Abstract
Brain serotonin (5-HT) neurotransmission plays a key role in the regulation of mood and has been implicated in a variety of neuropsychiatric conditions. Tryptophan hydroxylase (TPH) is the rate-limiting enzyme in the biosynthesis of 5-HT. Recently, we discovered a second TPH isoform (TPH2) in vertebrates, including man, which is predominantly expressed in brain, while the previously known TPH isoform (TPH1) is primarly a non-neuronal enzyme. Overwhelming evidence now points to TPH2 as a candidate gene for 5-HT-related psychiatric disorders. To assess the role of TPH2 gene variability in the etiology of psychiatric diseases we performed cDNA sequence analysis of TPH2 transcripts from human post mortem amygdala samples obtained from individuals with psychiatric disorders (drug abuse, schizophrenia, suicide) and controls. Here we show that TPH2 exists in two alternatively spliced variants in the coding region, denoted TPH2a and TPH2b. Moreover, we found evidence that the pre-mRNAs of both splice variants are dynamically RNA-edited in a mutually exclusive manner. Kinetic studies with cell lines expressing recombinant TPH2 variants revealed a higher activity of the novel TPH2B protein compared with the previously known TPH2A, whereas RNA editing was shown to inhibit the enzymatic activity of both TPH2 splice variants. Therefore, our results strongly suggest a complex fine-tuning of central nervous system 5-HT biosynthesis by TPH2 alternative splicing and RNA editing. Finally, we present molecular and large-scale linkage data evidencing that deregulated alternative splicing and RNA editing is involved in the etiology of psychiatric diseases, such as suicidal behaviour.
Introduction
Serotonin (5-hydroxytryptamine, 5-HT) is a monoaminergic neurotransmitter involved in multiple facets of behavioural control [1], and it has been known for more than four decades that tryptophan hydroxylase (TPH; EC 1.14.16.4) perfoms the first-step and rate-limiting step in its biosynthesis [1], [2]. The serotonergic projection system is the most extensive monoaminergic system in the brain of vertebrates, with its roots in a handful of 5-HT-synthesizing neurons within the midbrain, pons, and medulla oblongata, which altogether constitute the raphe nuclei B1–B9 [1]. These few serotonergic raphe neurons innervate most cortical and subcortical brain areas, including the amygdala, a brain structure critically involved in the modulation of emotional behaviour related to anxiety, fear and reward [3], [4]. Dysregulations in the serotonergic system in the brain have been implicated in a variety of psychiatric disorders, such as depression, suicide, schizophrenia and addiction, which are accompanied by abnormal amygdala function [5].
Recently, a second TPH gene (TPH2) was identified, which encodes for the main 5-HT-synthesizing enzyme in neurons, whereas the previously known TPH gene (TPH1) is predominantly expressed in peripheral tissues [1], [6]. While TPH1 is still intensively investigated with regard to its role in developmental processes in embryos and nourishing mothers [7], [8], cancer [9], [10], platelet functions in primary haemostasis [11], liver regeneration [12], insulin secretion [13], and pulmonary hypertension [14], psychiatric 5-HT research now mainly focuses on TPH2 [1], [6], [15], [16], [17], [18], [19]. Although TPH1 has a higher catalytic rate than TPH2 [20] and since in several human brain areas, TPH1 mRNA is more abundant than TPH2 [21], TPH1 protein was not detectable by immunohistochemistry [22]. Thus, TPH1 seems not to contribute to brain 5-HT biosynthesis but it cannot be ruled out that impaired TPH1 activity leads to psychiatric illness due to metabolic disorders, such as diabetes [13] or impaired liver function [12].
Numerous studies have identified associations of single nucleotide polymorphisms (SNPs) in the human TPH2 gene with psychiatric diseases [16], [17], [18], [19]. These studies mainly focused on non-coding SNPs and only few functional data exist. For example, we previously reported the association of a TPH2 promoter SNP (-614T>A; rs11178997) with reduced transcriptional activity [23], whereas another allelic TPH2 promoter variant (-844G>T; rs4570625) was shown to associate with amygdala hyperexcitabilty in reaction to emotional stimuli [24], [25]. A functional coding Tph2 SNP was first described in mouse, where the highly conserved proline447 is changed to arginine by the SNP C1473G resulting in a 55% decrease of 5-HT biosynthesis when expressed in PC12 cells [26]. Consistently, BALB/cJ and DBA/2J mouse strains, homozygous for the allele 1473G, showed substantially reduced 5-HT-synthesizing activity in the brain [26]. To date, three non-synonymous TPH2 SNPs have been associated with psychiatric disorders in humans, which severely impair TPH2 enzymatic activity by causing the amino acid substitutions p.P206S (c.757C>T; exon 6), p.W303R (c.907C>T; exon 7), and p.R441H (c.1322G>A; also known as 1463G>A; exon 11) [27], [28], [29]. Interestingly, we and others tried to confirm the SNP c.1322G>A at the genomic level in more than 5,000 patients of matched collectives without any success [15], [29], [30], [31], [32], [33]. In addition, splice variants in the non-coding 3′-UTR were described, but no functional effects were detected on 5-HT biosynthesis by them [34]. As brain 5-HT biosynthesis is regulated by TPH2 [6], [26], and TPH2 SNPs increase the risk for psychiatric disorders [27], [28], [29], we decided to further analyze the polymorphic variability of the human TPH2 gene and focused on its coding region.
Here we show by cDNA sequence analysis of post mortem RNA samples obtained from the human amygdala that TPH2 transcripts exist in at least two alternatively spliced variants in the coding region, namely TPH2a and TPH2b. Moreover, extensive RNA editing of both TPH2 isoforms leads to protein variants with distinct catalytic properties. Finally, our data indicate that drug abuse may disturb RNA editing and that imbalanced RNA editing might be involved in the pathogenesis of psychiatric disorders.
Results and Discussion
Human TPH2 Is Alternatively Spliced
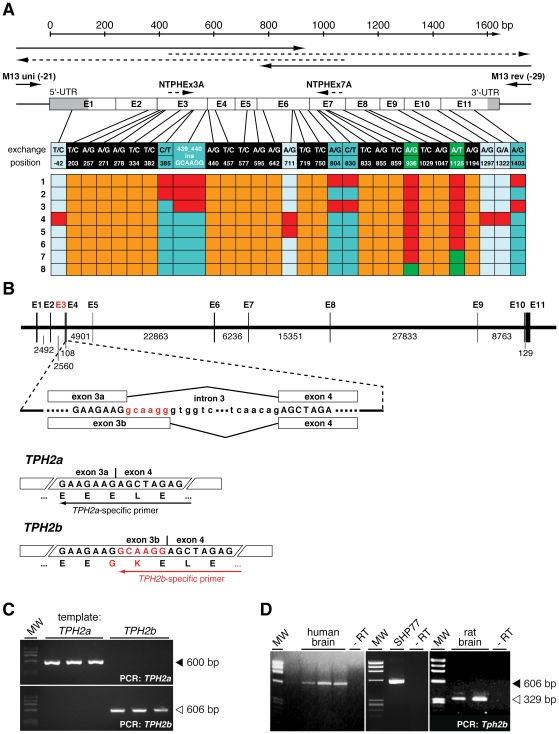
To identify functional SNPs in the human TPH2 gene, we analyzed post mortem brain samples from drug abuse and suicide victims, schizophrenic patients, and controls without a psychiatric history (a full description of the collectives is included in methods). RNA samples were obtained from the amygdala and transcribed into cDNA to amplify, clone and sequence the TPH2 open reading frame and parts of the untranslated regions. By alignment of the obtained sequences to the TPH2 mRNA reference sequence (GenBank NM_173353), we detected multiple SNPs in nearly all TPH2 exons (Figure 1A; Table S1). Among these, only three were known previously, namely the database SNPs rs7305115 (c.936A>G) and rs4290270 (c.1125A>T) and the recently reported SNP c.1322G>A (also known as G1463A) [29].
Figure 1. Human TPH2 exists in two splice variants.
