03 MB DOC)