(A) Sequencing strategy of TPH2 cDNA clones obtained from human amygdala of patients with psychopathological disorders and controls. Sequence alignments with the TPH2 mRNA reference sequence (GenBank NM_173353) led to the identification of 29 SNPs and a 6 bp insertion in exon 3 (n = 104 independent sequences). A compilation of representative TPH2 cDNA clones (1–8) and the positions of all found SNPs are shown; red boxes indicate the presence of a SNP in the corresponding clone. Eight SNPs were present in dependence of the insertion, forming two mutually exclusive polymorphism patterns. SNPs detectable in presence of the insertion (TPH2a) are indicated in dark blue, light blue SNPs were found only in their absence (TPH2b). The SNPs at the green positions correspond to the known SNPs rs7305115 and rs4290270. The insertion is a product of alternative splicing of intron 3. (B) Schematic representation of the alternative splicing of TPH2 pre-mRNA. In higher vertebrates splicing of intron 3 usually occurs at the highly conserved GC splicing donor site (SDS) resulting in the known TPH2, now called TPH2a. In humans, primates and rats a GT dinucleotide exists 6 bp downstream of the GC SDS, and acts an alternative SDS leading to the inclusion of two additional amino acids, Gly and Lys. This longer TPH2 isoform is now referred to as TPH2b. (C) Specificity of splice-specific TPH2 primers on plasmid DNA. (D) RT-PCR using splice-specific primers showed the presence of TPH2b transcripts in normal human and rat brain and also human neuroendocrine SHP77 cells.
Most notably, we detected a 6 bp insertion (c.439_440insGCAAGG) between exons 3 and 4 by this procedure, which corresponds to a novel splice isoform. Intron 3 starts with a non-canonical GC splicing donor site (SDS), leading to the fusion of exons 3a and 4 (Figure 1B). However, another GT dinucleotide exists 6 bp downstream of the GC SDS, which operates as an alternative SDS, leading to the inclusion of two additional triplets coding for the amino acids Gly and Lys (exon 3b; Figure 1B). We called this novel TPH2 splice isoform, TPH2b, to discern it from the known TPH2 reference sequence (GenBank NM_173353), which is now referred to as TPH2a.
Interestingly, intron 3 of all higher vertebrates starts with a non-canonical GC SDS (Table S2). Exceptions are fishes, which carry a canonical GT SDS at the same position, suggesting evolutionary conservation of this GC-AG intron for at least 450 million years. GC-AG introns occur with a frequency of only about 0.7% in the human genome, but 60% of them are involved in alternative splicing, especially during embryogenesis [35], [36]. Thus, our findings of alternative TPH2 splicing suggests that this intron might be important for developmental processes, as 5-HT is known to be involved even in pre-neuronal growth regulation [8], [37], [38]. Notably, the alternative GT SDS found in humans is also present in primates and rats, but not in mice (Table S2), excluding the latter as an animal model for the investigation of TPH2 alternative splicing.
At the protein level, the Gly-Lys insertion (p.146_147insGK) leads to the interruption of a negatively charged stretch of Glu residues by the positive Lys in the hinge structure between the regulatory and catalytic domains (Figure 1B). The hinge region is crucial for substrate accessibility in all aromatic amino acid hydroxylases [2] and it was recently shown that human TPH2 is no exception in this regard [39]. Hence, this structural feature of TPH2B predicted an impact on its hydroxylating activity (see below).
TPH2b transcripts can be found in every individual, even without a psychiatric history, using a splice-specific primer (Figure 1B–D). Moreover, TPH2b transcripts are also readily detectable in rat brain and human small cell lung carcinoma SHP77 cells, which express TPH2 (Figure 1D) together with other neuroendocrine markers. Therefore, our data indicate that TPH2B is an isoform that contributes to the brain 5-HT biosynthesis in a hitherto undefined manner.
TPH2a and TPH2b Pre-mRNAs Undergo Editing
Most notably, the analysis of TPH2 gene variability revealed two distinct patterns of synonymous and non-synonymous base exchanges in dependence of the c.439_440insGCAAGG insertion (Figure 1A; Table 1). Thus, the majority of TPH2a transcripts are characterized by the SNPs c.-42T>C (5′-UTR; exon 1), c.711A>G (p.R237; exon 6), c.1297A>G (p.R433G; exon 10), and c.1322G>A (p.R441H; exon 11), whereas TPH2b transcripts contained four other polymorphisms, namely c.385C>T (p.Q129X; exon 3), c.804A>G (p.K268; exon 6), c.830C>T (p.P277L; exon 7), and c.1403A>G (p.Q648R; exon 11). Both isoform-specific polymorphism patterns appeared almost without exception together with the database SNPs rs7305115 and rs4290270 (Figure 1A). Interestingly, the TPH2b polymorphism c.385C>T is a nonsense base exchange, which creates a premature stop codon upstream of the catalytic domain of TPH2 [39] and would represent a null mutation with regard to enzymatic activity. This variant would be expected to be a substrate for nonsense-mediated mRNA decay (NMD) [40], [41], but this is obviously not the case given the ease of its detection (Figure 1D).
Table 1. Amygdala-specific editing of TPH2a transcripts.
| TPH2 Isoform | Genotype rs4290270 | No. Clones | Editing Position and Percentual Distribution in the Respective Transcripts | |||
| 1 | 2 | 3 | 4 | |||
| Amygdala | ||||||
| TPH2a | A | 48 | 33% | 31% | 33% | 33% |
| T | 17 | 0% | 0% | 0% | 0% | |
| TPH2b | A | 27 | 78% | 96% | 96% | 96% |
| T | - | - | - | - | - | |
| other brain areasa | ||||||
| TPH2a | A | 8 | 0% | 0% | 0% | 0% |
| T | 19 | 0% | 0% | 0% | 0% | |
| TPH2b | A | 11 | 100% | 100% | 100% | 100% |
| T | - | - | - | - | - | |
The percentual distribution of edited positions in TPH2a/b transcripts revealed that TPH2a is only edited in the amygdala, while TPH2b editing is detectable in all investigated brain areas. Note that in presence of SNP rs4290270 A neither TPH2a editing could be observed nor expression of TPH2b.
Cortex, thalamus, hypothalamus, hippocampus, cerebellum, median raphe, pons, and striatum.
The isoform-specific polymorphism patterns were detected in the vast majority of TPH2a and TPH2b transcripts, but they cannot be explained by the presence of only two TPH2 alleles. Therefore, our results prompted us to hypothesize posttranscriptional modification of TPH2 transcripts by RNA editing, a mechanism known to regulate the activity of many neuronal proteins [42], [43], [44], [45].
Posttranscriptional RNA editing of primary transcripts alters genomically encoded sequences and enables multiple transcripts from a single gene, thereby generating proteomic diversity from a limited number of genes [46]. In humans, RNA editing was first described for the apolipoprotein B (APOB) [47], [48], [49]. A cytidine deaminase of the apolipoprotein B mRNA editing enzyme catalytic polypeptide (APOBEC) family converts a cytidine to uridine in the APOB primary transcripts by hydrolytic deamination [50]. This C-to-U editing (C>U) of APOB transcripts changes a glutamine codon to a premature stop codon in the intestine, giving rise to a functionally important, truncated 48 kDa protein, whereas the non-edited APOB100 is expressed in liver. In mammals, base exchanges from adenosine-to-inosine (A>I) represent the most common RNA editing mechanism, which influences neurotransmission by modulating the functional properties of glutamate receptors [44], serotonin receptors [43] and potassium channels [42]. The best studied example, the 5-HT2C receptor, is dynamically edited at five positions (A to E) in exon V, leading to multiple receptor isoforms, which differ in constitutive activity and intracellular signal transduction efficacy, thereby modulating the strength of 5-HT neurotransmission [43], [51], [52]. A>I editing is catalyzed by deaminases of the adenosine deaminase acting on RNA (ADAR) family and inosine is recognized as guanosine by the translation machinery [46]. Given that RNA editing mediates fine-regulation of central nervous system (CNS) neurotransmission, dysregulations of RNA editing have been associated with brain disorders [53].
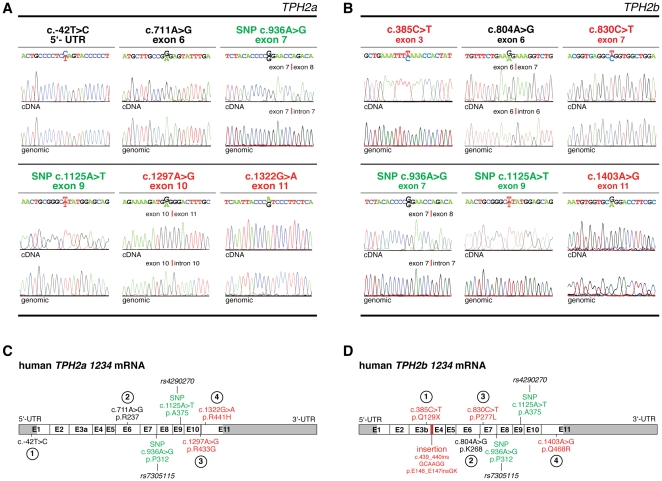
To assess for TPH2 editing, we cloned and sequenced the corresponding TPH2 exons from the genomic DNA of individuals, in whom we detected TPH2 polymorphisms. However, neither the polymorphisms c.-42T>C, c.711A>G, c.1297A>G and c.1322G>A (TPH2a) nor c.385C>T, c.804A>G, c.830C>T and c.1403A>G (TPH2b) were found in any of these samples. Only the database SNPs rs7305115 and rs4290270 could be detected at the genomic level and were confirmed as genuine SNPs (Figure 2A and 2B). Thus, we found that TPH2a and TPH2b pre-mRNAs are extensively RNA-edited by a yet unidentified mechanism, which involves mutually exclusive RNA editing patterns (indicated by Arabic numbers) and leads to the expression of the corresponding TPH2a 1234 and TPH2b 1234 transcripts, respectively (Figure 2C and 2D). Moreover, detection of partially edited TPH2a (TPH2a 2, TPH2a 134) and TPH2b (TPH2b 1, TPH2b 234) transcripts (Figure 1A) suggests a dynamic TPH2 RNA editing machinery in analogy to 5-HT2C receptor editing [43]. This results in a wide variety of different TPH2 isoforms, and increases the biochemical diversity and complexity of central 5-HT biosynthesis.
Figure 2. TPH2a and TPH2b undergo extensive mRNA editing.
(A, B) Alignment of genomic TPH2 sequences and corresponding cDNA traces of TPH2a and TPH2b transcripts revealed that neither the TPH2a SNPs c.-42T>C, c.711A>G, c.1297A>G and c.1322G>A (A) nor the TPH2b polymorphisms c.385C>T, c.804A>G, c.830C>T and c.1403A>G are encoded genomically (B), indicating posttranscriptional RNA editing for those positions. Only the SNPs rs7305115 (c.936A>G) and rs4290270 (c.1125A>T) could be verified as genuine SNPs at the genomic level. (C, D) The editing patterns for TPH2a and TPH2b transcripts are mutually exclusive. Four edited positions exist in each alternatively spliced variant, indicated by arabic numbers. Schematic representation of TPH2a 1234 (C) and TPH2b 1234 (D) transcripts. Synonymous and non-synonymous base substitutions are indicated in black and red, respectively; SNPs are shown in green.
Furthermore, we observed TPH2a editing exclusively in the amygdala, whereas TPH2b was edited in all brain regions analyzed (Table 1), and also in SHP77 cells (data not shown). Accordingly, our data underscore previous findings that RNA editing is often restricted to discrete brain regions [54].
RNA editing of human TPH2 transcripts is remarkably miscellaneous (Figure 2) and comprises all editing mechanisms known for mammals, including rare U>C editing [55], which we found for TPH2a position 1 (c.-42T>C; 5′-UTR). Interestingly, we also found RNA editing for the c.1322G>A (R441H) polymorphism, which has been associated with major depression [29]. This SNP is currently a matter of major debate, since it could not be confirmed at the genomic level neither in our study (Figure 2A), nor in laboratories worldwide [15], [29], [30], [31], [32], [33]. Thus, its tempting to speculate that the detection of c.1322G>A in blood DNA samples by Zhang et al. [29] might be due to a rare de novo mutation in elderly patients. It is conceivable that this G>A transition may have resulted from deamination of a methylated cytosine, as c.1322G is part of a CpG dinucleotide, the major target of DNA methyltransferases [56]. The transition c.1322G>A might also have resulted from somatic hypermutation, which was shown to modulate genomic DNA by edited RNA in B cells [57]. However, our results strongly favour RNA editing for c.1322G>A by an extremely rare mechanism, which thus far was only described for the proviral RNA of the human immunodeficiency virus [58]. Thus, TPH2a transcripts might offer a unique possibility to study G>A editing in a physiological context. Furthermore, to our knowledge, most mammalian pre-mRNAs are edited by only one mechanism [42], [43], [44], [45]. Thus, TPH2 transcripts might offer an interesting target for the investigation of how different editing machineries act in concert on a single transcript.
Editing Defines the Kinetics of TPH2A and TPH2B Isoforms
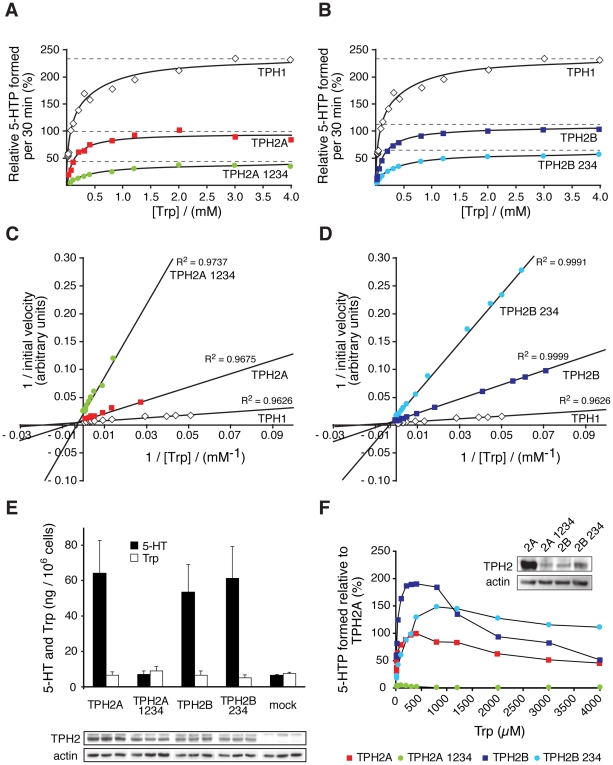
Alternative splicing and RNA editing of human TPH2 transcripts generate multiple protein variants with potentially different properties. To address this question, we stably expressed TPH2a and TPH2b and their edited isoforms in rat pheochromocytoma PC12 cells and performed kinetic studies with the cellular lysates (Figure 3A–D). Instead of TPH2b 1234 we analyzed a TPH2b 234 cDNA, which we also detected in our samples (Figure 1A, Table 1), but lacks nonsense editing at position 1 (c.385C>T; p.Q129X) and enables the expression of a full-length TPH2B 234 protein.
Figure 3. Kinetic properties of TPH2 variants are modulated by RNA editing.
(A, B) 5-hydroxytryptophan (5-HTP) formation of TPH2 containing cellular PC12 lysates in presence of the synthetic cofactor 6-methyl-tetrahydrobiopterin (6MPH4). (C, D) Double reciprocal Lineweaver-Burk plots for Km(W) determination of TPH2 variants. Enzymatic activities of TPH2A and TPH2B, and TPH2A 1234 and TPH2B 234 isoforms were similar, respectively, when using the synthetic cofactor 6MPH4. TPH1 served as a control. RNA editing decreased enzyme activity in both isoforms. Shown are combined data of 4–7 independent experiments. (E) 5-HT and Trp contents of stably transfected PC12 cells. Western blots of the TPH2 variants were used for normalization of 5-HT levels (n = 10 independent experiments). (F) Enzymatic activity of TPH2 variants expressed in non-neuronal HEK293 cells in presence of the natural cofactor tetrahydrobiopterin (BH4). At concentrations above the physiological Trp range of 30–50 µM, all variants except TPH2B 234, exhibit strong substrate inhibition. Shown are combined data of 6 independent experiments.
It has been helpful to use synthetic analogues of TPH cofactors to discern catalytic differences between TPH enzymes isolated from different tissues (Table S3), because the differences are rather small if the natural cofactor tetrahydrobiopterin (BH4) is used [2], [20]. Therefore, we used 6-methyl-tetrahydrobiopterin (6MPH4), which provides the highest resolution of kinetic differences, and obtained hyperbolic plots for TPH2A, TPH2B, TPH2A 1234 and TPH2B 234, according to the Michaelis-Menten equation (Figure 3A and 3B). The analysis of kinetics by double-reciprocal Lineweaver-Burk plots allowed determination of the Michaelis constants (Km) for Trp of the indicated TPH2 variants (Figure 3C and 3D). The Km(Trp) values for TPH2A and TPH2B were nearly similar (Table 2), but significantly higher for the edited variants, indicating a reduced affinity for the substrate Trp (Table 2). However, Km(Trp) values for TPH2A and TPH2B were close to the previously reported constants and consistently higher than for TPH1 (Table S3) [20], [39].
Table 2. Kinetic constants of TPH2 variants.
| Km, Trp (µM)6MPH4 (300 µM) | Km, Trp (µM)BH4 (300 µM) | |
| TPH2A | 126±9 (n = 6) | 16±11 (n = 6) |
| TPH2A 1234 | 292±97 (n = 7)* | 23±3 (n = 6) |
| TPH2B | 124±11 (n = 4) | 37±69 (n = 6)# |
| TPH2B 234 | 304±77 (n = 7)* | 116±18 (n = 6)* , # |
: p<0.05 versus non-edited;
: p<0.05 versus all others.
PC12 cells have been used to directly assess 5-HT synthesis of recombinant TPH2 mutants [29]. The expression of the four TPH2 variants in PC12 cells revealed equal 5-HT contents for TPH2A and TPH2B, and, unexpectedly, also for TPH2B 234, for which we had expected lower activity (Figure 3E). However, it was previously shown that PC12 cells can only store limited neurotransmitter amounts in their vesicles [59]. Thus, it is conceivable that the TPH2 activity in these three stable cell lines by far exceeded their vesicular 5-HT storage capacity. Interestingly, TPH2A 1234-expressing cells did not produce significantly elevated 5-HT levels compared to mock-transfected cells, which contained comparable 5-HT amounts of about 18% of the maximal levels detected (Figure 3E). Therefore, our data indicate a major loss of TPH2A 1234 enzymatic activity by RNA editing and support the recently reported 80% reduction of 5-HT synthesis in PC12 cells expressing the TPH2-R441H mutant [29]. This mutant corresponds to a TPH2A 4 protein, but cumulative effects of the remaining editing positions might also contribute to TPH2A 1234 inactivation. However, our results raise the question whether the 20% residual activity of TPH2-R441H might simply reflect a low intrinsic 5-HT synthesis capacity of PC12 cells, either due to a low endogenous TPH expression or the known substrate promiscuity of tyrosine hydroxylase [2], which is highly expressed in these catecholaminergic cells.
To circumvent any above-mentioned artefactual influences of PC12 cells, we determined the Km(Trp) values of the TPH2 variants also in non-neuronal HEK293 cells using the natural cofactor BH4 (Table 2). As expected, kinetics with BH4 resulted in a lower resolution of the Km(Trp) values of the four TPH2 variants (Table 2), but revealed major differences in the relative maximal velocities (Vmax) of the corresponding enzymes. At physiological Trp concentrations of 30 to 50 µM, all TPH2 variants obeyed the Michaelis-Menten equation, but showed significant substrate inhibition at higher concentrations (Figure 3F). TPH2A 1234 presented the lowest Vmax with 5% of TPH2A and confirmed its enzymatic inactivation by RNA editing, as shown by 5-HT measurements (Figure 3E). Interestingly, TPH2B presented the highest Vmax, which was twice as high as for TPH2A and well in accordance with the prediction that the GK insertion into the hinge region allows easier access of the substrates to the catalytic core. RNA editing of TPH2b resulted in a 50% decrease of Vmax of the corresponding TPH2B 234 protein, which exhibited the highest Km(Trp) value of all variants (Table 2). However, TPH2B 234 activity, which was still consistently higher than for TPH2A, can be totally abolished by RNA editing at position 1 (c.385C>T; p.Q129X), resulting in a premature stop codon upstream of the catalytic domain [39] and the expression of an inactive, truncated TPH2B 1 protein. Thus, TPH2B might be important for a rapid response in amygdala 5-HT synthesis, when 5-HT levels need to be increased. Furthermore, it possesses the possibility to be rapidly switched off by editing of its RNA at the position 1 (c.385C>T).
Although TPH2b 1 transcripts would be expected for degradation by NMD [40], [41], we detected them easily (Figure 1D). In this respect, APOB48 transcripts are protected from NMD by the C>U editing machinery, which allows for the expression of the truncated APOB form [47], [48], [49]. Accordingly, our data suggest that TPH2b 1 transcripts might also be protected from NMD by the same mechanism and point to a physiological role of the TPH2B truncation.
Our data show that the activities of both TPH2 isoforms are inhibited by RNA editing and can even be completely abolished by this mechanism (Figure 3F). Moreover, TPH2 physiologically acts as a tetramer [2], [39] and forms functional heteromers with its mutant variants [27], [29]. Coexpression studies revealed intermediate enzymatic activities of the resulting hybrids as compared with the corresponding wildtype and mutant homotetramers [27], [29]. Therefore, TPH2 proteomic diversity generated by alternative splicing and RNA editing suggests further control of 5-HT biosynthesis at the level of enzyme oligomerization.
In conclusion, our results underscore that human CNS 5-HT biosynthesis is a highly regulated process, which is based on the expression of a wide variety of functional TPH2 proteins with different properties. This should enable a complex fine-tuning of 5-HT biosynthesis in response to agonist stimulation in order to maintain optimal 5-HT neurotransmission.
TPH2 Editing Is Abnormal in Individuals with Psychiatric Disorders
Interestingly, TPH2a 1234 editing (Figure 2C) was found elevated by 20% in transcripts obtained from the amygdala of drug abuse and suicide victims compared to controls (Table S4). Thus, the known 5-HT hypofunction in psychiatric disorders may result at least in part from the expression of the low active TPH2A 1234 protein in these individuals. In contrast, no TPH2a editing could be detected in schizophrenic patients (Table S4). Moreover, high levels of TPH2b editing were found in all patients and controls, whereas TPH2b transcripts of suicides and schizophrenics showed a substantial decrease in editing at position 1 (c.385C>T) by 50% and 30%, respectively (Table S4). Thus, dysregulations in TPH2 editing could be involved in the pathogenesis of psychiatric diseases or may directly result from substance abuse.
For comparison, altered RNA editing of 5-HT2C receptor transcripts was found in depressed suicides and schizophrenics [60], [61], [62], [63], leading to distinct receptor isoforms with different activities. In rodents, changes in 5-HT2C receptor editing were evident in response to stress [51] and 5-HT availability [60], whereas antidepressants were found to antagonize these changes [51], [52], [64]. This complex fine-tuning of the 5-HT2C receptor sensitivity is considered to be a crucial mechanism to keep receptor activation within an optimal range for information processing in face of changing synaptic input [64]. Our findings suggest that brain 5-HT biosynthesis is also regulated by TPH2 pre-mRNA editing, which could be affected by drug abuse and environmental factors in analogy to the 5-HT2C receptor. However, our collective of post mortem brain samples is small and conclusions have to be drawn with care. Nonetheless, our findings invite large-scale follow up studies.
SNP rs4290270 Regulates TPH2 Splicing and Editing
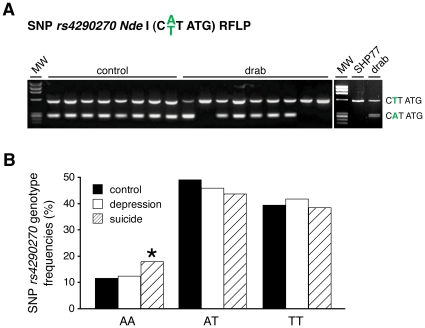
Most notably, we never detected TPH2b transcripts or editing in the presence of the SNP rs4290270 A (Figure 1; Table 1). This strongly resembles the recently found regulation of 5-HT2C receptor splicing by the small nucleolar RNA (snoRNA) HBII-52 and its deregulation by mutagenesis of the 5-HT2C receptor mRNA binding site [65]. However, we could not find a complementary snoRNA for the sequence context of the TPH2 rs4290270 SNP in the existing databases. Nonetheless, such trans-acting factors are only one possible explanation, as different expression or splicing efficacies due to rs4290270-mediated differences in TPH2 pre-mRNA secondary structure could be also responsible.
Fortunately, rs4290270 is part of the palindromic recognition sequence of Nde I, thus individual genotypes can be easily determined by restriction fragment length polymorphism (Figure 4A). In line with the finding that SHP77 cells exhibit alternative TPH2 splicing and editing of TPH2b, these cells are homozygous for rs4290270 T (Figure 4A). Thus, these cells may represent a suitable cell culture system to investigate the dynamics of splicing and editing.
Figure 4. Alternative splicing and editing only occurs in presence of SNP rs4290270 T.
(A) The individual genotype of SNP rs4290270 can be easily detected by restriction fragment length polymorphism (RFLP) analysis with Nde I in drug abusers (drab) and controls. (B) Large scale rs4290270 genotyping revealed a genetic predisposition for suicidality of homozygous A/A-carriers. The distribution was in the Hardy Weinberg equilibrium. *: p<0.05; controls: n = 373; major depression: n = 436; suicide: n = 369.
Importantly, we detected a significantly higher frequency of the A/A genotype of the SNP rs4290270 in a cohort of 369 suicides, as compared with 436 patients with major depression and 373 controls (Figure 4B). Thus, while we still have to elucidate the underlying mechanism of how rs4290270 A affects TPH2 alternative splicing and editing, the data demonstrate a genetic predisposition of homozygous A-allele carriers for suicide.
Implications for Psychiatric Research
Our functional data imply disturbed TPH2 activity in drug abuse, suicide, and schizophrenia. The regulation of TPH2 expression reveals an unprecedented mechanism of mutually exclusive editing of the alternatively spliced isoforms TPH2a and TPH2b. For this reason, our data establish TPH2 as an excellent subject for future investigations of the underlying RNA editing machineries.
Recent studies have tried to explain the reduced 5-HT neurotransmission in psychiatric disorders with disturbances in TPH2 expression [66], [67], [68], [69], [70]. However, based on our results that alternative splicing and RNA editing lead to TPH2 variants with different kinetic properties, we conclude, that neither the currently used RNA-based techniques, such as real time PCR or RNase protection assays, nor immunohistochemical protein methods allow a proper estimation of TPH2 enzymatic activity in psychiatric research. Based on the data presented here, careful re-examination of recent reports on TPH2 expression disturbances in neuropsychiatric diseases is mandatory. Moreover, since we are now in knowledge of the alternative splicing and editing that governs TPH2 activity, powerful new methods are eagerly awaited to assess for the editing status of TPH2 transcripts to gain insight into the regulation of 5-HT synthesis in the human brain.
Methods
Ethics Statement
All clinical investigations have been conducted according to the principles expressed in the Declaration of Helsinki and approved by the Ethics Committee of the Medical Faculty of the Ludwigs Maximilians University (LMU) Munich (Head: Prof. Dr. Gustav Paungartner, Members: Prof. Dr. Eckhard Held, Prof. Dr. Wolfgang Eisenmenger, PD Dr. Thomas Beinert, Prof. Dr. Hans Ulrich Gallwas, Prof. Dr. Detlef Kunze, Dr. Viktoria Mönch, Prof. Dr. Randolph Penning, Prof. Dr. Klaus Hahn, Prof. Dr. Klaus Jürgen Pfeifer, and Dr. Christian Zach). Ethikantrag, Projekt Nr. 213/00; positive vote from: 12.05.2005 “Genetische, biochemische und funktionelle Untersuchungen an depressiven Patienten und gesunden Kontrollpersonen”. Ethikantrag, Projekt Nr. 164/00; positive vote from: 14.04.2003 “Genetische Polymorphismen bei Suizidenten”. Written informed consent was given by the patients and healthy volunteers. Autopsy samples: The autopsies were court ordered from the state attorney. In that case informed consent from the next of kin is not required, because relatives have no possibility for intervention. Within these autopsies it is necessary to take routinely additional tissue probes for probable further investigations. The probes of the present study originate from these investigations. The Ethics Committee of the LMU Munich approved this procedure. All autopsies, including those of the control individuals were performed according to the legal requirements. They were court ordered according to the German legal situation from the state attorney due to unknown causes of death. For the control individuals the natural cause of death was verified finally by these autopsies. In all of these cases (patients and controls) informed consent from the next of kin is not required, because relatives have no possibility for intervention. Blood and brain samples were exclusively taken during the routine autopsies to perform the court ordered analysis. Furthermore post mortem material will be preserved for subsequently necessary investigations on behalf of the state attorney. Blood and brain samples were never taken for research. For research projects we use only remaining post mortem samples which have been released and approved for use in research by the Ethics Committee of the LMU Munich. As described, the consent for research use of autopsy tissues will be given by the localEthics Committees of the universities. This is the current procedure in legal medicine in Germany.
Brain Samples
Brain specimens (as indicated in the Ethics statement) were derived from 11 individuals, who died as a consequence of opiate addiction (8 males, 3 females, mean age 30.4±9.8 years; post mortem interval (PMI): 14.5±9.6 hours), 8 suicide victims (6 males, 2 females, mean age 42,8±9.4 years; PMI: 18.8±13.7 hours) and 7 schizophrenic patients (5 males, 2 females, mean age 43±13.5 years; PMI: 23.9±6.4 hours). The control tissues were obtained from 10 individuals, who died suddenly from CNS-unrelated diseases (5 males, 5 females, mean age 44.7±15.8 years; PMI: 19.9±7.6 hours). Causes of death were acute cardiac failure (n = 5), accident (n = 3), and homicide (n = 2). The clinical, respectively medical, data sheets of the control individuals were available and excluded any lifetime psychiatric or neurological disorders. According to the medical records, there was no history of psychopharmacological medication, alcohol or drug abuse. Additionally, a toxicological report for all individuals was provided and negative for additional drug intoxification, whereas information on pre-existing psychiatric disturbances was missing for the suicide victims. All individuals were Caucasians from the same geographical region in southern Germany.
Molecular Biological Methods
All cloning procedures, PCR (used primers are indicated in Table S5), and immunoblotting were conducted according to standard protocols or manufacture's instructions. Post mortem brain samples were collected using the RNAlater kit (Qiagen, Hilden, Germany) and immediately frozen at −80°C until RNA extraction. After homogenization, total RNA was extracted from tissues using the RNeasy Lipid Tissue Midi Kit (Qiagen), treated with DNase I (Invitrogen, Carlsbad, CA, USA) and dissolved in RNase-free water. cDNA was synthesized from 2 µg RNA using MMLV reverse transcriptase and random hexamer primers (Invitrogen). The TPH2 coding sequence was amplified with ORF-fw and ORF-rev primers and subcloned into pCR®-XL-TOPO (Invitrogen). The obtained clones were sequenced and aligned with the TPH2 mRNA reference sequence (GenBank NM_173353). TPH2 polymorphisms were analyzed genomically by amplification of the TPH2 exons from genomic DNA using intronic primers (Table S5), subcloning into pGEM®-T easy (Promega, Madison, WI, USA), and DNA sequencing.
Detection of TPH2 splice isoforms was performed using the TPH2SPL_fw forward primer together with the splice-specific TPH2a_rev and TPH2b_rev reverse primers and the following PCR conditions: 15 s denaturation at 95°C, 10 s annealing at 75°C/70°C, and 30 s elongation at 75°C/72°C for TPH2a/TPH2b, respectively. The detection of Tph2b in the rat brain was carried out using the primers rTPH2ex2A (forward) and TPH2SPLrat (reverse).
TPH2 expression constructs were generated from TPH2a/b cDNAs obtained from patients by reamplification with 6xHisTph2-fw (forward) primer containing an ATG with a Kozak consensus sequence and a 6xHis tag and supsequent cloning into pTargeT™ (Promega). Stable PC12 and HEK293 cell lines were obtained with linearized TPH2 constructs and DreamFect (OZ Biosciences, Marseille, France), followed by selection of transfected cells with 500 µg mL−1 for at least two weeks. Cell cultures were maintained under standard conditions in DMEM supplemented with 10% fetal bovine serum (FBS, HEK293) and 15% FBS/2.5% donor horse serum (PC12) and antibiotics.
TPH Activity Assay
The activity of cell homogenates was determined as described [6], [8], monitoring for 5-hydroxytryptophan (5-HTP) accumulation by HPLC in presence of the aromatic amino acid decarboxylase inhibitor 3-hydroxybenzylhydrazine hydrochloride (NSD1015). All reagents were purchased from Sigma-Aldrich (St. Louis, MO, USA). In brief, cells were harvested by scraping and washed twice with phosphate-buffered saline, resuspended in 75 mM tris-acetate buffer (pH 7.5), and lyzed by sonication. After withdrawal of an aliquot for protein determination, the homogenates were immediately preincubated in 100 µL buffer containing 2 mg/mL catalase, 25 mM DTT and 100 µM Fe(NH4)2(SO4)2 for 10 min at 30°C in the dark. The pre-incubated samples were incubated at 37°C for 30 min after addition of 400 µL 15 mM tris-acetate buffer (pH 6.4) containing the indicated concentrations of L-Trp, 300 µM 6-methyl-tetrahydrobiopterin (6MPH4) or tetrahydrobiopterin (BH4) and 2 mM NSD1015. The reaction was terminated by addition of 300 mM perchloric acid (final concentration) and centrifugation for deproteination. The cleared supernatants were directly analyzed using reverse phase HPLC with fluorometric detection (HPLC-FD) as previously described [8]. The measured 5-HTP levels were normalized to the amount of TPH2 protein in each lysate by immunodetection using mouse anti-WH3 (TPH2, Sigma-Aldrich) and goat anti-actin (Santa Cruz Biotechnology, Santa Cruz, CA, USA) antibodies.
5-HT Measurement
To determine 5-HT levels in stable TPH2-expressing PC12 cell lines, 1.7 million cells were homogenized in 100 µL buffer containing 5 mM sodium metabisulfite and 300 mM perchloric acid (Sigma-Aldrich). Cleared supernatants were directly used for HPLC-FD measurement as previously described [8]. Cell pellets were boiled in 100 µL SDS loading buffer for 5-HT normalization to the TPH2 protein expression levels by immunoblotting.
Subjects for Genotyping
The case sample of suicide victims consisted of 369 individuals (269 males, 100 females; mean age: 46.42 years±17.77 years). Of these, 290 committed violent suicides, as e.g. hanging (36%), shooting (18%), penetrating lesions (7%), jumping from height, drowning and lying under a train (17%). 79 employed soft suicide methods, such as intoxication with drugs or other substances (21%). Blood samples for DNA extraction were obtained in the course of autopsy at the Institute for Legal Medicine of the LMU Munich. There was no information on pre-existing psychiatric disturbances.
A total of 436 unrelated Caucasian patients with major depression (270 males, 166 females; mean age: 48.69±14.07 years), hospitalized in the Psychiatric Department of the LMU Munich and diagnosed according to the DSM-IV and ICD-10 criteria were included in the study. All patients were interviewed by experienced psychiatrists using the Structured Clinical Interview for DSM-IV disorders (SCID-I). Severity of depression was assessed using the 17-item Hamilton Rating Scale for Depression (HAMD-17) and the Clinical Global Impression Scale (CGI). Only subjects with a minimum score of 18 on the HAMD-17 scale were included in the study. Patients with severe organic disorders were excluded to avoid cases with secondary depression. Furthermore, all patients with comorbidity of other psychiatric disturbances (e.g. substance/alcohol dependence, personality disorders, anxiety disorders) were excluded. The patient sample contained significantly more females than males as compared with the control sample (62%/38% versus 48%/52%; p = 0.001, χ2 = 11.3, df = 1). Because of no significant differences concerning all other investigated variables (age, clinical variables such as CGI and HAMD-17 scores), males and females were not analyzed separately.
As control group, 373 ethnically matched subjects were selected from the general population (185 males, 188 females; mean age: 44.42±16.00 years). All probands were screened for psychiatric disturbances using personality questionnaires (MMPI, NEO-FI, TCI) and a short structured interview with a psychiatrist. Probands with known history of psychiatric disorders were excluded from the study. All patients and controls were of Caucasian origin from the German population and came from the same geographical area in southern Germany. Blood was collected from these subjects for DNA extraction; patients and controls participated after giving written informed consent. The study was approved by the ethics committee of the Medical Faculty of the LMU Munich (project number 213/00; positive vote from: 12.05.2005).
Genotyping of SNP rs4290270
Genomic DNA was isolated from whole blood according standard procedures. The SNP rs4290270 was genotyped applying the TaqMan® technology (Assay-on-Demand; assay-ID: C_26385365) on an ABI7000 system (Applied Biosystems, Foster City, CA, USA). The standard PCR reaction was carried out using TaqMan® Universal PCR Master Mix reagent kit according to the manufacture's instructions.
Statistics
All data are presented as means ± SEM and p-values are from two-tailed Student's t-tests type 3. Genotype frequencies were tested for Hardy-Weinberg equilibrium as described [71]. Values of p<0.05 were considered as statistically significant.
Supporting Information
Identified SNPs in the human TPH2 gene. The positions of base exchanges are indicated according the TPH2 mRNA reference sequence (GenBank NM_173353) and the described nomenclature system.
(0.07 MB DOC)
Exon-intron boundaries of Tph2 genes of higher vertebrates. The consensus sequence of the five species is indicated by ‘cons’. Alternative 3′-SDS in rats giving rise to four rTPH2 isoforms.
(0.05 MB DOC)
Compiled kinetic constants for mammalian TPH1/2 isoforms.
(0.05 MB DOC)
TPH2a and TPH2b editing in the amygdala of the tested individuals with psychiatric disorders.
(0.06 MB DOC)
TPH2 primers used for PCR amplification.
(0.05 MB DOC)
Acknowledgments
We thank Daniela Eser and Caroline Nothdurfter for patient recruitment, Hans-Hilger Ropers for discussions and Jakob Vowinckel for critical reading of the manuscript. The technical assistance of Sabine Otto and Monika Dopatka is highly appreciated.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: The study was funded in part by grants of the Deutsche Forschungsgemeinschaft, the Bundesministerium für Bildung und Forschung and the Max Planck Society. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Walther DJ, Bader M. A unique central tryptophan hydroxylase isoform. Biochemical Pharmacology. 2003;66:1673–1680. doi: 10.1016/s0006-2952(03)00556-2. [DOI] [PubMed] [Google Scholar]
- 2.Fitzpatrick PF. Tetrahydropterin-dependent amino acid hydroxylases. Annual Review of Biochemistry. 1999;68:355–381. doi: 10.1146/annurev.biochem.68.1.355. [DOI] [PubMed] [Google Scholar]
- 3.Rosen JB, Donley MP. Animal studies of amygdala function in fear and uncertainty: relevance to human research. Biological Psychiatry. 2006;73:49–60. doi: 10.1016/j.biopsycho.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 4.Baxter MG, Murray EA. The amygdala and reward. Nature Reviews Neuroscience. 2002;3:563–573. doi: 10.1038/nrn875. [DOI] [PubMed] [Google Scholar]
- 5.Talarovicova A, Krskova L, Kiss A. Some assessments of the amygdala role in suprahypothalamic neuroendocrine regulation: a minireview. Endocrine Regulations. 2007;41:155–162. [PubMed] [Google Scholar]
- 6.Walther DJ, Peter JU, Bashammakh S, Hortnagl H, Voits M, et al. Synthesis of serotonin by a second tryptophan hydroxylase isoform. Science. 2003;299:76. doi: 10.1126/science.1078197. [DOI] [PubMed] [Google Scholar]
- 7.Matsuda M, Imaoka T, Vomachka AJ, Gudelsky GA, Hou Z, et al. Serotonin regulates mammary gland development via an autocrine-paracrine loop. Developmental Cell. 2004;6:193–203. doi: 10.1016/s1534-5807(04)00022-x. [DOI] [PubMed] [Google Scholar]
- 8.Walther DJ, Bader M. Serotonin synthesis in murine embryonic stem cells. Brain Research Molecular Brain Research. 1999;68:55–63. doi: 10.1016/s0169-328x(99)00046-7. [DOI] [PubMed] [Google Scholar]
- 9.Cattaneo MG, Codignola A, Vicentini LM, Clementi F, Sher E. Nicotine stimulates a serotonergic autocrine loop in human small-cell lung carcinoma. Cancer Research. 1993;53:5566–5568. [PubMed] [Google Scholar]
- 10.Walther DJ, Peter JU, Bader M. 7-Hydroxytryptophan, a novel, specific, cytotoxic agent for carcinoids and other serotonin-producing tumors. Cancer. 2002;94:3135–3140. doi: 10.1002/cncr.10592. [DOI] [PubMed] [Google Scholar]
- 11.Walther DJ, Peter JU, Winter S, Holtje M, Paulmann N, et al. Serotonylation of small GTPases is a signal transduction pathway that triggers platelet alpha-granule release. Cell. 2003;115:851–862. doi: 10.1016/s0092-8674(03)01014-6. [DOI] [PubMed] [Google Scholar]
- 12.Lesurtel M, Graf R, Aleil B, Walther DJ, Tian Y, et al. Platelet-derived serotonin mediates liver regeneration. Science. 2006;312:104–107. doi: 10.1126/science.1123842. [DOI] [PubMed] [Google Scholar]
- 13.Paulmann N, Grohmann M, Voigt JP, Bert B, Vowinckel J, et al. Intracellular serotonin modulates insulin secretion from pancreatic beta-cells by protein serotonylation. PLoS biology. 2009;7:e1000229. doi: 10.1371/journal.pbio.1000229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morecroft I, Dempsie Y, Bader M, Walther DJ, Kotnik K, et al. Effect of tryptophan hydroxylase 1 deficiency on the development of hypoxia-induced pulmonary hypertension. Hypertension. 2007;49:232–236. doi: 10.1161/01.HYP.0000252210.58849.78. [DOI] [PubMed] [Google Scholar]
- 15.Bicalho MA, Pimenta GJ, Neves FS, Correa H, de Moraes EN, et al. Genotyping of the G1463A (Arg441His) TPH2 polymorphism in a geriatric population of patients with major depression. Molecular Psychiatry. 2006;11:799–800. doi: 10.1038/sj.mp.4001861. [DOI] [PubMed] [Google Scholar]
- 16.Harvey M, Shink E, Tremblay M, Gagne B, Raymond C, et al. Support for the involvement of TPH2 gene in affective disorders. Molecular Psychiatry. 2004;9:980–981. doi: 10.1038/sj.mp.4001557. [DOI] [PubMed] [Google Scholar]
- 17.Walitza S, Renner TJ, Dempfle A, Konrad K, Wewetzer C, et al. Transmission disequilibrium of polymorphic variants in the tryptophan hydroxylase-2 gene in attention-deficit/hyperactivity disorder. Molecular Psychiatry. 2005;10:1126–1132. doi: 10.1038/sj.mp.4001734. [DOI] [PubMed] [Google Scholar]
- 18.Kulikov AV, Osipova DV, Naumenko VS, Popova NK. Association between Tph2 gene polymorphism, brain tryptophan hydroxylase activity and aggressiveness in mouse strains. Genes, Brain, and Behavior. 2005;4:482–485. doi: 10.1111/j.1601-183X.2005.00145.x. [DOI] [PubMed] [Google Scholar]
- 19.Zill P, Buttner A, Eisenmenger W, Moller HJ, Bondy B, et al. Single nucleotide polymorphism and haplotype analysis of a novel tryptophan hydroxylase isoform (TPH2) gene in suicide victims. Biological Psychiatry. 2004;56:581–586. doi: 10.1016/j.biopsych.2004.07.015. [DOI] [PubMed] [Google Scholar]
- 20.McKinney J, Knappskog PM, Haavik J. Different properties of the central and peripheral forms of human tryptophan hydroxylase. Journal of Neurochemistry. 2005;92:311–320. doi: 10.1111/j.1471-4159.2004.02850.x. [DOI] [PubMed] [Google Scholar]
- 21.Zill P, Buttner A, Eisenmenger W, Moller HJ, Ackenheil M, et al. Analysis of tryptophan hydroxylase I and II mRNA expression in the human brain: a post-mortem study. Journal of Psychiatric Research. 2007;41:168–173. doi: 10.1016/j.jpsychires.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 22.Sakowski SA, Geddes TJ, Thomas DM, Levi E, Hatfield JS, et al. Differential tissue distribution of tryptophan hydroxylase isoforms 1 and 2 as revealed with monospecific antibodies. Brain Research. 2006;1085:11–18. doi: 10.1016/j.brainres.2006.02.047. [DOI] [PubMed] [Google Scholar]
- 23.Scheuch K, Lautenschlager M, Grohmann M, Stahlberg S, Kirchheiner J, et al. Characterization of a Functional Promoter Polymorphism of the Human Tryptophan Hydroxylase 2 Gene in Serotonergic Raphe Neurons. Biological Psychiatry. 2007;62:1288–1294. doi: 10.1016/j.biopsych.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 24.Brown SM, Peet E, Manuck SB, Williamson DE, Dahl RE, et al. A regulatory variant of the human tryptophan hydroxylase-2 gene biases amygdala reactivity. Molecular Psychiatry. 2005;10:884–888, 805. doi: 10.1038/sj.mp.4001716. [DOI] [PubMed] [Google Scholar]
- 25.Canli T, Congdon E, Gutknecht L, Constable RT, Lesch KP. Amygdala responsiveness is modulated by tryptophan hydroxylase-2 gene variation. Journal of Neural Transmission. 2005;112:1479–1485. doi: 10.1007/s00702-005-0391-4. [DOI] [PubMed] [Google Scholar]
- 26.Zhang X, Beaulieu JM, Sotnikova TD, Gainetdinov RR, Caron MG. Tryptophan hydroxylase-2 controls brain serotonin synthesis. Science. 2004;305:217. doi: 10.1126/science.1097540. [DOI] [PubMed] [Google Scholar]
- 27.Cichon S, Winge I, Mattheisen M, Georgi A, Karpushova A, et al. Brain-specific tryptophan hydroxylase 2 (TPH2): a functional Pro206Ser substitution and variation in the 5′-region are associated with bipolar affective disorder. Human Molecular Genetics. 2008;17:87–97. doi: 10.1093/hmg/ddm286. [DOI] [PubMed] [Google Scholar]
- 28.McKinney J, Johansson S, Halmoy A, Dramsdahl M, Winge I, et al. A loss-of-function mutation in tryptophan hydroxylase 2 segregating with attention-deficit/hyperactivity disorder. Molecular Psychiatry. 2008;13:365–367. doi: 10.1038/sj.mp.4002152. [DOI] [PubMed] [Google Scholar]
- 29.Zhang X, Gainetdinov RR, Beaulieu JM, Sotnikova TD, Burch LH, et al. Loss-of-function mutation in tryptophan hydroxylase-2 identified in unipolar major depression. Neuron. 2005;45:11–16. doi: 10.1016/j.neuron.2004.12.014. [DOI] [PubMed] [Google Scholar]
- 30.Garriock HA, Allen JJ, Delgado P, Nahaz Z, Kling MA, et al. Lack of association of TPH2 exon XI polymorphisms with major depression and treatment resistance. Molecular Psychiatry. 2005;10:976–977. doi: 10.1038/sj.mp.4001712. [DOI] [PubMed] [Google Scholar]
- 31.Glatt CE, Carlson E, Taylor TR, Risch N, Reus VI, et al. Response to Zhang et al. (2005): loss-of-function mutation in tryptophan hydroxylase-2 identified in unipolar major depression. Neuron. 2005. 45, 11-16. Neuron 48: 704-705; author reply 705-706. [DOI] [PubMed]
- 32.van den Bogaert A, de Zutter S, Heyrman L, Mendlewicz J, Adolfsson R, et al. Response to Zhang et al (2005): loss-of-function mutation in tryptophan hydroxylase-2 identified in unipolar major Depression. Neuron. 2005. 45, 11-16. Neuron 48: 704; author reply 705-706. [DOI] [PubMed]
- 33.Zhou Z, Peters EJ, Hamilton SP, McMahon F, Thomas C, et al. Response to Zhang et al. (2005): loss-of-function mutation in tryptophan hydroxylase-2 identified in unipolar major depression. Neuron. 2005. 45, 11-16. Neuron 48: 702-703; author reply 705-706. [DOI] [PubMed]
- 34.Abumaria N, Ribic A, Anacker C, Fuchs E, Flugge G. Stress upregulates TPH1 but not TPH2 mRNA in the rat dorsal raphe nucleus: identification of two TPH2 mRNA splice variants. Cellular and Molecular Neurobiology. 2008;28:331–342. doi: 10.1007/s10571-007-9259-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Farrer T, Roller AB, Kent WJ, Zahler AM. Analysis of the role of Caenorhabditis elegans GC-AG introns in regulated splicing. Nucleic Acids Research. 2002;30:3360–3367. doi: 10.1093/nar/gkf465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thanaraj TA, Clark F. Human GC-AG alternative intron isoforms with weak donor sites show enhanced consensus at acceptor exon positions. Nucleic Acids Research. 2001;29:2581–2593. doi: 10.1093/nar/29.12.2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Buznikov GA, Peterson RE, Nikitina LA, Bezuglov VV, Lauder JM. The pre-nervous serotonergic system of developing sea urchin embryos and larvae: pharmacologic and immunocytochemical evidence. Neurochemical Research. 2005;30:825–837. doi: 10.1007/s11064-005-6876-6. [DOI] [PubMed] [Google Scholar]
- 38.Lauder JM, Wallace JA, Krebs H. Roles for serotonin in neuroembryogenesis. Advances in Experimental Medicine and Biology. 1981;133:477–506. doi: 10.1007/978-1-4684-3860-4_28. [DOI] [PubMed] [Google Scholar]
- 39.Carkaci-Salli N, Flanagan JM, Martz MK, Salli U, Walther DJ, et al. Functional domains of human tryptophan hydroxylase 2 (hTPH2). Journal of Biological Chemistry. 2006;281:28105–28112. doi: 10.1074/jbc.M602817200. [DOI] [PubMed] [Google Scholar]
- 40.Hentze MW, Kulozik AE. A Perfect Message: RNA Surveillance and Nonsense-Mediated Decay. Cell. 1999;96:307–310. doi: 10.1016/s0092-8674(00)80542-5. [DOI] [PubMed] [Google Scholar]
- 41.Reed R, Hurt E. A Conserved mRNA Export Machinery Coupled to pre-mRNA Splicing. Cell. 2002;108:523. doi: 10.1016/s0092-8674(02)00627-x. [DOI] [PubMed] [Google Scholar]
- 42.Bhalla T, Rosenthal JJ, Holmgren M, Reenan R. Control of human potassium channel inactivation by editing of a small mRNA hairpin. Nature Structural & Molecular Biology. 2004;11:950–956. doi: 10.1038/nsmb825. [DOI] [PubMed] [Google Scholar]
- 43.Burns CM, Chu H, Rueter SM, Hutchinson LK, Canton H, et al. Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature. 1997;387:303–308. doi: 10.1038/387303a0. [DOI] [PubMed] [Google Scholar]
- 44.Higuchi M, Single FN, Kohler M, Sommer B, Sprengel R, et al. RNA editing of AMPA receptor subunit GluR-B: a base-paired intron-exon structure determines position and efficiency. Cell. 1993;75:1361–1370. doi: 10.1016/0092-8674(93)90622-w. [DOI] [PubMed] [Google Scholar]
- 45.Meier JC, Henneberger C, Melnick I, Racca C, Harvey RJ, et al. RNA editing produces glycine receptor alpha3(P185L), resulting in high agonist potency. Nature Neuroscience. 2005;8:736–744. doi: 10.1038/nn1467. [DOI] [PubMed] [Google Scholar]
- 46.Nishikura K. Editor meets silencer: crosstalk between RNA editing and RNA interference. Nature Reviews Molecular Cell Biology. 2006;7:919–931. doi: 10.1038/nrm2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blanc V, Davidson NO. C-to-U RNA editing: mechanisms leading to genetic diversity. Journal of Biological Chemistry. 2003;278:1395–1398. doi: 10.1074/jbc.R200024200. [DOI] [PubMed] [Google Scholar]
- 48.Chester A, Somasekaram A, Tzimina M, Jarmuz A, Gisbourne J, et al. The apolipoprotein B mRNA editing complex performs a multifunctional cycle and suppresses nonsense-mediated decay. EMBO Journal. 2003;22:3971–3982. doi: 10.1093/emboj/cdg369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wedekind JE, Dance GS, Sowden MP, Smith HC. Messenger RNA editing in mammals: new members of the APOBEC family seeking roles in the family business. Trends in Genetics. 2003;19:207–216. doi: 10.1016/S0168-9525(03)00054-4. [DOI] [PubMed] [Google Scholar]
- 51.Englander MT, Dulawa SC, Bhansali P, Schmauss C. How stress and fluoxetine modulate serotonin 2C receptor pre-mRNA editing. Journal of Neuroscience. 2005;25:648–651. doi: 10.1523/JNEUROSCI.3895-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Iwamoto K, Nakatani N, Bundo M, Yoshikawa T, Kato T. Altered RNA editing of serotonin 2C receptor in a rat model of depression. Neuroscience Research. 2005;53:69–76. doi: 10.1016/j.neures.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 53.Maas S, Kawahara Y, Tamburro KM, Nishikura K. A-to-I RNA editing and human disease. RNA Biology. 2006;3:1–9. doi: 10.4161/rna.3.1.2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Barlati S, Barbon A. RNA editing: a molecular mechanism for the fine modulation of neuronal transmission. Acta Neurochirurgica Supplementum. 2005;93:53–57. doi: 10.1007/3-211-27577-0_7. [DOI] [PubMed] [Google Scholar]
- 55.Sharma PM, Bowman M, Madden SL, Rauscher FJ, 3rd, Sukumar S. RNA editing in the Wilms' tumor susceptibility gene, WT1. Genes and Development. 1994;8:720–731. doi: 10.1101/gad.8.6.720. [DOI] [PubMed] [Google Scholar]
- 56.Bestor TH. The DNA methyltransferases of mammals. Human Molecular Genetics. 2000;9:2395–2402. doi: 10.1093/hmg/9.16.2395. [DOI] [PubMed] [Google Scholar]
- 57.Steele EJ, Lindley RA, Wen J, Weiller GF. Computational analyses show A-to-G mutations correlate with nascent mRNA hairpins at somatic hypermutation hotspots. DNA Repair (Amst) 2006;5:1346–1363. doi: 10.1016/j.dnarep.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 58.Bourara K, Litvak S, Araya A. Generation of G-to-A and C-to-U changes in HIV-1 transcripts by RNA editing. Science. 2000;289:1564–1566. doi: 10.1126/science.289.5484.1564. [DOI] [PubMed] [Google Scholar]
- 59.Schonn JS, Desnos C, Henry JP, Darchen F. Transmitter uptake and release in PC12 cells overexpressing plasma membrane monoamine transporters. Journal of Neurochemistry. 2003;84:669–677. doi: 10.1046/j.1471-4159.2003.01561.x. [DOI] [PubMed] [Google Scholar]
- 60.Gurevich I, Tamir H, Arango V, Dwork AJ, Mann JJ, et al. Altered editing of serotonin 2C receptor pre-mRNA in the prefrontal cortex of depressed suicide victims. Neuron. 2002;34:349–356. doi: 10.1016/s0896-6273(02)00660-8. [DOI] [PubMed] [Google Scholar]
- 61.Niswender CM, Herrick-Davis K, Dilley GE, Meltzer HY, Overholser JC, et al. RNA editing of the human serotonin 5-HT2C receptor. alterations in suicide and implications for serotonergic pharmacotherapy. Neuropsychopharmacology. 2001;24:478–491. doi: 10.1016/S0893-133X(00)00223-2. [DOI] [PubMed] [Google Scholar]
- 62.Iwamoto K, Kato T. RNA editing of serotonin 2C receptor in human postmortem brains of major mental disorders. Neuroscience Letters. 2003;346:169–172. doi: 10.1016/s0304-3940(03)00608-6. [DOI] [PubMed] [Google Scholar]
- 63.Sodhi MS, Burnet PW, Makoff AJ, Kerwin RW, Harrison PJ. RNA editing of the 5-HT(2C) receptor is reduced in schizophrenia. Molecular Psychiatry. 2001;6:373–379. doi: 10.1038/sj.mp.4000920. [DOI] [PubMed] [Google Scholar]
- 64.Gurevich I, Englander MT, Adlersberg M, Siegal NB, Schmauss C. Modulation of serotonin 2C receptor editing by sustained changes in serotonergic neurotransmission. Journal of Neuroscience. 2002;22:10529–10532. doi: 10.1523/JNEUROSCI.22-24-10529.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kishore S, Stamm S. The snoRNA HBII-52 regulates alternative splicing of the serotonin receptor 2C. Science. 2006;311:230–232. doi: 10.1126/science.1118265. [DOI] [PubMed] [Google Scholar]
- 66.Bach-Mizrachi H, Underwood MD, Kassir SA, Bakalian MJ, Sibille E, et al. Neuronal tryptophan hydroxylase mRNA expression in the human dorsal and median raphe nuclei: major depression and suicide. Neuropsychopharmacology. 2006;31:814–824. doi: 10.1038/sj.npp.1300897. [DOI] [PubMed] [Google Scholar]
- 67.Bach-Mizrachi H, Underwood MD, Tin A, Ellis SP, Mann JJ, et al. Elevated expression of tryptophan hydroxylase-2 mRNA at the neuronal level in the dorsal and median raphe nuclei of depressed suicides. Molecular Psychiatry. 2008;13:507–513. doi: 10.1038/sj.mp.4002143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.De Luca V, Likhodi O, Van Tol HH, Kennedy JL, Wong AH. Tryptophan hydroxylase 2 gene expression and promoter polymorphisms in bipolar disorder and schizophrenia. Psychopharmacology. 2005;183:378–382. doi: 10.1007/s00213-005-0191-4. [DOI] [PubMed] [Google Scholar]
- 69.De Luca V, Likhodi O, Van Tol HH, Kennedy JL, Wong AH. Gene expression of tryptophan hydroxylase 2 in post-mortem brain of suicide subjects. The International Journal of Neuropsychopharmacology. 2006;9:21–25. doi: 10.1017/S1461145705005572. [DOI] [PubMed] [Google Scholar]
- 70.Haghighi F, Bach-Mizrachi H, Huang YY, Arango V, Shi S, et al. Genetic architecture of the human tryptophan hydroxylase 2 Gene: existence of neural isoforms and relevance for major depression. Molecular Psychiatry. 2008;13:813–820. doi: 10.1038/sj.mp.4002127. [DOI] [PubMed] [Google Scholar]
- 71.Zill P, Baghai TC, Zwanzger P, Schule C, Eser D, et al. SNP and haplotype analysis of a novel tryptophan hydroxylase isoform (TPH2) gene provide evidence for association with major depression. Molecular Psychiatry. 2004;9:1030–1036. doi: 10.1038/sj.mp.4001525. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Identified SNPs in the human TPH2 gene. The positions of base exchanges are indicated according the TPH2 mRNA reference sequence (GenBank NM_173353) and the described nomenclature system.
(0.07 MB DOC)
Exon-intron boundaries of Tph2 genes of higher vertebrates. The consensus sequence of the five species is indicated by ‘cons’. Alternative 3′-SDS in rats giving rise to four rTPH2 isoforms.
(0.05 MB DOC)
Compiled kinetic constants for mammalian TPH1/2 isoforms.
(0.05 MB DOC)
TPH2a and TPH2b editing in the amygdala of the tested individuals with psychiatric disorders.
(0.06 MB DOC)
TPH2 primers used for PCR amplification.
(0.05 MB DOC)